Abstract
We have determined the crystal structure of the core (C) protein from the Kunjin subtype of West Nile virus (WNV), closely related to the NY99 strain of WNV, currently a major health threat in the U.S. WNV is a member of the Flaviviridae family of enveloped RNA viruses that contains many important human pathogens. The C protein is associated with the RNA genome and forms the internal core which is surrounded by the envelope in the virion. The C protein structure contains four α helices and forms dimers that are organized into tetramers. The tetramers form extended filamentous ribbons resembling the stacked α helices seen in HEAT protein structures.
Introduction
West Nile virus (WNV) is an emerging, mosquito-borne human pathogen that first appeared in the U.S. in 1999 (Brinton, 2002) and currently constitutes a serious health threat, with 264 registered deaths and 9858 reported cases in 2003 (CDC, 2004). Sporadic outbreaks have also occurred in Europe, and the virus has been isolated from birds in the UK (Buckley et al., 2003). Typical symptoms of WNV infection include fever, rashes, and myalgia; however, occasionally more severe and potentially fatal symptoms develop, including acute encephalitis and fulminant hepatitis (Petersen et al., 2003). WNV are divided into two lineages: lineage 1 includes the New York strain (NY99) responsible for the recent outbreaks in the U.S., as well as the closely related Kunjin virus, which was recently classified as an Australian subtype of WNV Lanciotti et al. 1999, Scherret et al. 2001. Unlike the highly pathogenic NY99 strain, humans infected with Kunjin virus do not develop the more severe symptoms (Hall et al., 2003). Lineage 2 consists of several African strains, which are apparently not involved in outbreaks of the disease (Lanciotti et al., 1999). There is currently no vaccine available, but inoculation with a cDNA for the whole genome of an attenuated form of Kunjin virus could confer resistance to infection by the NY99 strain of WNV in mice (Hall et al., 2003).
WNV is a member of the Flaviviridae family, which includes many important human pathogens, like dengue, yellow fever, tick-borne encephalitis (TBE), and hepatitis C viruses (Strauss and Strauss, 2002). The flaviviruses have a positive-sense, single-stranded RNA genome, containing one large open reading frame, which is translated as a single polyprotein (Lindenbach and Rice, 2002). The 3433 amino acid polyprotein of WNV is cleaved by host and viral proteases to the three structural proteins, C (core), prM (membrane), and E (envelope), and seven nonstructural (NS) proteins (Brinton, 2002). In the virion, the genome is contained within a spherical or isometric nucleocapsid or core made from the C protein. The 123 amino acid C precursor is targeted to the membrane by a carboxy-terminal hydrophobic sequence and is cleaved to its mature 105 residue form by the viral NS2B-NS3 protease. The core is surrounded by a lipid membrane containing the two membrane proteins prM and E (Mukhopadhyay et al., 2003). Particle maturation involves cleavage of pre-M to M and resulting reorganization of the E proteins from a dimeric to a trimeric arrangement Kuhn et al. 2002, Zhang et al. 2003.
Cryo-electron microscopy (EM) reconstruction of WNV (Mukhopadhyay et al., 2003) showed an outer envelope layer consisting of 180 copies of E protein, very similar to that of dengue virus Kuhn et al. 2002, Zhang et al. 2003. Consequently, the WNV E protein is presumed to have a similar structure to the E protein of dengue virus and TBE Modis et al. 2003, Rey et al. 1995. The EM reconstructions of WNV and dengue virus did not show any structure for the inner core, however, presumably due to its being disordered or having different symmetry from the envelope.
There are only four amino acid differences (out of 105) in the mature C protein between Kunjin virus and the NY99 strain of WNV and seven differences between Kunjin virus and a Nigerian strain, a member of WNV lineage 2 (Figure 1) . Dengue virus C protein shares about 57% identity with WNV. The WNV C protein contains a large number of positively charged residues, which are distributed throughout the protein, unlike the core proteins of togaviruses and arteriviruses, which have distinct, positively charged N-terminal regions Choi et al. 1991, Doan and Dokland 2003. The N-terminal and C-terminal charged regions have both been implicated in RNA binding (Khromykh and Westaway, 1996). The dengue virus C protein was recently solved by NMR methods (Ma et al., 2004). It forms a dimer in solution and contains four α helices. The N-terminal 20 or so residues were disordered, similar to the RNA binding regions of togaviruses and arteriviruses. Here, we report the crystal structure of the C protein from the Kunjin subtype of WNV. This is the first crystal structure of a flavivirus core protein. The C protein forms dimers, similar to those of dengue virus C, which are organized into tetramers with highly positively charged surfaces. The α helices in the tetramers stack up to form long filamentous ribbons in the crystal.
Figure 1.
Sequence Alignment of the C Proteins of Kunjin Virus, the NY99 and Nigerian Strains of WNV, and Dengue
The start and end of the trypsin-cleaved fragment (bullets) as well as the site of maturation cleavage (arrowhead) are indicated. The position of the α helices in the C structure are shown above the sequence (spirals) (Gouet et al., 1999).
Results and Discussion
The first 103 amino acids of the C protein of Kunjin virus (strain MRM61C) were cloned with an N-terminal 6×His tag and purified by affinity and ion exchange chromatography. Crystals of the full-length C protein were unstable and disordered and did not diffract X-rays to high resolution. Assuming that the crystal disorder was caused by flexible regions in the protein, we treated the full-length protein with trypsin, resulting in a stable fragment starting at Val23 and ending at Arg98 (Figure 1). This fragment yielded stable crystals which diffracted synchrotron radiation anisotropically to about 2.8 Å resolution. The crystals belong to space group I41 with a = 85.7 Å and c = 214.4 Å, with eight protein monomers in the asymmetric unit and VM = 2.8. The structure was solved to 3.2 Å resolution by MAD on a Se-Met derivative, giving a final Rcryst = 0.25 and Rfree = 0.31 (Table 1) and has been deposited in the Protein Data Bank with accession code 1SFK (Figures 2A and 2B) .
Table 1.
Crystallographic Data
| Data Set |
Kunc29 Native |
SF83 Se-Met |
||
|---|---|---|---|---|
| ESRF BM14 |
||||
| X-ray source | ESRF BM14 | Peak | Inflection | Remote |
| Wavelength (Å) | 0.8856 | 0.97956 | 0.97976 | 0.8856 |
| Oscillation angle (°) | 1 | 1 | 2 | 1 |
| Number of frames | 139 | 200 | 51 | 100 |
| Resolution range (Å) | 30.0–3.2 (3.31–3.2)a | 40.0–3.7 (3.83–3.7) | 30.0–3.9 (4.04–3.9) | 50.0–3.64 (3.77–3.64) |
| Total number of reflections | 72,872 | 70,802 | 30,605 | 36,861 |
| Number of unique reflections | 12,515 | 8,246 | 7,022 | 8,629 |
| Completeness (%) | 99.4 (95.3) | 100 (100) | 100 (100) | 99.9 (99.7) |
| Redundancy | 5.8 (4.8) | 8.6 (8.6) | 4.4 (4.4) | 4.3 (4.3) |
| Rmergeb | 0.075 (0.32) | 0.10 (0.45) | 0.087 (0.46) | 0.094 (0.51) |
| I/σ(I) | 21.2 (3.0) | 26.7 (4.2) | 18.8 (2.7) | 17.1 (2.2) |
| Refinement Parameters | ||||
| Resolution range (Å) | 10–3.2 | |||
| No of atoms | 4,447 | |||
| Rcrystc | 0.251 | |||
| Rfreec | 0.311 | |||
| Rmsd on bond lengths (Å) | 0.014 | |||
| Rmsd on bond angles (°) | 1.56 | |||
Values in parentheses are for the high-resolution shell.
Rmerge = ΣhΣi |Ih,i − <Ih>|/ΣhΣiIh,i.
Rcryst = Σh|Fobs,h − Fcalc,h|/Σh|Fobs,h|. Rfree is the same, calculated for a random 5% of the reflections not used in refinement.
Figure 2.
Stereoviews of the WNV C Protein Structure
(A) Detail of the weighted 2Fo − Fc electron density map, showing part of helix α4 from Asn72 to Ile94 (Esnouf, 1999).
(B) Cα backbone trace of CD dimer. C, blue; D, red.
(C) Superposition of subunit A (blue) with residues B1108 to B1206 of Pyrococcus woesei TFB (PDB code 1AIS; green), aligned using the helical segments identified by the DALI search (Holm and Sander, 1995).
(D) Superposition of the C protein CD dimer (purple) with the solution structure dimer of dengue C protein (1R6R) (green). The least mean square fit was calculated using residues 54–96.
The C protein structure consists of four α helices interspersed by short loops Figure 1, Figure 2, Figure 3 . The protein forms a tight 2-fold symmetric dimer, in which the helices α1, α2, and α4 form three distinct “layers” and α3 forms a short, connecting helix flanking the dimer (Figure 3A). A DALI search for structural homologs (Holm and Sander, 1995) revealed a topological resemblance with the cyclin A-like domain, like that of archeal transcription factor B (TFB; PDB code 1AIS; Z = 4.0) (Kosa et al., 1997), and with the HEAT repeat domain of human eIF4Gii (1HU3; Z = 3.9) (Marcotrigiano et al., 2001) (Figure 2C). Like the WNV C protein, both of these proteins are involved in nucleic acid interactions; however, unlike WNV C, they are monomeric. A comparison with the dengue virus C protein dimer (1R6R) (Ma et al., 2004) shows that while helices α2 to α4 superimpose relatively well, with an rmsd of 3.6 Å for the Cα atoms of residues 44–96, α1 is in a completely different orientation (Figure 2D).
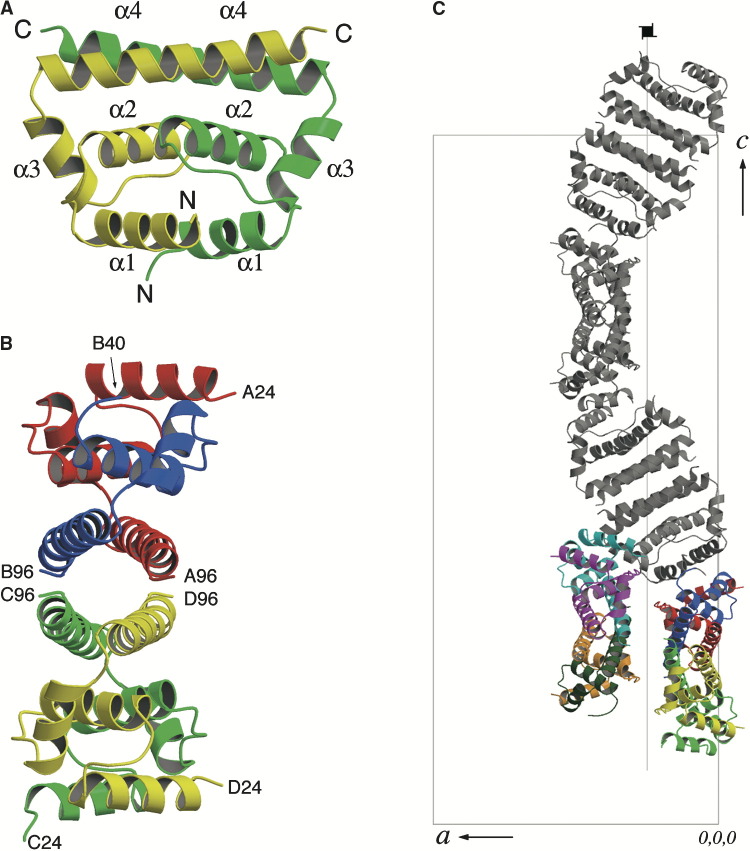
Figure 3.
Ribbon Diagrams of the WNV C Protein
The diagrams have been colored as follows: A, red; B, blue; C, green; D, yellow; E, cyan; F, magenta; G, orange; H, teal.
(A) Ribbon diagram of CD dimer (Esnouf, 1999). Secondary structure elements are indicated.
(B) Ribbon diagram of ABDC tetramer looking down the tunnel. N- and C-terminal residues are indicated.
(C) Arrangement of tetramers in the crystal, viewed down the b axis. The unit cell, the crystal a and c axes and the 4-fold screw axis are indicated. The ABCD and EFGH tetramers in the asymmetric unit are shown in color, while the symmetry-related ABCD tetramers that form one half of the tubule are shown in gray. The symmetry-related EFGH tetramers are omitted for clarity.
In the crystal, two dimers form a tetramer with 222 symmetry, with a cluster of four NCS-related α4 helices forming a tunnel-like cavity (Figure 3B). Two such tetramers, rotated by 58° around an axis parallel to the crystal a axis, comprise the asymmetric unit of the crystal (Figure 3C). In the crystal, symmetry-related tetramers form long, helical ribbons that spiral around the 4-fold screw axes and extend the length of the crystal c axis (Figure 3C). Two such ribbons together form a hollow tubule at each of the two 4-fold screw axes in the crystal. This leaves a large space between adjacent tubules, which is reflected in the relatively high solvent content (57%). Interestingly, the resulting repeated stacking of α helices in the tubules resembles the long, filamentous structures commonly formed by HEAT and ARM repeat proteins, multimeric structures typically involved in protein-protein interactions (Andrade et al., 2001).
The AB dimer encloses a hydrophobic pocket formed by Leu29, Leu36, Phe45, Leu49, Phe52, and Phe53 from the α1 and α2 helices (Figure 4A) . The symmetry related C′D′ dimer approaches the pocket, so that the α1 helices from A, C′, and D′ form a trimeric bundle. This would cause helix α1 from B and C′ to overlap; consequently, density for α1 in subunit B is missing, suggesting that this helix undergoes an order-disorder transition upon crystallization. Such order-disorder transitions are often of functional importance and may suggest a role in the conformational switching that is required during core assembly (Dokland, 2000). The density is very poor for the overlapping α helices in subunit C, perhaps due to two alternative orientations coexisting in the crystal. The GH dimer forms similar interactions with E′F′; in this case α1 is missing from H where it would overlap with F′. The different orientation of α1 in the dengue C protein (Ma et al., 2004) also suggests that this helix is flexible, perhaps reflecting a functional role in assembly or RNA binding (Figure 2D). The hydrophobic pocket contains additional density that was not interpretable at 3.2 Å resolution, but could conceivably represent part of the missing density for the B and H subunits, or perhaps a polyethylene glycol molecule from the crystallization solution. There is a cation, probably Ca2+, sitting on the crystallographic 2-fold axis between 2-fold related Thr43 residues and an anion, possibly Cl−, held between 2-fold related Arg31residues, about 9.5 Å from the cation, mediating the interactions between tubules in adjacent asymmetric units. The two ribbons within the tubule are held together by interactions between Arg69 residues in the A–G and B–E chains.
Figure 4.
Interactions and Surface Potential of the WNV Core Protein Tetramer
(A) View of the interface between the AB dimer (red, blue) and the symmetry-related C′D′ dimer (green, yellow) (Esnouf, 1999). The position that the missing helix α1 in the B subunit would have had where it overlaps with subunit C′ is shown in gray. The Tyr and Leu residues that comprise the hydrophobic pocket in AB are indicated, as is the density enclosed by the pocket in AB and C′D′.
(B) Electrostatic potential surface of C protein tetramer, viewed in the same orientation as Figure 3B.
The WNV C protein dimer presumably represents the building block for core assembly, which also involves interactions with the viral RNA. Both the N-terminal (1–32) and C-terminal (83–103) sequences of the Kunjin virus C protein are involved in RNA binding (Khromykh and Westaway, 1996). The N-terminal region corresponds to the disordered region (residues 1–22) that was cleaved off by the trypsin treatment, and the C-terminal region corresponds to helix α4, which forms an 11 Å wide tunnel in the tetramer (Figure 3B). The tunnel contains a number of positively charged residues, consistent with a role in RNA binding (Figure 4B). The N- and C-terminal RNA binding regions are located at opposite ends of the dimer. While a conformational switch in the N-terminal region, perhaps reflected in the order-disorder transition in helix α1, could bring the two termini together on one side, the organization would appear to be very different from that of the togaviruses or the arteriviruses, which have a clear “inside” or RNA binding face Choi et al. 1991, Doan and Dokland 2003. It is possible that the RNA instead associates on the surface of the C tetramer, in a way similar to that proposed for the tetrameric Borna virus nucleoprotein (Rudolph et al., 2003). Indeed, the C tetramer has a strongly positively charged surface (Figure 4B). In this model, the tetramers would form the building blocks for capsid assembly. The order-disorder transition in helix α1 (Figure 4A) might thus reflect tetramer-tetramer interactions in the nucleocapsid. However, the lack of a structured core in EM reconstructions of dengue or WNV could mean that there is no ordered shell, and that the RNA is instead packed together with protein in a nonspecific manner Kuhn et al. 2002, Mukhopadhyay et al. 2003, Zhang et al. 2003.
A hydrophobic region, corresponding to residues 45–65 in WNV, was proposed to localize the C protein to the membrane in dengue and TBE Kofler et al. 2002, Markoff et al. 1997. In the C structure, this region forms the hydrophobic pocket that is partially protected by helix α1. Movement of α1 might render the hydrophobic region in a position able to interact with the membrane. Part of the hydrophobic region of TBE C (residues 28–43), which corresponds to parts of helices α1 and α2 in WNV, could be removed while still retaining some viability (Kofler et al., 2002). This suggests that the RNA binding properties of the C-terminal domain are more important than the specific fold of the protein, or that RNA binding is more unspecific in nature. Furthermore, viability was partially restored by mutations that increased the hydrophobic nature of the remaining sequence, indicating that membrane localization is done through nonspecific hydrophobic interactions (Kofler et al., 2003).
In vivo, C is localized to the ER membrane through prM, prior to cleavage of C-prM by the NS2B-NS3 protease Lobigs and Lee 2004, Mackenzie and Westaway 2001. Incorporation of cores into virions occurs at the ER membrane and also involves nonstructural proteins NS2A and NS3. The EM structures of dengue and WNV Kuhn et al. 2002, Mukhopadhyay et al. 2003, Zhang et al. 2003 did not reveal cytoplasmic extensions of the M and E proteins, so there may not be a direct interaction between C and prM/E. Virion budding is not dependent on C, and VLPs could be produced by expression of TBE prM and E (Ferlenghi et al., 2001), while expression of dengue C, prM, and E led to the formation of VLPs that lacked C (Sugrue et al., 1997).
Based on differences in genomic organization as well as homologies in nonstructural proteins, the Flaviviridae are considered to belong to an evolutionarily distinct superfamily from Togaviridae and the Nidovirales, which includes coronaviruses and arteriviruses (Strauss and Strauss, 2002). Indeed, the structures of the core proteins from these three superfamilies are different. The Sindbis virus (Togaviridae) core protein has a chymotrypsin-like β-barrel fold (Choi et al., 1991). The recently determined structure of the core (N) protein of the arterivirus PRRSV displayed a novel motif based on an antiparallel β sheet flanked by α helices (Doan and Dokland, 2003). As already suspected from analysis of charge distribution and secondary structure predictions, the flavivirus core structure presented here has a different structure, based entirely on α-helical bundles. Yet, there are some similarities between the WNV C and PRRSV N structures: Both proteins are dimeric; the dimers have an antiparallel α helix pair and are flanked by connecting α helices; both structures form long filamentous ribbon-like or tubular structures that extend in the direction of the long crystal c axis. In WNV, these filaments are formed by stacking of α-helical domains (Figure 3C), similar to those formed by HEAT repeat proteins (Andrade et al., 2001). These similarities are probably based on similar propensities or requirements for forming multimeric shells with a range of different interactions, and may reflect subunit and RNA interactions in the virus.
Experimental Procedures
The 103 first amino acids of the C protein of Kunjin virus (strain MRM61C) were cloned with an N-terminal 6×His tag in the vector pET16b (Novagen), expressed in E. coli strain BL21(DE3) (Novagen) and purified by affinity and ion exchange chromatography. The protein was treated with low concentrations of trypsin, resulting in a stable 10.5 kDa fragment. N-terminal protein sequencing and mass spectrometry confirmed that the fragment started from Val23 and ended at Arg98. This 76 residue fragment was crystallized by vapor diffusion against 0.5%–2% PEG 3350 at pH 10.5 and yielded stable crystals of up to 0.2 mm length, which diffracted synchrotron radiation anisotropically to about 2.8 Å resolution. Se-Met derivative crystals were made by the same procedure by growing the cells in M9 minimal media supplemented with amino acids Lys, Phe, Thr, Ile, Leu, Val, and Se-Met and 0.4% glucose (Doublie, 1997).
Native and Se-Met derivative data were collected at the European Synchrotron Radiation Facility beamline BM14 using a marCCD detector (Mar USA, Inc.) and processed using HKL2000 (Otwinowski and Minor, 1997) to a resolution of 2.8 Å for the native data and 3.6 Å for the Se-Met data (Table 1). The crystals belonged to space group I41 with a = 85.7 Å and c = 214.4 Å, corresponding to a total of eight protein monomers in the asymmetric unit with VM = 2.8. Structure determination by Se-Met MAD methods was done using SOLVE and RESOLVE Terwilliger 2002, Terwilliger 2003. Starting from the initial RESOLVE trace, the noncrystallographic symmetry (NCS) that resulted in the generation of a total of four dimers in the asymmetric unit was identified and used for 8-fold NCS averaging and map improvement with DM (Cowtan and Main, 1998), as well as phase extension to 3.2 Å resolution using the native data. All eight subunits were built manually into the 2Fo − Fc map using the program O (Jones et al., 1991), and the sequence could be unambiguously assigned in residues 24–96 in most of the chains (Figure 2A). Density for residues 24–39 was missing in two chains, and was poor for another two chains. Refinement with REFMAC5 (Murshudov et al., 1997) yielded a final Rcryst = 0.25 and Rfree = 0.31 (Table 1).
Published: July 13, 2004
Accession Numbers
The structure has been deposited in the Protein Data Bank with accession code 1SFK.
References
- Andrade M.A., Petosa C., O'Donoghue S.I., Müller C.W., Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- Brinton M.A. The molecular biology of West Nile virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- Buckley A., Dawson A., Moss S.R., Hinsley S.A., Bellamy P.E., Gould E.A. Serological evidence of West Nile virus, Usuru virus and Sindbis virus infection of birds in the UK. J. Gen. Virol. 2003;84:2807–2817. doi: 10.1099/vir.0.19341-0. [DOI] [PubMed] [Google Scholar]
- CDC. (2004). West Nile Virus: Statistics, Surveillance and Control (Center for Disease Control). http://www.cdc.gov/ncidod/dvbid/westnile/surv&control.htm.
- Choi H.-K., Tong L., Minor W., Dumas P., Boege U., Rossmann M.G., Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine protease and the organization of the virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- Cowtan K., Main P. Miscellaneous algorithms for density modification. Acta Crystallogr. D Biol. Crystallogr. 1998;54:487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- Doan D.N.P., Dokland T. Structure of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus. Structure. 2003;11:1445–1451. doi: 10.1016/j.str.2003.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokland T. Freedom and restraint: themes in virus capsid assembly. Structure. 2000;8:R157–R167. doi: 10.1016/s0969-2126(00)00181-7. [DOI] [PubMed] [Google Scholar]
- Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- Esnouf R.M. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- Ferlenghi I., Clarke M., Ruttan T., Allison S.L., Schalich J., Heinz F.X., Harrison S.C., Rey F.A., Fuller S.D. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell. 2001;7:593–602. doi: 10.1016/s1097-2765(01)00206-4. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle E., Stuart D.I., Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Hall R.A., Nisbet D.J., Pham K.B., Pyke A.T., Smith G.A., Khromykh A.A. DNA vaccine coding for the full-length infectious Kunjin virus RNA protects mice against the New York strain of West Nile Virus. Proc. Natl. Acad. Sci. USA. 2003;100:10460–10464. doi: 10.1073/pnas.1834270100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou J.-Y., Cowan S.W., Kjeldgaard M. Improved methods for the building of protein models in electron density and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Khromykh A.A., Westaway E.G. RNA binding properties of core protein of the flavivirus Kunjin. Arch. Virol. 1996;141:685–699. doi: 10.1007/BF01718326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R.M., Heinz F.X., Mandl C.W. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 2002;76:3534–3543. doi: 10.1128/JVI.76.7.3534-3543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R.M., Leitner A., O'Riordan G., Heinz F.X., Mandl C.W. Spontaneous mutations restore the viability of tick-borne encephalitis virus mutants with large deletions in protein C. J. Virol. 2003;77:443–451. doi: 10.1128/JVI.77.1.443-451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosa P.F., Ghosh G., DeDecker B.S., Sigler P.B. The 2.1-Å crystal structure of an archeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc. Natl. Acad. Sci. USA. 1997;94:6042–6047. doi: 10.1073/pnas.94.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R.J., Zhang W., Rossmann M.G., Pletnev S.V., Corver J., Lenches E., Jones C.T., Mukhopadhyay S., Chipman P.R., Strauss E.G. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R.S., Roehrig J.T., Deubel V., Smith J., Parker M., Steele K., Crise B., Volpe K.E., Crabtree M.B., Scherret J.H. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science. 1999;286:2333–2337. doi: 10.1126/science.286.5448.2333. [DOI] [PubMed] [Google Scholar]
- Lindenbach B.D., Rice C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2002;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- Lobigs M., Lee E. Inefficient signalase cleavage promotes efficient nucleocapsid incorporation into budding flavivirus membranes. J. Virol. 2004;78:178–186. doi: 10.1128/JVI.78.1.178-186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Jones C.T., Groesch T.D., Kuhn R.J., Post C.B. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. USA. 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.M., Westaway E.G. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 2001;75:10787–10799. doi: 10.1128/JVI.75.22.10787-10799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J., Lomakin I.B., Sonenberg N., Pestova T.V., Hellen C.U.T., Burley S.K. A conserved HEAT domain within eIF4G directs assembly of the translation initation machinery. Mol. Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- Markoff L., Falgout B., Chang A. A conserved internal hydrophobic domain mediates the stable membrane integration of the dengue virus capsid protein. Virology. 1997;233:105–117. doi: 10.1006/viro.1997.8608. [DOI] [PubMed] [Google Scholar]
- Modis Y., Ogata S., Clements D., Harrison S.C. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Kim B.S., Chipman P.R., Rossmann M.G., Kuhn R.J. Structure of West Nile virus. Science. 2003;302:248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C.W. Jr., Sweet R.M., editors. Methods Enzymology. Academic Press; San Diego: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Petersen L.R., Marfin A.A., Gubler D.J. West Nile virus. JAMA. 2003;290:524–528. doi: 10.1001/jama.290.4.524. [DOI] [PubMed] [Google Scholar]
- Rey F.A., Heinz F.X., Mandl C., Kunz C., Harrison S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rudolph M.G., Kraus I., Dickmanns A., Eickmann M., Garten W., Ficner R. Crystal structure of the Borna virus nucleoprotein. Structure. 2003;11:1219–1226. doi: 10.1016/j.str.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Scherret J.H., Poidinger M., Mackenzie J.S., Broom A.K., Deubel V., Lipkin W.I., Briese T., Gould E.A., Hall R.A. The relationships between West Nile and Kunjin viruses. Emerg. Infect. Dis. 2001;7:697–705. doi: 10.3201/eid0704.010418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J.H., Strauss E.G. Viruses and Human Disease. Academic Press; San Diego: 2002. [Google Scholar]
- Sugrue R.J., Fu J., Howe J., Chan Y.C. Expression of the dengue virus structural proteins in Pichia pastoris leads to the generation of virus-like particles. J. Gen. Virol. 1997;78:1861–1866. doi: 10.1099/0022-1317-78-8-1861. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. Automated structure solution, density modification and model building. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1937–1940. doi: 10.1107/s0907444902016438. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Corver J., Chipman P.R., Zhang W., Pletnev S.V., Sedlak D., Baker T.S., Strauss J.H., Kuhn R.J., Rossmann M.G. Structures of immature flavivirus particles. EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]