Abstract
Ubiquitination and deubiquitination regulate several essential cellular processes such as protein degradation, cell-cycle progression, signaling, and DNA repair. Given the importance of these processes, it is not surprising that many microbes have developed the means to interfere with different stages of ubiquitin pathways to promote their survival and replication. This review focuses on virulence proteins of bacterial pathogens that mediate these effects and summarizes our current understanding of their actions.
Main Text
Introduction
Small reversible posttranslational modifications of proteins provide a widespread mechanism for regulating a broad range of cellular processes, enabling cells to respond rapidly to changes in their environment. Such modifications include phosphorylation, acetylation, and among eukaryotes, ubiquitination and ubiquitin-like modifications such as sumoylation and neddylation.
Ubiquitin (Ub) is a small protein of 76 amino acids (Goldstein et al., 1975). It is present in all eukaryotes and is highly conserved, with only three amino acid differences between the yeast and human proteins. Ubiquitination involves covalent addition of Ub monomers to lysine residues of target proteins or of Ub itself (to form poly-Ub chains), via the involvement of at least three enzymes: a Ub-activating enzyme (E1), a Ub-conjugating enzyme (E2), and a Ub ligase (E3) ( Figure 1). E1 enzymes are responsible for the ATP-dependent activation of the C terminus of Ub, which is then directed to the active site of an E2 enzyme. E3 ligases (of which there are over 1000 encoded by human genome) are a large and diverse group of proteins that have been subclassified by the presence of different motifs, including HECT, RING, and U box domains. HECT E3s transfer Ub from the E2 to a thioester linkage on the E3, and then to the substrate, while RING and U box E3s enable protein ubiquitination by acting as adaptors that bind to E2 proteins and substrates separately and facilitate direct transfer of Ub from the E2 protein to the substrate. A fourth group of E3 ligases, called the modulator of immune recognition (MIR) family, comprises membrane-bound proteins containing a specialized form of the RING finger, plant homeodomain (PHD) (Coscoy and Ganem, 2003, Ohmura-Hoshino et al., 2006).
Figure 1.
Schematic Figure of Ubiquitin Activation, Conjugation, and Deconjugation
E1 enzymes activate the C terminus of ubiquitin in an ATP-dependent manner, which directs ubiquitin to an E2 enzyme. E3 ligases transfer ubiquitin either from the E2 to a thioester linkage on the E3, and then to the substrate, or by acting as adaptors that bind to E2 proteins and substrates separately, and facilitate direct transfer of Ub from the E2 protein to the substrate. Chains of four or more Ub molecules linked through lysine 48 (K48) of ubiquitin target the modified protein for proteasomal degradation, whereas K63-linked polyubiquitin chains regulate several cellular processes, such as DNA repair, signaling, endocytosis, vesicular trafficking, and cell-cycle progression. Ubiquitin deconjugation is a reversible process catalyzed by deubiquitinating enzymes (DUBs). These play an important role in processing Ub precursors, proofreading Ub protein conjugates, and removing Ub from substrate proteins.
Several RING E3s appear to act independently, but others are components of larger multiprotein complexes. A good example of the latter are the cullin RING Ub ligases. The C-terminal regions of cullin proteins interact with the E2-interacting RING protein ROC-1. The N-terminal region of cullins interact with linker or adaptor proteins such as SCF (Skp-1, Cul-1, F box), which in turn bind to the substrate. Cullins are themselves positively regulated by the binding of the small Ub-like protein (Ubl) neural precursor cell-expressed developmentally downregulated 8 (NEDD8) (RUB1 in S. cerevisiae).
In addition to NEDD8, other small proteins with various degrees of sequence similarity to Ub have been identified that link to proteins in an analogous manner to ubiquitination. These include small ubiquitin-like modifier 1 (SUMO-1) also known as Ubl1, Sentrin, or PIC-1 (Jentsch and Pyrowolakis, 2000, Yeh et al., 2000). Sumoylation occurs on a wide range of substrates including signaling molecules and several transcriptional regulators (Muller et al., 2001, Sapetschnig et al., 2002, Seeler and Dejean, 2001). SUMO can also act by blocking Ub attachment sites (Hoege et al., 2002). A key distinction between SUMO and Ub is the inability of SUMO to conjugate to itself (Pickart, 1997). The ability of Ub to interact with a wide range of proteins, including itself, and to form a variety of different length and linkages of chains, increases its capacity to manipulate a larger array of signals compared to Ubls. For a comprehensive review of Ub and Ub-like protein modifications, the reader is referred to a recent review (Kerscher et al., 2006).
The fate of ubiquitinated proteins depends on the combination of interacting E2 and E3 enzymes and the length and linkage of the Ub chains. Chains of four or more Ub molecules linked through lysine 48 (K48) of Ub target the modified protein for proteasomal degradation (Thrower et al., 2000). K63-linked polyubiquitin chains regulate a wide range of important cellular processes such as DNA repair, signaling, endocytosis, vesicular trafficking, and cell-cycle progression (Haglund and Dikic, 2005, Hicke and Dunn, 2003, Huang and D'Andrea, 2006). Monoubiquitination (modification with one ubiquitin molecule) controls subcellular protein localization, endocytosis, and recruitment of Ub-binding proteins (Haglund and Dikic, 2005).
Deubiquitinating Enzymes
Protein ubiquitination is a reversible process, and Ub deconjugation, catalyzed by deubiquitinating enzymes (DUBs), plays an important role in processing Ub precursors, proofreading Ub protein conjugates, and removing Ub from target proteins (Figure 1). Major progress has been made in understanding pathways of ubiquitination, but our understanding of the functions of DUBs is much less advanced. DUBs are present in all eukaryotic cells, and approximately 100 are predicted to be encoded by the human genome. They include both metalloproteases (JAMM/MPN+) and cysteine proteases, with the majority being of the latter type. The cysteine protease DUBs have been grouped into four different classes based on their sequence and predicted activity: Ub-specific proteases (USPs, also referred to as Ub processing proteases, UBPs), Ub C-terminal hydrolases (UCHs), ovarian tumor-related proteases/Cezanne (OTUs), and Josephin domain proteases (JD) (Amerik and Hochstrasser, 2004, Nijman et al., 2005, Sulea et al., 2006). USPs represent the most diverse class, comprising proteases with two short conserved motifs, the Cys and His boxes, flanked by N- and C-terminal regions of differing length and sequence. The UCH class contains small proteins originally identified by their ability to hydrolyze small amides and esters at the C terminus of Ub. The other classes have only been recognized recently and contain only a few members. Proteases have also been divided into families and clans according to a hierarchical classification on the basis of degree of similarity in amino acid sequence. Families of proteases that are homologous are grouped together in a clan. According to this system, cysteine proteases have been divided into 9 clans and 47 families (http://merops.sanger.ac.uk/index.htm). Several cysteine proteases that are DUBs or Ub-like protein-specific proteases (ULPs) have been classified into the clans CA and CE. In clan CA, the Cys motif is N proximal to the His box, whereas the reverse is the case in clan CE.
In contrast to many other proteases, which are translated as inactive precursors, DUBs are generally produced as active enzymes, and structural analysis has revealed that the catalytic triad only assumes the active conformation upon binding of Ub, thereby preventing nonspecific protease activity (Nijman et al., 2005). Specificity of DUBs is also likely to be determined by regions outside the catalytic core of the enzyme, and both the target and the attached Ub monomer or polymer are likely to be important for recognition by the DUB.
Bacterial Intervention
Studies on the interactions between bacteria and host cells over the last decade have revealed a remarkable diversity of cellular processes that are modulated by Gram-positive and Gram-negative pathogens. Several Gram-negative bacterial pathogens use type three secretion (T3S) systems to deliver effector proteins into host cells. Components of the T3S apparatus (which resembles a needle-like structure in both form and function; Galan and Wolf-Watz, 2006) are conserved in different pathogens; however, each pathogen is endowed with a unique repertoire of effectors that contribute to the disease processes caused by that pathogen. These effectors interfere with the actin, microtubule, and intermediate filament cytoskeletons; microtubule motors; specific stages of the endocytic and secretory pathways; and a host of signaling pathways, affecting, for example, transcription, translation, cell division, cell migration, and immune responses. Given the importance of ubiquitination and deubiquitination in the regulation of many cellular processes, it is not surprising that pathogenic bacteria have developed the means to interfere extensively with different stages of Ub pathways. In this review we focus on the bacterial virulence proteins that mediate these effects (summarized in Table 1), and what is known of the mechanisms by which they interfere with Ub pathways.
Table 1.
Summary of Bacterial Effectors Directly Targeting Ub Pathways
| Pathogen | Virulence Protein | Biochemical Function | Target(s) | Physiological Effect(s) | Reference |
|---|---|---|---|---|---|
| C. trachomatis | ChlaDub1 | DUB and deneddylator | unknown | unknown | Misaghi et al., 2006 |
| ChlaDub2 | DUB and deneddylator | unknown | unknown | Misaghi et al., 2006 | |
| P. syringae | AvrPtoB | E3 ligase | unknown | suppression of plant immunity | Janjusevic et al., 2006 |
| S. Typhimurium | SseL | DUB | unknown | cytotoxicity | Rytkönen et al., 2007 |
| SopA | E3 ligase | induction of PMN trans-epithelial migration | Zhang et al., 2006 | ||
| S. flexneri | OspG | kinase | Ub E2 molecules | suppression of host innate response | Kim et al., 2005 |
| IpaH9.8 | E3 ligase | MAPKK Ste7 in yeast | suppression of inflammatory response | Rohde et al., 2007, Okuda et al., 2005 | |
| Yersinia | YopJ/YopP | DUB/acetyl-transferase | unknown/MKKs and IKKs | suppression of MAPK and NF-κB pathway, induction of apoptosis | Zhou et al., 2005, Mukherjee et al., 2006, Mittal et al., 2006 |
PMN, polymorphonuclear leukocytes.
Yersinia YopJ/P
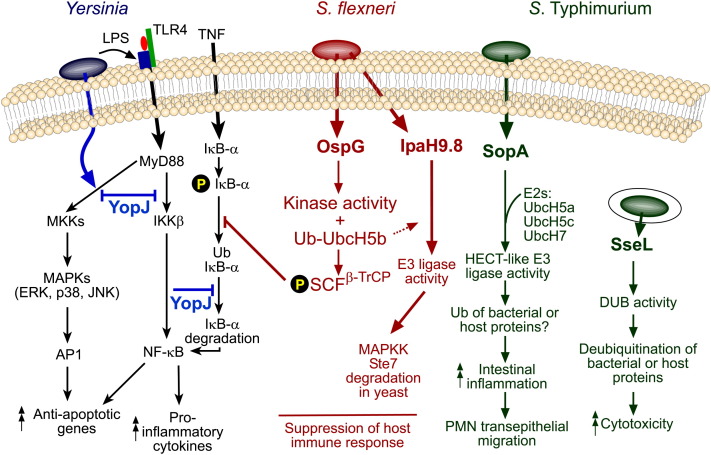
The first bacterial protein shown to have DUB activity was the Yersinia T3S effector YopJ/P (referred to as YopJ in Yersinia pseudotuberculosis and YopP in Yersinia enterocolitica) ( Figure 2). YopJ is present in all three Yersinia spp. pathogenic for humans; these bacteria are mainly extracellular pathogens causing both enteric diseases (Y. pseudotuberculosis and Y. enterocolitica) and plague (Y. pestis). YopJ plays an important role in inhibiting the inflammatory response and inducing apoptosis in macrophages. By interfering with the NF-κB pathway through inhibition of MAPK (mitogen-activated protein kinase) kinases and IKKβ, YopJ suppresses proinflammatory signaling via TNF and IL-8 production, as well as inducing apoptosis (Boland and Cornelis, 1998, Mills et al., 1997, Monack et al., 1997, Orth et al., 1999, Palmer et al., 1998, Yoon et al., 2003, Zhou et al., 2004). YopJ is a member of clan CE, family 55, and its substrate was originally thought to be SUMO-1, since expression of YopJ in mammalian cells decreased the cellular concentration of SUMO-1-conjugated proteins (Orth et al., 2000). However, a subsequent study showed that purified YopJ cleaves Ub-amino methyl coumarin (Ub-AMC) (a fluorogenic Ub conjugate) but not SUMO-1-AMC in vitro (Zhou et al., 2005). Furthermore, infection of macrophages with a Y. pseudotuberculosis strain expressing a mutant version of YopJ in which the catalytic cysteine was replaced by alanine resulted in the accumulation of ubiquitinated IκBα in the macrophage cytosol (Zhou et al., 2005). Normally, activation of the NF-κB pathway leads to phosphorylation, ubiquitination, and proteasomal degradation of IκBα, which allows the translocation of NF-κB to the nucleus, where it regulates transcription of genes involved in proinflammatory signaling. Translocated YopJ was therefore proposed to act as a DUB for ubiquitinated IκBα, thereby preventing its degradation. However, YopJ is evidently not able to hydrolyze ubiquitinated IκBα in vitro (Collier-Hyams et al., 2002). Furthermore, it has been shown recently by two groups that YopJ can function as an acetyltransferase in vitro by transferring acetyl groups to serine and threonine residues in the activation loop of MAPKK6, effectively preventing their phosphorylation and hence kinase activation (Bliska, 2006, Mittal et al., 2006, Mukherjee et al., 2006). It is important to know if YopJ can cause similar modifications in vivo, since it is not immediately obvious how a loss of acetyltransferase activity on MAPKK6 would result in an accumulation of ubiquitinated IκBα in infected cells (Zhou et al., 2005). However, these studies raise the possibility that YopJ is a bifunctional enzyme that acetylates one or more substrate and deubiquitinates another. Such relaxed substrate specificity of a bacterial enzyme would not be unprecedented: E. coli TAP is a multifunctional enzyme that has both protease and deacetylating activities (Lee et al., 2006).
Figure 2.
Examples of Interference of Mammalian Ub Pathways by Bacterial Pathogens
YopJ/P, a Yersinia T3S effector protein with DUB and acetyltransferase activity, is translocated into host cells, where it interferes with the NF-κB pathway through inhibition of MKKs (mitogen-activated protein kinase kinases) and IKKβ and possibly by deubiquitinating ubiquitinated IκB-α. YopJ plays an important role in inhibiting the inflammatory response and induces apoptosis in macrophages. Shigella also inhibits host immune responses. Following translocation into host cells, OspG acts as a kinase and blocks TNF-induced IκBα degradation, conceivably by phosphorylating a component of SCFβ-TrCP complex. IpaH9.8, another Shigella effector, functions as an E3 Ub ligase that interferes with the pheromone response signaling pathway in yeast by ubiquitinating the MAPKK Ste7. To facilitate invasion and intracellular survival, Salmonella translocates two sets of effector proteins into the host cells. The Salmonella pathogenicity island 1 (SPI-1) T3S system translocates effector proteins across the host-cell plasma membrane. Several of these effectors are ubiquitinated following translocation, but one, SopA, functions as a HECT-like E3 Ub ligase that preferentially uses the inflammation-associated E2s UbcH5a, UbcH5c, and UbcH7 for ubiquitination in vitro. SopA induces a host inflammatory response by promoting polymorphonuclear leukocyte transepithelial migration. Following uptake, Salmonella translocates another set of effectors across the Salmonella-containing vacuole via the SPI-2 T3S system. SseL is translocated approximately 6 hr postuptake and acts as a DUB in vivo to trigger a delayed cytotoxic effect in macrophages.
Salmonella
Salmonella enterica serovar Typhimurium is an invasive pathogen that colonizes the small intestine, and in the murine model of typhoid it penetrates the epithelium and transits from Peyer's patches and mesenteric lymph nodes to the spleen and liver, where it replicates in macrophages. Following invasion or phagocytosis by macrophages, the vast majority of intracellular S. enterica remain in a membrane-bound compartment, the Salmonella-containing vacuole (SCV), which the bacteria modify to provide an environment favorable for replication. To invade host cells, Salmonella uses a T3S system encoded by the Salmonella pathogenicity island 1 (SPI-1). This system translocates several effector proteins across the host-cell plasma membrane. Some of these remodel the actin cytoskeleton to cause localized membrane ruffling and bacterial uptake. Thereafter, another set of effectors is translocated across the vacuolar membrane. This is accomplished by the T3S system encoded by SPI-2, which translocates over 20 effectors, detectable approximately 4 hr following entry of bacteria. Together, these effector proteins enable bacterial replication in a variety of host-cell types and also induce a delayed cytotoxicity in macrophages (Kuhle and Hensel, 2004, Waterman and Holden, 2003).
Salmonellae that are both pathogenic and nonpathogenic for humans interfere with ubiquitination pathways. Whereas pathogenic Salmonella elicits an NF-κB-dependent proinflammatory response upon colonization of human intestinal epithelial cells in vitro, nonpathogenic strains are capable of suppressing this response. The inhibition does not affect the phosphorylation of IκBα but somehow affects its subsequent ubiquitination, and thereby prevents its degradation (Neish et al., 2000). The inhibition appears to be specific to the E3-SCFβ- TrCP substrates IκBα and β-catenin and caused by a rapid loss of neddylated Cullin-1, which is a subunit of the E3-SCF complex necessary for ubiquitination of IκBα and subsequent activation of NF-κB (Collier-Hyams et al., 2005). The Salmonella molecule(s) responsible and the mechanisms involved in this interesting activity are not yet known.
AvrA is predicted to be a clan CE protein translocated by the SPI-1 T3S system in a subset of strains of S. enterica (Hardt and Galan, 1997). AvrA has 56% identity to YopJ and also inhibits the NF-κB pathway. However, this seems to occur at a different stage than YopJ, because in transfected cells YopJ had little effect on NF-κB-dependent reporter gene activity when cells were stimulated with IKK-β, whereas AvrA effectively inhibited reporter gene activity (Collier-Hyams et al., 2002). However, like YopJ, AvrA contributes to apoptosis of host cells (Collier-Hyams et al., 2002). While the inhibitory activity of AvrA on the NF-κB pathway was shown to be dependent on a putative catalytic cysteine, it has yet to be established if AvrA is a DUB.
Several other effectors involved in Salmonella invasion are either ubiquitinated or modulate ubiquitination following their translocation to the host cells. SopE is a guanine nucleotide exchange factor for Rac1 that stimulates the actin polymerization required for membrane ruffling and bacterial uptake (Patel and Galan, 2006). SptP, which is codelivered into host cells in similar quantities to SopE, acts as a GTPase-activating protein and counteracts the activity of SopE (Fu and Galan, 1999). SopE is rapidly ubiquitinated and degraded, while the half-life of SptP is much longer. Domain-swapping experiments revealed that differential degradation kinetics is controlled by information in the N-terminal secretion and translocation domains of these proteins. The differential half-life of these proteins therefore accounts for the transient nature of the ruffling response induced by Salmonella (Kubori and Galan, 2003).
SopA is another SPI-1 effector that has an important role in the induction of host inflammatory responses by promoting polymorphonuclear (PMN) leukocyte transepithelial migration (Wood et al., 2000) (Figure 2). In epithelial cells, translocated SopA is ubiquitinated by an E3 ligase, HsRMA1, and subsequently degraded. While the majority of S. enterica serovar Typhimurium (S. Typhimurium) bacteria remain within membrane-bound vacuoles, a small proportion escape into the epithelial cell cytosol, where they replicate. Ubiquitinated SopA appears to have a minor role in promoting vacuolar escape, since mutation of sopA or RNAi of HsRMA1 reduced the numbers of bacteria that escaped their vacuoles (Zhang et al., 2005). In macrophages, cytosolic bacteria fail to replicate (Beuzon et al., 2002) and become surrounded by polyubiquitinated proteins and proteasomes, which might contribute to bacterial growth suppression (Perrin et al., 2004). Remarkably, recent work has shown that SopA is itself a HECT-like E3 Ub ligase (Zhang et al., 2006). Consistent with a regulatory role in inflammation, SopA preferentially used the inflammation-associated E2s UbcH5a, UbcH5c, and UbcH7 for ubiquitination in vitro, and a Salmonella strain expressing an E3 ligase-dead point mutant version of SopA induced less PMN transepithelial migration in vitro (Zhang et al., 2006). SopA could ubiquitinate bacterially derived or host proteins (or both), and identification of its physiological target(s) is now crucial in understanding its role in inflammation and enteritis.
SigD/SopB is a fourth SPI-1 effector that is ubiquitinated upon translocation to the host cell. This inositol phosphatase localizes to the surface of the nascent SCV, where it persists for several hours (Marcus et al., 2002). It acts by retaining high levels of phosphatidylinositol-3-phosphate [PtdIns(3)P] in the SCV membrane and has important roles in bacterial replication (Galan and Collmer, 1999, Hernandez et al., 2004) and suppression of apoptosis in epithelial cells by activation of Akt (Knodler et al., 2005), and membrane fission during bacterial invasion (Terebiznik et al., 2002). By 2 hr postinvasion, ubiquitin-conjugated SigD/SopB can be detected, but curiously this does not appear to lead to its degradation by the proteasome (Marcus et al., 2002), and the functional significance of its ubiquitination (if any) is not understood.
Interference with ubiquitination by Salmonella continues after the expression of the SPI-2 T3S system. SseL is a newly characterized effector of cysteine protease clan CE whose translocation onto the vacuolar membrane can be detected in epithelial cells and macrophages after approximately 6 hr following uptake of bacteria (Rytkönen et al., 2007) (Figure 2). A yeast two-hybrid screen with SseL and a HeLa cell cDNA library revealed that SseL interacts directly with Ub. A purified GST-SseL fusion protein specifically cleaved ubiquitin substrates, and infection of epithelial cells and macrophages with S. Typhimurium sseL mutant strains led to the accumulation of ubiquitin-modified proteins, indicating that SseL functions as a DUB in vivo. However, unlike YopJ, SseL did not influence cytokine production or interfere with activation or degradation of IκBα upon infection of macrophages. sseL mutant strains did not display an intracellular replication defect but were defective for a SPI-2-dependent late-stage cytotoxic effect on macrophages and were attenuated for virulence in the systemic phase of infection in mice (Rytkönen et al., 2007). This shows that ubiquitinated proteins of host or bacterial origin accumulate on (or in close proximity to) the SCV membrane, and these are deubiquitinated by SseL. How this leads to cytotoxicity remains to be established, but the study provides evidence that the cytotoxic activity of Salmonella is an important component of its virulence. It is tempting to speculate that the deubiquitinating activity of SseL might function to counteract ubiquitination and degradation of other translocated bacterial effectors by the host-cell machinery. However, this seems unlikely for two reasons. First, an important collective function of SPI-2 T3S system effectors is to enable intracellular growth (Waterman and Holden, 2003) and if SseL acted by protecting these effectors then an sseL mutant would be predicted to have an intracellular replication defect. Secondly, SseL has a clear preference for K63-linked Ub chains (Rytkönen et al., 2007), indicating that it is likely to be involved in signaling events rather than interfering directly with protein degradation.
Shigella
Shigella flexneri is an important cause of human bacillary dysentery. The morphological features of Shigella entry into epithelial cells are similar to those induced by S. Typhimurium, and although both require T3S systems, the effector protein repertoire and infection process following invasion are distinct. S. flexneri invades the colonic epithelium initially through M cells of Peyer's patches. Bacteria then interact with macrophages in the lymphoid follicle, and enterocytes via their basolateral membranes. These interactions result in a strong proinflammatory response that exacerbates infection of the intestinal epithelium. However, it has become clear that stimulation of proinflammatory responses is subsequently modulated by the action of several bacterial effectors (Arbibe et al., 2007, Kim et al., 2005). OspG is one of approximately 20 effectors delivered to the host cells by the Shigella T3S system. A yeast two-hybrid screen showed that OspG interacts with ubiquitinylated E2 molecules, including UbcH5b (Figure 2). OspG was shown to have kinase activity but evidently does not phosphorylate E2 proteins. Nevertheless, OspG can block TNF-induced IκBα degradation, a process that is dependent on UbcH5b (Kim et al., 2005). This effect requires a lysine residue important for kinase activity of OspG and suggests that OspG acts at the step of phospho-IκBα ubiquitination. Possibly, OspG acts by phosphorylating a component of SCFβ- TrCP in the context of OspG-UbcH5b. The biochemical evidence for dampening of the inflammatory response is supported by the phenotype of an ospG mutant, which induced a much stronger inflammatory response during infection of rabbit ileal loops (Kim et al., 2005).
IpaH9.8 is another effector delivered by Shigella into host cells, where it translocates to the nucleus (Haraga and Miller, 2003, Toyotome et al., 2001). Recently it has been shown that IpaH9.8 is an E3 Ub ligase (Rohde et al., 2007) (Figure 2). Its biochemical characterization was achieved by using Saccharomyces both as a host for an initial phenotypic screen, and then to identify its target and biochemical function. IpaH9.8 was shown to interfere with the pheromone response signaling pathway by ubiquitinating the MAPKK Ste7, which leads to its proteasome-dependent destruction (Rohde et al., 2007). Work from another group has shown that, in mammalian cells, IpaH9.8 interacts with U2AF35—a component of an essential pre-mRNA splicing factor complex (Okuda et al., 2005). Deletion of ipaH9.8 resulted in increased proinflammatory cytokine production and, consistent with the hypothesis that IpaH9.8 might interfere with the function of U2AF35, reduction of U2AF35 expression by siRNA-mediated knockdown caused a decrease in the expression of these cytokines (Okuda et al., 2005). It would be interesting to know if IpaH9.8 can ubiquitinate U2AF35, which might help to reconcile the results from these two studies. The finding that IpaH9.8 is an E3 ligase has broader implications, since homologous proteins with similar domain organization and containing a critical cysteine residue are found not only in Shigella but also in several other bacterial pathogens. These proteins include Salmonella SspH1, an effector of both the SPI-1 and SPI-2 T3S systems. Purified SspH1 was shown to have E3 ligase activity on PKN1 (Rohde et al., 2007), a host-cell kinase previously shown to interact with SspH1 (Haraga and Miller, 2006).
Chlamydia
The availability of ubiquitin-based probes has provided useful reagents to identify new DUBs from mammalian cells and viruses (Galardy et al., 2005, Ovaa et al., 2005). Recently, this approach was used to identify two DUBs from Chlamydia trachomatis (Misaghi et al., 2006). Lysates of infected HeLa cells were incubated with an epitope-tagged Ub-vinylmethylester conjugate, which forms a covalent adduct with the active site cysteine of DUBs (Borodovsky et al., 2001). Following immunoprecipitation, an interacting protein was fractionated by SDS-PAGE and identified by mass spectrometry. Genome sequence analysis revealed that the gene encoding this DUB is flanked by a homolog. Both corresponding proteins (named ChlaDub1 and ChlaDub2) were shown to have deubiquitinating and deneddylating activities. Although ChlaDub1 is clearly translocated into host cells by bacteria, its localization, biochemical target(s), and possible role during infection remain to be established.
Escherichia coli
CNF1 toxin is produced by uropathogenic E. coli, and related toxins have been found in Y. pseudotuberculosis and Bordetella pertussis. The toxin is taken up by mammalian cells by endocytosis and then passes into the cytosol by translocation through the endosomal membrane (Boquet, 2001). CNF1 functions by deamidating a specific glutamine residue of Rho GTPases to glutamate. This prevents endogenous or induced GTPase activity of these molecular switches, effectively locking them into a permanently active state. However, this effect is only transient, as the host cell responds by depleting the modified GTPases by ubiquitination and proteasome-mediated degradation (Doye et al., 2002). Hence, although the immediate effect of CNF1 deamidation of Rho proteins is their activation, the long-term consequence is ubiquitination and degradation. In the case of RhoA, this is carried out by an E3 ligase called Smurf1 (Boyer et al., 2006).
Listeria
Listeria monocytogenes is a Gram-positive bacterial pathogen that interacts with host-cell surface proteins to trigger signaling pathways leading to bacterial uptake. One such pathway involves a bacterial cell surface-localized protein, InlB, and the growth factor receptor Met (Shen et al., 2000). Interaction between Met and its ligand normally triggers the recruitment of the Ub ligase Cbl, which ubiquitinates Met and leads to its endocytosis. It was found recently that InlB can mediate the same process, and furthermore that the Ub-dependent endocytosis machinery is required for internalization of bacteria (Veiga and Cossart, 2005). While the interaction between InlB and ubiquitination machinery is indirect, this discovery reveals another means by which bacterial pathogens harness host Ub-dependent pathways to promote their virulence. Following uptake into host cells, L. monocytogenes escapes its vacuole and begins to replicate and spread in the host-cell cytosol. This is dependent on the action of the secreted cytolysin listeriolysin O (LLO). LLO is potentially damaging to the host-cell plasma membrane, and to ensure that bacteria remain intracellular, the activity of LLO must be restricted to the vacuolar membrane. By mechanisms that are still unclear, LLO is rapidly ubiquitinated and degraded by the proteasome machinery (Schnupf et al., 2006).
Plant Pathogens
Bacterial plant pathogens also use specialized secretion systems to deliver effector proteins into host cells. As with animal pathogens, the processes of activation, conjugation, and deconjugation of Ub and Ubls all appear to be targeted by bacterial effector proteins. VirF of Agrobacterium is one of several proteins involved in transfer of T-DNA from this bacterium into plant cells. It localizes to the plant cell nucleus, where it causes the degradation of a plant protein VIP1. This is thought to be mediated by an F box motif in VirF that binds the plant homolog of Skp1, a component of SCF in some E3 ligase complexes (Tzfira et al., 2004). AvrPtoB is a T3S effector translocated into tomato cells by Pseudomonas syringae, where it inhibits hypersensitivity—a plant immune response involving rapid localized host-cell death at the site of infection. The amino acid sequence of AvrPtoB provided no information as to its function, but the crystal structure of the C-terminal domain of AvrPtoB revealed striking similarity to U box and RING finger components of E3 ligases (Janjusevic et al., 2006). Purified AvrPtoB was shown to have E3 ligase activity in vitro, and putative E2 binding site residues were shown to be critical for suppression of the plant hypersensitive response (Janjusevic et al., 2006). Presumably, AvrPtoB recruits E2 partners and substrates and transfers Ub to host-cell proteins(s) that positively regulate the plant defense response. However, the cellular substrate(s) of this activity have not yet been identified. Xanthomonas campestris is a related pathogen of solanaceous plants; one of its T3S system effectors, XopD, has been characterized as a cysteine protease of clan CE, family 48. This effector localizes to the plant cell nucleus, where it cleaves (as yet uncharacterized) sumoylated proteins (Hotson et al., 2003). XopD appears to have a strict preference for plant SUMOs and was recently used to obtain insight into substrate specificity of ULPs (Chosed et al., 2007). Several other bacterial plant pathogens and symbionts are predicted to contain proteins that have been classified as members of clan CE, family 48 proteases. These include Acidovorax avenae, Bradyrhizobium japonicum, Mesorhizobium loti, and Rhizobium leguminosarum (http://merops.sanger.ac.uk/index.htm).
Viral Intervention
Although this review focuses on bacteria, several viruses are also known to interfere with different steps in the ubiquitin pathway. Some encode proteins that modify cullin-like RING Ub ligases by interacting with substrate adaptors, redirecting the Ub ligase complex to new targets, or protecting cellular proteins from ubiquitination (Barry and Fruh, 2006). Both the Herpes and Poxvirus families encode proteins with E3 ligase activity. These E3 ligases promote ubiquitination of MHC-1, B7.2, ICAM-1, and CD4, resulting in removal of these cell surface molecules and obstruction of host immune responses (Coscoy et al., 2001, Duncan et al., 2006, Hewitt et al., 2002, Mansouri et al., 2003). Other viruses produce E3 ligases that interfere with apoptosis and interferon (IFN) production (Halford et al., 2006, Melroe et al., 2004, Taylor and Barry, 2006). Recently, it has become clear that some viruses, including herpesvirus, cytomegalovirus, adenovirus, and coronaviruses also encode DUBs; however, the targets for most of these DUBs have not yet been identified (Kattenhorn et al., 2005, Balakirev et al., 2002, Barretto et al., 2005, Sulea et al., 2006, Wang et al., 2006).
Conclusions
From the work reviewed above and elsewhere (Boyer and Lemichez, 2004, Angot et al., 2007), it is clear that interference of ubiquitination and deubiquitination plays an important role in the virulence of several bacterial pathogens and frequently influences host immune responses. Although the current list of bacterial effectors involved in these processes is relatively short, they would appear to fall into three general categories: those that act directly on pathways by “mimicking” host-cell proteins (e.g., E3 ligases and DUBs), those that act indirectly (e.g., E. coli CNF1 and Shigella OspG), and those that are themselves subject to ubiquitination in ways that influence bacterial virulence (e.g., Salmonella SopE). Much of our (albeit limited) understanding of this aspect of bacterial virulence has been gained in the last few years, and it therefore seems likely that the number of bacterial proteins that either mimic or modulate host-cell proteins involved in Ub pathways will increase considerably in the future. Of the bacterial DUBs that have been identified to date, none have the same architecture as the “classic” eukaryotic DUBs (clan CA); they are all members of clan CE proteases. As our knowledge of the clan organization of DUBs and ULPs increases (Hotson and Mudgett, 2004), it will become easier to identify putative new Ub- or ULP-specific proteases among both plant and animal pathogens. However, identifying new modifiers of the Ub conjugation machinery is unlikely to be straightforward, since the bacterial E3 ligases identified to date do not display striking amino acid sequence similarities, either between themselves or with their mammalian counterparts. A major problem is that the targets of most of these bacterial effectors are currently unknown, and identifying them can be fraught with difficulty: many of these effectors are very potent, are produced transiently, and localize to specific regions of the infected cell. Specificity could therefore be achieved by precise spatiotemporal control of effector delivery rather than being an intrinsic property of the effectors. The controversial literature regarding the function of YopJ emphasizes the need for careful biochemical analysis of putative cysteine proteases to reveal their physiological functions in vivo. Ideally, this involves the study of the protein in the natural context of infection, as opposed to analysis following its overexpression either in vitro or in vivo, but obviously this is not feasible in many instances. Notwithstanding these concerns, the growing literature on bacterial manipulation of Ub pathways reflects a very important and widespread aspect of bacterial pathogenesis, and future research in this area will provide exciting new insights into the mechanisms underlying bacterial virulence.
Acknowledgments
A.R. is supported by a fellowship from the Swedish Research Council. Work in D.W.H.'s laboratory is funded by the Medical Research Council (UK), Wellcome Trust (UK), and European Commission. We thank Jaime Mota, Piotr Mazurkiewicz, and Christoph Tang for critical review of the manuscript.
References
- Amerik A.Y., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Angot A., Vergunst A., Genin S., Peeters N. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 2007;3:e3. doi: 10.1371/journal.ppat.0030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbibe L., Kim D.W., Batsche E., Pedron T., Mateescu B., Muchardt C., Parsot C., Sansonetti P.J. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat. Immunol. 2007;8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- Balakirev M.Y., Jaquinod M., Haas A.L., Chroboczek J. Deubiquitinating function of adenovirus proteinase. J. Virol. 2002;76:6323–6331. doi: 10.1128/JVI.76.12.6323-6331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A.D., Baker S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005;79:15189–15198. doi: 10.1128/JVI.79.24.15189-15198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry M., Fruh K. Viral modulators of cullin RING ubiquitin ligases: Culling the host defense. Sci. STKE. 2006;2006:pe21. doi: 10.1126/stke.3352006pe21. [DOI] [PubMed] [Google Scholar]
- Beuzon C.R., Salcedo S.P., Holden D.W. Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiol. 2002;148:2705–2715. doi: 10.1099/00221287-148-9-2705. [DOI] [PubMed] [Google Scholar]
- Bliska J.B. Yersinia inhibits host signaling by acetylating MAPK kinases. ACS Chem. Biol. 2006;1:349–351. doi: 10.1021/cb600261k. [DOI] [PubMed] [Google Scholar]
- Boland A., Cornelis G.R. Role of YopP in suppression of tumor necrosis factor alpha release by macrophages during Yersinia infection. Infect. Immun. 1998;66:1878–1884. doi: 10.1128/iai.66.5.1878-1884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boquet P. The cytotoxic necrotizing factor 1 (CNF1) from Escherichia coli. Toxicon. 2001;39:1673–1680. doi: 10.1016/s0041-0101(01)00154-4. [DOI] [PubMed] [Google Scholar]
- Borodovsky A., Kessler B.M., Casagrande R., Overkleeft H.S., Wilkinson K.D., Ploegh H.L. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L., Lemichez E. Targeting of host-cell ubiquitin and ubiquitin-like pathways by bacterial factors. Nat. Rev. Microbiol. 2004;2:779–788. doi: 10.1038/nrmicro1005. [DOI] [PubMed] [Google Scholar]
- Boyer L., Turchi L., Desnues B., Doye A., Ponzio G., Mege J.L., Yamashita M., Zhang Y.E., Bertoglio J., Flatau G. CNF1-induced ubiquitylation and proteasome destruction of activated RhoA is impaired in Smurf1−/− cells. Mol. Biol. Cell. 2006;17:2489–2497. doi: 10.1091/mbc.E05-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosed R., Tomchick D.R., Brautigam C.A., Mukherjee S., Negi V.S., Machius M., Orth K. Structural analysis of Xanthomonas XopD provides insights into substrate specificity of ULPs. J. Biol. Chem. 2007 doi: 10.1074/jbc.M608730200. in press. [DOI] [PubMed] [Google Scholar]
- Collier-Hyams L.S., Zeng H., Sun J., Tomlinson A.D., Bao Z.Q., Chen H., Madara J.L., Orth K., Neish A.S. Cutting edge: Salmonella AvrA effector inhibits the key proinflammatory, anti-apoptotic NF-kappa B pathway. J. Immunol. 2002;169:2846–2850. doi: 10.4049/jimmunol.169.6.2846. [DOI] [PubMed] [Google Scholar]
- Collier-Hyams L.S., Sloane V., Batten B.C., Neish A.S. Cutting edge: Bacterial modulation of epithelial signaling via changes in neddylation of cullin-1. J. Immunol. 2005;175:4194–4198. doi: 10.4049/jimmunol.175.7.4194. [DOI] [PubMed] [Google Scholar]
- Coscoy L., Ganem D. PHD domains and E3 ubiquitin ligases: Viruses make the connection. Trends Cell Biol. 2003;13:7–12. doi: 10.1016/s0962-8924(02)00005-3. [DOI] [PubMed] [Google Scholar]
- Coscoy L., Sanchez D.J., Ganem D. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 2001;155:1265–1273. doi: 10.1083/jcb.200111010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye A., Mettouchi A., Bossis G., Clement R., Buisson-Touati C., Flatau G., Gagnoux L., Piechaczyk M., Boquet P., Lemichez E. CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell. 2002;111:553–564. doi: 10.1016/s0092-8674(02)01132-7. [DOI] [PubMed] [Google Scholar]
- Duncan L.M., Piper S., Dodd R.B., Saville M.K., Sanderson C.M., Luzio J.P., Lehner P.J. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 2006;25:1635–1645. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Galan J.E. A salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature. 1999;401:293–297. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- Galan J.E., Collmer A. Type III secretion machines: Bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- Galan J.E., Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- Galardy P., Ploegh H.L., Ovaa H. Mechanism-based proteomics tools based on ubiquitin and ubiquitin-like proteins: Crystallography, activity profiling, and protease identification. Methods Enzymol. 2005;399:120–131. doi: 10.1016/S0076-6879(05)99008-3. [DOI] [PubMed] [Google Scholar]
- Goldstein G., Scheid M., Hammerling U., Schlesinger D.H., Niall H.D., Boyse E.A. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. USA. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K., Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford W.P., Weisend C., Grace J., Soboleski M., Carr D.J., Balliet J.W., Imai Y., Margolis T.P., Gebhardt B.M. ICP0 antagonizes Stat 1-dependent repression of herpes simplex virus: Implications for the regulation of viral latency. Virol. J. 2006;3:44. doi: 10.1186/1743-422X-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A., Miller S.I. A Salmonella enterica serovar typhimurium translocated leucine-rich repeat effector protein inhibits NF-kappa B-dependent gene expression. Infect. Immun. 2003;71:4052–4058. doi: 10.1128/IAI.71.7.4052-4058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A., Miller S.I. A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cell. Microbiol. 2006;8:837–846. doi: 10.1111/j.1462-5822.2005.00670.x. [DOI] [PubMed] [Google Scholar]
- Hardt W.D., Galan J.E. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc. Natl. Acad. Sci. USA. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L.D., Hueffer K., Wenk M.R., Galan J.E. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science. 2004;304:1805–1807. doi: 10.1126/science.1098188. [DOI] [PubMed] [Google Scholar]
- Hewitt E.W., Duncan L., Mufti D., Baker J., Stevenson P.G., Lehner P.J. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 2002;21:2418–2429. doi: 10.1093/emboj/21.10.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L., Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Hotson A., Mudgett M.B. Cysteine proteases in phytopathogenic bacteria: Identification of plant targets and activation of innate immunity. Curr. Opin. Plant Biol. 2004;7:384–390. doi: 10.1016/j.pbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Hotson A., Chosed R., Shu H., Orth K., Mudgett M.B. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 2003;50:377–389. doi: 10.1046/j.1365-2958.2003.03730.x. [DOI] [PubMed] [Google Scholar]
- Huang T.T., D'Andrea A.D. Regulation of DNA repair by ubiquitylation. Nat. Rev. Mol. Cell Biol. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- Janjusevic R., Abramovitch R.B., Martin G.B., Stebbins C.E. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science. 2006;311:222–226. doi: 10.1126/science.1120131. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Pyrowolakis G. Ubiquitin and its kin: How close are the family ties? Trends Cell Biol. 2000;10:335–342. doi: 10.1016/s0962-8924(00)01785-2. [DOI] [PubMed] [Google Scholar]
- Kattenhorn L.M., Korbel G.A., Kessler B.M., Spooner E., Ploegh H.L. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell. 2005;19:547–557. doi: 10.1016/j.molcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Kim D.W., Lenzen G., Page A.L., Legrain P., Sansonetti P.J., Parsot C. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. USA. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knodler L.A., Finlay B.B., Steele-Mortimer O. The Salmonella effector protein SopB protects epithelial cells from apoptosis by sustained activation of Akt. J. Biol. Chem. 2005;280:9058–9064. doi: 10.1074/jbc.M412588200. [DOI] [PubMed] [Google Scholar]
- Kubori T., Galan J.E. Temporal regulation of salmonella virulence effector function by proteasome-dependent protein degradation. Cell. 2003;115:333–342. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- Kuhle V., Hensel M. Cellular microbiology of intracellular Salmonella enterica: Functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell. Mol. Life Sci. 2004;61:2812–2826. doi: 10.1007/s00018-004-4248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L.C., Lee Y.L., Leu R.J., Shaw J.F. Functional role of catalytic triad and oxyanion hole-forming residues on enzyme activity of Escherichia coli thioesterase I/protease I/phospholipase L1. Biochem. J. 2006;397:69–76. doi: 10.1042/BJ20051645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri M., Bartee E., Gouveia K., Hovey Nerenberg B.T., Barrett J., Thomas L., Thomas G., McFadden G., Fruh K. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J. Virol. 2003;77:1427–1440. doi: 10.1128/JVI.77.2.1427-1440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S.L., Knodler L.A., Finlay B.B. Salmonella enterica serovar Typhimurium effector SigD/SopB is membrane-associated and ubiquitinated inside host cells. Cell. Microbiol. 2002;4:435–446. doi: 10.1046/j.1462-5822.2002.00202.x. [DOI] [PubMed] [Google Scholar]
- Melroe G.T., DeLuca N.A., Knipe D.M. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 2004;78:8411–8420. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S.D., Boland A., Sory M.P., van der Smissen P., Kerbourch C., Finlay B.B., Cornelis G.R. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc. Natl. Acad. Sci. USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaghi S., Balsara Z.R., Catic A., Spooner E., Ploegh H.L., Starnbach M.N. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol. Microbiol. 2006;61:142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- Mittal R., Peak-Chew S.Y., McMahon H.T. Acetylation of MEK2 and IkappaB kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc. Natl. Acad. Sci. USA. 2006;103:18574–18579. doi: 10.1073/pnas.0608995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack D.M., Mecsas J., Ghori N., Falkow S. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc. Natl. Acad. Sci. USA. 1997;94:10385–10390. doi: 10.1073/pnas.94.19.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Keitany G., Li Y., Wang Y., Ball H.L., Goldsmith E.J., Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- Muller S., Hoege C., Pyrowolakis G., Jentsch S. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- Neish A.S., Gewirtz A.T., Zeng H., Young A.N., Hobert M.E., Karmali V., Rao A.S., Madara J.L. Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alpha ubiquitination. Science. 2000;289:1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Ohmura-Hoshino M., Goto E., Matsuki Y., Aoki M., Mito M., Uematsu M., Hotta H., Ishido S. A novel family of membrane-bound E3 ubiquitin ligases. J. Biochem. (Tokyo) 2006;140:147–154. doi: 10.1093/jb/mvj160. [DOI] [PubMed] [Google Scholar]
- Okuda J., Toyotome T., Kataoka N., Ohno M., Abe H., Shimura Y., Seyedarabi A., Pickersgill R., Sasakawa C. Shigella effector IpaH9.8 binds to a splicing factor U2AF(35) to modulate host immune responses. Biochem. Biophys. Res. Commun. 2005;333:531–539. doi: 10.1016/j.bbrc.2005.05.145. [DOI] [PubMed] [Google Scholar]
- Orth K., Palmer L.E., Bao Z.Q., Stewart S., Rudolph A.E., Bliska J.B., Dixon J.E. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science. 1999;285:1920–1923. doi: 10.1126/science.285.5435.1920. [DOI] [PubMed] [Google Scholar]
- Orth K., Xu Z., Mudgett M.B., Bao Z.Q., Palmer L.E., Bliska J.B., Mangel W.F., Staskawicz B., Dixon J.E. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science. 2000;290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- Ovaa H., Galardy P.J., Ploegh H.L. Mechanism-based proteomics tools based on ubiquitin and ubiquitin-like proteins: Synthesis of active site-directed probes. Methods Enzymol. 2005;399:468–478. doi: 10.1016/S0076-6879(05)99032-0. [DOI] [PubMed] [Google Scholar]
- Palmer L.E., Hobbie S., Galan J.E., Bliska J.B. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-alpha production and downregulation of the MAP kinases p38 and JNK. Mol. Microbiol. 1998;27:953–965. doi: 10.1046/j.1365-2958.1998.00740.x. [DOI] [PubMed] [Google Scholar]
- Patel J.C., Galan J.E. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J. Cell Biol. 2006;175:453–463. doi: 10.1083/jcb.200605144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin A.J., Jiang X., Birmingham C.L., So N.S., Brumell J.H. Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr. Biol. 2004;14:806–811. doi: 10.1016/j.cub.2004.04.033. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. Targeting of substrates to the 26S proteasome. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- Rohde J.R., Breitkreutz A., Chenal A., Sansonetti P.J., Parsot C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host & Microbe. 2007;1:77–83. doi: 10.1016/j.chom.2007.02.002. this issue. [DOI] [PubMed] [Google Scholar]
- Rytkönen A., Poh P., Garmendia J., Boyle C., Thompson A., Liu M., Freemont P., Hinton J.C.D., Holden D.W. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc. Natl. Acad. Sci. USA. 2007;104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapetschnig A., Rischitor G., Braun H., Doll A., Schergaut M., Melchior F., Suske G. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 2002;21:5206–5215. doi: 10.1093/emboj/cdf510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnupf P., Portnoy D.A., Decatur A.L. Phosphorylation, ubiquitination and degradation of listeriolysin O in mammalian cells: Role of the PEST-like sequence. Cell. Microbiol. 2006;8:353–364. doi: 10.1111/j.1462-5822.2005.00631.x. [DOI] [PubMed] [Google Scholar]
- Seeler J.S., Dejean A. SUMO: Of branched proteins and nuclear bodies. Oncogene. 2001;20:7243–7249. doi: 10.1038/sj.onc.1204758. [DOI] [PubMed] [Google Scholar]
- Shen Y., Naujokas M., Park M., Ireton K. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell. 2000;103:501–510. doi: 10.1016/s0092-8674(00)00141-0. [DOI] [PubMed] [Google Scholar]
- Sulea T., Lindner H.A., Menard R. Structural aspects of recently discovered viral deubiquitinating activities. Biol. Chem. 2006;387:853–862. doi: 10.1515/BC.2006.108. [DOI] [PubMed] [Google Scholar]
- Taylor J.M., Barry M. Near death experiences: Poxvirus regulation of apoptotic death. Virology. 2006;344:139–150. doi: 10.1016/j.virol.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Terebiznik M.R., Vieira O.V., Marcus S.L., Slade A., Yip C.M., Trimble W.S., Meyer T., Finlay B.B., Grinstein S. Elimination of host cell PtdIns(4,5)P(2) by bacterial SigD promotes membrane fission during invasion by Salmonella. Nat. Cell Biol. 2002;4:766–773. doi: 10.1038/ncb854. [DOI] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman L., Rechsteiner M., Pickart C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyotome T., Suzuki T., Kuwae A., Nonaka T., Fukuda H., Imajoh-Ohmi S., Toyofuku T., Hori M., Sasakawa C. Shigella protein IpaH(9.8) is secreted from bacteria within mammalian cells and transported to the nucleus. J. Biol. Chem. 2001;276:32071–32079. doi: 10.1074/jbc.M101882200. [DOI] [PubMed] [Google Scholar]
- Tzfira T., Vaidya M., Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- Veiga E., Cossart P. Listeria hijacks the clathrin-dependent endocytic machinery to invade mammalian cells. Nat. Cell Biol. 2005;7:894–900. doi: 10.1038/ncb1292. [DOI] [PubMed] [Google Scholar]
- Wang J., Loveland A.N., Kattenhorn L.M., Ploegh H.L., Gibson W. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: Mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 2006;80:6003–6012. doi: 10.1128/JVI.00401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman S.R., Holden D.W. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 2003;5:501–511. doi: 10.1046/j.1462-5822.2003.00294.x. [DOI] [PubMed] [Google Scholar]
- Wood M.W., Jones M.A., Watson P.R., Siber A.M., McCormick B.A., Hedges S., Rosqvist R., Wallis T.S., Galyov E.E. The secreted effector protein of Salmonella dublin, SopA, is translocated into eukaryotic cells and influences the induction of enteritis. Cell. Microbiol. 2000;2:293–303. doi: 10.1046/j.1462-5822.2000.00054.x. [DOI] [PubMed] [Google Scholar]
- Yeh E.T., Gong L., Kamitani T. Ubiquitin-like proteins: New wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- Yoon S., Liu Z., Eyobo Y., Orth K. Yersinia effector YopJ inhibits yeast MAPK signaling pathways by an evolutionarily conserved mechanism. J. Biol. Chem. 2003;278:2131–2135. doi: 10.1074/jbc.M209905200. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Higashide W., Dai S., Sherman D.M., Zhou D. Recognition and ubiquitination of Salmonella type III effector SopA by a ubiquitin E3 ligase, HsRMA1. J. Biol. Chem. 2005;280:38682–38688. doi: 10.1074/jbc.M506309200. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Higashide W.M., McCormick B.A., Chen J., Zhou D. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol. Microbiol. 2006;62:786–793. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- Zhou L., Tan A., Hershenson M.B. Yersinia YopJ inhibits pro-inflammatory molecule expression in human bronchial epithelial cells. Respir Physiol Neurobiol. 2004;140:89–97. doi: 10.1016/j.resp.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Zhou H., Monack D.M., Kayagaki N., Wertz I., Yin J., Wolf B., Dixit V.M. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF-kappa B activation. J. Exp. Med. 2005;202:1327–1332. doi: 10.1084/jem.20051194. [DOI] [PMC free article] [PubMed] [Google Scholar]