To the Editor:
Rhinovirus infection is a common trigger of bronchiolitis and early wheezing in children.1 Its detection is clinically important because rhinovirus-induced bronchiolitis/early wheezing probably is an important risk factor for recurrent wheezing and childhood asthma.1, 2 The mechanisms behind the undesirable long-term sequela remain poorly understood, but potential factors include atopic inheritance, weak antiviral defense, and viral factors.3
Looking closely at previous reports on quantitative rhinovirus detection, we did not find any on the rhinovirus genomic load in bronchiolitis and short-term clinical outcomes, including the need for intensive care treatment. In other conditions, however, higher rhinovirus genomic load is related to the severity and/or duration of acute lower respiratory tract illness, and 1 study reported that it discriminated the response to systemic corticosteroids in terms of less recurrent wheezing.1, 4, 5 Data on the link between rhinovirus genomic load and clinical outcomes, however, are discordant because studies in subjects with asthma have not shown any clinical association.6 For these reasons and the relatively small samples in earlier studies, we examined the clinical significance of rhinovirus genomic load in bronchiolitis in 694 children with severe bronchiolitis. Our aim was to prospectively investigate whether rhinovirus genomic load in standardized nasopharyngeal aspirate (NPA) samples is associated with short-term outcomes of bronchiolitis. On the basis of previous literature, our hypothesis was that higher rhinovirus genomic load in bronchiolitis is associated with worse short-term outcomes.
For this analysis, we combined data from 2 multicenter prospective cohort studies of children younger than 2 years hospitalized for bronchiolitis; both studies used the same protocol. The US study7 was carried out at 16 sites across 12 US states during the 2007-2010 winter seasons (Multicenter Airway Research Collaboration [MARC]-30 USA) (see Table E1 in this article's Online Repository at www.jacionline.org), whereas the Finnish counterpart study8 was carried out in 3 Finnish sites during the 2008-2010 winter seasons (MARC-30 Finland). See more details of the MARC-30 and recruitment in this article's Online Repository at www.jacionline.org. The study protocol was approved by the ethics committees of participating hospitals, and the study was commenced only after obtaining written informed consent from the guardian.
Investigators interviewed a guardian using a standard questionnaire and conducted a hospital chart review for further clinical data. NPA sampling was performed using a standardized protocol. Samples were stored at −80°C for later virus diagnostics, which included real-time PCR for adenovirus, coronaviruses NL-63, HKU1, OC43, and 229E, enterovirus, human metapneumovirus, influenza virus types A and B, 2009 novel H1N1, parainfluenza virus types 1, 2, and 3, rhinovirus, respiratory syncytial virus (RSV) A and B, Bordetella pertussis, and Mycoplasma pneumonia, as previously described.7 Rhinovirus genomic load was quantified by using real-time RT-PCR as the number of amplification cycles needed for a positive PCR test result (cycle threshold [CT]). CT values provide a semi-quantitative measure of genomic load, with a highly significant inverse linear relationship between genomic load and CT values. See more details of the virus diagnostics in this article's Methods section in the Online Repository at www.jacionline.org.
Our primary outcome measure was hospital length of stay (LOS) of 3 days or more.7, 8 The secondary outcome measure was intensive care treatment, defined as use of mechanical ventilation (continuous positive airway pressure and/or intubation during inpatient stay regardless of location) and/or admission to the intensive care unit.9 Tertiles of rhinovirus CT values permitted classification into 3 rhinovirus genomic load groups: low (CT ≥ 32.7), intermediate (CT, 27.2-32.6), and high (CT < 27.2). The association between rhinovirus genomic load and the outcomes was analyzed using unadjusted and multivariable logistic regression models. Several sensitivity analyses were performed to assess the robustness of the findings. All analyses were performed using SAS 9.4 (SAS Institute, Cary, NC). See more details of the outcomes and statistical methods in the Online Repository at www.jacionline.org.
Of 2615 enrolled children with bronchiolitis from 19 sites, 694 children (27%) had rhinovirus and comprised the analytic cohort (564 US children and 130 Finnish children). Among these children, the median age was 6 months (interquartile range, 3-12 months), 63% were boys, and 46% were non-Hispanic white. Two hundred sixty (37%) children had an LOS of 3 days or more, and 102 (15%) required intensive care treatment. See more details of demographics and clinical course in Table E2, Table E3 in this article's Online Repository at www.jacionline.org.
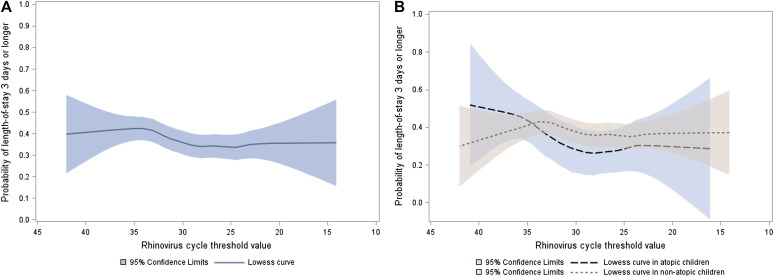
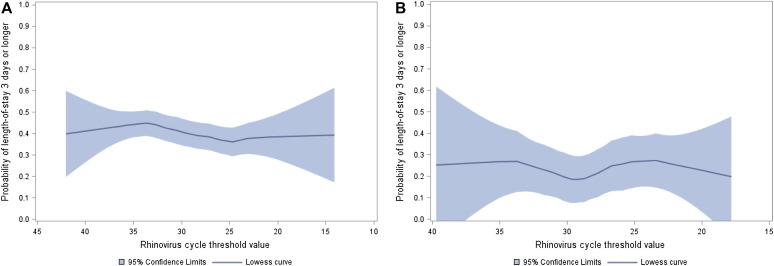
Overall, there was no significant association between rhinovirus genomic load (an inverse of the CT value) and risk of LOS of 3 days or more or risk of intensive care treatment, either in unadjusted analyses or in multivariable models adjusting for 8 patient-level variables and clustering of patients within sites (all P ≥ .40, Fig 1 , A; see Table E4 in this article's Online Repository at www.jacionline.org). Likewise, a sensitivity analysis focused on the first episode in infants younger than 12 months showed no significant associations (all P > .30; see Table E4). Similarly, rhinovirus genomic load had no significant associations with the outcomes, by country (see Fig E1, A and B, in this article's Online Repository at www.jacionline.org), coinfection (see Table E5 in this article's Online Repository at www.jacionline.org), atopy status (Fig 1, B; see Table E6, Table E7 in this article's Online Repository at www.jacionline.org), comorbid status (see Table E8 in this article's Online Repository at www.jacionline.org), or respiratory distress severity score (see Table E9 in this article's Online Repository at www.jacionline.org).
Fig 1.
The relation between rhinovirus CT value and hospital LOS overall (A) and by atopic status (B) in children hospitalized for bronchiolitis.
Fig E1.
The relation between rhinovirus CT value and hospital LOS in US (A) and Finnish (B) cohorts of children with bronchiolitis.
In summary, we found no association between rhinovirus genomic load and short-term outcomes of bronchiolitis. Our hypothesis was justified on the basis of previous clinical data,1, 4, 5 which were also supported by in vitro data.3 Although multiple viral infections are relatively common in severe bronchiolitis (15% to 30%),7, 8 the interplay between viruses is poorly understood. Coinfection with RSV and rhinovirus has been linked to more severe short-term outcomes of bronchiolitis compared with RSV alone,7 but we found no link between rhinovirus genomic load, coinfections, and these same outcomes. Even when examining the rhinovirus-only group, the association was null. Moreover, investigation of the interaction between the rhinovirus genomic load and atopic status was interesting because atopic children appear to be more susceptible than nonatopic children to rhinovirus-induced wheezing.2
Considering the large sample size, careful standardization of NPA sampling, and virus diagnostics done with the same protocol in a single laboratory, our results truly suggest no significant association between rhinovirus genomic load and an LOS of 3 days or more or need for intensive care treatment. Although 1 study suggested that rhinovirus genomic load has more clinical relevance in children older than 12 months,4 this association is not supported by our data or other reports.5 Because our results contrast the direct association between RSV genomic load and short-term outcomes of bronchiolitis,10 we speculate that a host response to infection may be more important than virus load in determining the short-term clinical course of rhinovirus-induced bronchiolitis.3
The study has potential limitations. First, bronchiolitis is a clinical diagnosis without a common international definition,11 so we included children up to age 2 years with recurrent wheezing. Results, however, remained consistent when the analysis was restricted to children experiencing their first episode of breathing difficulty during infancy (age <12 months). Second, clinical decisions (eg, hospital admission/discharge or intensive care treatment) were not based on standardized criteria, which may have caused further variability of care. However, the significant association persisted after adjusting for clustering at the hospital level. Third, one might argue that samples from the upper respiratory tract do not reflect conditions in the lower respiratory tract and that nasal airway epithelial cells may respond differently than bronchial epithelial cells to rhinovirus infections.12 To our knowledge, there are no data on the comparison of rhinovirus genomic load between upper and lower airway samples and their relation to symptoms. Fourth, one could also argue whether we measured the peak of rhinovirus replication due to lack of longitudinal sampling. A peak in virus concentration typically occurs at 48 to 72 hours after infection in experimental models.13 Because the duration of prehospital symptoms is typically 1 to 3 days in rhinovirus-induced bronchiolitis,2 our time window of the first 24 hours of the hospitalization may have been optimal. Fifth, we did not sequence rhinoviruses.14, 15 Last, the results may not be generalizable to outpatient clinics because all our study subjects were hospitalized.
Challenges in future studies include more careful standardization of analysis (ie, standardization to housekeeping gene), investigation of viremia (ie, links to more compromised clinical outcome), virus genotyping (ie, rhinovirus species and rapid evolution of the virus), and more careful analysis of the replication/transcription status of the virus (ie, separate analysis of positive- and negative-stranded virus RNA).5, 6 Our findings call attention to the need for more detailed analysis of virology, along with host response and genetics, when investigating predictors of short-term outcomes of severe rhinovirus-induced bronchiolitis.
Footnotes
This study was supported by the Academy of Finland (grant nos. 132595 and 114034), Helsinki, Finland; the Sigrid Juselius Foundation, Helsinki, Finland; the Foundation for Pediatric Research, Helsinki, Finland; and the National Institutes of Health, Bethesda, Md (grant no. U01 AI-67693). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, or the other institutes that provided support.
Disclosure of potential conflict of interest: T. Jartti has received research support from the Academy of Finland (grant nos. 132595 and 114034), the Sigrid Juselius Foundation, and the Foundation for Pediatric Research. K. Hasegawa has received research support from the Milton Fund. J. M. Mansbach has received research support from the National Institutes of Health and has consultant arrangements with Regeneron. The rest of the authors declare that they have no relevant conflicts of interest.
Methods
Study design, setting, and subjects
The present analysis combines data from 2 multicenter prospective cohort studies of children hospitalized for bronchiolitis. Using a similar protocol, one study was from the United StatesE1 and the other was from Finland.E2 Both studies were performed as part of MARC. MARC is a program of the Emergency Medicine Network (www.emnet-usa.org), a collaboration with more than 225 participating hospitals. The study design, setting, participants, and methods of data collection used in the studies have been reported previously.E1, E2 Using a standardized protocol, we enrolled children younger than 2 years hospitalized for an attending physician's diagnosis of bronchiolitis. The exclusion criteria consisted of previous enrollment and delay of more than 48 hours in transfer to a participating hospital after the original hospitalization. All patients were treated at the discretion of the treating physician. The institutional review board at each of the participating hospitals approved the study.
NPA collection and virology testing
For the collection of NPAs, the child was placed supine, 1 mL of normal saline was instilled into 1 naris, and an 8-F suction catheter was used to remove the mucus. This procedure was performed once on each nostril. After the sample collection from both nares, 2 mL of normal saline was suctioned through the catheter to clear the tubing and to ensure that a standard volume of aspirate was obtained. Once collected, the NPA sample was added to the transport medium. The samples were immediately placed on ice within 1 hour of collection, and then stored at −80°C within 24 hours of collection.
PCR assay
Real-time RT-PCR was used for the detection of RNA respiratory viruses, such as rhinovirus, RSV types A and B, parainfluenza virus types 1, 2, and 3, influenza virus types A and B, 2009 novel H1N1, human metapneumovirus, coronaviruses NL-63, HKU1, OC43, and 229E, and enterovirus. Real-time PCR was used for the detection of DNA pathogens, which included adenovirus, M pneumoniae, and B pertussis. These tests are routinely conducted in Baylor College of Medicine, and details of the primers and probes have been described previously.E3, E4, E5 The upper and lower limits of rhinovirus detection were 15 and 40 CT, respectively.E6
Statistical methods
For the purpose of our analyses, we focused on rhinovirus. We categorized CT values into tertiles to classify patients into 3 rhinovirus genomic load status groups: low (CT ≥ 32.7), intermediate (CT, 27.2-32.6), and high (CT < 27.2). We compared patients' demographic characteristics, medical history, and hospital course by rhinovirus genomic load status using chi-square or Kruskal-Wallis tests as appropriate. To examine the association of genomic load status with the outcomes, we constructed 2 logistic regression models. First, we fitted an unadjusted model that included only genomic load status as the independent variable. Second, we constructed a multivariable model adjusting for 8 patient-level variables (ie, age, sex, race, gestational age, history of wheezing, history of eczema, comorbid medical disorder, and viral coinfection status [rhinovirus plus RSV and rhinovirus plus non-RSV pathogens]). We chose these potential confounders on the basis of clinical plausibility and a priori knowledge.E1, E2, E7 We did not adjust for markers for acute severity (eg, vital signs and retractions) or duration of symptoms before bronchiolitis hospitalization because these were considered as intermediate factors in the association of interest. In both models, we used generalized estimating equations to account for patient clustering at the hospital level.
We performed a series of sensitivity analyses to assess the robustness of our findings. First, we examined the association of rhinovirus genomic load and the primary outcome, modeling the CT value as a continuous variable, in the US cohort and the Finnish cohort separately. Second, after confirming a similar association in both the cohorts, we combined the US and Finnish data set, and then repeated the analysis by using a more restrictive definition of children with bronchiolitis—that is, those younger than 12 months and without history of wheezing. Third, we stratified the analysis by coinfection status (rhinovirus only, rhinovirus plus RSV, and rhinovirus plus non-RSV pathogens). Last, we also stratified the analysis by children's atopic status (ie, history of eczema). All analyses were performed using SAS 9.4 (SAS Institute, Cary, NC). Results are presented as proportions with 95% CIs, medians with interquartile ranges, and odds ratios with 95% CIs. All P values were 2-tailed, with P < .05 considered statistically significant.
Results
Patients' characteristics
Among the analytic cohort of 694 children, 259 children (37%) had bronchiolitis with rhinovirus only and 435 (63%) had bronchiolitis with 2 or more viruses. More specifically, 297 (43%) had rhinovirus plus RSV and 138 (20%) had rhinovirus plus non-RSV pathogens. The median hospital LOS was 2 days (interquartile range, 1-4 days). Of the 694 children in the analytic cohort, 234 children (34%) were categorized into the low rhinovirus genomic load group, 230 children (33%) into the intermediate load group, and 230 children (33%) into the high load group.
Table E1.
Principal investigators at the 19 participating sites in MARC-30
| MARC-30 US sites | |
| Besh Barcega, MD | Loma Linda University Children's Hospital, Loma Linda, Calif |
| John Cheng, MD, and Carlos Delgado, MD | Children's Healthcare of Atlanta at Egleston, Atlanta, Ga |
| Dorothy Damore, MD, and Nikhil Shah, MD | New York Presbyterian Hospital, New York, NY |
| Haitham Haddad, MD | Rainbow Babies & Children's Hospital, Cleveland, Ohio |
| Paul Hain, MD, and Mark Riederer, MD | Monroe Carell Jr. Children's Hospital at Vanderbilt, Nashville, Tenn |
| Frank LoVecchio, DO | Maricopa Medical Center, Phoenix, Ariz |
| Charles Macias, MD, MPH | Texas Children's Hospital, Houston, Tex |
| Jonathan Mansbach, MD, MPH | Boston Children's Hospital, Boston, Mass |
| Eugene Mowad, MD | Akron Children's Hospital, Akron, Ohio |
| Brian Pate, MD | Children's Mercy Hospital & Clinics, Kansas City, Mo |
| M. Jason Sanders, MD | Children's Memorial Hermann Hospital, Houston, Tex |
| Alan Schroeder, MD | Santa Clara Valley Medical Center, San Jose, Calif |
| Michelle Stevenson, MD, MS | Kosair Children's Hospital, Louisville, Ky |
| Erin Stucky Fisher, MD | Rady Children's Hospital, San Diego, Calif |
| Stephen Teach, MD, MPH | Children's National Medical Center, Washington, DC |
| Lisa Zaoutis, MD | Children's Hospital of Philadelphia, Philadelphia, Pa |
| MARC-30 Finland sites | |
| Tuomas Jartti, MD | Turku University Hospital, Turku, Finland |
| Matti Korppi, MD | Tampere University Hospital, Tampere, Finland |
| Sami Remes, MD | Kuopio University Hospital, Kuopio, Finland |
Table E2.
Demographic characteristics and medical history of children hospitalized with rhinovirus bronchiolitis by genomic load category
| Characteristic | Virus genomic load∗ |
|||
|---|---|---|---|---|
| Low (n = 234) | Intermediate (n = 230) | High (n = 230) | P value | |
| Age (mo) | .15 | |||
| <2 | 35 (15) | 45 (20) | 42 (18) | |
| 2-5.9 | 79 (34) | 66 (29) | 66 (29) | |
| 6-11.9 | 73 (31) | 54 (24) | 71 (31) | |
| 12-23.9 | 47 (20) | 65 (28) | 51 (22) | |
| Sex: male | 144 (62) | 158 (69) | 136 (59) | .09 |
| Race/ethnicity | <.001 | |||
| Non-Hispanic white | 80 (34) | 124 (54) | 118 (51) | |
| Non-Hispanic black | 71 (30) | 42 (18) | 38 (17) | |
| Hispanic | 72 (31) | 58 (25) | 70 (30) | |
| Other | 11 (5) | 6 (3) | 4 (2) | |
| Insurance | .52 | |||
| Nonprivate | 154 (66) | 162 (70) | 160 (70) | |
| Private | 80 (34) | 68 (30) | 70 (30) | |
| Family history of asthma | .36 | |||
| Neither parent | 152 (65) | 153 (67) | 166 (72) | |
| Either mother or father | 66 (28) | 66 (29) | 56 (24) | |
| Both parents | 10 (4) | 8 (4) | 4 (2) | |
| Unknown/missing | 6 (3) | 3 (1) | 4 (2) | |
| Maternal smoking during pregnancy | 41 (18) | 38 (17) | 37 (16) | .91 |
| Gestational age | .84 | |||
| <32 wk | 16 (7) | 21 (9) | 16 (7) | |
| 32-36 wk | 41 (18) | 42 (18) | 41 (18) | |
| ≥37 wk or “full term” | 173 (74) | 160 (70) | 171 (74) | |
| Is or was breast-fed | 147 (63) | 149 (65) | 159 (69) | .34 |
| History of wheezing | 74 (32) | 82 (36) | 77 (34) | .66 |
| History of eczema | 62 (27) | 34 (15) | 52 (23) | .006 |
| History of intubation | 24 (10) | 22 (10) | 28 (12) | .64 |
| Major, relevant, comorbid medical disorder† | 65 (28) | 48 (21) | 48 (21) | .12 |
| Cohort | <.001 | |||
| United States | 211 (90) | 176 (77) | 177 (77) | |
| Finland | 23 (10) | 54 (23) | 53 (23) | |
Data are expressed as n (%) unless otherwise indicated.
Categorized CT values into tertiles to classify patients into 3 rhinovirus genomic load status groups: low (CT ≥ 32.7), intermediate (CT, 27.2-32.6), and high (CT < 27.2).
Defined by respiratory, cardiac, neurologic, gastrointestinal, and immunologic diseases.
Table E3.
Clinical course of children hospitalized with rhinovirus bronchiolitis by genomic load category
| Characteristic | Virus genomic load∗ |
P value | ||
|---|---|---|---|---|
| Low (n = 234) | Intermediate (n = 230) | High (n = 230) | ||
| When difficulty breathing began (prehospitalization) | .10 | |||
| ≥1 d | 66 (28) | 74 (32) | 87 (38) | |
| <1 d | 160 (68) | 153 (67) | 135 (59) | |
| No difficulty prehospitalization | 8 (3) | 3 (1) | 8 (3) | |
| Presence of apnea (chart) | 14 (6) | 13 (6) | 15 (7) | .92 |
| Weight (kg), median (IQR) | 7.3 (5.1-9.5) | 7.0 (4.7-10.0) | 7.3 (4.7-9.6) | .92 |
| Pulse (bpm), median (IQR) | 160 (144-176) | 160 (144-173) | 160 (147-176) | .94 |
| Respiratory rate per minute, median (IQR) | 48 (40-60) | 50 (40-60) | 48 (40-58) | .86 |
| Oxygen saturation by pulse oximetry or ABG | .81 | |||
| <90% | 32 (14) | 31 (13) | 24 (10) | |
| 90% to 93.9% | 40 (17) | 39 (17) | 41 (18) | |
| ≥94% | 155 (66) | 155 (68) | 163 (71) | |
| Retractions | .68 | |||
| None | 33 (14) | 44 (19) | 36 (16) | |
| Mild | 94 (40) | 83 (36) | 85 (40) | |
| Moderate or severe | 88 (38) | 91 (40) | 85 (37) | |
| Missing | 19 (8) | 12 (5) | 24 (10) | |
| Oral intake | .01 | |||
| Adequate | 102 (44) | 132 (57) | 123 (53) | |
| Inadequate | 96 (41) | 68 (30) | 82 (36) | |
| Missing | 36 (15) | 30 (13) | 25 (11) | |
| Coinfection | <.001 | |||
| Rhinovirus + RSV | 131 (56) | 97 (42) | 69 (30) | |
| Rhinovirus + non-RSV pathogens | 36 (15) | 47 (20) | 55 (24) | |
| Sole rhinovirus infection | 67 (29) | 86 (37) | 106 (46) | |
| Length of stay (d), median (IQR) | 2 (1-4) | 2 (1-4) | 2 (1-3) | .39 |
| ≥3 | 96 (41) | 85 (37) | 79 (34) | .33 |
| Intensive care treatment | 39 (17) | 30 (13) | 33 (14) | .71 |
| Intubation and/or CPAP | 20 (9) | 12 (5) | 11 (5) | .20 |
| Intensive care unit admission | 37 (16) | 29 (13) | 30 (13) | .65 |
Data are expressed as n (%) unless otherwise indicated.
ABG, Arterial blood gas; bpm, beats per minute; CPAP, continuous positive airway pressure; IQR, interquartile range.
Categorized CT values into tertiles to classify patients into 3 rhinovirus genomic load status groups: low (CT ≥ 32.7), intermediate (CT, 27.2-32.6), and high (CT < 27.2).
Table E4.
Unadjusted and multivariable associations of rhinovirus genomic load with bronchiolitis outcomes
| Outcome and rhinovirus genomic load category | Unadjusted model∗ |
Adjusted model† |
Sensitivity analysis‡ |
|||
|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| Length of stay ≥3 d | ||||||
| Low | Reference | — | Reference | — | Reference | — |
| Intermediate | 0.85 (0.56-1.29) | .78 | 1.07 (0.75-1.54) | .70 | 0.89 (0.58-1.37) | .60 |
| High | 0.96 (0.73-1.27) | .43 | 1.05 (0.65-1.68) | .85 | 0.92 (0.63-1.34) | .65 |
| Intensive care treatment | ||||||
| Low | Reference | — | Reference | — | Reference | — |
| Intermediate | 0.89 (0.58-1.37) | .60 | 0.97 (0.67-1.40) | .87 | 0.69 (0.30-1.54) | .36 |
| High | 0.92 (0.63-1.34) | .65 | 0.78 (0.43-1.40) | .40 | 0.84 (0.45-1.55) | .58 |
OR, Odds ratio.
Unadjusted model adjusting for clustering of patients within the sites using the generalized estimating equations.
Multivariable model adjusting for 8 patient-level variables (age, sex, race, gestational age, history of wheezing, history of eczema, comorbid medical disorder, and viral coinfection status [rhinovirus plus RSV and rhinovirus plus non-RSV pathogens]) and clustering of patients within the sites.
Multivariable model using a restrictive definition of children with bronchiolitis—ie, those younger than 12 months and without history of wheezing (n = 389).
Table E5.
Unadjusted and multivariable associations of rhinovirus genomic load with bronchiolitis outcomes, according to the coinfection status
| Outcome and rhinovirus genomic load category | Rhinovirus only |
Rhinovirus plus RSV |
Rhinovirus plus non-RSV pathogens |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unadjusted model∗ |
Adjusted model† |
Unadjusted model∗ |
Adjusted model† |
Unadjusted model∗ |
Adjusted model† |
|||||||
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | |
| Length of stay ≥3 d | ||||||||||||
| Low | Reference | — | Reference | — | Reference | — | Reference | — | Reference | — | Reference | — |
| Intermediate | 0.97 (0.52-1.82) | .92 | 1.24 (0.50-3.06) | .64 | 1.02 (0.68-1.52) | .94 | 1.12 (0.79-1.59) | .53 | 1.63 (0.80-3.33) | .18 | 2.44 (1.06-5.63) | .04 |
| High | 1.17 (0.61-2.25) | .63 | 1.27 (0.62-2.59) | .52 | 0.86 (0.52-1.43) | .56 | 0.83 (0.45-1.55) | .57 | 1.52 (0.54-4.26) | .43 | 2.25 (0.86-5.90) | .10 |
| Intensive care treatment | ||||||||||||
| Low | Reference | — | Reference | — | Reference | — | Reference | — | Reference | — | Reference | — |
| Intermediate | 0.98 (0.28-3.44) | .98 | 1.06 (0.22-5.12) | .94 | 0.84 (0.50-1.44) | .53 | 0.56 (0.32-0.98) | .04 | 0.69 (0.21-2.29) | .54 | 0.97 (0.40-2.43) | .95 |
| High | 1.06 (0.52-2.16) | .88 | 1.07 (0.37-3.07) | .90 | 0.90 (0.59-1.37) | .61 | 0.88 (0.54-1.43) | .60 | 0.77 (0.21-2.83) | .69 | 1.23 (0.43-3.51) | .70 |
OR, Odds ratio.
Unadjusted model adjusting for clustering of patients within the sites using the generalized estimating equations.
Multivariable model adjusting for 7 patient-level variables (age, sex, race, gestational age, history of wheezing, history of eczema, and comorbid medical disorder) and clustering of patients within the sites.
Table E6.
Unadjusted and multivariable associations of rhinovirus genomic load with bronchiolitis outcomes in atopic children∗ (n = 148)
| Outcome and rhinovirus genomic load category | Unadjusted model† |
Adjusted model‡ |
||
|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |
| Length of stay ≥3 d (n = 51 for outcome) | ||||
| Low | Reference | — | Reference | — |
| Intermediate | 0.50 (0.28-0.88) | .02 | 0.35 (0.13-0.91) | .03 |
| High | 0.57 (0.31-1.02) | .06 | 0.61 (0.26-1.40) | .24 |
| Intensive care treatment (n = 15 for outcome) | ||||
| Low | Reference | — | Reference | — |
| Intermediate | 0.39 (0.08-1.89) | .24 | 0.12 (0.02-0.81) | .03 |
| High | 0.67 (0.21-2.16) | .51 | 0.55 (0.12-2.60) | .45 |
OR, Odds ratio.
Children with history of eczema.
Unadjusted model adjusting for clustering of patients within the sites using the generalized estimating equations.
Multivariable model adjusting for 7 patient-level variables (age, sex, race, gestational age, history of wheezing, comorbid medical disorder, and viral coinfection status [rhinovirus plus RSV and rhinovirus plus non-RSV pathogens]) and clustering of patients within the sites.
Table E7.
Unadjusted and multivariable associations of rhinovirus genomic load with bronchiolitis outcomes in nonatopic children∗ (n = 546)
| Outcome and rhinovirus genomic load category | Unadjusted model† |
Adjusted model‡ |
||
|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |
| Length of stay ≥3 d (n = 209 for outcome) | ||||
| Low | Reference | — | Reference | — |
| Intermediate | 1.05 (0.74-1.50) | .78 | 1.18 (0.81-1.71) | .39 |
| High | 0.95 (0.56-1.60) | .84 | 1.12 (0.62-2.04) | .70 |
| Intensive care treatment (n = 87 for outcome) | ||||
| Low | Reference | — | Reference | — |
| Intermediate | 0.89 (0.53-1.50) | .67 | 0.74 (0.39-1.41) | .36 |
| High | 0.88 (0.60-1.29) | .51 | 0.86 (0.59-1.27) | .46 |
OR, Odds ratio.
Children without history of eczema.
Unadjusted model adjusting for clustering of patients within the sites using the generalized estimating equations.
Multivariable model adjusting for 7 patient-level variables (age, sex, race, gestational age, history of wheezing, comorbid medical disorder, and viral coinfection status [rhinovirus plus RSV and rhinovirus plus non-RSV pathogens]) and clustering of patients within the sites.
Table E8.
Unadjusted and multivariable associations of rhinovirus genomic load with bronchiolitis outcomes in children without comorbid medical disorder (n = 528)
| Outcome and rhinovirus genomic load category | Unadjusted model∗ |
Adjusted model† |
||
|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |
| Length of stay ≥3 d (n = 198 for outcome) | ||||
| Low | Reference | — | Reference | — |
| Intermediate | 0.88 (0.64-1.22) | .44 | 1.00 (0.69-1.44) | .99 |
| High | 0.82 (0.52-1.30) | .40 | 1.07 (0.68-1.70) | .77 |
| Intensive care treatment (n = 81 for outcome) | ||||
| Low | Reference | — | Reference | — |
| Intermediate | 0.88 (0.52-1.49) | .63 | 0.80 (0.37-1.73) | .57 |
| High | 0.93 (0.69-1.26) | .64 | 0.99 (0.60-1.63) | .98 |
OR, Odds ratio.
Unadjusted model adjusting for clustering of patients within the sites using the generalized estimating equations.
Multivariable model adjusting for 8 patient-level variables (age, sex, race, gestational age, history of wheezing, comorbid medical disorder, and viral coinfection status [rhinovirus plus RSV and rhinovirus plus non-RSV pathogens]) and clustering of patients within the sites.
Table E9.
Unadjusted and multivariable associations of rhinovirus genomic load with respiratory distress severity score∗† at presentation (n = 694)
| Rhinovirus genomic load category | Unadjusted model† |
Adjusted model‡ |
||
|---|---|---|---|---|
| β Coefficient (95% CI) | P value | β Coefficient (95% CI) | P value | |
| Low | Reference | — | Reference | — |
| Intermediate | 0.03 (0.47-0.40) | .88 | 0.02 (0.42-0.45) | .94 |
| High | 0.11 (0.55-0.34) | .64 | 0.14 (0.31-0.59) | .55 |
Bajaj L, Turner CG, Bothner J. A randomized trial of home oxygen therapy from the emergency department for acute bronchiolitis. Pediatrics 2006;117:633-40.
Linear regression model with respiratory distress severity score as the dependent variable.
Multivariable linear regression model adjusting for 8 patient-level variables (age, sex, race, gestational age, history of wheezing, history of eczema, comorbid medical disorder, and viral coinfection status [rhinovirus plus RSV and rhinovirus plus non-RSV pathogens]).
References
- 1.Jartti T., Nieminen R., Vuorinen T., Lehtinen P., Vahlberg T., Gern J. Short- and long-term efficacy of prednisolone for first acute rhinovirus-induced wheezing episode. J Allergy Clin Immunol. 2015;135:691–698. doi: 10.1016/j.jaci.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson D.J., Evans M.D., Gangnon R.E., Tisler C.J., Pappas T.E., Lee W.M. Evidence for a causal relationship between allergic sensitization and rhinovirus wheezing in early life. Am J Respir Crit Care Med. 2012;185:281–285. doi: 10.1164/rccm.201104-0660OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baraldo S., Contoli M., Bazzan E., Turato G., Padovani A., Marku B. Deficient antiviral immune responses in childhood: distinct roles of atopy and asthma. J Allergy Clin Immunol. 2012;130:1307–1314. doi: 10.1016/j.jaci.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Takeyama A., Hashimoto K., Sato M., Sato T., Kanno S., Takano K. Rhinovirus load and disease severity in children with lower respiratory tract infections. J Med Virol. 2012;84:1135–1142. doi: 10.1002/jmv.23306. [DOI] [PubMed] [Google Scholar]
- 5.Esposito S., Daleno C., Scala A., Castellazzi L., Terranova L., Sferrazza Papa S. Impact of rhinovirus nasopharyngeal viral load and viremia on severity of respiratory infections in children. Eur J Clin Microbiol Infect Dis. 2014;33:41–48. doi: 10.1007/s10096-013-1926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller E.K., Hernandez J.Z., Wimmenauer V., Shepherd B.E., Hijano D., Libster R. A mechanistic role for type III IFN-λ1 in asthma exacerbations mediated by human rhinoviruses. Am J Respir Crit Care Med. 2012;185:508–516. doi: 10.1164/rccm.201108-1462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansbach J.M., Piedra P.A., Teach S.J., Sullivan A.F., Forgey T., Clark S. Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis. Arch Pediatr Adolesc Med. 2012;166:700–706. doi: 10.1001/archpediatrics.2011.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jartti T., Aakula M., Mansbach J.M., Piedra P.A., Bergroth E., Koponen P. Hospital length-of-stay is associated with rhinovirus etiology of bronchiolitis. Pediatr Infect Dis J. 2014;33:829–834. doi: 10.1097/INF.0000000000000313. [DOI] [PubMed] [Google Scholar]
- 9.Hasegawa K., Pate B.M., Mansbach J.M., Macias C.G., Fisher E.S., Piedra P.A. Risk factors for requiring intensive care among children admitted to ward with bronchiolitis. Acad Pediatr. 2015;15:77–81. doi: 10.1016/j.acap.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasegawa K, Jartti T, Mansbach JM, Laham RF, Jewell AM, Espinola JA, et al. Respiratory syncytial virus genomic load and disease severity among children hospitalized with bronchiolitis: multicenter cohort studies in the US and Finland. J Infect 2014 [E-pub ahead of print]. [DOI] [PMC free article] [PubMed]
- 11.American Academy of Pediatrics Subcommittee on Diagnosis and Management of Bronchiolitis Diagnosis and management of bronchiolitis. Pediatrics. 2006;118:1774–1793. doi: 10.1542/peds.2006-2223. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Souza N., Favoreto S., Wong H., Ward T., Yagi S., Schnurr D. In vitro susceptibility to rhinovirus infection is greater for bronchial than for nasal airway epithelial cells in human subjects. J Allergy Clin Immunol. 2009;123:1384–1390. doi: 10.1016/j.jaci.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendley J.O., Gwaltney J.M., Jr. Viral titers in nasal lining fluid compared to viral titers in nasal washes during experimental rhinovirus infection. J Clin Virol. 2004;30:326–328. doi: 10.1016/j.jcv.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Cordey S., Junier T., Gerlach D., Gobbini F., Farinelli L., Zdobnov E.M. Rhinovirus genome evolution during experimental human infection. PLoS One. 2010;5:e10588. doi: 10.1371/journal.pone.0010588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bochkov Y.A., Grindle K., Vang F., Evans M.D., Gern J.E. Improved molecular typing assay for rhinovirus species A, B, and C. J Clin Microbiol. 2014;52:2461–2471. doi: 10.1128/JCM.00075-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- Mansbach J.M., Piedra P.A., Teach S.J., Sullivan A.F., Forgey T., Clark S. Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis. Arch Pediatr Adolesc Med. 2012;166:700–706. doi: 10.1001/archpediatrics.2011.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jartti T., Aakula M., Mansbach J.M., Piedra P.A., Bergroth E., Koponen P. Hospital length-of-stay is associated with rhinovirus etiology of bronchiolitis. Pedatr Infect Dis J. 2014;33:829–834. doi: 10.1097/INF.0000000000000313. [DOI] [PubMed] [Google Scholar]
- Beckham J.D., Cadena A., Lin J., Piedra P.A., Glezen W.P., Greenberg S.B. Respiratory viral infections in patients with chronic, obstructive pulmonary disease. J Infect. 2005;50:322–330. doi: 10.1016/j.jinf.2004.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr L., Fox J.D., Tilley P.A., Ahmed-Bentley J. Evaluation of real-time PCR for diagnosis of Bordetella pertussis infection. BMC Infect Dis. 2006;6:62. doi: 10.1186/1471-2334-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchell J.M., Thurman K.A., Mitchell S.L., Thacker W.L., Fields B.S. Evaluation of three real-time PCR assays for detection of Mycoplasma pneumoniae in an outbreak investigation. J Clin Microbiol. 2008;46:3116–3118. doi: 10.1128/JCM.00440-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Holloway B., Dare R.K., Kuypers J., Yagi S., Williams J.V. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46:533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansbach J.M., Piedra P.A., Stevenson M.D., Sullivan A.F., Forgey T.F., Clark S. Prospective multicenter study of children with bronchiolitis requiring mechanical ventilation. Pediatrics. 2012;130:e492–e500. doi: 10.1542/peds.2012-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]