Abstract
Small plus-stranded RNA viruses do not code for RNA helicases that would facilitate the proper folding of viral RNAs during replication. Instead, these viruses might use RNA chaperones as shown here for the essential p33 replication protein of Tomato bushy stunt virus (TBSV). In vitro experiments demonstrate that the purified recombinant p33 promotes strand separation of a DNA/RNA duplex. In addition, p33 renders dsRNA templates sensitive to single-strand specific S1 nuclease, suggesting that p33 can destabilize highly structured RNAs. We also demonstrate that the RNA chaperone activity of p33 facilitates self-cleavage by a ribozyme in vitro. In addition, purified p33 facilitates in vitro RNA synthesis on double-stranded (ds)RNA templates up to 5-fold by a viral RNA-dependent RNA polymerase. We propose that the RNA chaperone activity of p33 facilitates the initiation of plus-strand synthesis as well as affects RNA recombination. Altogether, the TBSV RNA chaperone might perform similar biological functions to the helicases of other RNA viruses with much larger coding capacity.
Keywords: Tomato bushy stunt virus, Replication, RNA chaperone, Duplex unwinding, Plus-strand synthesis, In vitro, RdRp, Virus replicase
Graphical Abstract
Research Highlights
►RNA chaperone activity of the small replication protein; enhanced plus-strand synthesis in vitro; highly disordered structural regions.
Introduction
RNA viruses utilize their genomes not only as templates to produce more viral RNA progeny during replication, but for many other processes, such as translation, regulation of replication, encapsidation and movement from cell to cell. The folding of viral RNAs in biologically relevant conformation is important because the RNA can be trapped in one of many incorrect/nonfunctional structures (Russell, 2008). However, RNA-binding proteins, such as helicases, RNA chaperones and cofactors, which facilitate conformational transitions of RNA, thus preventing the deleterious effect of RNA misfolding, have been proposed to play auxiliary roles in most steps during the viral infection process. Unlike larger RNA viruses, small plus-strand (+)RNA viruses with genomes less than 6000 nt do not code for RNA helicases (Koonin and Dolja, 1993). This prompted discussions if these small RNA viruses might not need helicases with robust unwinding functions for their replication or, alternatively, they might recruit RNA helicases(s) from the host. Another alternative possibility is that these RNA viruses might code for RNA chaperones, which are also capable of unwinding/destabilizing RNA structures (Cristofari and Darlix, 2002, Zuniga et al., 2009). Accordingly, viral-coded RNA chaperones have been shown to participate in the replication of picornaviruses, coronaviruses, flaviviruses and hepatitis delta virus (DeStefano and Titilope, 2006, Huang and Wu, 1998, Ivanyi-Nagy et al., 2008, Wang et al., 2003, Zuniga et al., 2010), or affect the packaging of coronaviruses, hepatitis C virus, retroviruses and minus-stranded RNA viruses (Cristofari et al., 2004, Cruceanu et al., 2006, Ivanyi-Nagy et al., 2006, Levin et al., 2005, Mir and Panganiban, 2006a, Mir and Panganiban, 2006b, Rein et al., 1998, Zuniga et al., 2007). Host-coded RNA chaperones have also been shown to affect calicivirus and viroid RNA replication (Daros and Flores, 2002, Karakasiliotis et al., 2006). The unique feature of RNA chaperones is that they do not require rNTPs for destabilizing RNA structures, but instead, they bind cooperatively to the RNA and leads to structural changes in the RNA. Thus, RNA chaperones will profoundly affect the folding process, interactions between RNAs and the accessibility of the RNA to other proteins (Russell, 2008, Zuniga et al., 2009).
One group of small plus-stranded RNA viruses, which does not code for RNA helicases, is tombusviruses. These viruses, including Tomato bushy stunt virus (TBSV) and Cucumber necrosis virus (CNV), code for an essential RNA-binding protein, termed p33, which is known to affect many steps during the infection process. For example, an essential function of p33 is to bind a C·C mismatch within an internal cis-acting stem-loop element that leads to selective recruitment of the viral RNA into replication (Monkewich et al., 2005, Pogany et al., 2005). p33 is also involved in the formation of spherule-like structures on the cytoplasmic interface of peroxisomal membranes, which represent the site of viral RNA replication (McCartney et al., 2005, Navarro et al., 2006, Panavas et al., 2005). Its role in viral pathogenesis has also been documented (Burgyan et al., 2000). In addition to the RNA-binding domain, p33 also contains two short p33:p33/p92pol interaction domains that are likely important for the assembly of the viral replicase and for RNA-binding, too (Panaviene et al., 2003, Pogany et al., 2005, Rajendran and Nagy, 2004). Phosphorylation close to the RNA-binding domain is thought to regulate the function of p33 during replication (Shapka et al., 2005, Stork et al., 2005). Overall, the essential p33 seems to be the master regulator of the tombusvirus replication process (Nagy, 2008, Nagy and Pogany, 2006, Nagy and Pogany, 2008).
Depending on the protein and RNA concentrations, mutations or the composition of the buffer, the tombusvirus p33 auxiliary protein has been shown to bind to the viral RNA either specifically or non-specifically (Pogany et al., 2005, Rajendran and Nagy, 2003). The specific interaction between p33 and the viral RNA is required for the recruitment of the TBSV (+)RNA to the membrane-bound replicase complex (Monkewich et al., 2005, Pogany et al., 2005). However, after the selective (+)RNA recruitment, the function of p33 could be different within the viral replicase complex, where p33 is present in high amount, reaching ~ 10–20-fold excess over the p92pol (Rajendran and Nagy, 2006, Serva and Nagy, 2006). The high local concentration of p33 within the replicase complex could facilitate non-specific binding of p33 to the viral RNA in a cooperative manner (Rajendran and Nagy, 2003), possibly allowing p33 to function as an RNA chaperone within the replicase complex. Therefore, based on cooperative RNA-binding ability of p33, we speculated that p33 might be able to perform many functions during infection by acting as an RNA chaperone that modifies the structure of the viral RNA to allow the participation of the viral RNA in different ribonucleoprotein complexes. To test this hypothesis, we performed biochemical experiments with purified recombinant p33 using in vitro assays. The purified p33 was shown to efficiently unwind short ssDNA/ssRNA duplexes, render double-stranded (ds)RNA templates sensitive to S1 nuclease, and stimulate self-cleavage by a ribozyme in vitro. We have demonstrated that p33 can promote initiation of RNA synthesis by up to 5-fold by a viral RNA-dependent RNA polymerase (RdRp) in vitro. Based on these data, we propose a role for the RNA chaperone activity of p33 during tombusvirus replication.
Results
In vitro strand separation activity of the recombinant p33 on DNA oligo — RNA template duplex
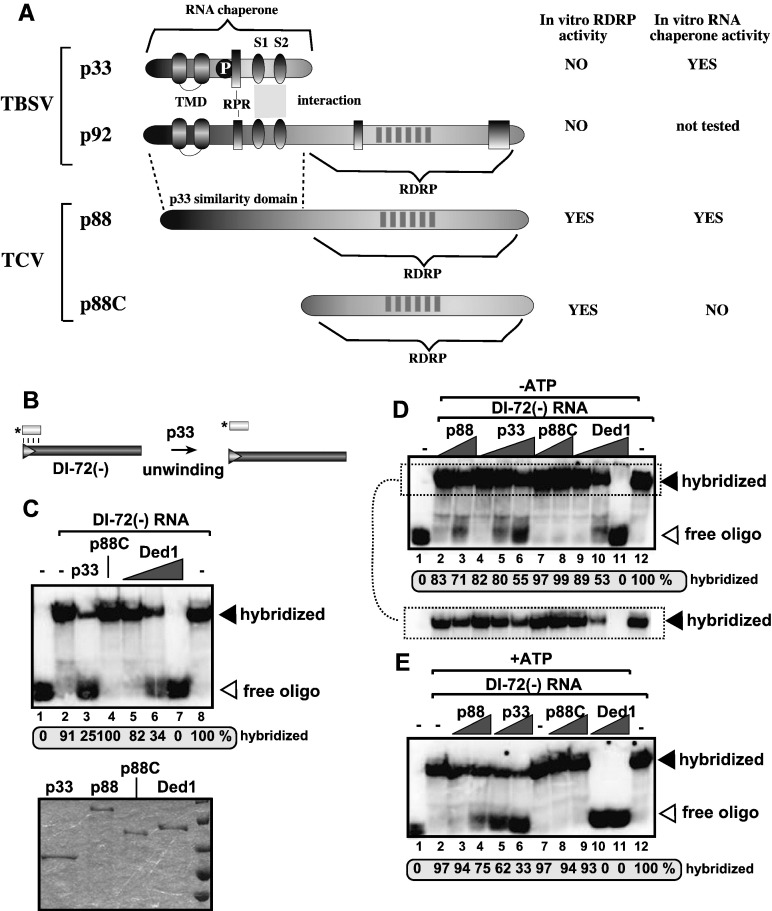
To test the putative RNA chaperone activity of p33, we used a strand separation assay with purified recombinant TBSV p33 and Turnip crinkle virus (TCV) RdRp proteins in vitro (Fig. 1 ). A 21-nt ssDNA oligo was 5′ end labeled and annealed with the unlabeled full-length DI-72(−)RNA, followed by the addition of purified TBSV p33 (Fig. 1B). We have found that 45–75% of ssDNA/ssRNA duplex was separated by p33 when present at high concentration (Fig. 1C, lane 3 and 1D, lane 6) in the absence of ATP. On the other hand, the N-terminally truncated TCV p88 RdRp, termed p88C, lacking the p33 similarity domain (Fig. 1A) did not show unwinding activity at a detectable level (Fig. 1C, lane 4), even at the highest concentration (Figs. 1D–E). Since p88C is an RNA-binding protein and it can efficiently use DI-72(−)RNA as a template (Cheng et al., 2005, Rajendran and Nagy, 2003, Rajendran et al., 2002), the lack of unwinding activity by p88C shows that RNA-binding to the template by an RNA-binding protein alone is not sufficient for strand separation in this assay. This is further supported by the lack of RNA chaperone activity by the N-terminally truncated p33, termed p33C (supplementary Fig. S1A–C), which has strong RNA-binding capacity (Pogany et al., 2005, Rajendran and Nagy, 2003).
Fig. 1.
Unwinding of the ssDNA/ssRNA duplex by p33 in an in vitro strand separation assay. (A) Schematic representation of the known domains in the tombusvirus replication proteins p33 and p92pol. Note that the N-terminal segment in p92pol contains the same sequence as in p33 due to the overlapping expression strategy of TBSV genome, while the C-terminal region of p92pol carries the RdRp domain. TMD, trans-membrane domains; P, phosphorylation sites; RPR, arginine-proline-rich RNA-binding domain; S1 and S2 are subdomains of the p33:p33/p92 interaction domain. The closely related TCV p88 is similar to p92pol and carries the p33 similarity domain as well as the RdRp domain. P88C is an N-terminally truncated p88 derivative, which has strong RdRp activity in vitro. (B) Scheme of the strand separation assay showing the hybridized ssDNA/ssRNA duplex. The 5′ end labeled, 21 base ssDNA oligo was annealed to the unlabeled DI-72(−)RNA prior to the addition of purified recombinant p33. (C) In vitro strand separation assay with p33 using an ssDNA oligo/ssRNA template duplex. Lane 1: 5′ end labeled-ssDNA oligo; Lanes 2 and 8: annealed ssDNA/ssRNA, no p33; Lane 3: annealed ssDNA/ssRNA template plus 4 pmol purified recombinant TBSV MBP-p33; Lane 4: annealed ssDNA/ssRNA plus 4 pmol purified recombinant TCV MBP-p88C; Lanes 5–7: annealed ssDNA/ssRNA plus 0.4, 1 and 4 pmol purified recombinant MBP-Ded1p, respectively. The assay was performed in the absence of ATP. The 32P-labeled free ssDNA and ssDNA/ssRNA duplex were separated on non-denaturing 5% acrylamide gels. Quantification of the ssDNA/ssRNA duplex was done with ImageQuant. Bottom image shows an SDS-PAGE analysis of purified recombinant proteins used in these assays. Each experiment was repeated at least three times. (D) In vitro strand separation assay with recombinant proteins using an ssDNA oligo/ssRNA template duplex in the absence of ATP. Lane 1: 5′ end labeled-ssDNA oligo; Lanes 2–3: annealed ssDNA/ssRNA, plus 1 and 4 pmol of p88; Lanes 4–6: annealed ssDNA/ssRNA template plus 0.4, 1 and 4 pmol purified recombinant TBSV MBP-p33; Lanes 7–8: annealed ssDNA/ssRNA plus 1 and 4 pmol purified recombinant TCV MBP-p88C; Lanes 9–11: annealed ssDNA/ssRNA plus 0.4, 1 and 4 pmol purified recombinant MBP-Ded1p, respectively; Lane 12: annealed ssDNA/ssRNA, no p33. The percentage of unwound ssDNA/ssRNA duplex is shown. The lower image shows a shorter exposure for the ssDNA/ssRNA duplex. (E) In vitro strand separation assay with recombinant proteins using an ssDNA oligo/ssRNA template duplex in the presence of ATP. Lane 1: 5′ end labeled-ssDNA oligo; Lane 2: annealed ssDNA/ssRNA, no p33; Lanes 3–4: annealed ssDNA/ssRNA, plus 1 and 4 pmol of p88; Lanes 5–6: annealed ssDNA/ssRNA template plus 1 and 4 pmol purified recombinant TBSV MBP-p33; Lane 7: annealed ssDNA/ssRNA, no p33; Lanes 8–9: annealed ssDNA/ssRNA plus 1 and 4 pmol purified recombinant TCV MBP-p88C; lanes 10–11: annealed ssDNA/ssRNA plus 1 and 4 pmol purified recombinant MBP-Ded1p, respectively; Lane 12: annealed ssDNA/ssRNA, no p33. The percentage of unwound ssDNA/ssRNA duplex is shown. See additional details in panel C.
When compared with the TBSV p33 (Fig. 1D, lanes 5–6), the purified p88 RdRp of the closely related TCV, which carries the p33 similarity domain (Fig. 1A), showed weak unwinding activity in vitro (Fig. 1D, lanes 2–3) at similar concentration in the absence of added ATP in vitro. However, the strand separation activity of p33 was much weaker than that of Ded1p DEAD-box RNA helicase, which was capable of complete separation of the ssDNA/ssRNA duplex (Fig. 1E, lanes 10–11). As expected, ATP stimulated the unwinding activity of Ded1p helicase (Fig. 1E, lane 10 versus Fig. 1D, lane 10), while ATP had no major effect on the strand separation activity of p33 (Fig. 1C, lane 3 and Fig. 1D, lanes 5–6 versus 1E, lanes 5–6) or p88 (1D, lanes 2–3 versus 1E, lanes 3–4). These data support that p33, and, to a lesser extent, p88, have RNA chaperone-like activities in vitro.
P33 replication protein renders a dsRNA template ribonuclease sensitive in vitro
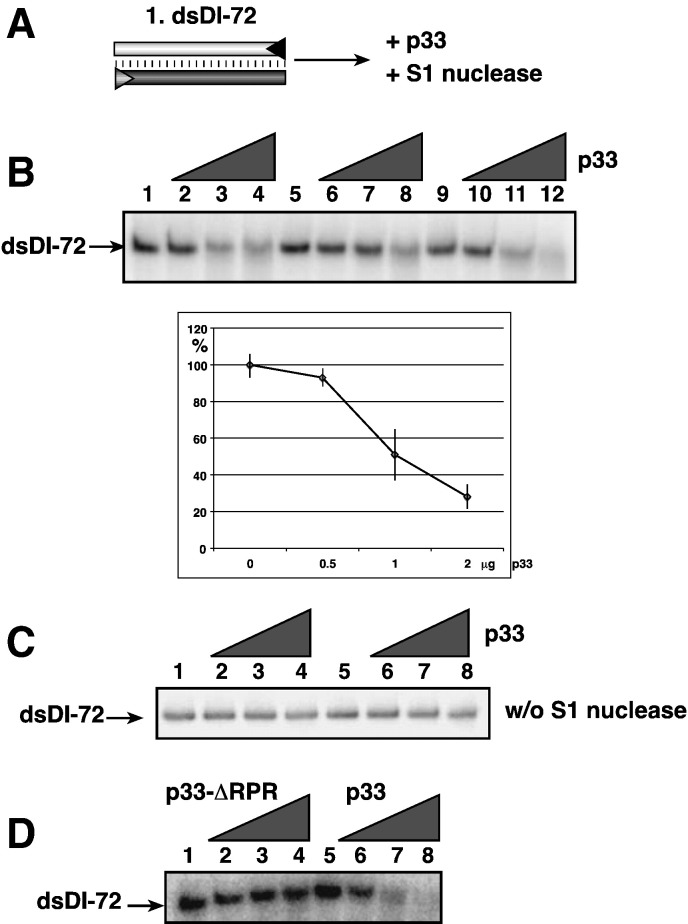
To test if p33 can open up dsRNA structures, thus rendering the dsRNA sensitive to nucleases, we developed an S1 nuclease-based assay (Fig. 2A). While dsDI-72 was completely resistant to S1 nuclease digestion under in vitro conditions (Fig. 2B, lanes 1, 5, and 9), addition of purified recombinant p33 made dsRNA S1 ribonuclease sensitive, leading to RNA degradation up to 75% (Fig. 2B, lanes 4, 8, and 12). The same p33 preparation when applied in the absence of S1 did not lead to degradation of dsRNA, excluding the possibility that the purified recombinant p33 preparation was contaminated by a bacterial ribonuclease (Fig. 2C). This experiment also showed that p33 could not efficiently separate 621 bp long fully base-paired RNA-RNA duplex in vitro (Fig. 2C), suggesting that the RNA chaperone activity of p33 is not robust.
Fig. 2.
Increased sensitivity of dsRNA to single-strand specific S1 nuclease due to the RNA chaperone activity of p33. (A) Schematic presentation of the dsDI-72 RNA template and the treatment applied. (B) Representative denaturing gel of radiolabeled RNA template (5 ng) that remained after the S1 nuclease treatment in the presence of 0, 0.5, 1 and 2 μg of purified recombinant p33 are shown. The samples were phenol-chloroform extracted prior to gel-analysis. Each experiment was repeated at least three times. (C) A control experiment showing that dsRNA template is insensitive to treatment with purified p33 in the absence of S1 nuclease. The condition during the experiments is comparable to that shown in panel B, except S1 was not added to the samples. (D) A control experiment showing that dsRNA template is insensitive to treatment with S1 nuclease in the presence of a p33 mutant lacking the RPR RNA-binding sequence. See further details in panel B.
In addition, p33 lacking the 6 amino acid RPR RNA-binding domain (construct p33-ΔRPR) (Rajendran and Nagy, 2003) did not render dsDI-72 RNA S1 nuclease sensitive (Fig. 2D, lanes 2–4). These data are consistent with the model that the RNA-binding ability of p33 is critical to render the dsRNA template S1 nuclease sensitive.
P33 replication protein facilitates self-cleavage by a ribozyme in vitro
Since RNA chaperones bind to the RNA non-specifically and cooperatively, like p33 when present in high concentration (Rajendran and Nagy, 2003), they can facilitate RNA cleavage by ribozymes (Cristofari and Darlix, 2002, Zuniga et al., 2007). This property of the RNA chaperones are due to refolding of the ribozyme sequence trapped in non-active conformations. To test the effect of p33 on ribozyme activity, we used the self-cleaving ribozyme of satellite Tobacco ringspot virus (Buzayan et al., 1986, Panavas and Nagy, 2003) in the presence of recombinant p33 (Fig. 3A). We have found that the purified p33 stimulated the ribozyme activity by ~ 4-fold, reducing the amount of uncleaved RNA and increasing the amount of self-cleaved RNAs (Fig. 3B, lane 3 versus lane 1). This finding supports the model that p33 could be involved in RNA-folding in a sequence non-specific manner.
Fig. 3.
Recombinant p33 promotes the self-cleavage activity of the satellite Tobacco ringspot virus ribozyme in vitro. (A) Schematic presentation of the RNA template carrying the ribozyme sequence before and after the self-cleavage. The satellite (−) ribozyme of TRSV (Buzayan et al., 1986, Fedor, 2000) forms an autocatalytic hairpin structure, which self-cleaves at the 3′ end TBSV RIV(+)RNA, releasing two RNA products as shown. (B) Representative denaturing gel of 32P-labeled RNA cleavage products, which are derived from the template RNA synthesized by in vitro transcription with T7 RNA polymerase in the presence of 0, 1 and 2 μg of purified recombinant p33 are shown. Each experiment was repeated at least three times.
Inhibition of the RdRp-driven RNA synthesis by ssDNA complementary to the promoter in the template RNA is neutralized by p33
Since the RNA chaperone activity of p33 replication protein is likely important within the viral replicase complex where the local p33 concentration is high, we wanted to test if p33 could change the activity of a viral RdRp on various RNA templates. We have chosen a heterologous RdRp derived from the closely related TCV for these studies since the TBSV or other tombusvirus p92pol RdRp is not active in vitro (Fig. 1). Since the full-length p88 RdRp protein of TCV functions as a weak RNA chaperone by itself (Fig. 1), we used the N-terminally-truncated p88C, which lacks RNA chaperone activity in vitro (Fig. 1), yet it is a highly active RdRp on ssRNA templates in vitro (Panavas et al., 2006, Rajendran et al., 2002). Altogether, a major advantage of using this heterologous system is that the tombusvirus p33 does not interact directly with TCV p88C (Rajendran and Nagy, 2004), thus making it unlikely that any stimulatory effect of p33 on p88C RdRp activity would be due to direct enhancement of RdRp activity via protein–protein interaction (i.e., due to p33 acting like a transcription factor). Rather, the most probable way for p33 to affect the RdRp activity of p88C is to alter the structure of the RNA template via the RNA chaperone activity of p33.
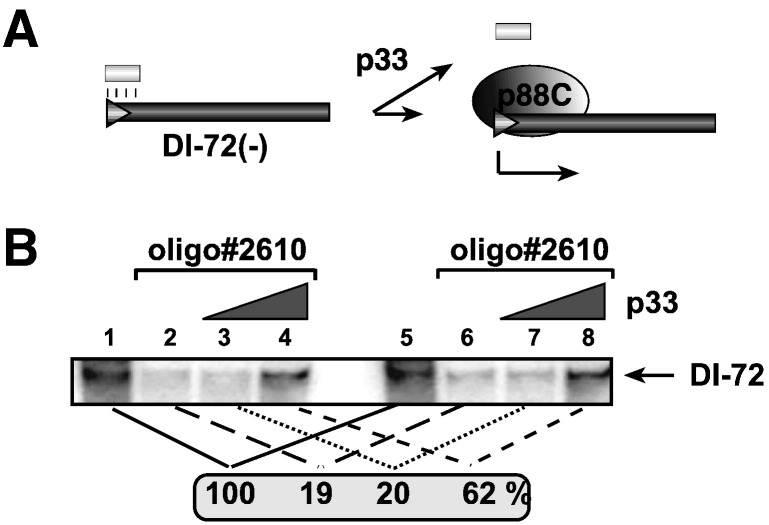
Accordingly, we set up an in vitro assay based on the p88C RdRp protein and a partial ssRNA/ssDNA duplex (Fig. 4A), to test for the RNA chaperone activity of p33 replication protein. Since previous studies have established that the accessibility of the terminal promoters by the tombusviral or TCV RdRps is a critical factor during initiation, we used a short ssDNA complementary to the very 3′ end of DI-72(−) to form partial DNA/RNA duplex within the complementary sequences. We have found that the ssDNA inhibited RNA synthesis by ~ 80% in the RdRp assay (Fig. 4B, lanes 2, 6 versus lanes 1, 5). This observation suggests that the initiation site was much less accessible to the p88C RdRp in the presence of ssDNA than on the free ssRNA template.
Fig. 4.
Recombinant p33 promotes plus-strand initiation on (−)RNA templates by the RdRp. (A) Schematic presentation of the ssDNA oligo/ssRNA template duplex used to program the TCV p88C RdRp preparation in vitro. The cPR promoter sequence used for plus-strand initiation by the TCV RdRp is shown with an empty arrowhead. (B) Representative denaturing gel of 32P-labeled RNA product synthesized by TCV p88C RdRp in vitro in the presence of 0, 1 and 2 μg of purified recombinant p33 is shown. The level of RNA synthesis was compared to that of the RdRp activity obtained in the absence of ssDNA and p33 (100%). The samples were treated with S1 nuclease to exclude terminal transferase-based labeling of DI-72(−)RNA, which might be present in the affinity-purified TCV p88C or p33 preparations. Each experiment was repeated three times.
Importantly, we have found that the addition of recombinant p33 enhanced the RdRp activity of p88C by ~ 3-fold on the partial ssRNA/ssDNA duplex (Fig. 4B, lane 4 and 8). This finding supports the idea that p33 facilitates the accessibility of the cPR promoter in DI-72(−) template by destabilizing the ssRNA/ssDNA duplex (Fig. 4A) as also shown in the strand separation assay (Fig. 1).
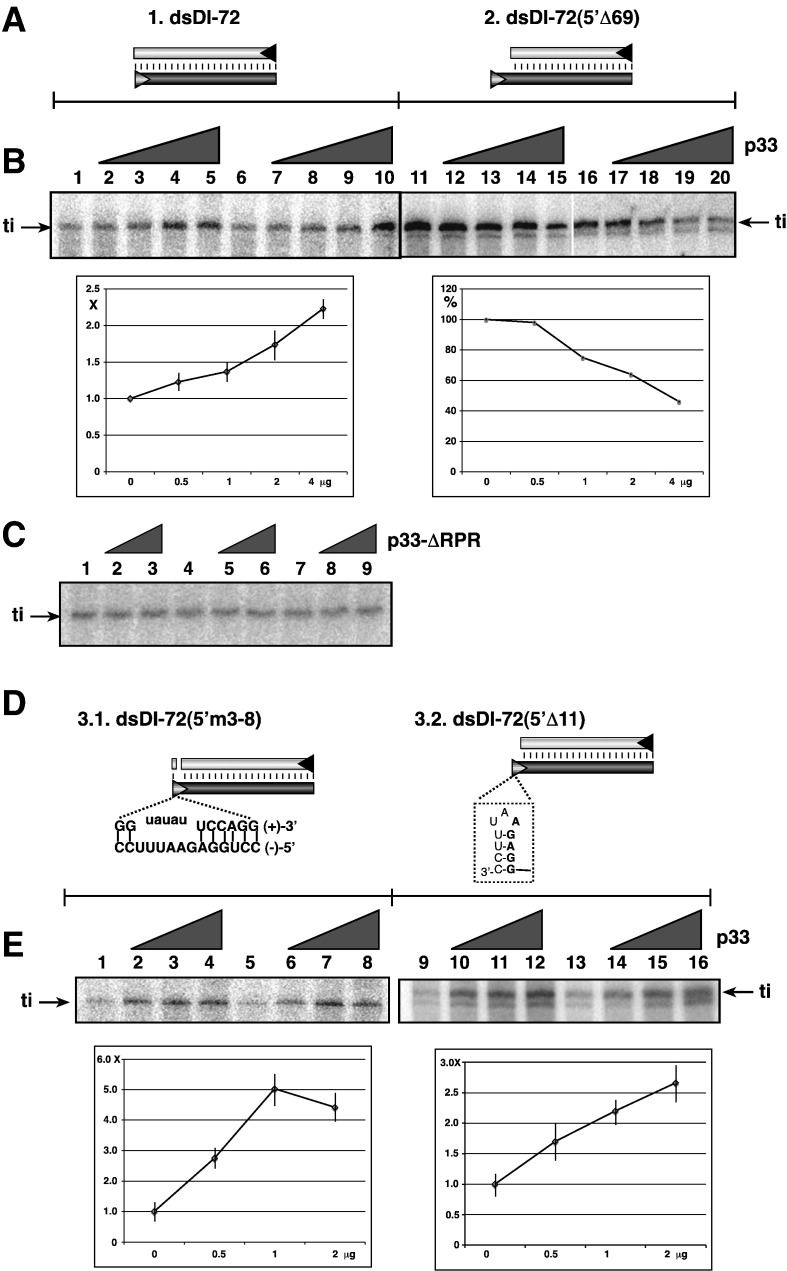
P33 facilitates initiation of RNA synthesis on dsRNA template by p88C RdRp
To further test the RNA chaperone activity of p33, we used additional dsRNA templates in the p88C RdRp assay. Previous studies have established that p88C was inefficient in initiation of RNA synthesis on TBSV dsRNA templates when compared to the ss(−)RNA template (Panavas et al., 2006). Inefficient initiation on dsRNA templates was likely due to poor accessibility of the promoter to the p88C RdRp in the dsRNA when compared to ssRNA. However, once initiation takes place, the RdRp can unwind the dsRNA structure during strand elongation (Panavas et al., 2006).
Using gel-isolated dsDI-72 RNA template in p88C RdRp assay, we found that addition of increasing amounts of recombinant p33 enhanced p88C activity by more than two-fold (Figs. 5A and B, lanes 1–10). This enhancement is not due to stabilization of p88C by p33 because the activity of p88C on template dsDI-72(5′Δ69) with an unbase-paired ssRNA tail including the cPR promoter was decreased by the addition of p33 (Fig. 5B, lanes 11–20). Moreover, deletion of the RNA-binding domain (construct p33-ΔRPR) of p33 eliminated the stimulatory effect of p33 on p88C with dsDI-72 template (Fig. 5C), suggesting that the ability of p33 to bind to the RNA is important for stimulation of the RdRp activity on dsRNA template.
Fig. 5.
Recombinant CNV p33 promotes initiation on dsRNA template by the recombinant TCV RdRp. (A) Schematic presentation of the two dsRNA templates used to program the TCV p88C RdRp preparation in vitro. (B) Representative denaturing gels of 32P-labeled RNA products synthesized by in vitro transcription with TCV p88C RdRp in the presence of 0, 0.5, 1, 2 and 4 μg of purified recombinant p33 are shown. The dsRNA templates were gel-isolated after annealing of plus- and minus-stranded RNAs (in 1:1 ratio), and they were used in equal amounts (0.5 μg per sample). The level of RNA synthesis was compared to that of the RdRp activity obtained in the absence of p33 (100%). The samples were treated with S1 nuclease to exclude terminal transferase-based labeling of dsDI-72, which might be present in the affinity-purified TCV p88C or CNV p33 preparations. Each experiment was repeated three times. “ti” represents terminally initiated template-sized RNA product. (C) Representative denaturing gels of radiolabeled RNA products synthesized by in vitro transcription with TCV p88C RdRps in the presence of 0, 1 and 2 μg of a p33 mutant are shown. The p33 mutant lacked the RPR sequence involved in RNA-binding. See Fig. 5B for further details. Each experiment was repeated at least three times. (D) Schematic presentation of the two RNA templates used to program the TCV p88C RdRp preparation in vitro. See Fig. 5A for further details. (E) Representative denaturing gels of radiolabeled RNA products synthesized by in vitro transcription with p88C RdRp in the presence of 0, 0.5, 1 and 2 μg of purified recombinant p33 are shown. The samples were treated with S1 nuclease. See Fig. 5B for further details.
To further test the possible RNA chaperone activity of p33, we used template dsDI72(5′m3-8) (Fig. 5D), which has a “bubble structure” that enhances the availability of cPR sequence by p88C RdRp by 25% (Panavas et al., 2006). Interestingly, addition of recombinant p33 to the assay led to ~ 5-fold increase in template activity of p88C (Fig. 5E, lanes 3–4 and 7–8). Similarly, dsDI-72(5′Δ11) (Fig. 5D), which forms a short inhibitory hairpin within the cPR sequence, was used ~ 2.5-fold more efficiently by p88C RdRp after addition of the recombinant p33 to the assay (Fig. 5E, lanes 9–16). The easiest interpretation of these data is that p33, due to its RNA chaperone activity, facilitates the initiation step for p88C RdRp by opening up the stable secondary structure within the cPR sequence in the dsRNA template, which is then used more efficiently by p88C that lacks RNA chaperone activity.
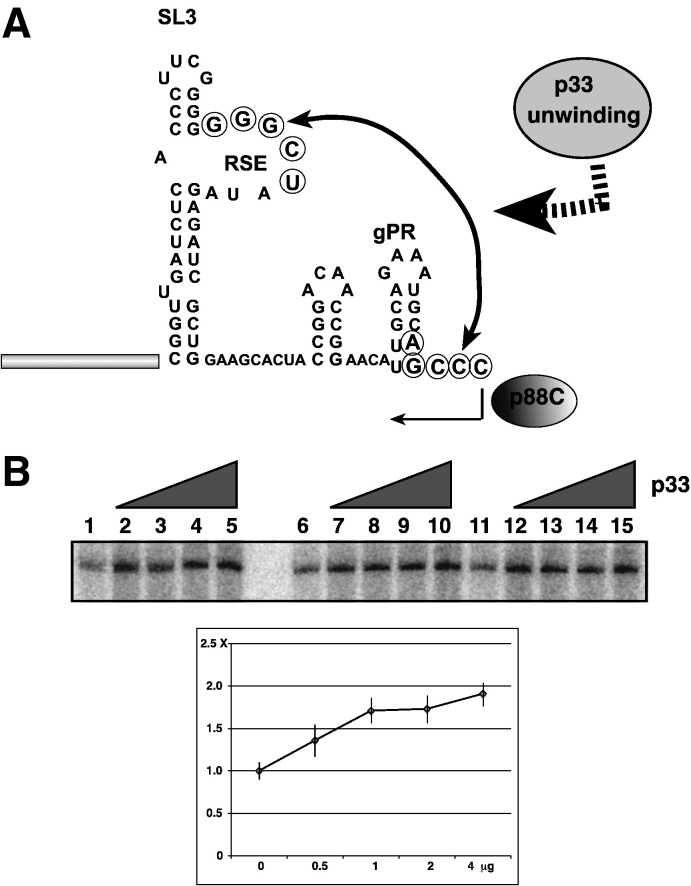
P33 promotes initiation on (+)RNA template by the p88C RdRp
The (+)RNA template in tombusviruses contains a silencer element that inhibits initiation of (−)RNA synthesis due to base-pairing between an internal loop sequence of a distant hairpin and the 5 nt 3′ terminal sequence, which makes the initiation site poorly accessible for the viral RdRp (Fig. 6A) (Pogany et al., 2003). To test if the chaperone activity of p33 could promote initiation on the (+)RNA template, we used DI-72(+) in our standard p88C RdRp assay in the presence of various amounts of recombinant p33. The RdRp assay revealed that p33 enhanced initiation on the (+)RNA template by ~ 2-fold (Fig. 6B), suggesting that p33 can facilitate opening up the base-pairs formed between the silencer and promoter, thus likely facilitating the loading of the viral RdRp on the (+)RNA template. However, the RNA chaperone activity of p33 is rather inefficient on the DI-72(+)RNA, thus it is possible that host factors, such as elongation factor 1A, which is recruited for tombusvirus replication, might also be involved in minus-strand initiation (Li et al., 2009).
Fig. 6.
Recombinant p33 promotes minus-strand initiation on (+)RNA templates by the RdRp. (A) Schematic presentation of the 3′ terminus of the (+)DI-72 RNA template used to program p88C RdRp in vitro. The base-pairing interaction between the replication silencer (RSE) and the minus-strand initiation promoter (gPR) is indicated with a double-headed arrow. The site of initiation of complementary RNA synthesis is marked below the sequence. (B) Representative denaturing gels of radiolabeled RNA products synthesized by p88C RdRp in the presence of 0, 0.5, 1, 2 and 4 μg of purified p33 are shown. The samples were treated with S1 nuclease. Each experiment was repeated at least three times.
Discussion
A large group of small RNA viruses codes for auxiliary replication proteins that lack helicase/ATPase motifs. Among these is the essential p33 replication protein of tombusviruses, which has essential functions in RNA replication (Nagy and Pogany, 2006, White and Nagy, 2004). The ability of p33 to bind to the viral RNA is critical during tombusvirus replication and it also affects viral RNA recombination (Jaag et al., 2007, Panaviene et al., 2003, Panaviene and Nagy, 2003). Interestingly, p33 can bind to the viral RNA in two different fashions. The first type of RNA-binding by p33 is specific to a region in the plus-stranded TBSV RNA termed RII(+)-SL with a signature C·C mismatch (also termed p33 recognition element) (Monkewich et al., 2005, Pogany et al., 2005). This specific p33-RNA interaction requires small amount of p33, likely in the form of p33 dimer (Pogany et al., 2005). The above specific p33-TBSV (+)RNA interaction is critical for the selective recognition of the TBSV RNA and TBSV replication both in vivo (Monkewich et al., 2005, Pogany et al., 2005) and in vitro in a cell-free extract prepared from yeast (Pogany and Nagy, 2008, Pogany et al., 2008). The second type of RNA-binding is non-specific binding of p33 to ssRNA and ssDNA, and to a lesser extent to dsRNA (Rajendran and Nagy, 2003). This non-specific nucleic acid binding by p33 requires high concentration of p33 and occurs in a cooperative manner (Rajendran and Nagy, 2003). We envision that the non-specific RNA-binding activity of p33 is important within the assembled, membrane-bound tombusvirus replicase complex, which contains large amount of p33 and only the selectively recruited TBSV (+)RNA in addition to p92pol and host factors (Nagy, 2008, Nagy and Pogany, 2010, Serva and Nagy, 2006). Within this context, the RNA chaperone activity of p33 could be important for tombusvirus replication (see below).
Based on in vitro approaches using purified recombinant p33, we have shown that p33 could function as an RNA chaperone. The supporting evidence for p33 RNA chaperone function includes: (i) a strand separation activity of p33 on a ssDNA/ssRNA duplex; (ii) p33 stimulated self-cleavage by a ribozyme in vitro by 4-fold, supporting the RNA-folding ability of p33; and (iii) increasing the sensitivity of dsRNA templates to the ssRNA-specific S1 nuclease, suggesting that p33 binding to the dsRNA leads to unwinding/opening up the dsRNA structure, and rendering dsRNA S1 nuclease sensitive. Further evidence for the RNA chaperone activity of the purified p33 is the requirement for the RNA-binding regions in p33 for RNA chaperone activity (Fig. 5C), suggesting that p33 must interact with the RNA template in order to function as a chaperone.
In addition to the above assays, the RNA chaperone activity of p33 is also likely responsible for stimulation of initiation of RNA synthesis by the heterologous TCV p88C RdRp (which, by itself, does not show RNA chaperone activity, Figs. 1C and D) on dsRNA templates or ssDNA/ssRNA duplex; as well as for enhancing the level of minus-strand RNA synthesis from the minus-strand initiation promoter (gPR), which is likely due to opening up the silencer–promoter interaction. The use of the heterologous p88C RdRp with the TBSV p33 [these proteins do not interact in vitro (Rajendran and Nagy, 2004)] in the above assays makes it unlikely that the stimulatory effect of p33 on TCV p88C RdRp activity using dsRNA templates is due to activation/stimulation of p88C RdRp via protein–protein interaction. Rather, the stimulatory effect of p33 is likely through RNA chaperoning activity that alters the RNA structures and leads to more accessible promoters for p88C-driven RNA synthesis.
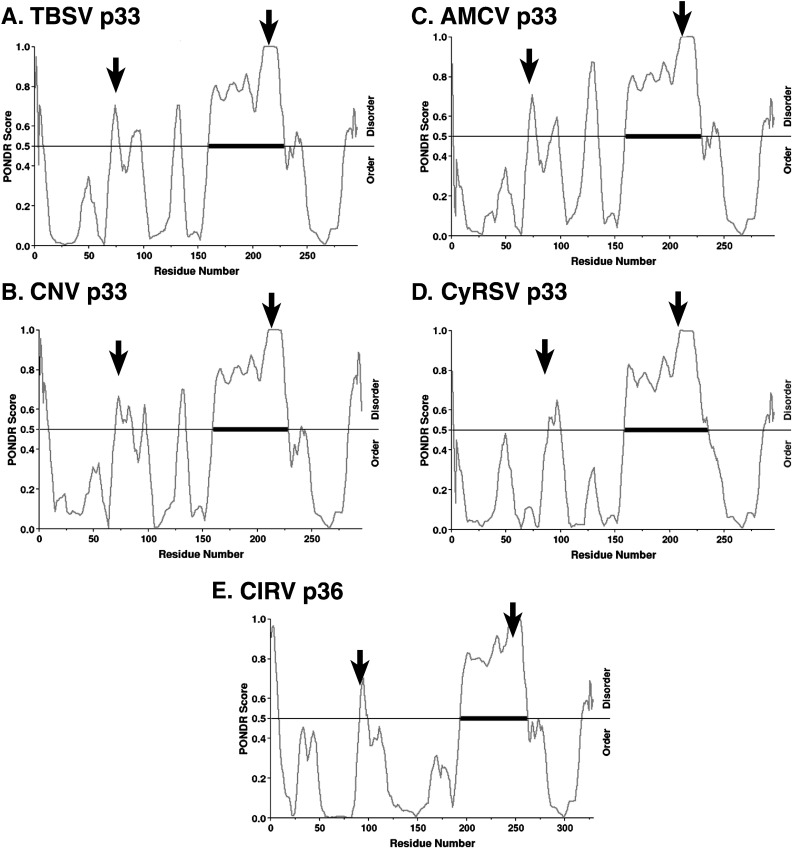
Many known RNA chaperones have long intrinsically disordered (unstructured) regions, which undergo disorder-to-order transitions upon binding to RNA (Ivanyi-Nagy et al., 2005, Ivanyi-Nagy et al., 2008, Zuniga et al., 2009). This leads to unwinding (unfolding) of the bound segment of the RNA, which then can refold to a new conformation (Tompa and Csermely, 2004). To predict if p33 replication protein contains naturally disordered regions, we used PONDR VL-XT (Bracken et al., 2004, Romero et al., 1997, Romero et al., 2004). As shown in Fig. 7 , the p33 replication proteins of five tombusviruses were predicted to contain a long and a shorter naturally disordered regions. These disordered regions contain two separate RNA-binding sequences (supplementary Fig. S1) (Panaviene et al., 2003, Rajendran and Nagy, 2003). Altogether, the presence of conserved naturally disordered regions in the p33 replication proteins further support the model that p33 functions as an RNA chaperone.
Fig. 7.
The RNA-binding sequences are predicted to be present within intrinsically disordered (unstructured) regions in p33 replication proteins of five tombusviruses. The probability of disorder is shown graphically in p33 based on analysis with PONDR-VL-XT. Amino acids with disorder value (PONDR score) of more than 0.5 are considered “disordered”, whereas below 0.5 are considered “ordered”. Arrows point at the two RNA-binding regions.
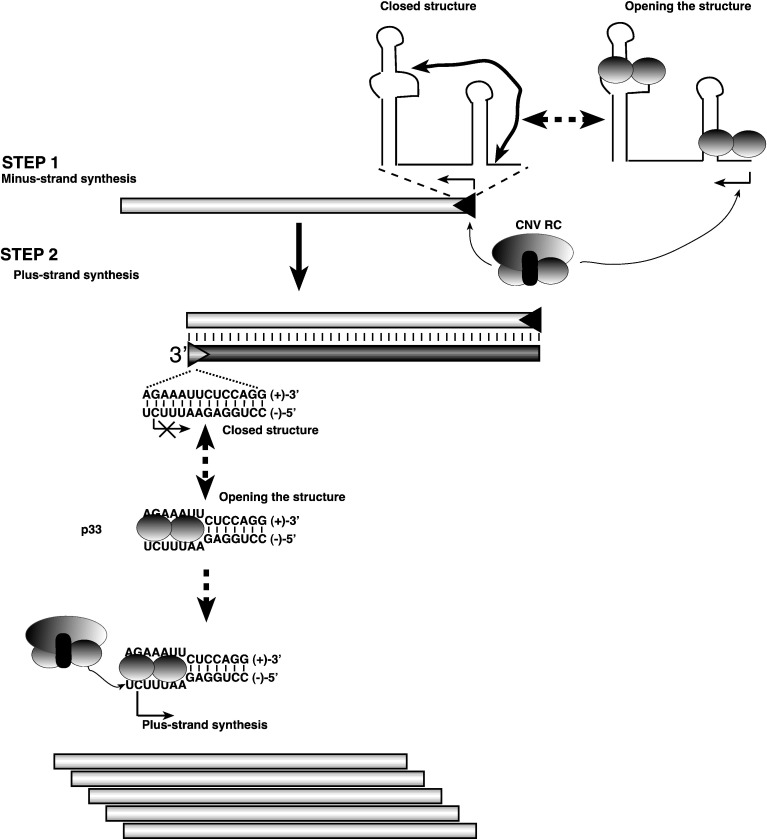
Based on previous results that p33 can bind to the viral RNA in a cooperative fashion and it can also bind to dsRNA, albeit less efficiently then to ssRNA (Rajendran and Nagy, 2003), we suggest that partial coating of the RNA by p33 might help unwinding secondary structures or dsRNA forms. Since p33 could not separate the dsRNA strands in the 621 bp long dsDI-72 RNA (not shown), we propose that p33 has a weak RNA chaperone activity, which might only be enough to open up dsRNA form from the end carrying the AU-rich stretch (within the plus-strand initiation promoter, Fig. 5) or the short silencer–promoter structure (Figs. 6A and B). Indeed, providing a weaker dsRNA structure in dsDI72(5′m3–8) (Fig. 5D), which has a “RNA-bubble structure” (Panavas et al., 2006), led to 5-fold enhanced RdRp activity by the TCV p88C due to the RNA chaperoning activity of p33. It is plausible that p88C cannot unwind the dsRNA regions due to lacking RNA chaperone activity (Fig. 1) and that results in inefficient loading of the RdRp on the dsRNA template. However, p33 might help opening up the dsRNA structure from the “weaker” AU-rich end, followed by loading the RdRp and then plus-strand synthesis (Fig. 8 ). Indeed, we have shown previously that both the tombusvirus replicase and p88 RdRp can only open up the “left-side” of the dsRNA carrying the AU-rich stretch. Thus, our in vitro data indicate that the primary function of p33 as an RNA chaperone is to facilitate initiation of RNA synthesis by making the promoter regions accessible to the viral RdRp. Unwinding of additional internal RNA structures might be accomplished by the viral RdRp during RNA synthesis as demonstrated earlier (Panavas et al., 2006). Interestingly, binding of p33 or p33C to ssRNA regions actually inhibited RdRp activity (Fig. 5B, lanes 11–20 and S1C). This suggests that p33 and especially p33C bound strongly to the ssRNA might hinder RdRp activity. Based on this and other data, we propose that the RNA chaperone activity of p33 affects plus-strand synthesis and possibly also contributes to minus-strand synthesis via facilitating initiation (Fig. 8). Future in vitro assembly approaches should help to address this and related questions.
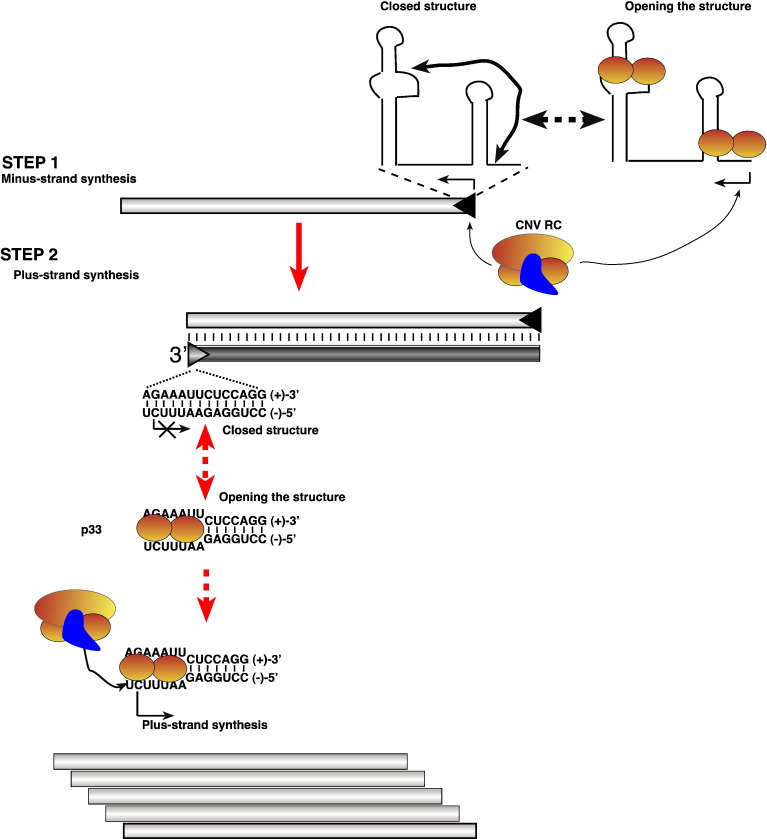
Fig. 8.
A model on the possible role of the RNA chaperone activity of p33 in tombusvirus replication. We predict that the RNA chaperone function of p33 replication protein is involved in opening the closed silencer–promoter structure, which then leads to minus-strand synthesis by the tombusvirus replicase (Step 1) However, the RNA chaperone activity of p33 is rather inefficient on the (+)RNA template, thus it is possible that host factors, such as elongation factor 1A, which is recruited for tombusvirus replication, might also be involved in minus-strand initiation. More importantly, p33 is likely involved in opening up the AU-rich terminus of the putative dsRNA replication intermediate (Step 2). We propose that binding of p33 to dsRNA could facilitate loading of the replicase to the open AU-rich end in the dsRNA replication intermediate, followed by initiation of plus-strand synthesis (bottom). It is likely that binding of p33 could stabilize the open structure within the AU-rich stretch in the cPR promoter as shown. After initiation, the tombusvirus replicase can efficiently unwind the remaining part of the dsRNA template during RNA synthesis as demonstrated earlier (Panavas et al., 2006).
Materials and methods
Purification of recombinant proteins
TCV p88C RdRp and the TBSV and CNV p33 were expressed as MBP-fusion proteins in E. coli and purified using affinity-based chromatography as described earlier (Rajendran and Nagy, 2003, Rajendran et al., 2002), except that 2 μg RNase A was added to the sonicated cell lysate and the solution was incubated for 15 min on ice to remove RNA. The protein concentrations were adjusted to 250 μg/ml with column buffer (Rajendran and Nagy, 2003, Rajendran et al., 2002). Note that these preparations lacked ribonuclease activity (not shown).
Preparation of dsRNA templates
First, single-stranded (ss)RNA templates were obtained by in vitro transcription with T7 RNA polymerase using PCR amplified DNA templates (Panavas and Nagy, 2005, Panavas et al., 2002). Second, to make the dsRNA constructs, we annealed the heat denatured ssRNA transcripts (94 °C for 5 min) in STE buffer (10 mM TRIS, pH 8.0, 1 mM EDTA, and 100 mM NaCl), followed by slow cooling (30 min) to 25 °C using a thermocycler (Panavas and Nagy, 2005, Panavas et al., 2006). The annealed RNAs were loaded onto 5% non-denaturing polyacrylamide gels, followed by staining with ethidium bromide. After cutting out the annealed RNA band, we eluted the dsRNA into 0.6 M ammonium acetate, followed by phenol-chloroform extraction and ethanol precipitation (Panavas et al., 2006). The quality of the obtained dsRNA was checked by non-denaturing polyacrylamide gels.
Strand separation (unwinding) assay
Oligo #20 (5′-GGAAATTCTCCAGGATTTCTC) was [32P]-labeled at the 5′ end with γATP (0.05 mCi) using polynucleotide kinase (Fermentas). The annealing of 1 pmol of oligo and 1 pmol of DI-72(−)RNA in STE buffer (10 mM TRIS, pH 8.0, 1 mM EDTA, and 100 mM NaCl) was done after heat denaturation of RNA transcript at 94 °C for 2 min followed by slowly cooling down the samples to 25 °C in 30 min. Different amounts of MBP-p33, MBP-p88, MBP-p88c or MBP-Ded1 (0.4, 1.0, and 2.0 pmol) were added separately to the annealed ssDNA/ssRNA duplex in the RdRp buffer, followed by incubation at room temperature for 20 min. The samples were then analyzed by electrophoresis on non-denaturing 5% acrylamide gels, followed by phosphoimaging (Rajendran and Nagy, 2003).
S1 nuclease sensitivity assay
The dsRNA was obtained using [32P]UTP-labeled (−)DI-72 RNA and unlabelled (+)DI-72 RNA in STE buffer. Different amounts (0, 0.5, 1, and 2 μg) of recombinant MBP-p33 were added to the dsRNA (approximately 15 ng) in RdRp buffer in 10 μl final volume. The samples were incubated for 15 min at room temperature, then 85U of S1 nuclease were added in 1.2 μl 10× S1 buffer (Promega) followed by 15 min incubation at 37 °C. The samples were loaded on a non-denaturing 8% acrylamide gel containing 0.1% SDS.
Ribozyme self-cleavage assay
The satellite (−) ribozyme of TRSV (Buzayan et al., 1986) fused to the 3′ end TBSV RIV RNA was made by T7 polymerase. The purified recombinant MBP-p33 was included in the 20 μl T7 reaction. The DNA template for the T7 polymerase was obtained by PCR using oligos #19 (GTAATACGACTCACTATAGGAATTCCTGTTTACGAAAG) and #1069 (CCGGTCGAGCTCTACCAGGTAATATACCACAACGTGTGT) with plasmid pYC2/CT DI-72-Rz (Panavas and Nagy, 2003). Incubation was at 37 °C for 30 min, followed by a phenol-chloroform extraction and ethanol precipitation. The radioactively labeled RNA was loaded to a denaturing 5% acrylamide gel/8 M urea.
In vitro replicase assays
TCV RdRp reactions were carried out for 2 h at 25 °C in the RdRp buffer consisting of 50 mM Tris-HCl (pH 8.2), 10 mM MgCl2, 10 mM dithiothreitol, 100 mM potassium glutamate, ATP, CTP, and GTP (1.0 mM each), and 0.3 μl of [32P]UTP (0.1 mCi/ml) in 50 μl volume (Rajendran et al., 2002). Each RdRp reaction mixture contained 0.5 μg template (ds or ssRNA), 1.5 μg purified p88C RdRp enzyme (Panavas et al., 2006) and p33. After phenol-chloroform extraction and ammonium acetate-isopropanol precipitation, the RNA products were treated with S1 nuclease followed by another phenol-chloroform extraction and ammonium acetate-isopropanol precipitation. The samples were analyzed by electrophoresis on denaturing 5% acrylamide gels. The gels were dried, exposed to a phosphor screen and analyzed using Typhoon phosphorimager and ImageQuant (GE Healthcare).
Acknowledgments
We thank Drs. Judit Pogany and Hannah Jaag for critical reading of the manuscript and for very helpful suggestions. The authors appreciate Dr. Tadas Panavas' effort at the beginning of this work. This work was supported by the National Institute of Allergy and Infectious Diseases (NIH-NIAID AI05767001A1) and by the Kentucky Tobacco Research and Development Center at the University of Kentucky, awarded to PDN and by a Philip Morris fellowship to JS.
Footnotes
Supplementary data to this article can be found online at doi:10.1016/j.virol.2010.10.015.
Appendix A. Supplementary data
Supplementary materials.
References
- Bracken C., Iakoucheva L.M., Romero P.R., Dunker A.K. Combining prediction, computation and experiment for the characterization of protein disorder. Curr. Opin. Struct. Biol. 2004;14(5):570–576. doi: 10.1016/j.sbi.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Burgyan J., Hornyik C., Szittya G., Silhavy D., Bisztray G. The ORF1 products of tombusviruses play a crucial role in lethal necrosis of virus-infected plants. J. Virol. 2000;74(23):10873–10881. doi: 10.1128/jvi.74.23.10873-10881.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzayan J.M., Hampel A., Bruening G. Nucleotide sequence and newly formed phosphodiester bond of spontaneously ligated satellite tobacco ringspot virus RNA. Nucleic Acids Res. 1986;14(24):9729–9743. doi: 10.1093/nar/14.24.9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.P., Panavas T., Luo G., Nagy P.D. Heterologous RNA replication enhancer stimulates in vitro RNA synthesis and template-switching by the carmovirus, but not by the tombusvirus, RNA-dependent RNA polymerase: implication for modular evolution of RNA viruses. Virology. 2005;341(1):107–121. doi: 10.1016/j.virol.2005.06.042. [DOI] [PubMed] [Google Scholar]
- Cristofari G., Darlix J.L. The ubiquitous nature of RNA chaperone proteins. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:223–268. doi: 10.1016/s0079-6603(02)72071-0. [DOI] [PubMed] [Google Scholar]
- Cristofari G., Ivanyi-Nagy R., Gabus C., Boulant S., Lavergne J.P., Penin F., Darlix J.L. The hepatitis C virus core protein is a potent nucleic acid chaperone that directs dimerization of the viral (+) strand RNA in vitro. Nucleic Acids Res. 2004;32(8):2623–2631. doi: 10.1093/nar/gkh579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruceanu M., Urbaneja M.A., Hixson C.V., Johnson D.G., Datta S.A., Fivash M.J., Stephen A.G., Fisher R.J., Gorelick R.J., Casas-Finet J.R., Rein A., Rouzina I., Williams M.C. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006;34(2):593–605. doi: 10.1093/nar/gkj458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daros J.A., Flores R. A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J. 2002;21(4):749–759. doi: 10.1093/emboj/21.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeStefano J.J., Titilope O. Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities. J. Virol. 2006;80(4):1662–1671. doi: 10.1128/JVI.80.4.1662-1671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor M.J. Structure and function of the hairpin ribozyme. J. Mol. Biol. 2000;297(2):269–291. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- Huang Z.S., Wu H.N. Identification and characterization of the RNA chaperone activity of hepatitis delta antigen peptides. J. Biol. Chem. 1998;273(41):26455–26461. doi: 10.1074/jbc.273.41.26455. [DOI] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Davidovic L., Khandjian E.W., Darlix J.L. Disordered RNA chaperone proteins: from functions to disease. Cell. Mol. Life Sci. 2005;62(13):1409–1417. doi: 10.1007/s00018-005-5100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Kanevsky I., Gabus C., Lavergne J.P., Ficheux D., Penin F., Fosse P., Darlix J.L. Analysis of hepatitis C virus RNA dimerization and core-RNA interactions. Nucleic Acids Res. 2006;34(9):2618–2633. doi: 10.1093/nar/gkl240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R., Lavergne J.P., Gabus C., Ficheux D., Darlix J.L. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 2008;36(3):712–725. doi: 10.1093/nar/gkm1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag H.M., Stork J., Nagy P.D. Host transcription factor Rpb11p affects tombusvirus replication and recombination via regulating the accumulation of viral replication proteins. Virology. 2007;368(2):388–404. doi: 10.1016/j.virol.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Karakasiliotis I., Chaudhry Y., Roberts L.O., Goodfellow I.G. Feline calicivirus replication: requirement for polypyrimidine tract-binding protein is temperature-dependent. J. Gen. Virol. 2006;87(Pt 11):3339–3347. doi: 10.1099/vir.0.82153-0. [DOI] [PubMed] [Google Scholar]
- Koonin E.V., Dolja V.V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 1993;28(5):375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- Levin J.G., Guo J., Rouzina I., Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:217–286. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- Li Z., Pogany J., Panavas T., Xu K., Esposito A.M., Kinzy T.G., Nagy P.D. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology. 2009;385(1):245–260. doi: 10.1016/j.virol.2008.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney A.W., Greenwood J.S., Fabian M.R., White K.A., Mullen R.T. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell. 2005;17(12):3513–3531. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Panganiban A.T. The bunyavirus nucleocapsid protein is an RNA chaperone: possible roles in viral RNA panhandle formation and genome replication. RNA. 2006;12(2):272–282. doi: 10.1261/rna.2101906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Panganiban A.T. Characterization of the RNA chaperone activity of hantavirus nucleocapsid protein. J. Virol. 2006;80(13):6276–6285. doi: 10.1128/JVI.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monkewich S., Lin H.X., Fabian M.R., Xu W., Na H., Ray D., Chernysheva O.A., Nagy P.D., White K.A. The p92 polymerase coding region contains an internal RNA element required at an early step in Tombusvirus genome replication. J. Virol. 2005;79(8):4848–4858. doi: 10.1128/JVI.79.8.4848-4858.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D. Yeast as a model host to explore plant virus–host interactions. Annu. Rev. Phytopathol. 2008;46:217–242. doi: 10.1146/annurev.phyto.121407.093958. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology. 2006;344(1):211–220. doi: 10.1016/j.virol.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. Multiple roles of viral replication proteins in plant RNA virus replication. Methods Mol. Biol. 2008;451:55–68. doi: 10.1007/978-1-59745-102-4_4. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Pogany J. Global genomics and proteomics approaches to identify host factors as targets to induce resistance against tomato bushy stunt virus. Adv. Virus Res. 2010;76:123–177. doi: 10.1016/S0065-3527(10)76004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro B., Russo M., Pantaleo V., Rubino L. Cytological analysis of Saccharomyces cerevisiae cells supporting cymbidium ringspot virus defective interfering RNA replication. J. Gen. Virol. 2006;87(Pt 3):705–714. doi: 10.1099/vir.0.81325-0. [DOI] [PubMed] [Google Scholar]
- Panavas T., Hawkins C.M., Panaviene Z., Nagy P.D. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology. 2005;338(1):81–95. doi: 10.1016/j.virol.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Panavas T., Nagy P.D. Yeast as a model host to study replication and recombination of defective interfering RNA of Tomato bushy stunt virus. Virology. 2003;314(1):315–325. doi: 10.1016/s0042-6822(03)00436-7. [DOI] [PubMed] [Google Scholar]
- Panavas T., Nagy P.D. Mechanism of stimulation of plus-strand synthesis by an RNA replication enhancer in a tombusvirus. J. Virol. 2005;79(15):9777–9785. doi: 10.1128/JVI.79.15.9777-9785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T., Pogany J., Nagy P.D. Internal initiation by the cucumber necrosis virus RNA-dependent RNA polymerase is facilitated by promoter-like sequences. Virology. 2002;296(2):275–287. doi: 10.1006/viro.2002.1422. [DOI] [PubMed] [Google Scholar]
- Panavas T., Stork J., Nagy P.D. Use of double-stranded RNA templates by the tombusvirus replicase in vitro: implications for the mechanism of plus-strand initiation. Virology. 2006;352(1):110–120. doi: 10.1016/j.virol.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Panaviene Z., Baker J.M., Nagy P.D. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology. 2003;308(1):191–205. doi: 10.1016/s0042-6822(02)00132-0. [DOI] [PubMed] [Google Scholar]
- Panaviene Z., Nagy P.D. Mutations in the RNA-binding domains of tombusvirus replicase proteins affect RNA recombination in vivo. Virology. 2003;317(2):359–372. doi: 10.1016/j.virol.2003.08.039. [DOI] [PubMed] [Google Scholar]
- Pogany J., Fabian M.R., White K.A., Nagy P.D. A replication silencer element in a plus-strand RNA virus. EMBO J. 2003;22(20):5602–5611. doi: 10.1093/emboj/cdg523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J., Nagy P.D. Authentic replication and recombination of Tomato bushy stunt virus RNA in a cell-free extract from yeast. J. Virol. 2008;82(12):5967–5980. doi: 10.1128/JVI.02737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J., Stork J., Li Z., Nagy P.D. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proc. Natl. Acad. Sci. U. S. A. 2008;105(50):19956–19961. doi: 10.1073/pnas.0810851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogany J., White K.A., Nagy P.D. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 2005;79(8):4859–4869. doi: 10.1128/JVI.79.8.4859-4869.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran K.S., Nagy P.D. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J. Virol. 2003;77(17):9244–9258. doi: 10.1128/JVI.77.17.9244-9258.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran K.S., Nagy P.D. Interaction between the replicase proteins of Tomato bushy stunt virus in vitro and in vivo. Virology. 2004;326(2):250–261. doi: 10.1016/j.virol.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Rajendran K.S., Nagy P.D. Kinetics and functional studies on interaction between the replicase proteins of Tomato bushy stunt virus: requirement of p33:p92 interaction for replicase assembly. Virology. 2006;345(1):270–279. doi: 10.1016/j.virol.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Rajendran K.S., Pogany J., Nagy P.D. Comparison of turnip crinkle virus RNA-dependent RNA polymerase preparations expressed in Escherichia coli or derived from infected plants. J. Virol. 2002;76(4):1707–1717. doi: 10.1128/JVI.76.4.1707-1717.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A., Henderson L.E., Levin J.G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23(8):297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- Romero, Obradovic, Dunker K. Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inform Ser. Workshop Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- Romero P., Obradovic Z., Dunker A.K. Natively disordered proteins: functions and predictions. Appl. Bioinform. 2004;3(2–3):105–113. doi: 10.2165/00822942-200403020-00005. [DOI] [PubMed] [Google Scholar]
- Russell R. RNA misfolding and the action of chaperones. Front. Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serva S., Nagy P.D. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 2006;80(5):2162–2169. doi: 10.1128/JVI.80.5.2162-2169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapka N., Stork J., Nagy P.D. Phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus adjacent to the RNA-binding site affects viral RNA replication. Virology. 2005;343(1):65–78. doi: 10.1016/j.virol.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Stork J., Panaviene Z., Nagy P.D. Inhibition of in vitro RNA-binding and replicase activity by phosphorylation of the p33 replication protein of Cucumber necrosis tombusvirus. Virology. 2005;343(1):79–92. doi: 10.1016/j.virol.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Tompa P., Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18(11):1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- Wang C.C., Chang T.C., Lin C.W., Tsui H.L., Chu P.B., Chen B.S., Huang Z.S., Wu H.N. Nucleic acid binding properties of the nucleic acid chaperone domain of hepatitis delta antigen. Nucleic Acids Res. 2003;31(22):6481–6492. doi: 10.1093/nar/gkg857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K.A., Nagy P.D. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- Zuniga S., Cruz J.L.G., Sola I., Mateos-Gomez P.A., Palacio L., Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J. Virol. 2010;84(4):2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S., Sola I., Cruz J.L.G., Enjuanes L. Role of RNA chaperones in virus replication. Virus Res. 2009;139(2):253–266. doi: 10.1016/j.virusres.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S., Sola I., Moreno J.L., Sabella P., Plana-Duran J., Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357(2):215–227. doi: 10.1016/j.virol.2006.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials.