Abstract
The orthohepadnaviruses, which include the major human pathogen hepatitis B virus, exist in a wide range of hosts. Since 2013, a large group of orthohepadnaviruses has been identified in bats worldwide and classified as 4 species within the genus Orthohepadnavirus. To further investigate orthohepadnaviruses in the Chinese bat population, 554 archived bat samples from 20 colonies covering 3 southern provinces were screened with results showing that 9 (1.6%) were positive. A systematic phylogenetic analysis has indicated the need for a new nomenclature for bat hepatitis B virus-like viruses: BtHBV, with the addition of 3 new species, one being divided into 6 genotypes. Viruses identified here shared 9.0–19.2% full genome divergence and classified into 3 different genotypes. This study illustrates the genetic diversity of orthohepadnaviruses in the Chinese bat population, and emphasizes need for further investigation of their public health significance.
Keywords: Bats, Orthohepadnavirus, Genetic diversity, Nomenclature
Highlights
-
•
Three new orthohepadnaviral lineages were identified in Chinese bats.
-
•
A new nomenclature was proposed for bat hepatitis B virus-like viruses.
-
•
This study indicates genetic diversity of orthohepadnaviruses in Chinese bats.
1. Introduction
Hepatitis B virus (HBV), the prototype member of the genus Orthohepadnavirus within the family Hepadnaviridae, is one of the most infectious human pathogens, responsible for >250 million chronic infections worldwide and around 887,000 human deaths annually (ICTV, 2017; Mason et al., 2011; Ott et al., 2012; Seeger et al., 2013; WHO, 2017). Orthohepadnaviruses have also been characterized in other primates and rodents (Seeger et al., 2013). Additionally, 14 species of bats from the Old World (Myanmar, China and Gabon) and New World (Panama) have recently been found to harbor a variety of orthohepadnaviruses (Drexler et al., 2013; He et al., 2013, 2015; Nie et al., 2018; Wang et al., 2017), of which 4 have been approved in the latest report of the International Committee on Taxonomy of Viruses (ICTV) as new species within the genus Orthohepadnavirus: Long-fingered bat hepatitis B virus, Pomona bat hepatitis B virus, Roundleaf bat hepatitis B virus, and Tent-making bat hepatitis B virus (ICTV, 2017). Molecular studies have shown that the Panamanian tent-making bat hepatitis B virus (TBHBV) is capable of infecting primary human hepatocytes through binding to the same receptor as human HBV, thereby suggesting that bat orthohepadnaviruses are ancestors of primate HBVs (Drexler et al., 2013; Rasche et al., 2016).
Orthohepadnaviruses are much more diverse in bats than in other host species. A single bat species can harbor diverse strains of these viruses, and a single orthohepadnavirus species can infect different bat species: e.g., Miniopterus schreibersi (M. schreibersi) bats have been found carrying both Neixiang-Ms69 and Anlong-Ms258 viruses, two viruses showing enough genetic diversity to be considered as two distinct species (Nie et al., 2018), whereas Neixiang-Ms69 and Neixiang-Rpu92, sharing almost 100% full genome nucleotide (nt) similarity, have been found in two distinct bat species of two separate families, M. schreibersi and Rhinolophus pusillus (R. pusillus) (Nie et al., 2018). Currently nomenclature of a bat orthohepadnavirus species uses HBV after the common name of the host in which they have been first detected; e.g., “Long-fingered bat hepatitis B virus” representing a virus species identified in Miniopterus bats, but this could refer to either strain 776 from Myanmar or Neixiang-Ms-69 from China — two different viruses (He et al., 2013; Nie et al., 2018). Such a mismatch can cause confusion. Accordingly, a new species nomenclature for bat HBV-like viruses (BtHBV) is necessary.
With the exception of humans and possibly rodents, bats are the most abundant as well as the most widely distributed mammals (Wilson and Reeder, 2005). They provide a huge natural virus bank from which >200 viruses have been isolated or identified, including such lethal agents as lyssaviruses, henipaviruses, ebolaviruses, and SARS-related coronaviruses (Moratelli and Calisher, 2015). Apart from orthohepadnaviruses in bats in America, Asia and Africa, homologues of hepatitis A, C and E virus have also been sporadically identified in these animals (Drexler et al., 2015; Drexler et al., 2012; Quan et al., 2013). To further understand the genetic diversity of HBV-like virus, we screened hepatitis viruses from bats collected in south China, and found another three variants. Systematic and phylogenetic analyses of the BtHBVs identified 7 species, for which we propose a new nomenclature that will be more helpful for understanding host species specificity and evolution of these viruses.
2. Materials and methods
2.1. Sample collection
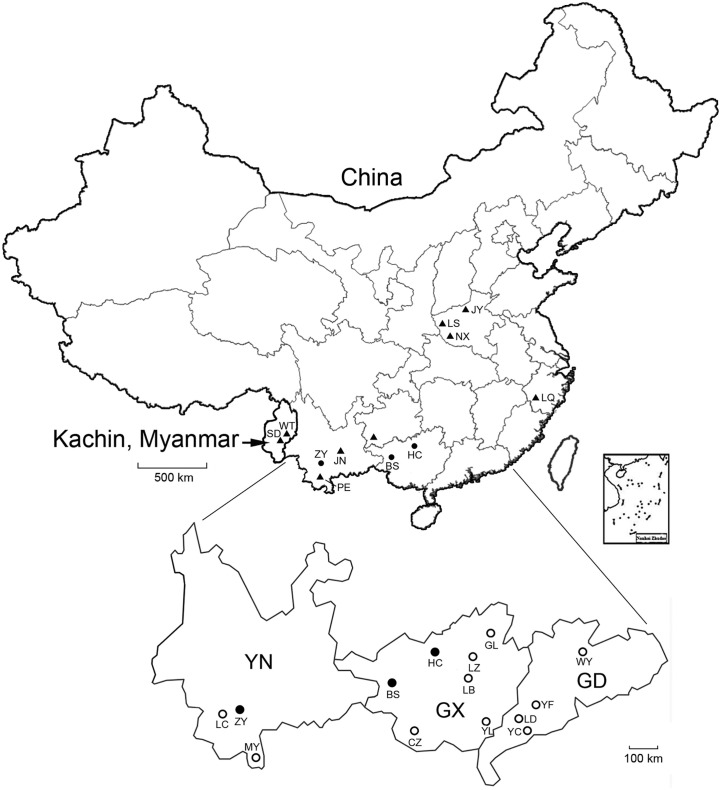
The procedures for sampling of bats in this study were reviewed and approved by the Administrative Committee on Animal Welfare of the Institute of Military Veterinary, Academy of Military Medical Sciences, China (Laboratory Animal Care and Use Committee Authorization, permit numbers: JSY-DW-2010-02 and JSY-DW-2015-01). All live bats were maintained and handled according to the Principles and Guidelines for Laboratory Animal Medicine (2006), Ministry of Science and Technology, China. To investigate orthohepadnaviral ecology in Chinese bats, animals were collected between 2005 and 2015 with nets near or in human-inhabited communities in Yunnan, Guangxi and Guangdong provinces. All bats were apparently healthy at capture, and were initially identified by morphological examination by a trained field biologist and confirmed by sequence analysis of the mt-cyt b gene (Wang et al., 2003). Following euthanization their livers and other organs were immediately collected and stored at −80 °C (Fig. 1 and Table 1 ).
Fig. 1.
Map showing the locations of bats collected in this study (circles), and previously reported BtHBVs (triangles) in China and Myanmar. Filled circles: bats positive for HBV; open circles: bats negative for HBV.
Table 1.
Details of bat samples and numbers of positive bat samples detected by PCR.a
| Province | Location | Colony | Bat species (English name) | Year | Diet | PCR P/T (%) |
|---|---|---|---|---|---|---|
| Yunnan | Zhenyuan | C1 | Hipposideros pomona (Pomona roundleaf bat) | 2013 | I | 4/40(10) |
| Mengyuan | C2 | Hipposideros armiger (Great roundleaf bat) | 2013 | I | 0/40(0) | |
| Lincang | C3 | Rousettus leschenaultia (Leschenault's rousette) | 2013 | F | 0/42(0) | |
| Subtotal | 4/122(3.3) | |||||
| Guangxi | Baise | C4 | Hipposideros pomona (Pomona roundleaf bat) | 2014 | I | 3/32(9.4) |
| Guilin | C5 | Hipposideros cineraceus (Ashy roundleaf bat) | 2015 | I | 0/55(0) | |
| C6 | Hipposideros pomona (Pomona roundleaf bat) | 2015 | I | 0/15(0) | ||
| Hipposideros pratti (Pratt's roundleaf bat) | 2015 | I | 0/1(0) | |||
| Liuzhou | C7 | Hipposideros pomona (Pomona roundleaf bat) | 2015 | I | 0/18(0) | |
| Laibin | C8 | Hipposideros pomona (Pomona roundleaf bat) | 2015 | I | 0/25(0) | |
| Hechi | C9 | Rhinolophus pearsonii (Pearson's horseshoe bat) | 2015 | I | 0/6(0) | |
| Rhinolophus pusillus (Least horseshoe bat) | 2015 | I | 0/9(0) | |||
| Rhinolophus thomasi (Thomas's horseshoe bat) | 2015 | I | 0/5(0) | |||
| C10 | Rhinolophus hipposideros (Lesser horseshoe bat) | 2015 | I | 0/15(0) | ||
| C11 | Hipposideros armiger (Great roundleaf bat) | 2015 | I | 2/3(66.7) | ||
| Hipposideros pomona (Pomona roundleaf bat) | 2015 | I | 0/39(0) | |||
| Congzuo | C12 | Hipposideros larvatus (Intermediate roundleaf bat) | 2015 | I | 0/22(0) | |
| Hipposideros armiger (Great roundleaf bat) | 2015 | I | 0/2(0) | |||
| C13 | Miniopterus schreibersi (Schreibers' long-fingered bat) | 2015 | I | 0/10(0) | ||
| Yulin | C14 | Miniopterus australis (Little long-fingered bat) | 2015 | I | 0/30(0) | |
| C15 | Miniopterus schreibersi (Schreibers' long-fingered bat) | 2015 | I | 0/29(0) | ||
| Subtotal | 5/277(1.8) | |||||
| Guangdong | Luoding | C16 | Rousettus lechenaultia (Leschenault's rousette) | 2005 | F | 0/38(0) |
| C17 | Cynopteru sphinx (Short-nosed fruit bat) | 2005 | F | 0/18(0) | ||
| Wengyuan | C18 | Hipposideros larvatus (Intermediate roundleaf bat) | 2005 | I | 0/40(0) | |
| Yunfu | C19 | Hipposideros larvatus (Intermediate roundleaf bat) | 2005 | I | 0/38(0) | |
| Yangchun | C20 | Hipposideros larvatus (Intermediate roundleaf bat) | 2005 | I | 0/21(0) | |
| Subtotal | 0/155(0) | |||||
| Total | 9/554 (1.6) |
Abbreviations: P/T (%), numbers of positive/tested (percentage); F, frugivorous; I, insectivorous.
2.2. Virus detection and full genome amplification
To screen for orthohepadnaviruses, viral DNA was extracted from liver tissue of each of the 554 bats using the QIAamp DNA Mini Kit (Qiagen) in a QIAcube (Qiagen).Virus detection was conducted using the TaKaRa PCR Kit (TaKaRa) as previously described (He et al., 2013). Double-distilled water was used as a negative control. Two samples of each lineage were chosen for amplification of their complete genomes using LA Taq polymerase (TaKaRa) and previously described primers (He et al., 2013). Positive PCR amplicons were ligated into pMD18T vector (TaKaRa) and used to transfect competent Escherichia coli DH5α cells (Tiangen). >3 clones of each amplicon were randomly picked for sequencing by the Sanger method in an ABI 3730 sequencer (ComateBio). Overlapping amplicons were assembled with SeqMan v7.0 into full genomic sequences.
2.3. Genomic characterization and phylogenetic characterization
Genomic structures were determined by Vector NTI v.8, followed by comparison with those of other orthohepadnaviruses. Full genomes and predicted open-reading frames (ORF) Pol, preS1/S2/S, preC/C, and X (only for orthohepadnaviruses) of representatives of hepadnaviruses, including unclassified ones, retrieved from Genbank were aligned with counterparts of viruses identified in this study using MAFFT v7.394 (Katoh and Standley, 2013), with nt identities being calculated using MegAlign v7.1 (DNASTAR, Inc., Madison, WI). Evolutionary models were determined using ModelGenerator v0.85 with Akaike Information Criterion 1(AIC1) (Keane et al., 2006). Phylogenetic and molecular evolutionary analyses were achieved using PhyML v3.3 by the maximum likelihood and best-fit substitution models under evaluation of 100 bootstraps (Guindon et al., 2010). The generated tree files were visualized with FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree).
2.4. Genbank accession numbers
Full genomic sequences obtained here were deposited in Genbank under the accession numbers KY905324-KY905329.
3. Results
3.1. Sample collection and viral detection
During 2005–2015, 554 frugivorous and insectivorous bats representing 13 species were collected from 1 to 3 adjacent colonies in 11 locations in Yunnan, Guangxi and Guangdong provinces (Fig. 1). Collection details are shown in Table 1. Pan-orthohepadnavirus PCR screening gave positive results in 9 bat samples from 3 colonies. Of these, 10.0% (4/40) and 9.4% (3/32) were from Hipposideros pomona (H. pomona) bats in Zhenyuan, Yunnan province, 2013, and Baise, Guangxi province, 2014 respectively. The remainder (2/3) came from H. armiger bats in Hechi, Guangxi province, 2015 (Table 1). The amplicons were sequenced, and phylogenetic analysis revealed that they formed three distinct lineages. Of note is that the three lineages were from three different bat colonies. All BtHBVs in this study were named as described previously (He et al., 2014), the first two letters representing the sampling location, with the remaining letters identifying the host species and numbers referring to the sampling order.
3.2. Full genomic characterization
Full genomes of six samples with two of each lineage were successfully sequenced. Analyses showed that the 6 full genomes ranged from 3254 to 3275 nt in length, consistent with previously published BtHBVs (3230–3287 nt) in Asia, including PEPRs in Pu′er and Rs3364 in Jinning, Yunnan, China, and 776 in Kachin state, Myanmar (He et al., 2013). As in all orthohepadnaviruses, the viruses identified here contained four ORFs: polymerase (Pol), surface (S), core (C), and X. Positions of all ORFs in these bat orthohepadnaviruses were similar to ORFs of currently reported members of the Orthohepadnavirus genus but were clearly distinct from the ORFs of Avihepadnavirus. The S protein genes encoded in the large ORF contained preS1, preS2, and S domains. The preS1 domain contained a signal of N-myristoylation at glycine-2. Additional to their ORF organization, human HBV and the BtHBVs shared a similar location for the direct repeat (DR) sequences DR1 and DR2 involved in genome replication. Secondary structure prediction highlighted the structural similarities between BtHBVs and HBVs of other animal origin in their ε-loops, which serve as templates for the priming of reverse transcription of pre-genomic RNA in all hepadnaviruses (Seeger et al., 2013).
3.3. Phylogenetic analysis and sequence comparison
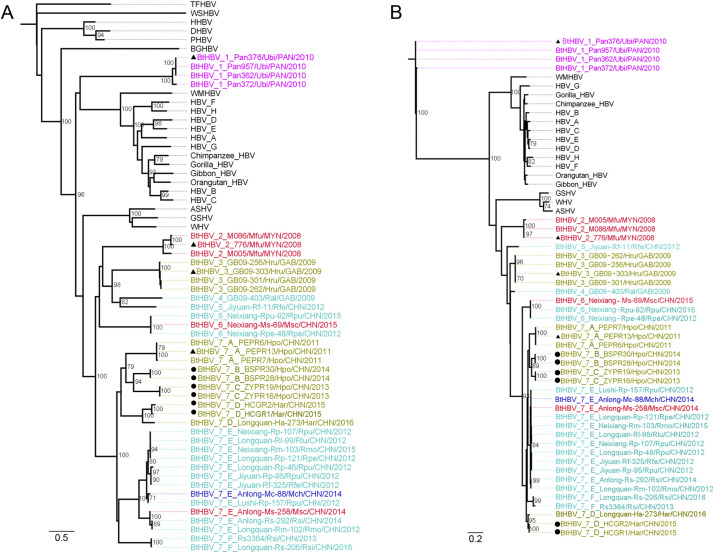
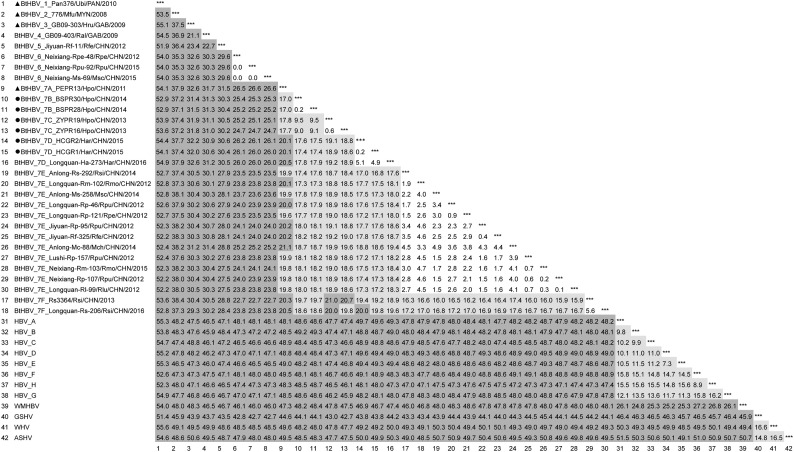
All representatives of hepadnaviruses available in Genbank were retrieved and aligned with the sequences obtained here. The best-fit substitution models, under AIC1, for full genomes, ORFs Pol, preS1/S2/S, preC1/C and X were selected as GTR + G [Gamma distribution parameter alpha (a) =0.61], TVM + G (a = 0.61), GTR + G (a = 0.62), GTR + I + G (I = 0.05, a = 0.63), and TVM + G (a = 0.45), respectively. Phylogenetic trees based on the full genomic and ORF nucleotide sequences are shown in Fig. 2, Fig. 3, Fig. 4 , with the calculated full genomic divergence shown in Fig. 5 . Full genomic analyses show that all BtHBVs are classified within the genus Orthohepadnavirus, and cluster within two groups, small one consisting of a clade of viruses (BtHBV 1) found in Panamanian bats Uroderma bilobatum (U. bilobatum) that show a similar relationship (51.4–55.6% divergence) with rodent and primate orthohepadnaviruses, and with the larger group (52.2–55.1%) consisting of BtHBVs from Myanmar, Gabon and China (Drexler et al., 2013; He et al., 2013, 2015; Nie et al., 2018; Wang et al., 2017) (Fig. 2, Fig. 5). The larger group can be divided into 6 clades (BtHBVs 2–7) according to divergence limits used by ICTV as species demarcation criteria (Mason et al., 2011) (Fig. 2, Fig. 5). BtHBV 2 consists of a lineage of viruses from Myanmar M. fuliginosus bats (He et al., 2013) (Fig. 2). BtHBV 3 and 4 comprise viruses with a divergence of 21.1% isolated from Gabonese H. cf. ruber and R. alcyone bats, respectively (Drexler et al., 2013) (Fig. 2, Fig. 5). BtHBV 5 consists of one isolate, Jiyuan-Rf-11 from Chinese R. ferrumequinum bats, showing 22.7–23.4% divergence with Gabonese viruses (Drexler et al., 2013; Wang et al., 2017) (Fig. 2, Fig. 5). BtHBV 6 is composed of viruses from a single location in China, but from two bat species, R. pearsonii and R. pusillus (Nie et al., 2018) (Fig. 2). BtHBV 7 has diverse members from a variety of bat species in widely spread geographic locations in China, such as the PEPRs from H. pomona in Yunnan, Longquan-Ha-273 from H. armiger in Zhejiang, and Neixiang-Rm-103 from R. monoceros in Henan (He et al., 2015; Nie et al., 2018) (Fig. 2).This clade can be further divided into 6 lineages (A to F) according to 6% divergence that has been used to differentiate genotypes of human HBVs (Norder et al., 1994) (Fig. 2, Fig. 5). The six full genomes of bat viruses described here clustered within three different lineages (B to D) with <06% intra- and 9.0–19.1% inter-lineages divergence within the clade BtHBV 7. The full genomes of the BSPRs (lineage B) from Baise, Guangxi, were much closer (~9.0% divergence) to the ZYPRs (lineage C) from Zhenyuan, Yunnan, than to the HCGRs (lineage D, 17.5% divergence) from Hechi, Guangxi, which showed ~19% divergence from the ZYPRs (Fig. 5). Compared with other viruses, the BSPRs and the ZYPRs were very similar, showing 17.3–19.7% divergences with other Chinese BtHBVs within the clade BtHBV 7, such as the PEPRs (lineage A), Anlong-Ms-258 (lineage E), and Rs3364 (lineage F) (Fig. 5), followed by 25.3–52.9% with members of BtHBV 1–6 (Fig. 5), then >44.6% with rodent and primate orthohepadnaviruses (Fig. 5). The HCGRs formed lineage D with Longquan-Ha-273, with the least divergence of 5% between each other (Fig. 2, Fig. 5), and also shared the same host species, H. armiger, but in two locations >1300 km apart (Nie et al., 2018) (Fig. 1), then 16.8–19.8% with other lineages within the BtHBV7 (Fig. 2, Fig. 5). The ORFs of Pol, preS1/S2/S, preC/C, and X were also used to validate the phylogenetic grouping. Generally, the Pol, preS1/S2/S, preC/C genes showed very similar topologies to the full genomes, and all BtHBVs can clearly be divided into 7 clades with BtHBV 7 containing 6 lineages (Fig. 2, Fig. 3, Fig. 4A). Due to the lack of X gene in other hepadnaviruses, the topology of orthohepadnaviruses differed from other trees, with BtHBV1s placing as an outgroup from other orthohepadnaviruses, and with the remaining BtHBVs forming 6 clades (Fig. 4B). Unlike other trees within which BtHBV 4 and 5 shared an evolutionary root, BtHBV 5 in the X gene tree formed an outgroup to BtHBVs 3, 4, 6 and 7, indicating a different evolutionary rate of X gene (Fig. 4B).
Fig. 2.
Phylogenetic analysis of BtHBVs and representatives of other hepadnaviruses based on their full genomic nucleotide sequences. According to the divergence of 20% used by ICTV as a species demarcation criterion, all BtHBVs can be divided into 7 clades (BtHBVs 1–7) with BtHBV 7 being classified into 6 lineages. Viruses isolated from the same host genus are highlighted in the same color. Circles: viruses identified in this study; triangles: phenotypes of 4 bat HBV species approved by the ICTV. The scale bar indicates nucleotide substitutions per site.
Fig. 3.
Topological comparison of phylogenetic trees of Pol (left) and preS1/S2/S (right) genes. Viruses isolated from the same host genus are highlighted in the same color. Circles: viruses identified in this study; triangles: phenotypes of 4 bat HBV species approved by the ICTV. The scale bar indicates nucleotide substitutions per site.
Fig. 4.
Phylogenetic analyses of preC/C (A) and X (B) genes. Viruses isolated from the same host genus are highlighted in the same color. Circles: viruses identified in this study; triangles: phenotypes of 4 bat HBV species approved by the ICTV. The scale bar indicates nucleotide substitutions per site.
Fig. 5.
Full genomic identity comparison of BtHBVs, and human- and rodent HBVs. Strains that showed little divergence (<1%) with phenotypes of 4 bat HBV species (triangles) are excluded. Dark gray: species with divergences within the ≥20% cutoff. Light gray: species with divergences of between 6% and 20%. Circles: viruses identified in this study. ***: not available.
4. Discussion
Until very recently, the family Hepadnaviridae contains two genera, Avihepadnavirus and Orthohepadnavirus, the former comprising three species and the latter having 8 members (ICTV, 2017). The species demarcation criteria in the genus Orthohepadnavirus are somewhat indefinite: for example, the full genome sequence divergence of WHV/HBV is 40%, GHSV/WHV 15%, WMHBV/HBV 20%, and WMHBV/WHV 30%, but the genetic divergences between different species have tentatively been set at ≥20% (ICTV, 2017; Mason et al., 2011). Recently a virus found in the white sucker fish (White sucker hepatitis B virus, WSHBV) has also been approved as a new species (White sucker hepatitis B virus), but unassigned to any genus (Hahn et al., 2015). Additionally, two other hepadnaviruses have been identified in fish (Bluegill hepadnavirus, BGHBV) and amphibians (Tibetan frog hepadnavirus, TFHBV), with both showing a high genetic diversity compared with other prototypes, and might represent totally new genus candidates (Dill et al., 2016). Currently viral nomenclature within the family follows the conventional tradition: hepatitis B virus (for avian, primate and chiropteran viruses) or hepatitis virus (for rodent viruses) prefixed by the host English name, e.g., duck hepatitis B virus (DHBV), woolly monkey hepatitis B virus (WMHBV), pomona bat hepatitis B virus (PBHBV), and woodchuck hepatitis virus (WHV). With increasing numbers of BtHBVs being reported, the current nomenclature cannot sustain the expansion of diversity of BtHBVs. Long-fingered bats and roundleaf bats are members of two genera, Miniopterus and Hipposideros, with each having >10 species (Wilson and Reeder, 2005). Within the Miniopterus spp. bats, not only do M. fuliginosus harbor orthohepadnaviruses (e.g. 776) (He et al., 2013), but M. schreibersi from China also carry such a virus – Neixiang-Ms-69 (Nie et al., 2018). However, Neixiang-Ms-69 only shows a distant relationship with isolate 776, with 35.3% divergence (Fig. 5); i.e., with a sufficient difference to separate them into two species. Similarly, GB09–303, the prototype of RBHBV, shares the same host genus (Hipposideros spp, roundleaf bats) as the HCGRs, but with ~32% divergence –i.e., much lower than the species demarcation criterion (20%) – indicating that they are members of two different species. The same situation has also been observed in Rhinolophus spp. bats (horseshoe bats) (Fig. 2), which harbor diverse orthohepadnaviruses showing 22.7%–31.7% divergences that have been grouped into 4 clades, BtHBV 4–7. Accordingly, unlike non-bat HBVs that can use the nomenclature of HBV or HV specified by host common English name to clearly distinguish the virus species, use of names such as LBHBV, RBHBV or HBHBV cannot precisely provide a match to a single viral species, which results in confusion in our understanding of the evolution and taxonomy of BtHBVs. Here we propose – and have used – a new nomenclature: BtHBV for bat-associated hepatitis B virus-like viruses, with different species marked by number, and with the strain name containing the species abbreviation, identifier, host species abbreviation, country, and collection time, so that all essential information about a virus can be clearly read from its name. Accordingly, presently named species have been changed to BtHBVs 1–3 and 7 (Table 2 , Fig. 2). With increasing numbers of orthohepadnaviruses being discovered in Chinese bats, 3 additional clades can be assigned showing >20% divergence: namely BtHBV 4–6, containing new species candidates. The 3 lineages of viruses identified here all fell into BtHBV 7 as variants.
Table 2.
Proposed species names and details of their corresponding strains.a
| Proposed species name | Strain | Previous species name | Genbank accession # | Reference |
|---|---|---|---|---|
| Bat hepatitis B virus 1 | BtHBV1/Pan372/Ubi/PAN/2010 | Tent-making bat hepatitis B virus | NC024445 | Drexler et al., 2013 |
| Bat hepatitis B virus 1 | BtHBV1/Pan362/PAN/2010 | Tent-making bat hepatitis B virus | KC790380 | Drexler et al., 2013 |
| Bat hepatitis B virus 1 | BtHBV1/Pan376/Ubi/PAN/2010 | Tent-making bat hepatitis B virus | KC790379 | Drexler et al., 2013 |
| Bat hepatitis B virus 1 | BtHBV1/Pan957/Ubi/PAN/2010 | Tent-making bat hepatitis B virus | KC790381 | Drexler et al., 2013 |
| Bat hepatitis B virus 2 | BtHBV2/M086/Mfu/MYN/2008 | Long-fingered bat hepatitis B virus | JX941468 | He et al., 2013 |
| Bat hepatitis B virus 2 | BtHBV2/776/Mfu/MYN/2008 | Long-fingered bat hepatitis B virus | NC020881 | He et al., 2013 |
| Bat hepatitis B virus 2 | BtHBV2/M005/Mfu/MYN/2008 | Long-fingered bat hepatitis B virus | JX941467 | He et al., 2013 |
| Bat hepatitis B virus 3 | BtHBV3/GB09–301/Hru/GAB/2009 | Roundleaf bat hepatitis B virus | KC790375 | Drexler et al., 2013 |
| Bat hepatitis B virus 3 | BtHBV3/GB09–303/Hru/GAB/2009 | Roundleaf bat hepatitis B virus | KC790376 | Drexler et al., 2013 |
| Bat hepatitis B virus 3 | BtHBV3/GB09–262/Hru/GAB/2009 | Roundleaf bat hepatitis B virus | KC790374 | Drexler et al., 2013 |
| Bat hepatitis B virus 3 | BtHBV3/GB09–256/Hru/GAB/2009 | Roundleaf bat hepatitis B virus | NC024443 | Drexler et al., 2013 |
| Bat hepatitis B virus 4 | BtHBV4/GB09–403/Ral/GAB/2009 | NA | NC024444 | Drexler et al., 2013 |
| Bat hepatitis B virus 5 | BtHBV5/Jiyuan-Rf-11/Rfe/CHN/2012 | NA | KY962687 | Nie et al., 2018 |
| Bat hepatitis B virus 6 | BtHBV6/Neixiang-Rpe-48/Rpe/CHN/2012 | NA | MG457483 | Nie et al., 2018 |
| Bat hepatitis B virus 6 | BtHBV6/Neixiang-Rpu-92/Rpu/CHN/2015 | NA | MG457484 | Nie et al., 2018 |
| Bat hepatitis B virus 6 | BtHBV6/Neixiang-Ms-69/Msc/CHN/2015 | NA | MG457480 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/PEPR6/Hpo/CHN/2011 | Pomona bat hepatitis B virus | KF939648 | He et al., 2015 |
| Bat hepatitis B virus 7 | BtHBV7/PEPR13/Hpo/CHN/2011 | Pomona bat hepatitis B virus | KF939650 | He et al., 2015 |
| Bat hepatitis B virus 7 | BtHBV7/PEPR7/Hpo/CHN/2011 | Pomona bat hepatitis B virus | KF939649 | He et al., 2015 |
| Bat hepatitis B virus 7 | BtHBV7/BSPR30/Hpo/CHN/2014 | NA | KY905329 | In this study |
| Bat hepatitis B virus 7 | BtHBV7/BSPR28/Hpo/CHN/2014 | NA | KY905328 | In this study |
| Bat hepatitis B virus 7 | BtHBV7/ZYPR19/Hpo/CHN/2013 | NA | KY905327 | In this study |
| Bat hepatitis B virus 7 | BtHBV7/ZYPR16/Hpo/CHN/2013 | NA | KY905326 | In this study |
| Bat hepatitis B virus 7 | BtHBV7/HCGR2/Har/CHN/2015 | NA | KY905325 | In this study |
| Bat hepatitis B virus 7 | BtHBV7/HCGR1/Har/CHN/2015 | NA | KY905324 | In this study |
| Bat hepatitis B virus 7 | BtHBV7/Longquan-Ha-273/Har/CHN/2016 | NA | MG457473 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/BtHBVRs3364/Rsi/CHN/2013 | NA | KX513949 | Wang et al., 2017 |
| Bat hepatitis B virus 7 | BtHBV7/Longquan-Rs-2064/Rsi/CHN/2016 | NA | MG457479 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Anlong-Rs-292/Rsi/CHN/2014 | NA | MG457469 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Longquan-Rm-102/Rmo/CHN/2012 | NA | KY962697 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Anlong-Ms-258/Msc/CHN/2014 | NA | MG457468 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Longquan-Rp-46/Rpu/CHN/2012 | NA | KY962700 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Longquan-Rp-121/Rpe/CHN/2012 | NA | KY962699 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Jiyuan-Rp-95/Rpu/CHN/2012 | NA | KY962695 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Jiyuan-Rf-325/Rfe/CHN/2012 | NA | KY962693 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Anlong-Mc-88/Mch/CHN/2014 | NA | MG457467 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Lushi-Rp-157/Rpu/CHN/2012 | NA | KY962701 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Neixiang-Rm-103/Rmo/CHN/2015 | NA | MG457481 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Neixiang-Rp-107/Rpu/CHN/2012 | NA | KY962704 | Nie et al., 2018 |
| Bat hepatitis B virus 7 | BtHBV7/Longquan-Rl-99/Rlu/CHN/2012 | NA | KY962696 | Nie et al., 2018 |
NA: not approved as species currently; Genbank accession #: Genbank accession number.
Hepadnaviruses have long been limited to primates, rodents and birds, but diverse hepadnaviruses have recently been found in bats from both Old and New Worlds. With orthohepadnaviruses increasingly being reported in bats in China The increasing numbers of Chinese BtHBVs being reported (He et al., 2015; Nie et al., 2018; Wang et al., 2017), are highly divergent genetically and distributed within all 3 of the proposed species, BtHBVs 5–7 (Fig. 2, Fig. 3, Fig. 4, Fig. 5). Such divergence is well reflected in BtHBV 7. To better understand the evolutionary dynamics of human HBVs, HBVs have been classified into genotypes A-J according to a divergence cutoff of 6% (Miyakawa and Mizokami, 2003; Olinger et al., 2008; Tatematsu et al., 2009). Similarly, BtHBV 7 can also be divided into 6 lineages based on the same cutoff. Accordingly here we propose 6 genotypes within BtHBV 7, and viruses found here constitute genotypes B, C and D (Fig. 2). All these findings indicate that orthohepadnaviruses in Chinese bats are much more diverse than has been expected and, undoubtedly, more new viruses or variants will be discovered in these creatures, which will be helpful in understanding their origin and evolution.
HBVs are transmissible not only among humans but can also infect certain other primates, such as chimpanzees and macaques. Laboratory infection of the tree shrew (Tupaia belangeri) has also been reported (Seeger and Mason, 2015), which suggests possible inter-species infection and transmission. It appears that this is also the case for BtHBVs, since some viruses within a species have been detected in distinctly different host species. For example, members of BtHBV 7 have been detected in bats of 10 species within 4 families (Fig. 2), and even within BtHBV 6 there are 3 members with almost no difference but isolated from R. perarsonii and R. pusillus of the genus Rhinopophus, family Rhinolophidae, and M. schreibersi, genus Miniopterus, family Miniopteridae (Fig. 2), which indicates that BtHBVs can jump the host barrier to circulate between different bat species. However, the HCGRs of H. armiger bats in Hechi do not appear to have infected cave-sharing H. pomona bats that are hosts to a number of BtHBVs, such as the PEPRs, BSPRs, and ZYPRs, even though both bat species are members of the genus Hipposideros. This may reflect potential host/genetic species barriers, although we cannot exclude the possibility that ecological reasons may prevent this cross-species transmission and/or that the limited sample size and survey duration has prevented the identification of positive individuals. All these observations suggested that the circulation of orthohepadnaviruses in bat population is very intricate. Understanding its complexity will require a study of viruses rescued from different infected hosts and epidemiological investigations of orthohepadnaviruses over a broader range of bat species, wider areas and of longer duration.
Acknowledgments
The study was supported by the General Program of the National Natural Science Foundation of China (31572529), the National Key Basic Research and Development Program of China (2016YFC1200100, 2017YFD0501800).
The funders played no role in study design, data collection and interpretation, or the decision to submit the work for publication.
References
- Dill J.A., Camus A.C., Leary J.H., Giallonardo F.D., Holmes E.C., Ng T.F.F. Distinct viral lingeages from fish and amphilians reveal the complex evolutionary history of hepadnaviruses. J. Virol. 2016;90:7920–7933. doi: 10.1128/JVI.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Seelen A., Corman V.M., Fumie Tateno A., Cottontail V., Melim Zerbinati R., Gloza-Rausch F., Klose S.M., Adu-Sarkodie Y., Oppong S.K., Kalko E.K.V., Osterman A., Rasche A., Adam A., Müller M.A., Ulrich R.G., Leroy E.M., Lukashev A.N., Drosten C. Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae. J. Virol. 2012;86:9134–9147. doi: 10.1128/JVI.00800-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Geipel A., Konig A., Corman V.M., van Riel D., Leijten L.M., Bremer C.M., Rasche A., Cottontail V.M., Maganga G.D., Schlegel M., Muller M.A., Adam A., Klose S.M., Carneiro A.J., Stocker A., Franke C.R., Gloza-Rausch F., Geyer J., Annan A., Adu-Sarkodie Y., Oppong S., Binger T., Vallo P., Tschapka M., Ulrich R.G., Gerlich W.H., Leroy E., Kuiken T., Glebe D., Drosten C. Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 2013;110:16151–16156. doi: 10.1073/pnas.1308049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Lukashev A.N., van den Brand J.M.A., Gmyl A.P., Brünink S., Rasche A., Seggewiβ N., Feng H., Leijten L.M., Vallo P., Kuiken T., Dotzauer A., Ulrich R.G., Lemon S.M., Drosten C., The Hepatovirus Ecology Consortium Evolutionary origins of hepatitis A virus in small mammals. Proc. Natl. Acad. Sci. U. S. A. 2015;112:15190–15195. doi: 10.1073/pnas.1516992112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimates maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hahn C.M., Iwanowicz L.R., Cornman R.S., Conway C.M., Winton J.R., Blazer V.S. Characterization of a novel hepadnavirus in the white sucker (Catostomus commersonii) from the great lakes region of the United States. J. Virol. 2015;89:11801–11811. doi: 10.1128/JVI.01278-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Fan Q., Yang F., Hu T., Qiu W., Feng Y., Li Z., Li Y., Zhang F., Guo H., Zou X., Tu C. Hepatitis virus in long-fingered bats, Myanmar. Emerg. Infect. Dis. 2013;19:638–640. doi: 10.3201/eid1904.121655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zhang Y., Xu L., Yang W., Yang F., Feng Y., Xia L., Zhou J., Zhen W., Feng Y., Guo H., Zhang H., Tu C. Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China. J. Virol. 2014;88:7070–7082. doi: 10.1128/JVI.00631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Zhang F., Xia L., Hu T., Chen G., Qiu W., Fan Q., Feng Y., Guo H., Tu C. Identification of a novel Orthohepadnavirus in pomona roundleaf bats in China. Arch. Virol. 2015;160:335–337. doi: 10.1007/s00705-014-2222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICTV The 10th report of International Committee on Taxonomy of Viruses. 2017. https://talk.ictvonline.org/taxonomy/
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T.M., Creevey C.J., Pentony M.M., Naughton T.J., McLnerney J.O. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol. Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason W.S., Gerlich W.H., Taylor J.M., Kann M., Mizokami T., Loeb D., Sureau C., Magnius L., Norder H. Family Hepadnaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth report of the International Committee on Taxonomy of Viruses. Academic Press; London, U. K.: 2011. pp. 445–455. [Google Scholar]
- Miyakawa Y., Mizokami M. Classifying hepatitis B virus genotypes. Intervirology. 2003;46:329–338. doi: 10.1159/000074988. [DOI] [PubMed] [Google Scholar]
- Moratelli R., Calisher C.H. Bats and zoonotic viruses: can we confidentily link bats with emerging deadly viruses? Mem. Inst. Oswaldo Cruz. 2015;110:1–22. doi: 10.1590/0074-02760150048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie F.-Y., Lin X.-D., Hao Z.-Y., Chen X.-N., Wang Z.-X., Wang M.-R., Wu J., Wang H.-W., Zhao G., Ma R.Z., Holmes E.C., Zhang Y.-Z. Extensive diversity and evolution of hepadnaviruses in bats in China. Virology. 2018;514:88–97. doi: 10.1016/j.virol.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norder H., Courouce A.M., Magnius L.O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- Olinger C.M., Jutavijittum P., Hubschen J.M., Yousukh A., Samountry B., Thammavong T., Toriyama K., Muller C.P. Possible new hepatitis B virus genotype, southeast Asia. Emerg. Infect. Dis. 2008;14:1777–1780. doi: 10.3201/eid1411.080437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott J.J., Stevens G.A., Groeger J., Wiersma S.T. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- Quan P.L., Firth C., Conte J.M., Williams S.H., Zambrana-Torrelio C.M., Anthony S.J., Ellison J.A., Gilbert A.T., Kuzmin I.V., Niezgoda M., Osinubi M.O., Recuenco S., Markotter W., Breiman R.F., Kalemba L., Malekani J., Lindblade K.A., Rostal M.K., Ojeda-Flores R., Suzan G., Davis L.B., Blau D.M., Ogunkoya A.B., Alvarez Castillo D.A., Moran D., Ngam S., Akaibe D., Agwanda B., Briese T., Epstein J.H., Daszak P., Rupprecht C.E., Holmes E.C., Lipkin W.I. Bats are a major natural reservoir for hepaciviruses and pegiviruses. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8194–8199. doi: 10.1073/pnas.1303037110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasche A., Souza B.F., de C.D., Drexler J.F. Bat hepadnaviruses and the origins of primate hepatitis B viruses. Curr. Opin. Virol. 2016;16:86–94. doi: 10.1016/j.coviro.2016.01.015. [DOI] [PubMed] [Google Scholar]
- Seeger C., Mason W.S. Molecular biology of hepatitis B virus infection. Virology. 2015;479-480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C., Zoulim F., Mason W.S., Knipe D.M., Howley P.M. Fields Virology. 6 ed. Lippincott Williams & Wilkins; Philadelphia,PA: 2013. Hepadnaviruses; pp. 2185–2221. [Google Scholar]
- Tatematsu K., Tanaka Y., Kurbanov F., Sugauchi F., Mano S., Maeshiro T., Nakayoshi T., Wakuta M., Miyakawa Y., Mizokami M. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J. Virol. 2009;83:10538–10547. doi: 10.1128/JVI.00462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liang B., Feng J., Sheng L., Zhang S. Molecular phylogenetic of Hipposiderids (Chiroptera: Hipposideridae) and Rhinolophids (Chiroptera: Rhinolophidae) in China based on mitochondrial cytochrome b sequences. Folia Zool. 2003;52:259–268. [Google Scholar]
- Wang B., Yang X.L., Li W., Zhu Y., Ge X.Y., Zhang L.B., Zhang Y.Z., Bock C.T., Shi Z.L. Detection and genome characterization of four novel bat hepadnaviruses and a hepevirus in China. Virol. J. 2017;14:40. doi: 10.1186/s12985-017-0706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Hepatitis B. 2017. http://www.who.int/mediacentre/factsheets/fs204/en/
- Wilson, D.E., Reeder, D.M., 2005. Mammal Species of the World. A Taxomomic and Geographic Reference, 3 ed. Johns Hopkins University Press, Batlimore, Maryland.