Highlights
-
•
Two large scale molecular epidemiology studies performed in Italy and Spain.
-
•
The population dynamics trends differed between the two countries.
-
•
Relevant association between changes in vaccination strategies and viral population.
-
•
Strong association between viral population dynamics and outbreak frequency.
-
•
Hill’s criteria for causation were accomplished.
Keywords: Infectious bronchitis virus, Vaccination strategies, Phylodynamics, Epidemiology
Abstract
The extreme variability and rapid evolution of Infectious bronchitis virus (IBV) has always represented the key challenge for its control because of the limited cross-protection among different strains. Several experimental trials have proven a broadening of the protection spectrum when animals are vaccinated with multiple genotypes. Nevertheless, the conditions of vaccine administration in field are so different that the generalization of experimental results is, at least, questionable. In the present study a large scale epidemiological-phylodynamic approach was used to reconstruct the demographic history of the major field genotype (i.e. the QX one) circulating in Italy and Spain. These two countries were selected because, even if they share a comparable epidemiological scenario, the implemented vaccination protocols did not vary in Spain while changed dramatically in Italy over the time period considered. One hundred and ninety-five Italian and 98 Spanish non-recombinant sequences of the hyper-variable region of the S1 gene obtained between 2012 and 2016 were analyzed using a serial coalescent-based approach to reconstruct viral population history over time. While the IBV QX population dynamics remained constant in Spain, a much more complex pattern was evidenced in Italy; both in terms of viral population size and clinical outbreak frequency. Remarkably, a strong association with changes in vaccination strategies was recognized. This allowed demonstrating, by accomplishing all Hill’s criteria for causation, the cause-effect relationship between the vaccine administration/withdrawal and the variation in viral population dynamics and, above all, IBV related outbreaks. Thus, a robust confirmation about the efficacy of IBV vaccination in field conditions was provided. Additionally, the history herein reported testifies the primary importance of rigorously planning not only the intervention strategies but also their monitoring and evaluation.
1. Introduction
Infectious bronchitis virus (IBV) represents one of the most relevant infectious diseases of poultry, causing severe economic losses mainly associated to respiratory and reproductive syndromes, decreased productive performances and increased mortality. Even if biosecurity and good management practices are fundamental in the disease containment, widespread vaccination is also essential to control the disease. Like other coronaviruses and ssRNA+ viruses, IBV displays a high mutation and recombination rate [1], [2]. This has caused the crowding of a huge genetic and phenotypic heterogeneity and the emergence of plenty of IBV variants whose cross-protection has been proven poor [3], [4]. In Italy, IBV is still the major threat for poultry farms and displays a high prevalence and variety of circulating genotypes. Although five genotypes are currently detected (i.e. QX (72%), followed by 793B (16%), Mass (8%), Q1 (3%) and D274 (1%)) [5], the QX genotype is by far the most commonly associated to clinical outbreaks and, in all probability, the only relevant field genotype. Despite the development of homologous vaccines was often impractical or impossible, the immunization using multiple vaccines, based on different genotypes, has been proven to be beneficial, broadening the protection spectrum (i.e. “protectotype concept”) [6]. Consequently, vaccination schedules typically involve a combination of two genotypes, with acertain variability among countries and/or regions. In Italy, broiler vaccination protocols conform to this theory: animals were routinely immunized with a Mass-based spray vaccine at hatchery and a 793B-based one in field via drinking water. Recently, a trend toward administration of the two vaccines at 1 day of life has been observed. However, the costs associated with the vaccination as well as the risk of rolling-reactions, have persuaded many companies to suspend or modify vaccination protocols against IBV. In a recent study, we demonstrated that such change had a noteworthy impact on IBV genotypes circulation in Italy, leading to the substantial disappearance of the 793B genotype, suggesting that its presence was due to the spreading of vaccine strains or vaccine-related ones [5]. However, the effects of this strategy on field strain epidemiology and clinical outbreak frequency has been never evaluated in detail. Despite experimental trials demonstrated that the Mass vaccination is not protective against a QX challenge when administered alone while it is effective in combination with a 793B vaccine strain [7], their generalization to field conditions is often hazardous due to the extreme differences between the two settings. On the other hand, the collection of the huge amount of data necessary for rigorous large-scale field studies is often impossible due to economical and practical constraints. The development of a theoretical framework and mathematical models to reconstruct evolutionary features and population history of rapidly evolving viruses offers a great opportunity to reconcile and investigate viral epidemiology and evolution [8], [9], [10], [11], an approach that has been proven effective for the study of many human [12], [13], [14], [15], [16] and animal [17], [18], [19], [20] diseases.
In the present study, we set up a systematic analysis of the effect that the stopping and re-introduction of IBV vaccination with the 793B genotype had on the population dynamics of the QX genotype in Italy and on the occurrence of episodes of overt disease. For comparison, a similar analysis was performed based on Spanish IBV sequence dataset, a country with a comparable epidemiological scenario and major genotype (QX and 793B) prevalence [21], where no changes in the vaccination strategies (i.e. Mass plus 793B at one day of age) have been implemented.
2. Material and methods
2.1. Samples
Italian samples (i.e. tracheal or cloacal swabs or tissues) were collected from 562 broilers farms located in Northern Italy, between November 2012 and April 2016, in response to clinical signs attributable to IBV. Ten swabs (tracheal or cloacal) were collected from each flock, air dried, and merged in a pool. Similarly, tissues (trachea or cecal tonsils) were collected from ten animals per flock, pooled and refrigerated until delivery to laboratory. At the time of sampling, age, clinical status and geographical location of flocks were recorded. This time period can be divided in 5 phases according to the different vaccination strategies implemented by the companies providing samples to our laboratory:
-
(1)
The first phase started in November 2012 until late spring 2013, when animals were vaccinated with a combination of Mass (1 day of life) and 793B (14 days of life).
-
(2)
During the second phase (early summer 2013-November 2013) field vaccination with the 793B genotype was discontinued.
-
(3)
During the third phase, between December 2013 and June 2014, a broad trial with a newly registered 793B vaccine (in association with Mass) was performed.
-
(4)
In the period July 2014-September 2015, the 793B vaccine was withdrawn again.
-
(5)
Finally, since November 2014 (fifth phase), all animals have been vaccinated with Mass plus 793B or QX based vaccine, typically administered at 1 day of age.
For the whole considered time period (phase 1–5) the Mass based vaccine was provided to all animals at 1 day of life. These managerial changes affected the vast majority, if not all, the poultry population.
Spanish samples were collected during the same time period from farms located in different Spanish regions (data not shown). Along this period the vaccination program in most of the broiler farms did not change and consisted in a Mass plus a 793B based vaccine administered at 1 day of age. Spanish samples were collected from each flock according to the same protocol (i.e. pools of ten samples of tracheal or cecal swabs or tissues) and delivered to reference laboratories.
2.2. Samples processing and sequencing
A common processing approach was arranged by Italian and Spanish laboratories participating to the present study: for all samples, pools from the same flock were vortexed in 1 mL of PBS while tissues were mechanically homogenated in 5 mL of PBS/g tissue. The solution was used for RNA extraction using NucleoSpin ® 8/96 RNA (Macherey-Nagel, Düren, Germany). Diagnosis of IBV infection was routinely performed using a real-time RT-PCR commercial kit (Quantification of Avian Infectious Bronchitis Virus-IBV-kit; Genesig, Southampton, UK). The hyper-variable region of the S1 gene of all positive samples was amplified using the primer XCE-1 and XCE-2 as described by Cavanagh et al. [22]. Sanger sequencing was performed on both strands using the same primers. Chromatograms were evaluated with FinchTV (http://www.geospiza.com) and consensus sequences were obtained using CromasPro (CromasPro Version 1.5). The same procedure was followed to obtain the Spanish sequences from samples collected between 2011 and 2015.
2.3. Sequence dataset
All Italian and Spanish sequences of the hyper-variable region of the S1 gene were aligned to the reference dataset proposed by Valastro et al. [23] using the MAFFT method [24]. Recombination analyses were performed with RDP4 (No Reference Selected) adjusting the settings for each method according to the RDP [25] manual recommendations. In particular RDP, GENECONV, Chimaera and 3Seq were used in a primary scan while the full set of available methods was used for the secondary scan. Only recombination events detected by more than 2 methods with a significance value lower than 10−5 (p-value <10−5) and Bonferroni correction were accepted. After exclusion of recombinant sequences, a phylogenetic tree was reconstructed for classification purpose using PhyML [26]. The substitution model was selected based on the Bayesian information criteria (BIC) calculated using Jmodeltest2.1.2 [27]. Italian and Spanish QX strains were then extracted and two independent datasets were constructed.
2.4. tMRCA, evolutionary rate and population dynamics
Population parameters were estimated independently for the two datasets using the Bayesian serial-coalescent based method implemented in BEAST 1.8.2 [28]. Substitution models were selected as previously described while the best fit between strict and relaxed clock models was chosen based on the Bayesian factor (BF) scores, calculated by estimating of the marginal likelihood of different models using both Path Sampling (PS) and Stepping Stone (SS) methods as proposed by Baele et al. [29]. A logBF greater than 10 was considered a good support in favor of the most complex model. The same approach was used for selecting among different parametric (i.e. constant, exponential growth, expansion growth and logistic growth) and non-parametric (i.e. Bayesian skyline, Bayesian skyride and Bayesian skygrid) demographic models [30] using the molecular clock model previously selected. Each dataset was analyzed performing at least two independent runs of 200 million generations each. Population parameters and trees were sampled every 20 thousand generations. Results of the two runs were merged using LogCombiner and accepted only if estimated sample size (ESS), after removal of a 10% burn-in, was greater than 200 and the trace plot, displayed using Tracer 1.5, demonstrated a good convergence and mixing. Parameter estimations were summarized in terms of mean, median and 95% Highest Posterior Density (95HPD).
2.5. Clinical outbreaks
In Italy, the number of IBV related clinical outbreaks was strictly related to the number of available sequences, since all positive samples were collected during clinical outbreaks and were processed for sequencing and genotyping. To evaluate the relationship between changes in viral population size and clinical episodes a correlation analysis was carried out. Accounting for a possible lag phase between QX population rise and its consequences, the IBV QX relative genetic diversity (i.e. effective population size (Ne) × generation time (τ)) was correlated with the corresponding monthly outbreak number shifted by 0 ± 8 months. Similarly, the relationship between age of outbreak occurrence and QX relative genetic diversity was evaluated using the same approach. All statistical analyses were performed using R [31].
3. Results
3.1. Dataset and model parameters
A total of 195 Italian and 98 Spanish non-recombinant sequences were obtained. For both alignments, the strict molecular clock was proven to poorly fit the data compared to “relaxed” models. Particularly, a log-normal uncorrelated relaxed molecular clock was preferred for the Spanish dataset (BF > 10) while both log-normal uncorrelated relaxed molecular clock and the random local clock displayed comparable performances for the Italian one (Table 1 ). Consequently, in absence of any strong evidence for preferring one against the other, a log-normal relaxed molecular clock [32] was implemented for both analyses. When different demographic models were compared the non-parametric skyline model [33] significantly outperformed other non-parametric and parametric ones (BF > 10). On the other hand, a parametric model imposing a constant population size over time explained the Spanish data comparably well with respect to other, more complex, scenarios and was also selected for further analysis (Table 1).
Table 1.
Logarithm of marginal likelihood estimation (logMLE) calculated using path sampling (PS) and stepping-stone (SS) methods for different molecular clock models (upper table) and demographic models (lower table).
| Country | Clock model | PS | SS |
|---|---|---|---|
| Italy | Strict | −1230,32 | −1231,11 |
| Lognormal | −1207,79 | −1205,78 | |
| Exponential | −1221,44 | −1221,76 | |
| Random | −1227,27 | −1228,19 | |
| Spain | Strict | −1712,84 | −1714,11 |
| Lognormal | −1676,83 | −1677,37 | |
| Exponential | −1689,11 | −1689,97 | |
| Random | −1676,44 | −1677,03 | |
| Country | Demographic model | PS | SS |
|---|---|---|---|
| Italy | Constant | −1228,76 | −1229,36 |
| Exponential | −1225,50 | −1226,50 | |
| Logistic | −1227,22 | −1228,33 | |
| Expansion | −1225,51 | −1226,13 | |
| Skyline | −1207,79 | −1205,78 | |
| Skyride | −1225,17 | −1226,74 | |
| Skygrid | −1225,17 | −1226,74 | |
| Spain | Constant | −1677,57 | −1677,38 |
| Exponential | −1683,56 | −1682,68 | |
| Logistic | −1682,22 | −1683,41 | |
| Expansion | −1688,64 | −1688,39 | |
| Skyline | −1676,83 | −1677,37 | |
| Skyride | −1681,82 | −1680,82 | |
| Skygrid | −1685,46 | −1684,49 | |
3.2. Viral population parameters
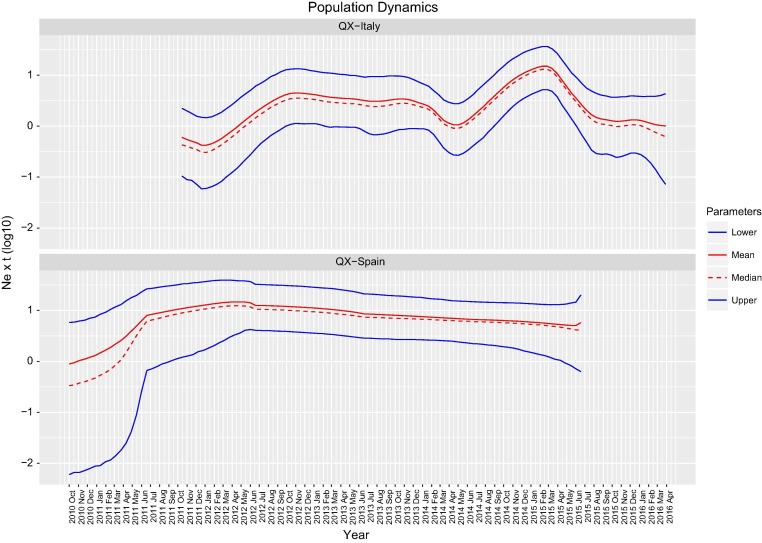
The analysis of QX genotype population dynamics over time revealed remarkable differences between the two countries (Fig. 1 ). Spanish relative genetic diversity remained constant (see previous paragraph) from its most recent common ancestor (MRCA) (i.e. October 2010; 95HPD March 2007– March 2011) to the end of the sampling period. This evidence was also confirmed performing the reconstruction of population history using a skyline model (Fig. 1). Differently, the estimation of the dynamics of Italian QX strains demonstrated a much more complex scenario. After the MRCA of sample strains (i.e. December 2011; 95HPD September 2011-January 2012) the viral population rapidly rose until late 2012 when it became substantially stable. A transitory decrease was observed between December 2013 and June 2014. Since late spring 2014, a new sharp increase, peaking in February 2015 was observed. The following trend was featured by a continuous viral population size reduction, initially steep and gradually more smooth, which reached a value comparable with the initial one by the end of the study.
Fig. 1.
Bayesian skyline plot of population dynamics estimated for Italian (top) and Spanish (bottom) IBV QX strains. Mean, median and upper and lower 95HPD values of relative genetic diversity (i.e. Ne · τ) are plotted over time. The time window considered ranges from the MRCA (mean value) to the most recent sample.
3.3. Clinical outbreaks
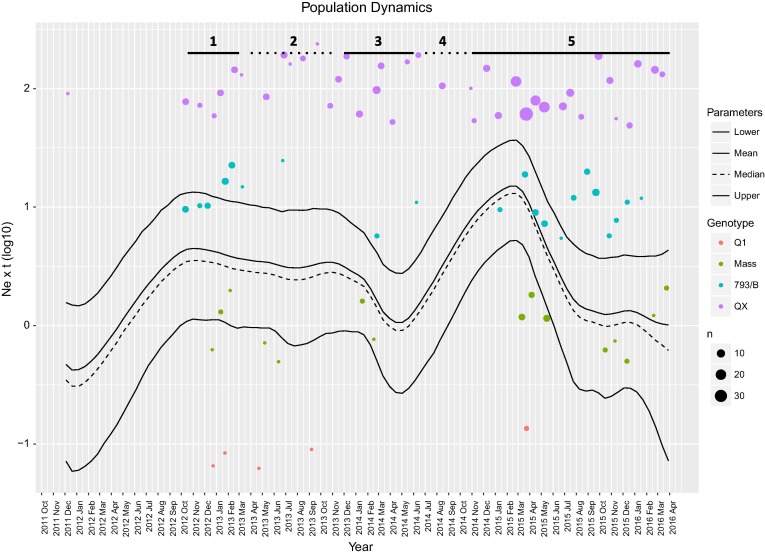
A total of 527 episodes of overt clinical signs ascribable to Infectious bronchitis were submitted to routinely differential diagnosis through PCR based assays and for 356 samples the presence of IBV was demonstrated. Four genotypes were detected during these outbreaks with QX being by far the most relevant (Fig. 2 ). Briefly, 253 (71, 06%) QX, 65 (18, 25%) 793B, 32 (8, 98%) Mass and 5 (1, 40%) Q1 strains were identified in this study.
Fig. 2.
Bayesian skyline plot of population dynamics estimated for Italian IBV QX strains. Mean, median and upper and lower 95HPD values of relative genetic diversity (i.e. Ne · τ) are plotted over time. The number of monthly clinical outbreaks associated to each genotype (color-coded) is reported as circles (randomly jittered to avoid graphical overlapping), whose radius is proportional to the number of events. The different phases (i.e. vaccination strategies) have been represented as solid (double vaccination) or dotted (Mass only vaccination) lines and numbered accordingly to the order reported in the manuscript. The time window considered ranges from the MRCA (mean value) to the most recent sample. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
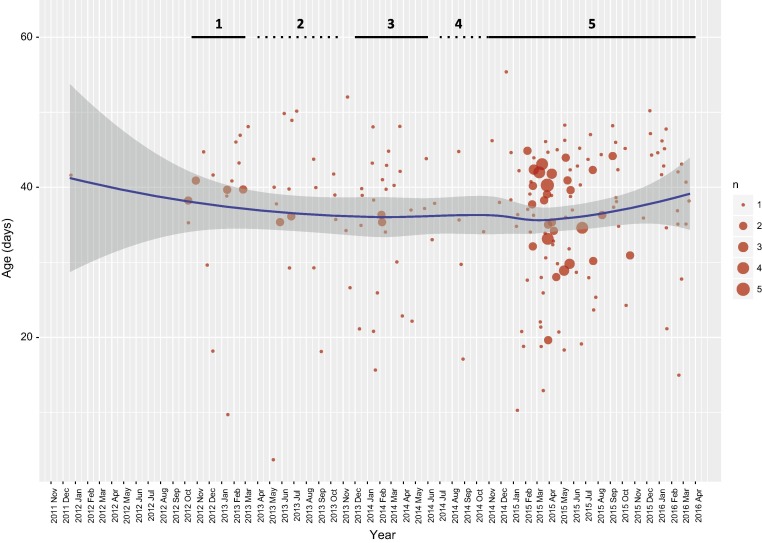
The number of QX-induced outbreaks appeared to be highly correlated with the viral population size. More specifically, the highest correlation was observed when the relative genetic diversity was related with the number of outbreaks occurring 3 months later. Namely, a correlation of 0,78 was observed with the total number of outbreaks while different correlation coefficients were reported for the 793B (0,39), Mass (0,61), Q1 (0,21) and QX (0,78) induced outbreaks. No association was evident between the geographic origin of the samples (which were substantially uniformly distributed) and outbreak occurrence. Similarly, no relationship was evidenced between clinical signs (the vast majority outbreaks were characterized by respiratory signs) and the specific QX strain involved. On the other hand, a statistically significant relationship (p-value = 0,045) was proven between animal age and relative genetic diversity: a negative correlation (−0,32) was observed when the relative genetic diversity was related with the animal age during outbreaks occurring 3 months later. Accordingly, the age of outbreak occurrence slightly fluctuated over time (Fig. 3 ) evidencing a tendency toward a more precocious emergence of clinical signs when vaccination was withdrawn.
Fig. 3.
The age of outbreak occurrence associated to QX genotype is plotted over time as circles (randomly jittered to avoid graphical overlapping), whose radius is proportional to the number of events. A smoothed tendency line with confidence intervals is reported to summarize the general trend. The different phases (i.e. vaccination strategies) have been represented as solid (double vaccination) or dotted (Mass only vaccination) lines and numbered accordingly to the order reported in the manuscript. The time window considered ranges from the most ancient to the most recent sample.
4. Discussion
Infectious bronchitis represents the most relevant poultry infectious disease in many developed countries, imposing huge managerial and economical costs for its control. Particularly, QX genotype is probably one of the most relevant genotype around the world [34], [3]. Indeed, local studies have confirmed this evidence in Italy and Spain where it is by far the most widespread IBV field genotype [21], [35]. Experimental trials have evidenced that the administration of a Mass-based (1 day of life) and 793B-based (14 day of life) vaccine combination provides a good protection against this challenge [7]. Nevertheless, the limited availability of accurate field data and trials hampered the definition of the actual efficacy of these vaccination strategies under field conditions. This, combined with the occurrence of sporadic vaccine-reactions and the absence of proper cost-benefit estimation, has sometimes caused, like in Italy, a certain skepticism about vaccination and its withdrawal, in particular of the 793B dose provided at farm level.
Paradoxically, this situation generated the opportunity to systematically prove, using a large scale epidemiological and phylodynamic approach, the paramount importance of proper vaccination strategies. The use of coalescent based modeling approaches was proven useful and accurate in many studies dealing with the estimation and comprehension of epidemiological dynamics of several human and animal diseases. Above all, these methods allow to reconstruct the demographic history by jointly modeling population parameters and genealogy shape [33]. Consequently, the estimations do not directly depend on the sampling activity, allowing the study of time periods for which no samples are available and to minimize the unavoidable bias due to sampling intensity.
For a vaccination protocol to be considered effective, it is expected to control both the infection spread and the appearance of clinical outbreaks. At first, a comparison between countries with similar epidemiological status but pursuing different vaccination plans was performed based on the coalescent-based analysis of the hyper-variable region of S1 gene. In Spain the same approach based on a combination of Mass plus 793B vaccination was applied over time. Accordingly, and differently from Italy, the IBV QX population dynamics remained constant during the whole study period, already suggesting that, despite not being able to eradicate the pathogen, this strategy was effective in controlling it in field conditions. Nevertheless, it should be also noted that comparison between independent populations could be biased by unconsidered confounding factors and the results should be compared with caution.
On the other hand, the particular scenario occurred in Italy allowed to test this hypothesis by comparing, at different time points, the same viral population in different condition of vaccine exposure (i.e. exposed, non-exposed). It seems clear that vaccination with Mass plus 793B genotypes was able to at least partially control QX spread during the first epidemiological phase, confirming the experimental evidences. While the persistence of a viral shedding was reported also under experimental conditions, the report of a certain frequency of clinical outbreaks in vaccinated flocks clashes with the absence of clinical signs reported by Terregino et al., (2008). Nevertheless, it is well known that vaccine administration is often sub-optimal in field conditions and the presence of several co-factors can easily explain the partial protection induced by these vaccination strategies. After vaccination via drinking water with the 793B genotype was suspended in spring 2013, the epidemiological situation remained substantially stable for almost a year. The transitory decrease in relative genetic diversity, which occurred between December 2013 and June 2014 (Fig. 1), mirrors the implementation of a large-scale trial with a newly registered 793B-based vaccine. Therefore, it can be speculated that this new strategy provided a higher protection against the circulating strains. Nevertheless, the companies decided to stop the trial because the reduction in the number of outbreaks was considered unsatisfactory. This decision was followed by a dramatic increase in viral population size, which preceded in about 2–3 months a peak in the number of clinical outbreaks (Fig. 2). This evidence is quite expected since the sole Mass vaccine was already reported to be ineffective against QX challenge [36]. The delay between the change in viral population and clinical manifestation is probably ascribable to the need of reaching an adequate infectious pressure plus the disease incubation time, a hypothesis that could also be adduced to explain the apparent limited practical effect of the introduction of the new 793B vaccine. In fact, it is possible to speculate that a relatively low viral burden can be tolerated without evident clinical problems thus limiting the benefits of a further reduction. Nevertheless, the economical impact of sub-clinical infections, demonstrated for several livestock diseases [37], [38], [39], [3], [35], was not accounted in the present study. The inverse relationship between relative genetic diversity as well as vaccination period and the age of outbreaks occurrence (Fig. 3) further supports this hypothesis. The higher and long lasting protection induced by double vaccination protocols can have both increased the individual resistance to infection and decreased the infectious pressure (of which the relative genetic diversity can be considered as a proxy) leading to a delayed disease outbreaks. More challenging is to explain the long latency observed between vaccination withdrawal and epidemiological changes. Other concomitant and unconsidered changes like co-infection, climate changes or other managerial choices, etc. could have also played a role and will deserve further investigation. The scenario herein described and the relative lack of information stress once more the need for a systematic and rigorous evaluation of the long term effects of managerial policies, since extemporaneous and subjective judgments can lead to a dramatic bias in disease epidemiology comprehension.
When vaccination was reintroduced, the effects on both viral circulation and clinical signs were immediate, restoring a situation comparable with the initial one, strongly supporting the cause-effect relationship between these two events. Among the factors that could have contributed to this impressive effectiveness, two changes could have played a major role. Firstly, part (less than 20%) of the Italian poultry population started to be vaccinated with QX-based vaccines, which can be reasonably assumed to provide an optimal protection against the homologous strains [40]. Secondly, a strong trend toward the administration of both Mass and 793B vaccine at 1 day of life emerged during the last year in Italy. Traditionally, the induced protection is thought to be higher when vaccines are administered two weeks apart compared with their combination on the same day [3]. Consequently, the benefits of the new strategy could be due to the higher standardization, vaccine coverage and to the decrease of vaccine-reactions compared with field vaccination via drinking water.
Even if what reported is extremely suggestive, to demonstrate a causation instead of a mere association is probably one of the major challenges of epidemiology, in particular when dealing with observation studies based on convenience sampling. A set of criteria for judging if the available information is enough to support a conclusion of causality were proposed by Sir Austin Bradford Hill [41] and are here briefly summarized: (I) Strength of Association. The stronger the relationship between the independent variable and the dependent variable, the less likely the relationship is due to an extraneous variable. (II) Temporality. A cause has to precede an effect in time. (III) Consistency. Multiple observations under different circumstances and study settings increase the credibility of a finding. (IV) Theoretical Plausibility. It is easier to accept an association as causal when there is a theoretical basis for such a conclusion. (V) Coherence. The association must be coherent with other knowledge. (VI) Specificity in the causes. In the ideal situation, the effect has only one cause. (VII) Dose Response Relationship. There should be a relationship between the risk factor exposure and the disease. (VIII) Experimental Evidence. Related researches based on experiments support a causal inference. (IX) Analogy. A commonly accepted phenomenon in one area can be applied to another area.
As we have reported in the previous paragraphs, all these criteria have been met in the present work. Criteria I and II are clearly accomplished by the Italian scenario where vaccine administration changes were invariably followed by a decrease on IBV population. Similarly, the reversibility of this effect was proven after vaccine withdrawal which led to an increase in viral population size and outbreak frequency in absence of other reasonable causes (point VI). The vaccination trial performed between December 2013 and June 2014, affecting only a limited portion of the poultry flocks, caused a less marked decrease in viral population size compared with the outstanding one experienced after November 2015, when the whole poultry population was massively vaccinated. Consequently, even if it is hard to be confidently proven in the present study, a dose response relationship (point VII) can be at least hypothesized.
Obtaining concordant results using a different study approach (i.e. the comparison between two countries) is a good proof in favor of point III. Finally, the biological mechanism of action of vaccination (point IV and V) is out of discussion and its efficacy has already been proven for IBV in experimental settings (point VIII) [40], [42], [43] and for other animal diseases using an approach comparable with that of the current work (point IX) [17], [20], [44]. Thus, the accomplishment of all Hill’s criteria provides strong background to demonstrate the causal nexus between selection of field vaccination protocols and IBV epidemiology.
As a secondary result, this study provides further confirmation to the “vaccine origin” hypothesis of the 793B strains circulating in Italy. In a previous study, we demonstrated the disappearance of this genotype after vaccination withdrawal in late spring 2013 [5]. During this follow-up study, the 793B genotype was only detected sporadically (4 strains) during the non-vaccination period and originated from broiler farms with nearby layers flocks (where the administration of vaccines containing this genotype was not discontinued). On the other hand, the detection of 793B strains indistinguishable from the vaccine (at least in the genomic region considered), became a common event after its reintroduction.
Globally, the present study provides an extensive evaluation of vaccination efficacy against the IBV QX genotype in field conditions. The comparison of two independent populations and of the same population at different time points, while exposed and not to changes in vaccination protocols, provided concordant and robust evidences about the efficacy of vaccination in controlling IBV field strain circulation and clinical outbreaks. The relevant lag phase between managerial changes and practical consequences highlights the relevance of other factors (biological, epidemiological, random, etc.) in mediating or delaying the effect of our control strategies.
This study testifies the primary importance of rigorously planning not only the intervention strategies but also their monitoring and evaluation, shunning the temptation of overtrusting in subjective and short term feelings.
References
- 1.Duffy S., Shackelton L.A., Holmes E.C. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9(4):267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 2.Simon-Loriere E., Holmes E.C. Why do RNA viruses recombine? Nat Rev Microbiol. 2011;9(8):617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sjaak de Wit J.J., Cook J.K., van der Heijden H.M. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40(3):223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanagh D., Elus M., Cook J. Relationship between sequence variation in the S1 spike protein of infectious bronchitis virus and the extent of cross-protection in vivo. Avian Pathol. 1997;26(1):63–74. doi: 10.1080/03079459708419194. [DOI] [PubMed] [Google Scholar]
- 5.Franzo G., Naylor C.J., Lupini C., Drigo M., Catelli E., Listorti V. Continued use of IBV 793B vaccine needs reassessment after its withdrawal led to the genotype’s disappearance. Vaccine. 2014;32(50):6765–6767. doi: 10.1016/j.vaccine.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook J.K., Orbell S.J., Woods M.A., Huggins M.B. Breadth of protection of the respiratory tract provided by different live-attenuated infectious bronchitis vaccines against challenge with infectious bronchitis viruses of heterologous serotypes. Avian Pathol. 1999;28(5):477–485. doi: 10.1080/03079459994506. [DOI] [PubMed] [Google Scholar]
- 7.Terregino C., Toffan A., Serena Beato M., De Nardi R., Vascellari M., Meini A. Pathogenicity of a QX strain of infectious bronchitis virus in specific pathogen free and commercial broiler chickens, and evaluation of protection induced by a vaccination programme based on the Ma5 and 4/91 serotypes. Avian Pathol. 2008;37(5):487–493. doi: 10.1080/03079450802356938. [DOI] [PubMed] [Google Scholar]
- 8.Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faria N.R., Suchard M.A., Rambaut A., Lemey P. Toward a quantitative understanding of viral phylogeography. Curr Opin Virol. 2011;1(5):423–429. doi: 10.1016/j.coviro.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grenfell B.T., Pybus O.G., Gog J.R., Wood J.L., Daly J.M., Mumford J.A. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303(5656):327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 11.Holmes E.C., Grenfell B.T. Discovering the phylodynamics of RNA viruses. PLoS Comput Biol. 2009;5(10):e1000505. doi: 10.1371/journal.pcbi.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stadler T., Kuhnert D., Bonhoeffer S., Drummond A.J. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV) Proc Natl Acad Sci USA. 2013;110(1):228–233. doi: 10.1073/pnas.1207965110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodrigo AG, Felsenstein J. Coalescent approaches to HIV population genetics. In: The evolution of HIV; 1999. p. 233–72.
- 14.Kuhnert D., Stadler T., Vaughan T.G., Drummond A.J. Simultaneous reconstruction of evolutionary history and epidemiological dynamics from viral sequences with the birth-death SIR model. J R Soc Interface. 2014;11(94):20131106. doi: 10.1098/rsif.2013.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su Y.C., Bahl J., Joseph U., Butt K.M., Peck H.A., Koay E.S. Phylodynamics of H1N1/2009 influenza reveals the transition from host adaptation to immune-driven selection. Nat Commun. 2015;6 doi: 10.1038/ncomms8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemey P., Suchard M., Rambaut A. Reconstructing the initial global spread of a human influenza pandemic: a Bayesian spatial-temporal model for the global spread of H1N1pdm. PLoS Curr. 2009;1 doi: 10.1371/currents.RRN1031. RRN1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alfonso-Morales A., Martinez-Perez O., Dolz R., Valle R., Perera C.L., Bertran K. Spatiotemporal phylogenetic analysis and molecular characterisation of infectious bursal disease viruses based on the VP2 hyper-variable region. PLoS ONE. 2013;8(6):e65999. doi: 10.1371/journal.pone.0065999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franzo G., Cortey M., Segalés J., Hughes J., Drigo M. Phylodynamic analysis of porcine circovirus type 2 reveals global waves of emerging genotypes and the circulation of recombinant forms. Mol Phylogenet Evol. 2016;100:269–280. doi: 10.1016/j.ympev.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 19.Franzo G., Dotto G., Cecchinato M., Pasotto D., Martini M., Drigo M. Phylodynamic analysis of porcine reproductive and respiratory syndrome virus (PRRSV) in Italy: action of selective pressures and interactions between different clades. Infect Genet Evol. 2015;31:149–157. doi: 10.1016/j.meegid.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 20.Murcia P.R., Wood J.L., Holmes E.C. Genome-scale evolution and phylodynamics of equine H3N8 influenza A virus. J Virol. 2011;85(11):5312–5322. doi: 10.1128/JVI.02619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanco A., Antillés N., Camprubí Q., Jové R., Biarnes M. LII Simposio cientifico de Avicultura; Malaga (Spain): 2011. Bronquitis aviar: epidemiología molecular y evolución delos diferentes genotipos predominantes en España desde 2011, 28–30 October. [Google Scholar]
- 22.Cavanagh D., Mawditt K., Britton P., Naylor C. Longitudinal field studies of infectious bronchitis virus and avian pneumovirus in broilers using type-specific polymerase chain reactions. Avian Pathol. 1999;28(6):593–605. doi: 10.1080/03079459994399. [DOI] [PubMed] [Google Scholar]
- 23.Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect Genet Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin D.P., Lemey P., Lott M., Moulton V., Posada D., Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26(19):2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guindon S., Dufayard J-., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 27.Darriba D., Taboada G.L., Doallo R., Posada D. JModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baele GF, Lemey PF, Bedford TF, Rambaut A, Suchard, MA, Alekseyenko AV. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 2012. JID – 8501455. [DOI] [PMC free article] [PubMed]
- 30.Ho S.Y., Shapiro B. Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol Ecol Resour. 2011;11(3):423–434. doi: 10.1111/j.1755-0998.2011.02988.x. [DOI] [PubMed] [Google Scholar]
- 31.Team RC. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013–2014.
- 32.Drummond A.J., Ho S.Y., Phillips M.J., Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4(5):e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drummond A.J., Rambaut A., Shapiro B., Pybus O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22(5):1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 34.Jackwood M.W. Review of infectious bronchitis virus around the world. Avian Dis. 2012;56(4):634–641. doi: 10.1637/10227-043012-Review.1. [DOI] [PubMed] [Google Scholar]
- 35.Moreno A., Franzo G., Massi P., Tosi G., Blanco A., Antilles N. A novel variant of the infectious bronchitis virus resulting from recombination events in Italy and Spain. Avian Pathol. 2016:1–28. doi: 10.1080/03079457.2016.1200011. [just-accepted] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun C., Han Z., Ma H., Zhang Q., Yan B., Shao Y. Phylogenetic analysis of infectious bronchitis coronaviruses newly isolated in China, and pathogenicity and evaluation of protection induced by Massachusetts serotype H120 vaccine against QX-like strains. Avian Pathol. 2011;40(1):43–54. doi: 10.1080/03079457.2010.538037. [DOI] [PubMed] [Google Scholar]
- 37.Alarcon P., Rushton J., Wieland B. Cost of post-weaning multi-systemic wasting syndrome and porcine circovirus type-2 subclinical infection in England – an economic disease model. Prev Vet Med. 2013;110(2):88–102. doi: 10.1016/j.prevetmed.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennett R.M., Christiansen K., Clifton-Hadley R.S. Estimating the costs associated with endemic diseases of dairy cattle. J Dairy Res. 1999;66(03):455–459. doi: 10.1017/s0022029999003684. [DOI] [PubMed] [Google Scholar]
- 39.Bennett R. The ‘direct costs’ of livestock disease: the development of a system of models for the analysis of 30 endemic livestock diseases in Great Britain. J Agric Econ. 2003;54(1):55–71. [Google Scholar]
- 40.Geerligs H., Boelm G., Meinders C., Stuurman B., Symons J., Tarres-Call J. Efficacy and safety of an attenuated live QX-like infectious bronchitis virus strain as a vaccine for chickens. Avian Pathol. 2011;40(1):93–102. doi: 10.1080/03079457.2010.542742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill A.B. The environment and disease: association or causation? Proc R Soc Med. 1965;58:295–300. doi: 10.1177/003591576505800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones R.C., Worthington K.J., Capua I., Naylor C.J. Efficacy of live infectious bronchitis vaccines against a novel European genotype, Italy 02. Vet Rec. 2005;156(20):646–647. doi: 10.1136/vr.156.20.646. [DOI] [PubMed] [Google Scholar]
- 43.Lim T.H., Kim M.S., Jang J.H., Lee D.H., Park J.K., Youn H.N. Live attenuated nephropathogenic infectious bronchitis virus vaccine provides broad cross protection against new variant strains. Poult Sci. 2012;91(1):89–94. doi: 10.3382/ps.2011-01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pereira C.A., Leal E.S., Durigon E.L. Selective regimen shift and demographic growth increase associated with the emergence of high-fitness variants of canine parvovirus. Infect Genet Evol. 2007;7(3):399–409. doi: 10.1016/j.meegid.2006.03.007. [DOI] [PubMed] [Google Scholar]