Highlights
-
•
Click chemistry describes reactions used to generate substances by joining small units together with heteroatom linkages.
-
•
Biosensors combined with analytical devices is convenient strategy for viral detection.
-
•
We reviewed the recent applications of click reactions in virus-related research.
-
•
This review provides an overview of the general principles and applications of click chemistry in virus-related research.
Keywords: Click chemistry, Virus, Labeling, Biosensor, Delivery, Diagnosis
Abstract
Click chemistry involves reactions that were originally introduced and used in organic chemistry to generate substances by joining small units together with heteroatom linkages (C-X-C). Over the last few decades, click chemistry has been widely used in virus-related research. Using click chemistry, the virus particle as well as viral protein and nucleic acids can be labeled. Subsequently, the labeled virions or molecules can be tracked in real time. Here, we reviewed the recent applications of click reactions in virus-related research, including viral tracking, the design of antiviral agents, the diagnosis of viral infection, and virus-based delivery systems. This review provides an overview of the general principles and applications of click chemistry in virus-related research.
1. Introduction
Click chemistry describes reactions originally introduced and used in organic chemistry to generate substances by joining small units together with heteroatom linkages (C-X-C) (Kolb et al., 2001). Over the last few decades, click chemistry has been widely used in bioscience fields, such as chemical biology, drug development and bionanoparticles, as a promising tool to modify biomolecules, such as DNA, protein and virions (Ahmad Fuaad et al., 2013; Avti et al., 2013; Jewett and Bertozzi, 2010; Kolb et al., 2001; Li et al., 2013; Liu et al., 2017; Moses and Moorhouse, 2007; Thirumurugan et al., 2013; Yi et al., 2018).
There are two main steps in the reaction to efficiently synthesize biomolecules or substances by joining substrates of small molecules with specific biomolecules: 1) biomolecules A and B are labeled with azide and alkyne functional groups via click chemistry, respectively, and 2) azide-labeled molecule A and alkyne-labeled molecule B are conjugated to form a stable conjugate (Himo et al., 2005; Kolb et al., 2001; Liu et al., 2017; Moses and Moorhouse, 2007; Salic and Mitchison, 2008). Copper-catalyzed alkyne-azide cycloaddition (CuAAC) is one of the most widely used click reactions, which reacts efficiently at room temperature and is stable to most functional groups to generate stable products (Kolb et al., 2001; Moses and Moorhouse, 2007). To label cells in situ using CuAAC, an azide or alkyne is conjugated to a biomolecule in the cell, such as a nucleic acid, nucleoside, amino acid, monosaccharide or fatty acid, which is termed the biosynthetic incorporating reaction (Amblard et al., 2009; Salic and Mitchison, 2008). Subsequently, the complementary alkyne or azide labeled with the reporter group is linked with the biomolecules via click chemistry in the presence of catalytic copper(I) (Amblard et al., 2009; Salic and Mitchison, 2008). However, although the CuAAC reaction is effective for bioconjugation, the reaction is limited in live cells due to that the copper, as a catalyst, is cytotoxic. Thus, some copper-free click chemistry methods have been developed, such as Cu-free alkyne-azide cycloaddition (Jewett et al., 2010), strain-promoted alkyne-azide cycloaddition (SPAAC) (Sachin et al., 2012), strain-promoted inverse-electron-demand Diels-Alder cycloaddition (SPIEDAC) (Nikic et al., 2014), the thiol–ene reaction (Hoyle and Bowman, 2010), etc.
Using click chemistry, fluorophores or other reporter molecules attach to the specific biomolecules, allowing the biomolecules to be identified, located and characterized. Recently, click chemistry has been widely used in virus-related research (Best, 2009; Bruckman et al., 2008; Cowan et al., 2012; Gao et al., 2012; Generous et al., 2014; Kalveram et al., 2013; Lu et al., 2015; Nwe and Brechbiel, 2009; Rubino et al., 2012; Wang et al., 2018). In this review, we will introduce the application of click chemistry in virus-related research, including viral tracking, the design of antiviral agents, the diagnosis of viral infection, and virus-based delivery systems. Furthermore, the advantages and disadvantages of click chemistry for virus-related research were also discussed. This review provides an overview of the general principles and applications of click chemistry in virus-related research.
2. Click chemistry in virus-related research
2.1. Viral tracking
To date, due to the lack of knowledge of viral infection, the pathogenesis of most viruses as well as the interaction between the virus and host cells is still unclear (Liu et al., 2017). Using click chemistry, viral protein and the virion can be clicked rapidly and quantitatively with small and highly stable tags, allowing the visualization of dynamic changes in labeled viral proteins or virions in cells and providing more knowledge of the interactions between the host cells and virus.
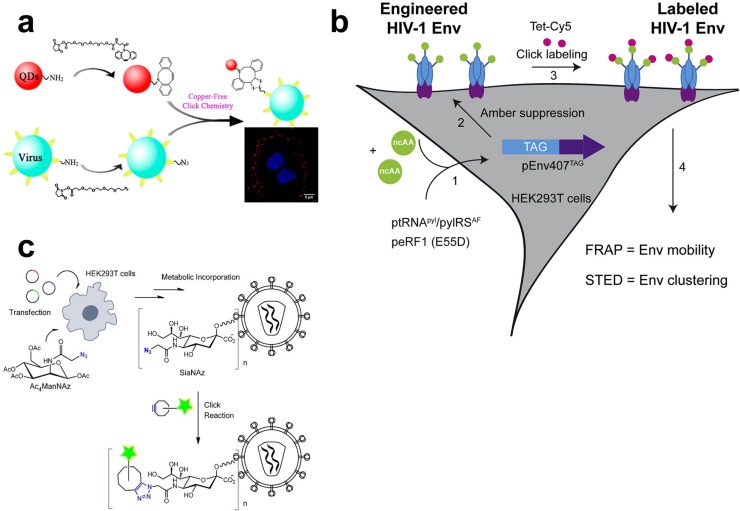
A classical click reaction for viral labeling is a protocol described by Bruckman (Bruckman et al., 2008). In this system, an alkyne group was quantitatively attached to the tyrosine residues on the surface of tobacco mosaic virus (TMV) via diazonium coupling and the CuAAC reaction (Bruckman et al., 2008). This study also proved that the CuAAC reaction did not depend on the structure of the starting materials and will not affect the function of the azide (Bruckman et al., 2008). However, copper-catalyzed ligation between Cu(II) and 1,2,3-triazoles has cytotoxicity and might affect the viral structure (Liu et al., 2017; Wang et al., 2003). Moreover, Cu(II) can greatly decrease the fluorescence of the quantum dots (QDs) (Beaune et al., 2011; Bernardin et al., 2010). Therefore, a copper-free reaction was used to label the vaccinia virus and avian influenza A virus in live cells with QDs by linking azide-clicked virions to the dibenzocyclooctyne-derived QDs, resulting in intact, fluorescence-labeled and infectious virions for single-virion tracking, with an 80% labeling efficiency (Fig. 1 a) (Hao et al., 2012). The fluorescence of labeled virions was strong enough for single-virion imaging and tracking (Hao et al., 2012).
Fig. 1.
Application of click chemistry in viral tracking. (a) Virions were labeled with quantum dots (QDs) via copper-free click chemistry by linking azide-clicked virions to the dibenzocyclooctyne-derived QDs, resulting in intact, fluorescence-labeled and infectious virions (Hao et al., 2012). (b) An amber (UAG) stop codon was inserted into the HIV Env protein by site-directed mutagenesis, followed by coexpression of the protein with an aminoacyl tRNA synthase/tRNA and noncanonical amino acid (ncAA) in the target cell (Fernandez and Freed, 2017; Sakin et al., 2017). Subsequently, the ncAA was incorporated at the amber stop codon, allowing the continuous translation through the amber stop codon (Fernandez and Freed, 2017; Sakin et al., 2017). When H-Tet-Cy5 was added, the Env was labeled between ncAA and tetrazine via a click reaction (Fernandez and Freed, 2017; Sakin et al., 2017). (c) Viral glycoprotein was labeled with a metabolically incorporated unnatural sugar (Ac4ManNAz), followed by a click reaction with organic fluorescent dyes, allowing virus-cell fusion to be visualized during the infection (Oum et al., 2016).
Recently, metabolic incorporation of a clickable group into viral protein was reported as another strategy to label a virus or viral protein for investigating the behavior of viral proteins (Fernandez and Freed, 2017; Lin et al., 2013; Nikic et al., 2014; Plass et al., 2012; Sakin et al., 2017). One protocol is based on genetic code expansion. First, an amber stop codon (UAG) was introduced into the target protein by site-directed mutagenesis (Fernandez and Freed, 2017; Nikic et al., 2014). Then, a noncanonical amino acid (ncAA) could be incorporated into the amber stop codon when the engineered protein was coexpressed with the aminoacyl tRNA synthase/tRNA pair in the host cell, resulting in translation continuing through the amber stop codon, which was designated as amber suppression (Fernandez and Freed, 2017; Nikic et al., 2014). As the ncAA bears a ring-strained alkyne or alkene, the ncAA is clicked to azide- or tetrazine-containing dyes via SPAAC or SPIEDAC reactions, and thus, the mobility of the target viral protein, such as HIV-1 Env, can be measured in a real-time manner (Fig. 1b) (Fernandez and Freed, 2017; Nikic et al., 2014; Sakin et al., 2017). Moreover, HIV-1 particles can be generated by adding eGFP-tagged Gag to the system, and therefore, the Env accumulating sites of particle budding can be visualized by stimulated emission depletion (STED) nanoscopy (Fernandez and Freed, 2017; Sakin et al., 2017). However, the efficiency of amber suppression used in this strategy was low (Fernandez and Freed, 2017; Plass et al., 2012; Sakin et al., 2017), which limits the incorporation of ncAA into the target protein. Another metabolic incorporation strategy describes the labeling of the viral membrane glycoprotein via metabolic incorporation of unnatural sugars (Ac4ManNAz) followed by a click-reaction with organic fluorescent dyes, allowing the visualization of the fusion of a single virus with cells during the infection (Fig. 1c) (Oum et al., 2016). The result showed that unnatural sugars did not affect membrane fusion, while it can interfere with the function of the Env (Oum et al., 2016). Moreover, the methionine analogue homopropargylglycine (HPG) was also used for incorporation into newly synthesized protein of the herpes simplex virus (HSV) using click chemistry (Su Hui Teo et al., 2016). Due to that the HPG can be selectively coupled by fluorochrome-capture reagents, the newly synthesized proteins can be tracked by a fluorescence microscope. The results showed that the newly synthesized proteins were rapidly accumulated in the nucleus and formed numerous subcompartments (Su Hui Teo et al., 2016). The immediate-early proteins ICP4 and ICP0 of HSV were excluded from these newly synthesized protein domains (NPDs), whereas ICP22 was selectively recruited in the NPDs, and the NPDs formed adjacent to promyelocytic leukemia (PML) domains (Su Hui Teo et al., 2016). These results indicated that NPDs recruit host and newly translated viral protein during the early stage of infection (Su Hui Teo et al., 2016).
In addition to viral protein, viral nucleic acids can also be labeled using click chemistry to track the dynamics of the nucleic acids. Newly generated viral RNA can be labeled by incorporating an uridine analogue, such as 5-ethynyluridine (EU), followed by clicking with a fluorescent azide (Hagemeijer et al., 2012; Kalveram et al., 2013). Thereafter, viral transcription can be visualized and quantified in the virus-infected cells through fluorescence microscopy and flow cytometry, respectively (Kalveram et al., 2013). It was found that newly synthesized viral RNA and nonstructural protein (NSP) were colocalized with dsRNA in the early stages of infection of Coronaviruses but not in the late stages of infection, indicating that dsRNA does not play a necessary role in activating viral RNA synthesis (Hagemeijer et al., 2012). However, both nascent viral RNA and host transcripts can be labeled using this protocol (Kalveram et al., 2013). To solve this problem, virus-infected cells can be treated with actinomycin D (ActD) to suppress host cell transcription, as ActD can inhibit DNA-dependent transcription by binding to dsDNA but has no effect on viral RNA-dependent RNA polymerases (Kalveram et al., 2013; Reich and Goldberg, 1964). It is noteworthy that the labeled viral RNA is sensitive to RNase, and therefore, it is critical to avoid contamination with nucleases throughout the entire process (Jao and Salic, 2008; Kalveram et al., 2013).
The protocols to label viral DNA are similar to the viral RNA labeling strategies. Using 5-ethynyl-2′-deoxyuridine (EdU)-labeled HSV-1 DNA, it was found that promyelocytic leukemia-nuclear bodies (PML-NBs) blocked viral replication by entrapping viral DNA during the entry of the viral genome into the nucleus (Alandijany et al., 2018). However, when the viral PML-NB antagonist ICP0 was added, interferon-stimulated gene (ISG) expression was induced in a PML-, interferon gamma inducible protein 16 (IFI16)-, and Janus-associated kinase (JAK)-dependent manner, while the ISG expression was inhibited by phosphonoacetic acid, indicating that viral DNA polymerase was involved in the induction of ISG expression (Alandijany et al., 2018). These results demonstrated that PML played dual roles in the regulation of intracellular host immunity, which depended on viral genome delivery and viral DNA polymerase, respectively (Alandijany et al., 2018). Senkevich and colleagues incorporated EdU into the nascent DNA of the vaccinia virus (VACV) in the virus-infected cells and found that the newly synthesized DNA was colocalized with the viral single-stranded DNA binding protein I3 in virus factories surrounded by the endoplasmic reticulum (Senkevich et al., 2017). Further research demonstrated that nuclear DNA synthesis stopped immediately after VACV infection, whereas the synthesis of viral DNA was enhanced in the virus factories of the cytoplasm (Senkevich et al., 2017). Viral proteins, host proteins (including proliferating cell nuclear antigen (PCNA) as well as topoisomerases IIα and IIβ) as well as nascent viral DNA were identified in the complexes, indicating that these proteins were required for poxvirus DNA replication (Senkevich et al., 2017). Unexpectedly, it was demonstrated that EdU has cytotoxic properties, which may be responsible for cell death and/or apoptosis (Cieslar-Pobuda and Los, 2013; Kohlmeier et al., 2013). Using a nucleoside analogue, the Komatsu group found a specific adenoviral DNA replication complex during the late phases of infection, which produced nascent viral genomes in virus-induced post-replication (ViPR) bodies (Komatsu et al., 2016). In addition, nucleolar protein Mybbp1a was identified as a molecular marker of ViPR bodies, which specifically reacted with viral core protein VII (Komatsu et al., 2016).
Moreover, a dual-labeled virus can be generated by labeling both viral nucleic acid and viral protein with different azides and alkynes using click chemistry. Huang and colleagues developed a universal and efficient dual labeling strategy for envelope viruses and capsid viruses, in which L-azidohomoalanine (AHA) and 5-vinyl-2′-deoxyuridine (VdU) were incorporated into the viral proteins and viral nucleic acids, respectively, followed by a copper-free click reaction or inverse electron demand Diels-Alder reaction (iEDDA) during the assembly of the progeny virions (Huang et al., 2017). The results showed that 80% of the progeny virions were dual-labeled and fully infectious, and their fluorescence was strong enough for tracking (Huang et al., 2017).
2.2. Development of an antiviral agent
Antiviral agents play important roles in inhibiting viral infection and curing the disease. However, many examples have proven that most viruses are increasingly resistant to the currently used antiviral agents. Therefore, the development of novel highly efficient antiviral agents is one of the urgent tasks for virologists and chemists.
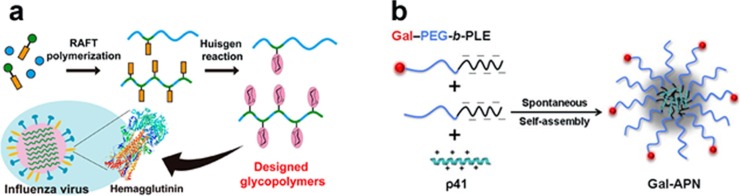
Based on click chemistry, many types of anti-influenza agents were developed with reliable effectiveness. Hemagglutinin (HA) and neuraminidase (NA) are important antigenic determinants on the surface of the influenza virus. Therefore, HA and NA are key targets for the development of anti-influenza agents. Nagao and coworkers designed glycopolymers carrying sialyl oligosaccharides by click chemistry and found that the glycopolymers can specifically recognize different types of influenza viruses (Fig. 2 a) (Nagao et al., 2017). Among the glycopolymers, GP4-6′ can significantly inhibit viral infection by interacting with viral hemagglutinin (Nagao et al., 2017). Excitingly, many groups reported that multivalent antibodies or ligands generated by click chemistry exhibited high anti-influenza activity. Multivalent interactions with multiple binding sites of the virus were found to be more efficient in inhibiting viral infection, and the lipidation of the polymer structures played an important role in inhibiting the viral infection (Nagao et al., 2017). Similarly, human serum albumin (HSA) and bovine serum albumin (BSA) conjugated with S-sialoside showed moderate NA inhibitory activity and higher affinity to HA due to the multivalent effect as well as optimized three-dimensional presentation of sialosides on the protein (Yang et al., 2016). To overcome the intrinsic limitations of the conventional influenza vaccine, gold nanoparticles (AuNPs) were conjugated with HA and flagellin (TLR5 agonist) by click chemistry, resulting in specific and stable conjugated proteins (Wang et al., 2017). The dual-conjugated AuNPs are effective in inducing a strong immune response in vitro and in mice, indicating that the proteins have effective immunogenicity and immunostimulation (Wang et al., 2017). Furthermore, per-O-methylated cyclodextrin (CD) derivatives functionalized with multivalent pentacyclic triterpene were synthesized using a 1,3-dipolar cycloaddition click reaction (Tian et al., 2017). Compound 28 showed the most potent anti-influenza activity by being tightly bound to influenza HA protein to inhibit the entry of the influenza virus into the host cell (Tian et al., 2017). Moreover, bispecific antibody conjugated with sortase via click chemistry has broad anti-influenza virus activity (Wagner et al., 2014). In addition, triazole derivatives synthesized by clicking a hydroxyl group or keto-group onto camphecene showed low cytotoxicity and high antiviral activity against influenza virus A/Puerto Rico/8/34 (H1N1) (Artyushin et al., 2017).
Fig. 2.
Application of click chemistry to evaluate antiviral agents. (a) A spacer (acrylamide, AAm) and alkyne (4-trimethylsilyl-3-butynyl acrylamide, TMS BtnAAm) were copolymerized by RAFT polymerization, followed by adding azidated sialyl oligosaccharides via "postclick" chemistry (Nagao et al., 2017). (b) Gal-APN was prepared by mixing cationic p41 and anionic copolymers (Gal-PEG-b-PLE and propargyl-PEG-b-PLE) (Zhang et al., 2015, 2013).
Hepatitis C virus (HCV) entry is a main target for the treatment of chronic HCV infection (McGowan et al., 2017; Xiao et al., 2014, 2016). To obtain an anti-HCV entry agent, a series of α- or β-cyclodextrin-pentacyclic triterpene conjugates labeled with water soluble triazole were synthesized via click chemistry (Xiao et al., 2014, 2016). The results showed that compounds α-cyclodextrin 15 and 18 displayed promising anti-HCV entry activities at the postbinding step, with average IC50 values of 1.18 μM and 0.25 μM, respectively (Xiao et al., 2016). To pursue an economical agent for HCV therapy, antiviral peptide nanocomplexes (APN) were galactosylated by click chemistry to generate Gal-APN (Zhang et al., 2015). Gal-APN exhibited prominent advantages to prevent HCV and suppress the intracellular expression of HCV proteins in vitro as well as preferential liver accumulation in vivo, indicating that Gal-APN is a potential liver-targeting therapeutic agent against HCV (Fig. 2b) (Zhang et al., 2015, 2013). To enhance cellular uptake and intracellular delivery, oligonucleotides were conjugated with lipid via click chemistry (Godeau et al., 2008). Further investigation showed that these lipid-conjugated oligonucleotides can efficiently inhibit the translation of the hepatitis C virus (Godeau et al., 2008).
The development of novel anti-HIV agents remains a tough challenge for medicinal chemistry, partly due to the rise in the nonnucleoside reverse transcriptase inhibitor (NNRTI) resistance of HIV (Pribut et al., 2016). Using 1,2,3-triazole as a new scaffold for novel NNRTIs, it was found that compound B5b7, a S-dihydro-alkoxy-benzyl-oxopyrimidine (DABO) derivative with a substituted 1,2,3-triazole moiety on the C-2 side chain, exhibited potent HIV-1 inhibitory activity with an EC50 value of 3.22 μM (Fang et al., 2015), whereas only two 1,2,3-triazole compounds displayed moderate activity by clicking an alkyl group onto lersivirine (LSV), a potent NNRTI that is effective against wild type HIV as well as several problematic mutant strains of the virus (Pribut et al., 2016). Moreover, compound 2, a copper(I)-catalyzed 1,2,3-triazole-derived compound, exhibited potent effectiveness against HIV-1 protease (Giffin et al., 2008). Furthermore, a specific HIV fusion inhibitor conjugated with sapogenin, a nonspecific antiviral agent, showed a strong cooperative effect against HIV-1 activity (Wang et al., 2014).
In summary, these results indicated that the unique bioorthogonality of click chemistry renders antiviral agents with high effectiveness to inhibit viruses.
2.3. Diagnosis of viral infection
Diagnosis at an early stage of viral infection is critical for antiviral therapy, especially for high-risk groups. Among the diagnostic strategies, biosensors combined with analytical devices is a convenient strategy for viral detection, which can sense the presence of a virus or viral genome with high sensitivity and good reproducibility.
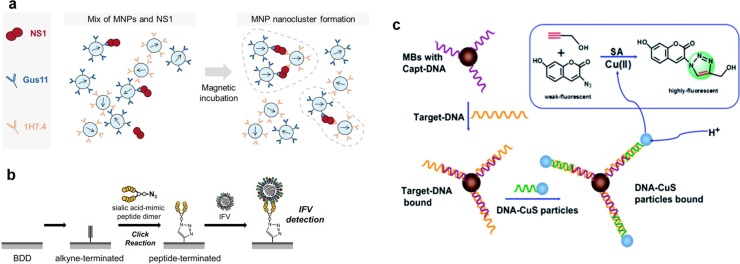
The conjugation of antibody or peptide on the biosensor via click chemistry is widely used for the early detection of infection. It was reported that tetraphenylethylene (TPE) clicked with a fluorescent 6′-sialyllactose moiety can be used as a "turn-on" fluorescent sensor to detect the influenza virus (Kato et al., 2010). A cocktail of thermally stable capture agents generated using click chemistry can be used to detect the anti-gp41 antibodies of HIV-1 from human sera with high sensitivity and specificity (Pfeilsticker et al., 2013). To increase the sensitivity of the biosensor, a variable domain of llama heavy-chain antibodies, which can specifically recognize the foot-and-mouth disease virus, was clicked with azides to generate bioorthogonal reactive groups (Trilling et al., 2014). The sensitivity of the resulting biosensor increased up to 800-fold compared with that of randomly oriented bioreceptors, indicating that the device may facilitate early diagnosis (Trilling et al., 2014). To detect infection by the dengue fever virus (DENV), an automated biosensing platform was designed by coating high-affinity monoclonal antibodies of DENV biomarker NS1 protein via bio-orthogonal Cu-free click chemistry (Fig. 3 a) (Antunes et al., 2015). The resulting biosensor takes only 8 min for DENV detection using a 6 μL serum sample, and the detection limit of the method was 25 ng/mL (Antunes et al., 2015). Recently, a boron-doped diamond (BBD) electrode clicked with sialic acid-mimic peptide, a common receptor of influenza viruses, was designed to capture influenza virions during the early stages of infection (Fig. 3b) (Matsubara et al., 2016). Using the BBD, 20–500 pfu of H1N1 and H3N2 virions can be detected, indicating that the BDD device integrated with the receptor-mimic peptide has high sensitivity in the early phase of infection (Matsubara et al., 2016). The great advantage of this BBD is that the sialic acid-mimic peptide is free from the risk of NA digestion (Matsubara et al., 2016).
Fig. 3.
Application of click chemistry to detect viral infection. (a) To detect DENV, magnetic nanoparticles (MNPs) are functionalized with capture (Gus11) and high-affinity monoclonal antibodies (1H7.4) against DENV NS1 protein via bio-orthogonal Cu-free click chemistry (Antunes et al., 2015). The presence of the target antigen NS1 triggers MNP agglutination and the formation of nanoclusters with rapid kinetics enhanced by external magnetic actuation (Antunes et al., 2015). The amount and size of the nanoclusters correlate with the target concentration and can be quantified using an optomagnetic readout method (Antunes et al., 2015). (b) The sialic acid-mimic peptide dimer was immobilized on the alkyne-terminated BDD electrode via a click reaction, and the capture of the influenza virus was detected electrochemically (Matsubara et al., 2016). (c) Biotin-modified capture DNA was linked on Streptavidin MagneSphere Paramagnetic Particles and hybridized with hepatitis B virus DNA, followed by hybridizing the target DNA with DNA-CuS particles to form a sandwich like structure (Yue et al., 2014). Subsequently, CuS particles on the sandwich structures were destroyed by acid to form Cu(II) that was reduced to Cu(I) by sodium ascorbate, which in turn catalyzes the reaction between a weakly fluorescent 3-azido-7-hydroxycoumarin and propargyl alcohol to form a fluorescent 1,2,3-triazole compound (Yue et al., 2014).
A biosensor clicked with a viral probe is a promising strategy for viral detection and quantification. To detect the viral genome, a HCV-specific acetylene-terminated DNA probe was clicked onto azido-derivatized poly(3,4-ethylenedioxythiophene) (PEDOT) electrodes, resulting in a label-free electrochemical DNA sensor for the detection of HCV (Galan et al., 2015). The result showed that the detection limit was 0.13 nM using the PEDOT-based biosensor (Galan et al., 2015). To quantify viral DNA, biotin-labeled capture DNA was immobilized onto Streptavidin MagneSphere Paramagnetic Particles and hybridized with hepatitis B viral DNA, followed by hybridizing the target DNA with DNA-CuS particles to form a sandwich like structure (Fig. 3c) (Yue et al., 2014). Then, the CuS particles were acidized to form Cu(II) and later reduced to Cu(I) by sodium ascorbate. The resulting Cu(I) catalyzed the formation of a fluorescent 1,2,3-triazole compound between a weakly fluorescent 3-azido-7-hydroxycoumarin and propargyl alcohol (Yue et al., 2014). It was found that there was a direct linear relationship between the fluorescence intensity and the target DNA concentrations in the range of 0.1–100 nM, and the detection limit is 0.04 nM (Yue et al., 2014). Thomson et al. reported an amplification-free HSV assay, in which magnetic nanoparticles were linked with a copolymer of hydroxyl- and azide-terminated oligoethylene glycol methacrylate, followed by the conjugation of the alkyne-labeled HS-specific DNA capture-probes (Thomson and Cooper, 2013; Thomson et al., 2012). Thereafter, the magnetic and fluorescent nanoparticles were linked by target DNA during the assay, and thus, the target DNA concentration can be quantified by the fluorescence intensity (Thomson and Cooper, 2013; Thomson et al., 2012). Following the assay and reagent optimization, the detection limit was 25 amol using undiluted serum, indicating a 160-fold improvement compared to that of conventional detection (Thomson and Cooper, 2013; Thomson et al., 2012).
2.4. Virus-based delivery system
Viral vectors, including those of the lentivirus and adeno-associated virus, as well as some nonviral strategies, such as cationic polymer and liposome, are limited by their packaging capacity, poor delivery, toxicity, and immunogenicity (Kretzmann et al., 2017). Therefore, improving the delivery of viral vectors remains a challenge for medical chemistry.
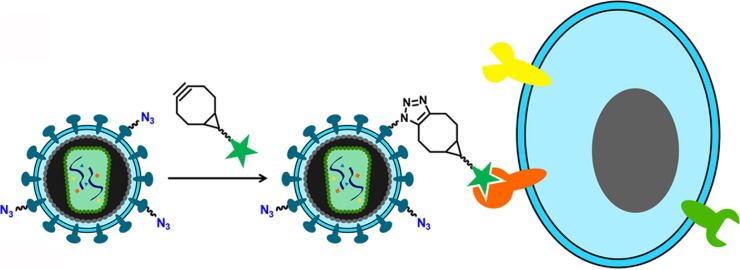
Functionalized viruses or viral vectors targeting specific cells via click chemistry are promising applications for gene or drug delivery into specific cells, especially cancer cells. For drug delivery, functionalized liposomes were generated by clicking the membrane-interacting domain (gH625) peptide of HSV-1 on the 1,2-dioleoyl-sn-glycero-3-phosphocholine-based liposomes, which may act as intracellular targeting carriers for the efficient delivery of drugs into tumor cells (Tarallo et al., 2011). Similarly, poly(amide)-based dendrimer functionalized with the gH625 allows the conjugate to rapidly penetrate the cells, suggesting that the peptidodendrimeric scaffold may be a promising material for efficient drug delivery (Carberry et al., 2012). A marked increase in gene delivery to a murine cancer cell line can achieved by the functionalization of human adenoviruses with unnatural azido sugars, followed by chemoselective modification with the small molecule effector folate, a known cancer-targeting motif (Banerjee et al., 2010). Furthermore, surface modification of unnatural glycans on lentiviral surfaces with cancer specific ligands via a strain-promoted click reaction also facilitates lentiviral transduction to target cancer cells (Fig. 4 ) (Chu et al., 2016).
Fig. 4.
Application of click chemistry in a virus-based delivery system. During the generation of lentivirus, peracetylated N-α-azidoacetylmannosamine (Ac4ManNAz), a sialic acid analog, can be directly incorporated onto viral surface glycans (Chu et al., 2016). Thereafter, functional molecules for targeting specific cells can be clicked onto the labeled virions via SPAAC (Chu et al., 2016).
3. Advantages and disadvantages of click chemistry for virus-related research
For bioconjugation, click chemistry has several advantages including the following: 1) there are no azides and alkynes in native biomolecules, which could be specifically introduced into biomolecules; 2) the reaction is very selective and only takes place between an azide and alkyne, without interacting with other biomolecules; 3) the click reaction takes place in water or aqueous solutions, indicating that the reaction can take place in living cells; 4) the reaction is fast, quantitative and irreversible, and the protocol is simple; 5) the reaction is pH-insensitive, and thus, there is no need to add any special buffer, acid or base; and 6) most importantly, the product is single and stable (Amblard et al., 2009; Best, 2009; Kolb et al., 2001). These advantages make click chemistry available for virus-related research in live cells.
However, the click chemistry is not perfect. There are a few limitations that need to be considered (Table 1 ). The most significant is that copper, as a catalyst in the reaction, is cytotoxic in vivo and in vitro, due in part to its capacity for sensitizing oxidative damage to proteins and nucleic acids (Hein et al., 2008; Meghani et al., 2017). Therefore, copper-free click reactions are ideal reactions for viral tracking and virus-based delivery systems in live cells or in vivo. Moreover, numerous stable triazoles are produced via click reactions, and thus, whether these triazoles are safe for animals or humans as well as the metabolic pathways of triazoles in biological systems need to be further investigated (Li et al., 2013). Furthermore, as proteins or nucleic acids can be easily digested by protease or nuclease, respectively (Picard-Lafond and Morin, 2017; Qi et al., 2017; Thomson and Cooper, 2013; Thomson et al., 2012; Yue et al., 2014), protease degradation needs to be considered during the modification of peptide or protein, while DNase or RNase inhibitors are needed for nucleotide-related reactions. For a more detailed explanation of the limitations of click chemistry, please see the review by Pickens et al. (Pickens et al., 2018).
Table 1.
Comparison of different usages of click chemistry in virus-related research.
| Usage | Advantages | Disadvantages | Reference |
|---|---|---|---|
| Viral tracking |
|
|
Liu et al. (2017) |
| Antiviral agent |
|
|
Ahmad Fuaad et al. (2013), McGowan et al. (2017), Tian et al. (2017), Xiao et al. (2014), (2016) |
| Diagnosis of viral infection |
|
|
Qi et al. (2017), Thomson and Cooper (2013), Thomson et al. (2012); Yue et al. (2014) |
| Virus-based delivery system |
|
|
Li et al. (2013), Picard-Lafond and Morin (2017) |
4. Conclusions and future perspectives
In summary, the high reaction yield, simple reaction and purification conditions, wide range of solvent and pH stabilities, and functional group tolerance make click chemistry ideal for virus-related research (Bruckman et al., 2008). Based on click chemistry, much progress was achieved to understand the molecular mechanisms of viral replication and pathogenesis as well as viral interactions with host cells. Although this review highlighted some of the currently knowledge based on click chemistry, we still have much to explore with regard to viral infection. It also provides platforms for viral detection, drug development and targeted delivery. To date, more products generated via click chemistry are likely to become commercially available in the field of biotechnology, especially for visualization, drug discovery, diagnosis, and therapeutics (Mandhare et al., 2014; Xu and Jones, 2015). Efforts are still needed to resolve the limitations of click chemistry in live cells and animals/humans, especially the cytotoxic activities of the compounds generated from click reactions. Hopefully, as time goes on, it can be anticipated that in the future, click reactions will contribute great advances in the research and applications of viral tracking, the design of antiviral agents, the diagnosis of viral infection, and virus-based delivery systems.
Author contributions
Conceptualization, R.L.Z.; writing original draft preparation, O.T. and O.H.; writing review & editing, R.L.Z.; figure editing, L. × .H and R.L.Z.
Conflict of interest
The authors declare that they have no conflict of interest. Permission has been obtained for the use of figures in this review.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Acknowledgments
This work was financially supported by the National Key Research and Development Program of China (No. 2017YFD0500103), the National Natural Science Foundation of China (No. 31772747), the Program for JLU Science and Technology Innovative Research Team (JLUSTIRT, No. 2017TD-28), the Graduate Innovation Fund of Jilin University, and the Fundamental Research Funds for the CentralUniversities.
References
- Ahmad Fuaad A.A., Azmi F., Skwarczynski M., Toth I. Peptide conjugation via CuAAC’ click’ chemistry. Molecules. 2013;18(11):13148–13174. doi: 10.3390/molecules181113148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alandijany T., Roberts A.P.E., Conn K.L., Loney C., McFarlane S., Orr A., Boutell C. Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PLoS Pathog. 2018;14(1):e1006769. doi: 10.1371/journal.ppat.1006769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amblard F., Cho J.H., Schinazi R.F. Cu(I)-catalyzed Huisgen azide-alkyne 1,3-dipolar cycloaddition reaction in nucleoside, nucleotide, and oligonucleotide chemistry. Chem. Rev. 2009;109(9):4207–4220. doi: 10.1021/cr9001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes P., Watterson D., Parmvi M., Burger R., Boisen A., Young P., Cooper M.A., Hansen M.F., Ranzoni A., Donolato M. Quantification of NS1 dengue biomarker in serum via optomagnetic nanocluster detection. Sci. Rep. 2015;5:16145. doi: 10.1038/srep16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artyushin O.I., Sharova E.V., Vinogradova N.M., Genkina G.K., Moiseeva A.A., Klemenkova Z.S., Orshanskaya I.R., Shtro A.A., Kadyrova R.A., Zarubaev V.V., Yarovaya O.I., Salakhutdinov N.F., Brel V.K. Synthesis of camphecene derivatives using click chemistry methodology and study of their antiviral activity. Bioorg. Med. Chem. Lett. 2017;27(10):2181–2184. doi: 10.1016/j.bmcl.2017.03.051. [DOI] [PubMed] [Google Scholar]
- Avti P.K., Maysinger D., Kakkar A. Alkyne-azide "click" chemistry in designing nanocarriers for applications in biology. Molecules. 2013;18(8):9531–9549. doi: 10.3390/molecules18089531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P.S., Ostapchuk P., Hearing P., Carrico I. Chemoselective attachment of small molecule effector functionality to human adenoviruses facilitates gene delivery to cancer cells. J. Am. Chem. Soc. 2010;132(39):13615–13617. doi: 10.1021/ja104547x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaune G., Tamang S., Bernardin A., Bayle-Guillemaud P., Fenel D., Schoehn G., Vinet F., Reiss P., Texier I. Luminescence of polyethylene glycol coated CdSeTe/ZnS and InP/ZnS nanoparticles in the presence of copper cations. Chemphyschem. 2011;12(12):2247–2254. doi: 10.1002/cphc.201100266. [DOI] [PubMed] [Google Scholar]
- Bernardin A., Cazet A., Guyon L., Delannoy P., Vinet F., Bonnaffe D., Texier I. Copper-free click chemistry for highly luminescent quantum dot conjugates: application to in vivo metabolic imaging. Bioconjug. Chem. 2010;21(4):583–588. doi: 10.1021/bc900564w. [DOI] [PubMed] [Google Scholar]
- Best M.D. Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry. 2009;48(28):6571–6584. doi: 10.1021/bi9007726. [DOI] [PubMed] [Google Scholar]
- Bruckman M.A., Kaur G., Lee L.A., Xie F., Sepulveda J., Breitenkamp R., Zhang X., Joralemon M., Russell T.P., Emrick T., Wang Q. Surface modification of tobacco mosaic virus with "click" chemistry. Chembiochem. 2008;9(4):519–523. doi: 10.1002/cbic.200700559. [DOI] [PubMed] [Google Scholar]
- Carberry T.P., Tarallo R., Falanga A., Finamore E., Galdiero M., Weck M., Galdiero S. Dendrimer functionalization with a membrane-interacting domain of herpes simplex virus type 1: towards intracellular delivery. Chem. Eur. J. 2012;18(43):13678–13685. doi: 10.1002/chem.201202358. [DOI] [PubMed] [Google Scholar]
- Chu Y.J., Oum Y.H., Carrico I.S. Surface modification via strain-promoted click reaction facilitates targeted lentiviral transduction. Virology. 2016;487:95–103. doi: 10.1016/j.virol.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Cieslar-Pobuda A., Los M.J. Prospects and limitations of "Click-Chemistry"-based DNA labeling technique employing 5-ethynyl-2’deoxyuridine (EdU) Cytometry A. 2013;83(11):977–978. doi: 10.1002/cyto.a.22394. [DOI] [PubMed] [Google Scholar]
- Cowan G.H., Roberts A.G., Chapman S.N., Ziegler A., Savenkov E.I., Torrance L. The potato mop-top virus TGB2 protein and viral RNA associate with chloroplasts and viral infection induces inclusions in the plastids. Front. Plant Sci. 2012;3:290. doi: 10.3389/fpls.2012.00290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Kang D., Zhang L., Huang B., Liu H., Pannecouque C., De Clercq E., Zhan P., Liu X. Synthesis and biological evaluation of a series of 2-((1-substituted-1H-1,2,3-triazol-4-yl)methylthio)-6-(naphthalen-1-ylmethyl)pyri midin-4(3 H)-one As potential HIV-1 inhibitors. Chem. Biol. Drug Des. 2015;86(4):614–618. doi: 10.1111/cbdd.12524. [DOI] [PubMed] [Google Scholar]
- Fernandez M.V., Freed E.O. "Expand and click": a new method for labeling HIV-1 envelope glycoproteins. Cell Chem. Biol. 2017;24(5):548–550. doi: 10.1016/j.chembiol.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan T., Prieto-Simon B., Alvira M., Eritja R., Gotz G., Bauerle P., Samitier J. Label-free electrochemical DNA sensor using "click"-functionalized PEDOT electrodes. Biosens. Bioelectron. 2015;74:751–756. doi: 10.1016/j.bios.2015.07.037. [DOI] [PubMed] [Google Scholar]
- Gao R., Liu P., Wong S.M. Identification of a plant viral RNA genome in the nucleus. PLoS One. 2012;7(11):e48736. doi: 10.1371/journal.pone.0048736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Generous A., Thorson M., Barcus J., Jacher J., Busch M., Sleister H. Identification of putative interactions between swine and human influenza A virus nucleoprotein and human host proteins. Virol. J. 2014;11:228. doi: 10.1186/s12985-014-0228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffin M.J., Heaslet H., Brik A., Lin Y.C., Cauvi G., Wong C.H., McRee D.E., Elder J.H., Stout C.D., Torbett B.E. A copper(I)-catalyzed 1,2,3-triazole azide-alkyne click compound is a potent inhibitor of a multidrug-resistant HIV-1 protease variant. J. Med. Chem. 2008;51(20):6263–6270. doi: 10.1021/jm800149m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeau G., Staedel C., Barthelemy P. Lipid-conjugated oligonucleotides via "click chemistry" efficiently inhibit hepatitis C virus translation. J. Med. Chem. 2008;51(15):4374–4376. doi: 10.1021/jm800518u. [DOI] [PubMed] [Google Scholar]
- Hagemeijer M.C., Vonk A.M., Monastyrska I., Rottier P.J.M., de Haan C.A.M. Visualizing coronavirus RNA synthesis in time by using click chemistry. J. Virol. 2012;86(10):5808–5816. doi: 10.1128/JVI.07207-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J., Huang L.L., Zhang R., Wang H.Z., Xie H.Y. A mild and reliable method to label enveloped virus with quantum dots by copper-free click chemistry. Anal. Chem. 2012;84(19):8364–8370. doi: 10.1021/ac301918t. [DOI] [PubMed] [Google Scholar]
- Hein C.D., Liu X.M., Wang D. Click chemistry, a powerful tool for pharmaceutical sciences. Pharm. Res. 2008;25(10):2216–2230. doi: 10.1007/s11095-008-9616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himo F., Lovell T., Hilgraf R., Rostovtsev V.V., Noodleman L., Sharpless K.B., Fokin V.V. Copper(I)-catalyzed synthesis of azoles. DFT study predicts unprecedented reactivity and intermediates. J. Am. Chem. Soc. 2005;127(1):210–216. doi: 10.1021/ja0471525. [DOI] [PubMed] [Google Scholar]
- Hoyle C.E., Bowman C.N. Thiol-ene click chemistry. Angew. Chem. Int. Ed. Engl. 2010;49(9):1540–1573. doi: 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
- Huang L.L., Liu K., Zhang Q., Xu J., Zhao D., Zhu H., Xie H.Y. Integrating two efficient and specific bioorthogonal ligation reactions with natural metabolic incorporation in one cell for virus dual labeling. Anal. Chem. 2017;89(21):11620–11627. doi: 10.1021/acs.analchem.7b03043. [DOI] [PubMed] [Google Scholar]
- Jao C.Y., Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U. S. A. 2008;105(41):15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett J.C., Bertozzi C.R. Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev. 2010;39(4):1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett J.C., Sletten E.M., Bertozzi C.R. Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc. 2010;132(11):3688–3690. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalveram B., Lihoradova O., Indran S.V., Head J.A., Ikegami T. Using click chemistry to measure the effect of viral infection on host-cell RNA synthesis. J. Vis. Exp. 2013;(78) doi: 10.3791/50809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Kawaguchi A., Nagata K., Hatanaka K. Development of tetraphenylethylene-based fluorescent oligosaccharide probes for detection of influenza virus. Biochem. Biophys. Res. Commun. 2010;394(1):200–204. doi: 10.1016/j.bbrc.2010.02.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmeier F., Maya-Mendoza A., Jackson D.A. EdU induces DNA damage response and cell death in mESC in culture. Chromosome Res. 2013;21(1):87–100. doi: 10.1007/s10577-013-9340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H.C., Finn M.G., Sharpless K.B. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Komatsu T., Robinson D.R., Hisaoka M., Ueshima S., Okuwaki M., Nagata K., Wodrich H. Tracking adenovirus genomes identifies morphologically distinct late DNA replication compartments. Traffic. 2016;17(11):1168–1180. doi: 10.1111/tra.12429. [DOI] [PubMed] [Google Scholar]
- Kretzmann J.A., Ho D., Evans C.W., Plani-Lam J.H.C., Garcia-Bloj B., Mohamed A.E., O’Mara M.L., Ford E., Tan D.E.K., Lister R., Blancafort P., Norret M., Iyer K.S. Synthetically controlling dendrimer flexibility improves delivery of large plasmid DNA. Chem. Sci. 2017;8(4):2923–2930. doi: 10.1039/c7sc00097a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Aneja R., Chaiken I. Click chemistry in peptide-based drug design. Molecules. 2013;18(8):9797–9817. doi: 10.3390/molecules18089797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Yan H., Li L., Yang M., Peng B., Chen S., Li W., Chen P.R. Site-specific engineering of chemical functionalities on the surface of live hepatitis D virus. Angew. Chem. Int. Ed. Engl. 2013;52(52):13970–13974. doi: 10.1002/anie.201305787. [DOI] [PubMed] [Google Scholar]
- Liu X., Ouyang T., Ouyang H., Ren L. Single particle labeling of RNA virus in live cells. Virus Res. 2017;237:14–21. doi: 10.1016/j.virusres.2017.05.007. [DOI] [PubMed] [Google Scholar]
- Lu Z.P., Xiao Z.L., Yang Z., Li J., Feng G.X., Chen F.Q., Li Y.H., Feng J.Y., Gao Y.E., Ye L.H., Zhang X.D. Hepatitis B virus X protein promotes human hepatoma cell growth via upregulation of transcription factor AP2alpha and sphingosine kinase 1. Acta Pharmacol. Sin. 2015;36(10):1228–1236. doi: 10.1038/aps.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandhare A., Banerjee P., Bhutkar S., Hirwani R. Click chemistry' for diagnosis: a patent review on exploitation of its emerging trends. Expert Opin. Ther. Pat. 2014;24(12):1287–1310. doi: 10.1517/13543776.2014.977864. [DOI] [PubMed] [Google Scholar]
- Matsubara T., Ujie M., Yamamoto T., Akahori M., Einaga Y., Sato T. Highly sensitive detection of influenza virus by boron-doped diamond electrode terminated with sialic acid-mimic peptide. Proc. Natl. Acad. Sci. U. S. A. 2016;113(32):8981–8984. doi: 10.1073/pnas.1603609113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan D.C., Khamlichi M.D., De Groot A., Pauwels F., Delouvroy F., Van Emelen K., Simmen K., Raboisson P. Synthesis and evaluation of novel HCV replication inhibitors. Mol. Divers. 2017;21(2):475–481. doi: 10.1007/s11030-017-9733-z. [DOI] [PubMed] [Google Scholar]
- Meghani N.M., Amin H.H., Leel B.J. Mechanistic applications of click chemistry for pharmaceutical drug discovery and drug delivery. Drug Discov. Today. 2017;22(11):1604–1619. doi: 10.1016/j.drudis.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Moses J.E., Moorhouse A.D. The growing applications of click chemistry. Chem. Soc. Rev. 2007;36(8):1249–1262. doi: 10.1039/b613014n. [DOI] [PubMed] [Google Scholar]
- Nagao M., Fujiwara Y., Matsubara T., Hoshino Y., Sato T., Miura Y. Design of glycopolymers carrying sialyl oligosaccharides for controlling the interaction with the influenza virus. Biomacromolecules. 2017;18(12):4385–4392. doi: 10.1021/acs.biomac.7b01426. [DOI] [PubMed] [Google Scholar]
- Nikic I., Plass T., Schraidt O., Szymanski J., Briggs J.A., Schultz C., Lemke E.A. Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew. Chem. Int. Ed. Engl. 2014;53(8):2245–2249. doi: 10.1002/anie.201309847. [DOI] [PubMed] [Google Scholar]
- Nwe K., Brechbiel M.W. Growing applications of "click chemistry" for bioconjugation in contemporary biomedical research. Cancer Biother. Radiopharm. 2009;24(3):289–302. doi: 10.1089/cbr.2008.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oum Y.H., Desai T.M., Marin M., Melikyan G.B. Click labeling of unnatural sugars metabolically incorporated into viral envelope glycoproteins enables visualization of single particle fusion. J. Virol. Methods. 2016;233:62–71. doi: 10.1016/j.jviromet.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeilsticker J.A., Umeda A., Farrow B., Hsueh C.L., Deyle K.M., Kim J.T., Lai B.T., Heath J.R. A cocktail of thermally stable, chemically synthesized capture agents for the efficient detection of anti-gp41 antibodies from human sera. PLoS One. 2013;8(10):e76224. doi: 10.1371/journal.pone.0076224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard-Lafond A., Morin J.F. Low-temperature synthesis of carbon-rich nanoparticles with a clickable surface for functionalization. Langmuir. 2017;33(22):5385–5392. doi: 10.1021/acs.langmuir.7b00135. [DOI] [PubMed] [Google Scholar]
- Pickens C.J., Johnson S.N., Pressnall M.M., Leon M.A., Berkland C.J. Practical considerations, challenges, and limitations of bioconjugation via Azide-Alkyne Cycloaddition. Bioconjug. Chem. 2018;29(3):686–701. doi: 10.1021/acs.bioconjchem.7b00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass T., Milles S., Koehler C., Szymanski J., Mueller R., Wiessler M., Schultz C., Lemke E.A. Amino acids for Diels-Alder reactions in living cells. Angew. Chem. Int. Ed. Engl. 2012;51(17):4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- Pribut N., Veale C.G., Basson A.E., van Otterlo W.A., Pelly S.C. Application of the Huisgen cycloaddition and’ click’ reaction toward various 1,2,3-triazoles as HIV non-nucleoside reverse transcriptase inhibitors. Bioorg. Med. Chem. Lett. 2016;26(15):3700–3704. doi: 10.1016/j.bmcl.2016.05.082. [DOI] [PubMed] [Google Scholar]
- Qi Y., Qiu L., Fan W., Liu C., Li Z. An enzyme-free flow cytometric bead assay for the sensitive detection of microRNAs based on click nucleic acid ligation-mediated signal amplification. Analyst. 2017;142(16):2967–2973. doi: 10.1039/c7an00989e. [DOI] [PubMed] [Google Scholar]
- Reich E., Goldberg I.H. Actinomycin and nucleic acid function. Prog. Nucleic Acid Res. Mol. Biol. 1964;3:183–234. doi: 10.1016/s0079-6603(08)60742-4. [DOI] [PubMed] [Google Scholar]
- Rubino F.A., Oum Y.H., Rajaram L., Chu Y., Carrico I.S. Chemoselective modification of viral surfaces via bioorthogonal click chemistry. J. Vis. Exp. 2012;(66):e4246. doi: 10.3791/4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachin K., Jadhav V.H., Kim E.M., Kim H.L., Lee S.B., Jeong H.J., Lim S.T., Sohn M.H., Kim D.W. F-18 labeling protocol of peptides based on chemically orthogonal strain-promoted cycloaddition under physiologically friendly reaction conditions. Bioconjug. Chem. 2012;23(8):1680–1686. doi: 10.1021/bc3002425. [DOI] [PubMed] [Google Scholar]
- Sakin V., Hanne J., Dunder J., Anders-Osswein M., Laketa V., Nikic I., Krausslich H.G., Lemke E.A., Muller B. A versatile tool for live-cell imaging and super-resolution nanoscopy studies of HIV-1 env distribution and mobility. Cell Chem. Biol. 2017;24(5) doi: 10.1016/j.chembiol.2017.04.007. 635-645. [DOI] [PubMed] [Google Scholar]
- Salic A., Mitchison T.J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105(7):2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich T.G., Katsafanas G.C., Weisberg A., Olano L.R., Moss B. Identification of vaccinia virus replisome and transcriptome proteins by isolation of proteins on nascent DNA coupled with mass spectrometry. J. Virol. 2017;91(19) doi: 10.1128/JVI.01015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Hui Teo C., Serwa R.A., O’Hare P. Spatial and temporal resolution of global protein synthesis during HSV infection using bioorthogonal precursors and click chemistry. PLoS Pathog. 2016;12(10) doi: 10.1371/journal.ppat.1005927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarallo R., Accardo A., Falanga A., Guarnieri D., Vitiello G., Netti P., D’Errico G., Morelli G., Galdiero S. Clickable functionalization of liposomes with the gH625 peptide from herpes simplex virus type I for intracellular drug delivery. Chem. Eur. J. 2011;17(45):12659–12668. doi: 10.1002/chem.201101425. [DOI] [PubMed] [Google Scholar]
- Thirumurugan P., Matosiuk D., Jozwiak K. Click chemistry for drug development and diverse chemical-biology applications. Chem. Rev. 2013;113(7):4905–4979. doi: 10.1021/cr200409f. [DOI] [PubMed] [Google Scholar]
- Thomson D.A., Cooper M.A. A paramagnetic-reporter two-particle system for amplification-free detection of DNA in serum. Biosens. Bioelectron. 2013;50:499–501. doi: 10.1016/j.bios.2013.06.062. [DOI] [PubMed] [Google Scholar]
- Thomson D.A., Tee E.H., Tran N.T., Monteiro M.J., Cooper M.A. Oligonucleotide and polymer functionalized nanoparticles for amplification-free detection of DNA. Biomacromolecules. 2012;13(6):1981–1989. doi: 10.1021/bm300717f. [DOI] [PubMed] [Google Scholar]
- Tian Z., Si L., Meng K., Zhou X., Zhang Y., Zhou D., Xiao S. Inhibition of influenza virus infection by multivalent pentacyclic triterpene-functionalized per-O-methylated cyclodextrin conjugates. Eur. J. Med. Chem. 2017;134:133–139. doi: 10.1016/j.ejmech.2017.03.087. [DOI] [PubMed] [Google Scholar]
- Trilling A.K., Hesselink T., van Houwelingen A., Cordewener J.H., Jongsma M.A., Schoffelen S., van Hest J.C., Zuilhof H., Beekwilder J. Orientation of llama antibodies strongly increases sensitivity of biosensors. Biosens. Bioelectron. 2014;60:130–136. doi: 10.1016/j.bios.2014.04.017. [DOI] [PubMed] [Google Scholar]
- Wagner K., Kwakkenbos M.J., Claassen Y.B., Maijoor K., Bohne M., van der Sluijs K.F., Witte M.D., van Zoelen D.J., Cornelissen L.A., Beaumont T., Bakker A.Q., Ploegh H.L., Spits H. Bispecific antibody generated with sortase and click chemistry has broad antiinfluenza virus activity. Proc. Natl. Acad. Sci. U. S. A. 2014;111(47):16820–16825. doi: 10.1073/pnas.1408605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Chan T.R., Hilgraf R., Fokin V.V., Sharpless K.B., Finn M.G. Bioconjugation by copper(I)-catalyzed azide-alkyne [3+2] cycloaddition. J. Am. Chem. Soc. 2003;125(11):3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- Wang C., Lu L., Na H., Li X., Wang Q., Jiang X., Xu X., Yu F., Zhang T., Li J., Zhang Z., Zheng B., Liang G., Cai L., Jiang S., Liu K. Conjugation of a nonspecific antiviral sapogenin with a specific HIV fusion inhibitor: a promising strategy for discovering new antiviral therapeutics. J. Med. Chem. 2014;57(17):7342–7354. doi: 10.1021/jm500763m. [DOI] [PubMed] [Google Scholar]
- Wang C., Zhu W., Wang B.Z. Dual-linker gold nanoparticles as adjuvanting carriers for multivalent display of recombinant influenza hemagglutinin trimers and flagellin improve the immunological responses in vivo and in vitro. Int. J. Nanomed. 2017;12:4747–4762. doi: 10.2147/IJN.S137222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang I.H., Burckhardt C.J., Yakimovich A., Greber U.F. Imaging, tracking and computational analyses of virus entry and egress with the cytoskeleton. Viruses. 2018;10(4) doi: 10.3390/v10040166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Wang Q., Si L., Shi Y., Wang H., Yu F., Zhang Y., Li Y., Zheng Y., Zhang C., Wang C., Zhang L., Zhou D. Synthesis and anti-HCV entry activity studies of beta-cyclodextrin-pentacyclic triterpene conjugates. ChemMedChem. 2014;9(5):1060–1070. doi: 10.1002/cmdc.201300545. [DOI] [PubMed] [Google Scholar]
- Xiao S., Wang Q., Si L., Zhou X., Zhang Y., Zhang L., Zhou D. Synthesis and biological evaluation of novel pentacyclic triterpene alpha-cyclodextrin conjugates as HCV entry inhibitors. Eur. J. Med. Chem. 2016;124:1–9. doi: 10.1016/j.ejmech.2016.08.020. [DOI] [PubMed] [Google Scholar]
- Xu H., Jones L.H. Click chemistry patents and their impact on drug discovery and chemical biology. Pharm. Pat. Anal. 2015;4(2):109–119. doi: 10.4155/ppa.14.59. [DOI] [PubMed] [Google Scholar]
- Yang Y., Liu H.P., Yu Q., Yang M.B., Wang D.M., Jia T.W., He H.J., He Y., Xiao H.X., Iyer S.S., Fan Z.C., Meng X., Yu P. Multivalent S-sialoside protein conjugates block influenza hemagglutinin and neuraminidase. Carbohydr. Res. 2016;435:68–75. doi: 10.1016/j.carres.2016.09.017. [DOI] [PubMed] [Google Scholar]
- Yi G., Son J., Yoo J., Park C., Koo H. Application of click chemistry in nanoparticle modification and its targeted delivery. Biomater. Res. 2018;22:13. doi: 10.1186/s40824-018-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue G., Ye H., Huang X., Ye W., Qiu S., Qiu B., Lin Z., Chen G. Quantification of DNA through a fluorescence biosensor based on click chemistry. Analyst. 2014;139(22):5669–5673. doi: 10.1039/c4an01438c. [DOI] [PubMed] [Google Scholar]
- Zhang J.J., Mulvenon A., Makarov E., Wagoner J., Knibbe J., Kim J.O., Osna N., Bronich T.K., Poluektova L.Y. Antiviral peptide nanocomplexes as a potential therapeutic modality for HIV/HCV co-infection. Biomaterials. 2013;34(15):3846–3857. doi: 10.1016/j.biomaterials.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Garrison J.C., Poluektova L.Y., Bronich T.K., Osna N.A. Liver-targeted antiviral peptide nanocomplexes as potential anti-HCV therapeutics. Biomaterials. 2015;70:37–47. doi: 10.1016/j.biomaterials.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]