Introduction
Polymerase chain reaction (PCR) is a technology for exponential amplification of a fragment of DNA. (The PCR is covered by patents owned by Hoffman-La Roche. A license is required to use the PCR process.) The limit of its sensitivity is a single molecule, making PCR a superb qualitative tool for the specific detection of rare DNA sequences. Under proper conditions, the yield of amplified DNA is proportional to the initial number of target molecules, rendering it a quantitative analytical tool as well. Since its original description in 1985, PCR has evolved into an assemblage of varied methodologies almost universally used in basic biological research, biotechnology, clinical research, clinical diagnostics, forensics, food technology, environmental testing, archaeology and anthropology, and other fields. Even though other nucleic acid amplification technologies have been described, PCR remains by far the most widely used.
Biochemical Basis of PCR
PCR involves the enzymatic synthesis of millions of copies of a specific DNA segment. The exponential amplification of a very small amount of template DNA is achieved using a heat-stable DNA polymerase and an automated heat block that is capable of rapid changes of temperature. The template DNA molecule is initially denatured to two single strands by heating to high temperature (typically 90–95°C) in the first stage of the PCR cycle (Figure 1A ). Two small oligonucleotides that are complementary to sequences on opposite strands of the template molecule and that flank the DNA segment to be amplified are used as primers for the DNA polymerase. The second stage of the PCR cycle consists of cooling the reaction, which permits the annealing of the single-stranded oligonucleotide primers to the denatured template molecule (Figure 1B). The third stage of the reaction is the extension of the new strand from the annealed primer in a 5′ to 3′ direction by the heat-stable polymerase (Figure 1C). This is performed at the optimum temperature for the polymerase (68–72°C). The most commonly used polymerase is the enzyme isolated from Thermus aquaticus (Taq DNA polymerase). After the first cycle, each template molecule has been amplified to two molecules. These in turn are denatured in the next cycle and amplified to produce four molecules. The four molecules are amplified to eight in the third cycle, and so on. Each successive cycle effectively doubles the amount of DNA product. The three-stage cycle of denaturation, annealing, and primer extension is repeated 25–40 times in a typical PCR procedure. The product of such a reaction is a large quantity (10−9 to 10−8 mol l−1 in a 10–100 μl volume) of a double-stranded DNA molecule whose length is determined by the distance between the primer sites on the original template molecule. Typically, PCR products or amplicons are 100–3000 bp in length, although much longer (up to ∼50000 bp) amplifications are possible under specific conditions. PCR products are visualized by electrophoresis followed by staining with fluorescent dyes or by hybridization to labeled oligonucleotide probes.
Figure 1.
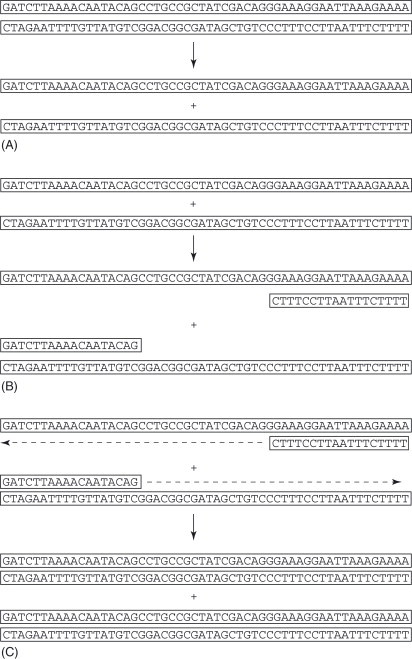
One cycle of a PCR. (A) The template is shown as a double-stranded DNA molecule, which is denatured to two single strands. (B) The short oligonucleotide primers anneal to the complementary regions of the single strands. (C) The polymerase generates a new strand from each single-stranded molecule, creating two double-stranded molecules.
Technical Aspects of PCR
One of the advantages of PCR technology is the speed and simplicity with which the technique can be performed. A PCR reaction can be set up in a short time, and multiple samples can be handled easily. Components of the amplification reaction (template DNA, DNA polymerase, oligonucleotide primers, buffers) are added to the amplification vessel. The most common containers for PCR are microcentrifuge tubes (0.5–0.2 ml) with thin walls to facilitate rapid heat transfer or multiwell plates (96- or 384-well plates being the most common). Some commercially available systems utilize specially designed vessels such as capillary tubes. The reaction tubes are then placed into an automated programmable thermal cycler for controlled heating and cooling for denaturation, annealing, and extension reactions. Following amplification, some type of DNA sequence analysis is typically performed, although technology for real-time monitoring of amplification obviates the need even to open the PCR tube after cycling is complete.
Template DNA
One of the advantages of PCR is that DNA from relatively clean samples need not be highly purified to be used as a template for amplification. It is possible to simply heat samples to near boiling to release DNA, then to add the resulting lysate to the PCR reaction tube. Such simplicity is the hallmark of PCR. With some types of biological samples, inhibitors of PCR such as heme or polyanionic mucopolysaccharides must be removed by DNA purification.
Virtually any form of DNA can be used as a template in a PCR reaction. Plasmids, cosmids, phagemids, lambda phage, M13 phage, genomic DNA, and many other sources of DNA have all been used successfully. A PCR can be performed directly on colonies or plaques. Although it is possible to amplify a single DNA molecule, a typical PCR reaction uses 105–106 copies of the target DNA as template. In practice, this means adding ∼1 μg of eukaryotic genomic DNA, 10 ng of yeast DNA, or 1 ng of bacterial DNA. The concentration of DNA is not critical for most applications, however, and a PCR reaction will work with a wide range of template concentrations. Often the concentration of template DNA is unknown and does not need to be determined. If necessary, a series of control reactions, each containing a different amount of template, can be performed.
Biochemical Components
DNA polymerase
A wide range of DNA polymerases is available for PCR. A cloned heat-stable DNA polymerase from Thermus aquaticus (Taq DNA polymerase) is the most commonly used. Other enzymes such as DNA polymerase from Pyrococcus furiosus (Pfu DNA polymerase) or Thermoccocus litoralis (Vent™ DNA polymerase, New England Biolabs) have been reported to have a lower error rate than Taq polymerase and may be advantageous for some applications. Typically, 0.5–2.5 units of enzyme are used per 50 μl reaction.
Oligonucleotide primers
Oligonucleotide primers are usually used at a concentration of 1 μmol l−1 in PCR reactions (50 pmol per 50 μl reaction). Concentrations between 0.1 and 1 μmol l−1 can be used. Higher concentrations may increase nonspecific annealing of primers and thus to nonspecific amplification products. PCR primers are normally between 18 and 30 nucleotides in length and should preferably have a guanine+cytosine (G+C) content of ∼50%. The temperature at which half the DNA molecules will be double-stranded, T m, in degree celsius, for a primer is estimated by the rule-of-thumb equation: 2×(number of As and Ts)+4×number of Gs and Cs) (A=adenine, T=thymine). However, more accurate calculation of oligonucleotide T m requires consideration of the effects of ionic strength and neighboring bases in the DNA strand (nearest neighbor T m calculation methods). Software is available for performing these calculations. The T m values for the two primers in a reaction should be similar, and the annealing temperature used is normally ∼5°C below the T m. As annealing temperature approaches T m, more specific amplifications are achieved, but overall yields may decrease. Despite careful calculations, empirical testing of annealing temperatures is essential for a well-optimized PCR assay. Complementarity at the 3′ ends of primer pairs should be avoided as this promotes the formation of primer oligomers (primer dimers). Such artifactual products are themselves templates for PCR amplification and compete with the desired products for deoxynucleotide triphosphates (dNTPs), polymerase, and primers. This leads to a decreased yield of the desired product and the presence of nonspecific products that may complicate analysis.
A primer oligomer forms when two or more primers anneal to each other and are extended during PCR. The double-stranded product acts as a template during subsequent rounds of amplification and competes with the desired product. In general, low annealing temperatures, high enzyme concentrations, and high primer concentrations increase the probability of primer oligomerization. However, it is stringency during the initial cycle of PCR that is most critical for primer oligomer formation, as well as for nonspecific PCR products in general. Various techniques for maintaining high stringency up until the point when primers and enzyme are mixed together have been described (generally called Hot Start). Hot Start methods involve mixing of enzyme and primers only after reactions have reached a stringent temperature. A simple technique for Hot Start is manual addition of a small volume (typically 5–10 μl) of reaction buffer containing the enzyme to the other components maintained in the thermal cycler at ∼80°C, then continuing with standard PCR amplification. Another method involves sealing a portion of the amplification reaction under wax, then adding the remaining reactants above the wax barrier. Enzymatic methods to accomplish the same goal include the use of a modified DNA polymerase that is blocked with a thermolabile group or the use of uracil DNA glycosylase (uracil-N-glycosylase, UNG), deoxyuridine triphosphate (dUTP), and a brief initial incubation at 50°C. Typically, dUTP is substituted on an equimolar basis for deoxythymidine triphosphate (dTTP) (200 μmol l−1), but higher concentrations of dUTP (125–300 μmol l−1) may be beneficial in some assays. Uracil DNA glycosylase is used at 1–2 Units per reaction. This method ensures that any nonspecific product or primer oligomer that is generated during the initial low-stringency conditions will contain uracil (U) and therefore be susceptible to cleavage by UNG. Uracil is excised during the 50°C incubation, and strand breakage occurs at these abasic sites during the initial denaturation step. In addition to inactivating any contaminating amplicons, this method reduces nonspecific products and primer oligomer. Because residual UNG can degrade the U-containing amplicons, PCR products must be analyzed immediately or stored at 4°C for short periods of time or frozen for longer times. Alternately, UNG can be removed by extraction with organic solvents or digested with proteases.
The 3′ end of a PCR primer should be perfectly complementary to the template DNA. Failure of the 3′ end to hybridize results in inefficient amplification. Internal sequences are less critical. Provided a suitable annealing temperature is used, degenerate primers (primers that consist of a pool of different but closely related oligonucleotides) may be used to ensure that primers will anneal to highly variable sequences or sequences that are only partially known. These primers are designed from amino acid sequences or from a comparison of similar genes from other organisms. Sequences at the 5′ end of the primer are least critical. Providing the hybridizing portion of the primer is long enough to ensure annealing to the template DNA, nonhomologous 5′ extensions (regions that do not correspond to the sequence of the template) can be used to introduce restriction enzyme sites into PCR products to be cloned or to add other sequences such as phage RNA polymerase promoters.
Multiplex PCR enables the detection of multiple gene sequences within the same reaction. Because several sets of PCR primers must function in the same reaction without interference, primer design and optimization of reaction conditions are critical.
Primers are often labeled with detectable groups to facilitate post-PCR analysis. Radioactive isotopes, haptens such as biotin, or fluorescent dyes are the most widely used. Labels attached to the 5′ end of a primer have little or no effect on amplification.
Many heat-stable polymerases used for PCR exhibit a template-independent activity that adds deoxynucleosides (predominantly deoxyadenosine) to the 3′ ends of all double-stranded molecules in the reaction. These A-overhangs must be filled in prior to blunt-end cloning of PCR products. Alternatively, special vectors are available that have single 3′ T-overhangs at the cloning site ready for insertion of the PCR product. Choosing a polymerase without this activity or adjusting reaction conditions to either minimize or maximize the extent of A-overhangs may produce sharper peaks in high-resolution electrophoretic analysis.
Deoxynucleoside triphosphates
dNTPs are typically used at a concentration of ∼200 μmol l−1 each dNTP in a PCR reaction. Excessively high concentrations promote nonspecific product formation. Modified dNTPs are sometimes used to label PCR products with radioactive or fluorescent markers or with haptens such as biotin, fluorescein, or digoxygenin. The modified dNTP is typically used at a concentration (0.1–1 μmol l−1) much lower than the unmodified dNTP (∼200 μmol l−1). Probes can be easily generated using PCR amplification with a labeled nucleotide, followed by removal of the unincorporated label.
Buffer components
Several reagents containing buffer ions, monovalent salts, and divalent cations required for polymerase activity, have been described for use in PCR amplification. The most widely used buffer consists of 10 mmol l−1 Tris–HCl (pH 8.3 at room temperature) and 50 mmol l−1 KCl. At the extension temperature (72°C), the pH of this Tris buffer falls to 7.2, near the optimum for Taq DNA polymerase. Sulfate-containing buffers such as 20 mmol l−1 Tris–SO4 (pH 8.5–9.0 at room temperature) and 20 mmol l−1 (NH4)2SO4 are also widely used. Monovalent cations (K+ or NH4 +) are included to adjust ionic strength. DNA polymerases require divalent cations for activity, and PCR reactions contain MgCl2 (1–5 mmol l−1). In general, higher MgCl2 concentrations promote nonspecific primer annealing and nonspecific product amplification. Reaction buffers usually include a low percentage (0.1–0.5%) of nonionic detergent such as Igepal CA-630, Tween 20, or Triton X-100 to minimize adsorption of polymerase to the surfaces of the PCR tube. Proteins (gelatin or bovine serum albumin) are sometimes included at similar concentrations as nonionic detergents for the same reason. Other components that have been reported to increase PCR product yields or specificity on specific templates include betaine, dimethylsulfoxide, formamide, and glycerol.
Thermal Cycling Equipment
The PCR process involves thermal cycling between denaturation, annealing, and extension temperatures (Figure 1). Each incubation step is a segment, and a complete round of denaturation, annealing, and extension is a cycle. First, samples are heated to a temperature adequate to melt the double-stranded DNA template and any secondary structure in the primers. Denaturation is most commonly performed at 94°C, although temperatures of 97–99°C may be required for templates with high GC content. After the first several cycles, denaturation temperature can be lowered to 90–92°C to reduce loss of polymerase activity through heat denaturation during the course of PCR. Next, samples are cooled to an annealing temperature at which the primers will hybridize to their target sequences. The choice of annealing temperature depends on primer T m, but is usually in the range of 50–60°C. Finally, the samples are heated to the extension temperature (68–72°C). If primers are designed to permit efficient annealing at 60–68°C, the annealing and extension steps can be combined into one step. Typical PCR amplification is performed over 20–30 cycles, although 40 or more cycles may be used with template DNA at very low initial concentration or with otherwise marginal amplifications involving, for example, degenerate primers or poor-quality template DNA.
Although it is possible to perform 20 or even 40 cycles of PCR manually using sandbaths, waterbaths, or heat blocks and physically moving the samples from one to the other at timed intervals, the tedium involved makes this approach impractical. Many different types of machines are commercially available to perform thermal cycling. Some cyclers use robotic arms to transfer a rack of PCR samples from one heat block to another. Other machines use resistive heating elements, compressors, water circulators, fan-forced air, high-intensity lamps, Peltier elements, or combinations of these heating and cooling devices. Microprocessors enable the user to program the instrument for time and temperature of each segment, and the number of cycles to be performed.
Thermal cyclers that can monitor progress of the amplification reaction while it is taking place are called real-time thermal cyclers. Real-time instruments are designed to monitor fluorescence from labels whose emission intensity is proportional to the amount of amplified DNA. Flexibility varies greatly among the available machines, but all instruments can measure fluorescence at least once during the annealing segment of every cycle. Intercalating dyes such as ethidium bromide or SYBR Green that are selective for double-stranded DNA provide the simplest method. Probes that hybridize adjacent to one another on one strand of the amplicon can be designed to have a fluorescence energy transfer donor and acceptor that will be close enough for energy transfer if and only if hybridization occurs. Emission from the hybridized donor- and acceptor-labeled probes is monitored during each cycle. A third approach is to use the 5′ nuclease activity of Taq DNA polymerase to cleave a probe labeled with a fluorophore and a quencher. When Taq DNA polymerase (and some, but not all, other heat-stable polymerases) encounters the probe, it cuts it, dissociating the fluorophore and quencher. As with energy transfer methods, amplification is detectable as an increase in fluorescence. In addition to monitoring fluorescence, real-time thermal cyclers provide data analysis for quantitation of initial template concentration. Real-time PCR has become the standard for quantitative PCR.
Post-PCR Analysis
Unless real-time thermal cyclers are used, some form of manipulation or analysis of PCR products must be performed. In preparative applications, amplicons are inserted into a suitable vector and propagated by standard molecular biological methods. In analytical applications, some form of DNA size or sequence analysis is performed. In many cases, high-performance liquid chromatography or electrophoresis on agarose or polyacrylamide gels to confirm that the amplicon is the expected size is sufficient. In clinical testing, hybridization of an oligonucleotide probe complementary to a sequence between the PCR primers is more commonly used to confirm specificity of the amplified DNA. Probe hybridization can be combined with DNA size analysis by electrophoresis or can be performed using methods analogous to immunoassays.
Considerations
Fidelity
For preparative applications and the analysis of single-base changes, fidelity of the DNA polymerase is important. The fidelity of the DNA polymerase in a PCR reaction determines the similarity between the sequence of the PCR product and the original template. Taq DNA polymerase sometimes incorporates an incorrect base in the growing DNA strand, and this error is propagated during subsequent rounds of cycling. The higher the fidelity, the more closely the sequence of the PCR product reflects the sequence of the original DNA template. A range of DNA polymerases is commercially available, and certain enzymes have demonstrably higher fidelity. In addition to intrinsic differences among the various heat-stable polymerases, fidelity is influenced by a number of factors. It is therefore possible to increase or decrease the number of mismatches between the product and initial template. Conditions of low fidelity may be chosen to introduce random point mutations into a PCR product. More general applications benefit from accurate amplification. Fidelity is improved by keeping polymerase, dNTP, and MgCl2 concentrations as low as practical, using the same concentrations of each of the four dNTPs, maintaining an annealing temperature near the T m of the primers, and programming a short extension segment and as few cycles as practical.
Cross-Contamination
Because PCR primers are incorporated into each molecule of PCR product, amplicons are themselves suitable templates for amplification. Re-amplification of previously amplified DNA is a major problem for PCR. To reduce the likelihood of contamination of reagents, separate laboratory work areas should be designated for reagent preparation, sample preparation, reaction assembly and thermal cycling, and post-PCR analysis. Depending on the analytical rigor required and the resources available, well-isolated rooms for each step of the process will be beneficial. In the research laboratory, however, it is more common to rely on separate work areas and equipment for pre- and postamplification work and gowning of laboratory personnel in cleanroom garments. Laboratory equipment, especially pipettors and shared equipment, and laboratory personnel are the most common sources of PCR product contamination. Clinical laboratories or other analytical laboratories now employ physical separation of work areas and equipment in addition to a biochemical method using the enzyme uracil DNA glycosylase (UNG) to prevent amplicon contamination. If PCR reactions are performed with dUTP rather than dTTP, amplicons will be susceptible to digestion by UNG. This method has been called PCR sterilization. The need to avoid contamination with amplified DNA constitutes an incentive to use real-time PCR for qualitative as well as quantitative assays. With real-time PCR, tubes containing amplified DNA need never be opened. No matter what precautions are taken, negative controls containing no DNA template must always be included with each batch of PCR reactions. If low-copy detection is the assay goal, then multiple negative controls are required to adequately test for amplicon contamination.
Reverse Transcription PCR
PCR can be used to detect and quantify RNA if RNA is first reverse-transcribed to complementary DNA (cDNA). Reverse transcriptases from Maloney murine leukemia virus (MMLV-RT) or avian myeloblastoma virus (AMV-RT) are commonly used. Either the buffer optimal for the reverse transcription (RT) or the PCR buffer is suitable in most cases, and reverse transcription is typically performed at 37–42°C for 15–60 min. Because these enzymes are heat-labile, they are inactivated during the first denaturation segment of PCR. Heat-stable reverse transcriptases enable reverse transcription at higher temperatures to denature RNA secondary structure. In the presence of Mn2+, some DNA-dependent DNA polymerases such as the enzyme from Thermus thermophilus (Tth DNA polymerase) have activity on RNA templates. Using MnCl2 for reverse transcription can enable cDNA synthesis with the same enzyme used for PCR. Unless a balance between MnCl2 and MgCl2 is carefully identified, Mn2+ must be removed or chelated, and Mg2+ must be added prior to PCR. RT-PCR can be performed within intact cells (in situ PCR) to identify cells expressing particular genes or to assess the presence of disease-related genes. Thermal cyclers are available to automate the process on microscope slides.
Applications
Clinical Diagnostics
The main clinical application of PCR technology is in the diagnosis of disease, and in many cases the speed and simplicity of PCRs have revolutionized clinical diagnosis. Bacteria that are difficult or impossible to culture on artificial media can now be detected by PCR. The causative organism of syphilis, Treponema pallidum, can be detected by a PCR assay in which a gene encoding a specific membrane protein is amplified. PCR is being employed to investigate the pathogenicity of Mycoplasma genitalium. As few as four organisms can be detected by this method. PCR-based detection assays have also been developed for Mycobacterium tuberculosis, an organism that takes up to several weeks to identify by conventional means owing to its slow growth on artificial media. M. tuberculosis, the bacterium that causes tuberculosis, infects about one-third of the world's population and is most prevalent in developing countries. Recently, this organism has re-emerged as a significant pathogen in the developed world, especially among the poor and homeless, and antibiotic-resistant strains are developing. PCR tests have become essential for rapid diagnosis and epidemiology of diseases such as tuberculosis.
Amplification of viral genes forms the basis of PCR-based detection systems for viral pathogens. The human immunodeficiency virus (HIV), which is responsible for AIDS, is currently detected primarily by immunological means. However, as with most viral infections, it takes some time following infection for antibodies to develop, and there is therefore a period when a patient infected with the virus would appear negative by conventional tests. A PCR assay can be used to detect the presence of the virus in this case. Proviral DNA that has integrated into the genomes of leucocytes can be detected, or the viral RNA genome can be detected in plasma by RT-PCR. Identification of HIV by PCR methods has played an important role in AIDS diagnostics and research, as well as improved safety testing in blood banks. PCR assays have also been used in epidemiological studies and in the identification of other retroviruses such as human T-lymphotropic virus types 1 and 2 (HTLV-1 and HTLV-2). PCR procedures have also been developed to detect human papillomaviruses. This diverse group of viruses can cause a number of diseases. A PCR assay can distinguish the relatively harmless strains from those that cause serious diseases such as cervical cancer. Current laboratory methods for the diagnosis of human enteroviruses, which cause infection in children, are slow and lack sensitivity. Detection is hindered by the low level of viruses present in clinical specimens. PCR technology, however, provides a rapid and accurate diagnostic test. PCR assays have also been used to detect coronaviruses, cytomegalovirus, human herpesvirus type 6, adenovirus, Epstein–Barr virus, rotaviruses, human parvovirus, and herpes simplex viruses, among many others. In addition to detection, PCR assays can be designed to differentiate between different strains or serotypes of a particular virus.
Parasitic infections cause important human diseases such as leishmaniasis and malaria. About 40% of the world's population is at risk from malaria, and drug-resistant strains have emerged and are spreading rapidly. As well as being helpful in diagnosis of disease, a PCR assay can distinguish between drug-resistant and drug-sensitive strains, thus enabling effective treatment to be prescribed.
Many PCR diagnostic kits are now commercially available. Hoffman-La Roche has introduced kits for Chlamydia, M. tuberculosis, HIV, hepatitis C virus, and others. It is likely that an even wider range of PCR diagnostic kits will become available, and PCR testing will continue to have a huge impact in the clinical diagnostic laboratory. Despite the simplicity and rapidity of PCR from the perspective of the research laboratory, it is viewed as a complex test in the clinical laboratory. The expenses of highly trained personnel and commercial PCR kits coupled with concerns about false positive test results due to contamination with previously amplified DNA have so far confined this promising technology to clinical research and specialty testing labs. Eventually, improved technology and assay automation will enable PCR testing to be as common in diagnostic laboratories as immunoassays.
Molecular Genetics
In addition to being an invaluable tool for the detection and diagnosis of infectious diseases, PCR tests have also proved useful in the diagnosis and analysis of genetic diseases. Many genetic disorders are caused by specific mutations, and the fragment of DNA involved can be amplified by PCR. DNA sequencing, high-resolution size analysis on sequencing gels, single-strand conformation polymorphism analysis, Southern blotting and probe hybridization, allele-specific oligonucleotide probes, and other techniques are used to test for the presence of mutations. This approach has been used to detect mutations causing diseases such as sickle cell anemia, beta-thalassemia, cystic fibrosis, and many others. Genetic diseases caused by trinucleotide repeat expansion can be tested using PCR followed by high-resolution electrophoretic analysis on sequencing gels. A commercial test for fragile X syndrome employs novel PCR reagents and thermal cycling conditions to enable amplification of (CGG) repeats up to 800 repeat units in length in the same reaction containing a normal allele (usually 29–31 repeat units). In general, PCR should be considered a versatile template onto which can be superimposed known DNA sequences for any disorder, whether genetic or infectious in nature, to rapidly configure a high-performance test.
PCR is routinely used in the analysis of human leucocyte antigen (HLA) polymorphisms (HLA typing) to assess donor compatibility prior to bone marrow or organ transplantation. The use of PCR in transfusion medicine is also being investigated, and the basis of a system for ABO blood typing using PCR has been described.
Prenatal Diagnostics
Prenatal diagnosis of genetic disease is normally carried out on a sample of amniotic fluid obtained by amniocentesis or by chorionic villus sampling (CVS). Conventional diagnostic tests for diseases such as cystic fibrosis can take as long as 2 weeks. Results of a PCR test can be available in 1 day. If the option to terminate a pregnancy is taken, this time savings can be of enormous benefit. The availability of PCR technology creates an incentive for the development of noninvasive prenatal sampling techniques. Because amniocentesis and CVS carry a certain amount of risk, less invasive sampling procedures are desirable. A small number of cells of fetal origin circulate in the maternal blood. Methods to purify these cells have been described. Mutations and polymorphisms on the Y chromosome already can be tested using maternal blood samples without the need to purify fetal cells. It seems likely that in the future PCR tests will be used for prenatal diagnosis of a whole range of genetic diseases from a sample of maternal blood.
PCR testing is currently available for preimplantation genetic diagnosis for in vitro fertilization. One cell is removed from an eight-cell zygote for PCR testing.
Basic Research
PCR has become an indispensable tool in the molecular biology laboratory. Many traditional methods and protocols have now been replaced by PCR-based techniques. One of the most common techniques employed by molecular biologists involves the cloning of DNA fragments into plasmid vectors. The plasmids are maintained in bacterial strains. To determine which bacterial colony contains the plasmid of interest, transformant colonies can be analyzed by PCR using primers designed from the vector (plasmid) DNA, bracketing the insert. In this way, colonies can be screened to find one containing a plasmid with an insert of a particular size, obviating the need to screen transformants by plasmid DNA preparation followed by restriction enzyme digestion. PCR can also be used to screen libraries for clones containing a particular DNA fragment or gene. This strategy dispenses with the filter hybridization step and can be both quicker and more sensitive than traditional methods. PCR is also invaluable in mutagenesis, radioactive, and nonradioactive labeling of DNA fragments, and many other applications. In addition, the technology developed for PCR amplification has been applied to the extremely important techniques of DNA sequencing and gene expression analysis.
PCR is being used in phylogenetic and evolutionary studies. PCR amplification and screening of ribosomal RNA (rRNA) genes and mitochondrial DNA have been used to estimate relationships between species and subspecies. Besides studies on the relationships of contemporary species, PCR allows studies on extinct species because it can be performed on material in museum collections. Amplification and sequencing of mitochondrial DNA are also used in evolutionary studies.
Forensics
PCR has become an essential tool in forensic science. Many methods that exploit differences in DNA to identify individuals or to distinguish between individuals require large amounts of DNA (>100 ng). However, PCR permits the amplification of minute amounts of DNA from biological evidence found at the scene of a crime and has greatly facilitated the identification of individuals. Now, sufficient DNA for analysis can be obtained from very small amounts of biological material, such as a single hair or a small blood stain.
Other Applications
Monitoring of foods for pathogens is increasingly performed using PCR analysis. Direct detection of microbial DNA or RNA can enable detection of contamination before visible signs appear. Assays for enterotoxigenic E. coli (E. coli 0157:H7), Listeria monocytogenes, Salmonella, and other pathogens are increasingly a component of food safety testing. Food products and ingredients can be tested for the presence of genetically modified organisms using PCR primers specific for the inserted gene sequence. Environmental samples can be tested for coliform and other bacteria or for biowarfare agents such as smallpox or anthrax. Because of its exquisite sensitivity, PCR amplification seems assured to find even more applications across research and analytical disciplines wherever there is a need to detect small amounts of genetic material.
See also:
AMPLIFICATION REACTIONS; DNA SEQUENCING; ELECTROPHORESIS | Nucleic Acids; FORENSIC SCIENCES | DNA Profiling; NUCLEIC ACIDS | Chromatographic and Electrophoretic Methods; NUCLEIC ACIDS | Immunoassays.
Further Reading
- Arnheim N., Erlich H. Polymerase Chain Reaction Strategy. Annual Reviews in Biochemistry. 1992;61:131–156. doi: 10.1146/annurev.bi.61.070192.001023. [DOI] [PubMed] [Google Scholar]
- Bloch W. A biochemical perspective of the polymerase chain reaction. Biochemistry. 1991;30:2735–2747. doi: 10.1021/bi00225a001. [DOI] [PubMed] [Google Scholar]
- Goodman M.F. DNA polymerase fidelity: Misinsertions and mismatched extensions. In: Innis M.A., Gelfand D.H., Sninsky J.J., editors. PCR Strategies. Academic Press; San Diego: 1995. pp. 130–139. [Google Scholar]
- Griffin H.G., Griffin A.M. PCR Technology, Current Innovations. CRC Press; Boca Raton, FL: 1994. [Google Scholar]
- Innis M.A., Gelfand D.H., Sninsky J.J. PCR Applications: Protocols for Functional Genomics. Academic Press; San Diego: 1999. [Google Scholar]
- Innis M.A., Gelfand D.H., Sninsky J.J., White T.J. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego: 1990. [Google Scholar]
- Markham A.F. The polymerase chain reaction: a tool for molecular medicine. British Medical Journal. 1993;306:441–446. doi: 10.1136/bmj.306.6875.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson M.J., Hames B.D., Taylor G.R. PCR 2: A Practical Approach. Oxford University Press; Oxford: 1995. [Google Scholar]
- Mullis K.B., Faloona F.A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods in Enzymology. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Mullis K.B., Ferré F., Gibbs R.A. The Polymerase Chain Reaction. Birkhäuser; Boston: 1994. [Google Scholar]


