Abstract
In general terms, virus replication involves three broad stages which are present in all viruses: initiation of infection, replication and expression of the genome, and the release of mature virions from the infected cell. At a more detailed level, virus replication can be broken down into the stages of attachment, entry, uncoating, transcription and genome replication, assembly, maturation, and release. However, there are many subtle differences between the replication processes of different viruses, imposed by the biology of the host cell and the nature of the virus genome.
Keywords: Assembly, Attachment, Entry, Genome replication, Maturation, Release, Replication, Transcription, Uncoating
Unlike cellular organisms, which ‘grow’ from an increase in the integrated sum of their components and reproduce by division, virus particles are produced from the assembly of preformed components. Once manufactured, virus particles (virions) do not grow or undergo division. This alone makes the process of virus replication distinct from the growth of all other biological agents, and although the term ‘grow’ is sometimes used in the vernacular to refer to propagation of viruses, it is best to avoid this word when referring to the processes of virus replication.
Although this article will attempt to paint a general picture of the process of virus replication, the type of host cell infected by the virus has a profound effect on the replication process. There are many examples of viruses undergoing different replicative cycles in different cell types. However, the coding capacity of the genome determines the basic replication strategy used by different viruses. This strategy may involve heavy reliance on the host cell, in which case the virus genome can be very compact and need only encode the essential information for a few proteins, for instance, in parvoviruses. Alternatively, large and complex virus genomes, such as those of poxviruses, encode most of the information necessary for replication, and the virus is only reliant on the cell for the provision of energy and the apparatus for macromolecular synthesis, such as ribosomes. Viruses with RNA genomes have no apparent need to enter the nucleus, although during the course of replication, some do. DNA viruses, as might be expected, mostly replicate in the nucleus where host cell DNA is replicated and the biochemical apparatus necessary for this process is located. However, some viruses with DNA genomes (e.g., poxviruses) have evolved to contain sufficient biochemical capacity to be able to replicate in the cytoplasm, with minimal requirement for host cell functions.
Virus replication can be divided into eight stages, as shown in Figure 1 . It should be emphasized that these are arbitrary divisions, used here for convenience in explaining the replication cycle of a theoretical, ‘typical’ virus. Regardless of their hosts, all viruses must undergo each of these stages in some form to successfully complete their replication cycle. Not all the steps described here are detectable as distinct stages for all viruses; often they blur together and appear to occur almost simultaneously. Some of the individual stages have been studied in great detail and a considerable amount of information is known about them. Other stages have been much harder to study, and less information is available.
Figure 1.
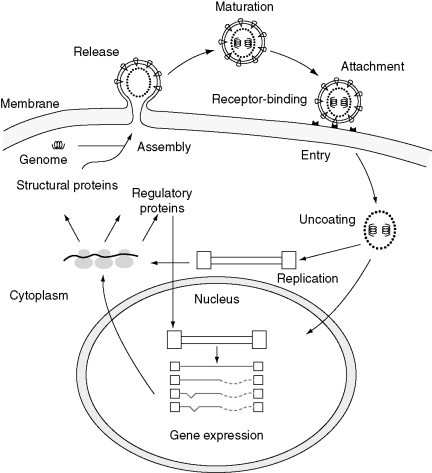
Schematic overview of a generalized scheme of virus replication. Reproduced from Cann AJ (2004)Principles of Molecular Virology, 4th edn. Amsterdam: Elsevier, with permission from Elsevier.
Attachment
The attachment phase of replication comprises specific binding of a virus-attachment protein (or ‘antireceptor’) to a cellular receptor molecule. Virus receptors on cell surfaces may be proteins (usually glycoproteins) or carbohydrate residues present on glycoproteins or glycolipids. Some complex viruses (e.g., in the Poxviridae or Herpesviridae) use more than one receptor and therefore have alternative routes of uptake into cells. Most bacteriophage receptors are on the bacterial cell wall, although certain phages use cellular appendages (pili, flagella) as primary adsorption sites. Attachment is an automatic docking process and the kinetics of receptor binding are controlled by the chemical and thermodynamic characteristics of the molecules involved, that is, their relative concentrations and availability.
In most cases, the expression (or absence) of receptors on the surface of host cells determines the tropism of a particular virus, that is, the types of cell in which it is able to replicate. The attachment phase of infection therefore has a major influence on viral pathogenesis and in determining the course of a virus infection. Plant viruses must overcome different problems to animal viruses in initiating infection. The outer surfaces of plants are composed of protective layers of waxes and pectin, and each cell is surrounded by a thick wall of cellulose overlying the cytoplasmic membrane. No known plant virus uses a specific cellular receptor of the type that animal and bacterial viruses use to attach to cells and plant viruses must rely on mechanical breaks in the cell wall to directly introduce a virus particle into a cell.
Some virus receptors consist of more than one protein and multiple interactions are required for virus entry. An example of this is human immunodeficiency virus-1 (HIV-1), the primary receptor for which is the T-cell antigen, CD4. The binding site for the HIV-1 attachment protein (antireceptor), gp 120, has been mapped to the first variable region of CD4, although additional amino acids of the second variable domain also contribute toward binding. The sequences important for CD4 binding have also been mapped in gp120. Deletions in this region or site substitutions abolish CD4 binding. In addition to CD4, there is at least one accessory factor which is necessary to form a functional HIV-1 receptor. These factors have now been identified as a family of proteins known as β-chemokine receptors. Multiple members of this class of proteins have been shown to play a role in the entry of HIV-1 into cells, and their distribution in the body is the primary control for the tropism of HIV-1 for different cell types.
Occasionally, the specificity of receptor binding can be subverted by nonspecific interactions between virus particles and host cells. Virus particles may be taken up by cells by pinocytosis or phagocytosis. However, without some form of physical interaction which holds the virus particle in close association with the cell surface, the frequency of these events would be very low. In addition, the fate of viruses absorbed into endocytic vacuoles is usually to be degraded, except in cases where the virus particle enters cells by this route. On occasion, binding of antibody-coated virus particles to Fc receptor molecules on the surface of monocytes and other blood cells can result in virus uptake. The presence of antiviral antibodies can result in increased virus uptake by cells and increased pathogenicity, rather than virus neutralization, as would normally be expected. The significance of such mechanisms in vivo is not known.
Entry
Entry of the virus particle into the host cell normally occurs a short time after attachment of the virus to the receptor. Unlike attachment, cell entry is generally an energy-dependent process, that is, the cell must be metabolically active for this to occur. Three main mechanisms are observed:
-
1.
Translocation of the entire virus particle across the cytoplasmic membrane of the cell. This process is relatively rare among viruses and is poorly understood. It is mediated by proteins in the virus capsid and specific membrane receptors.
-
2.
Endocytosis of the virus into intracellular vacuoles. This is probably the most common mechanism of virus entry into cells. It does not require any specific virus proteins (other than those already utilized for receptor binding) but relies on the normal formation and internalization of coated pits (term to be explained) at the cell membrane. Receptor-mediated endocytosis is an efficient process for taking up and concentrating extracellular macromolecules.
-
3.
Fusion of the virus envelope (where present) with the cell membrane, either directly at the cell surface or following endocytosis in a cytoplasmic vesicle. Fusion requires the presence of a specific fusion protein in the virus envelope, for example, influenza A virus hemagglutinin or the transmembrane glycoproteins of retroviruses. These proteins promote the joining of the cellular and virus membranes which results in the nucleocapsid being deposited directly in the cytoplasm. There are two types of virus-driven membrane fusions: pH-dependent and pH-independent.
The process of endocytosis is almost universal in animal cells and requires the formation of clathrin-coated pits which results in the engulfment of a membrane-bounded vesicle by the cytoplasm of the cell. At this point, any virus contained within these structures is still cut off from the cytoplasm by a lipid bilayer and therefore has not strictly entered the cell. As endosomes fuse with lysosomes, the environment inside these vessels becomes progressively more hostile as they are acidified and the pH falls, while the concentration of degradative enzymes rises. This means that the virus must leave the vesicle and enter the cytoplasm before it is degraded. There are a number of mechanisms by which this occurs, including membrane fusion and rescue by transcytosis. The release of virus particles from endosomes and their passage into the cytoplasm is intimately connected with (and often impossible to separate from) the process of uncoating.
Uncoating
Uncoating describes the events which occur after host cell entry, during which the virus capsid is partially or completely degraded or removed and the virus genome exposed, usually still in the form of a nucleic acid–protein complex. Uncoating occurs simultaneously with or immediately after entry and is thus difficult to study. In bacteriophages which inject their genome directly into the cell, entry and uncoating are the same process.
The removal of a virus envelope during membrane fusion is the initial stage of the uncoating process for enveloped viruses. Uncoating may occur inside endosomes, being triggered by the change in pH as the endosome is acidified, or directly in the cytoplasm. Entry into the endocytic pathway is a hazardous process for viruses because if they remain in the vesicle too long, they will be irreversibly damaged by low pH or lysosomal enzymes. Hence, some viruses have evolved proteins to control this process; for example, the influenza A virus M2 protein is a membrane channel which allows entry of hydrogen ions into the nucleocapsid, facilitating uncoating. The M2 protein is multifunctional, and also has a role in virus uncoating. In the picornaviruses, penetration of the cytoplasm by exit of virus from endosomes is tightly linked to uncoating. The acidic environment of the endosome causes a conformational change in the particle at around pH 5 that reveals hydrophobic domains not present on the surface of mature virus capsids. These hydrophobic patches interact with the endosomal membrane and form pores through which the RNA genome passes into the cytoplasm of the host cell.
The ultimate product of uncoating depends on the structure of the virus genome/nucleocapsid. In some cases, the resulting structure is relatively simple; for example, picornaviruses have only a small basic protein of approximately 23 amino acids covalently attached to the 5′ end of the RNA genome. In other cases, the virus core which remains is highly complex; for example, in the poxviruses uncoating occurs in two stages – removal of the outer membrane as the particle enters the cell and in the cytoplasm, followed by further uncoating as the core passes into the cytoplasm. In this case, the core still contains dozens of proteins and at least 10 distinct enzymes.
The structure and chemistry of the nucleocapsid determines the subsequent steps in replication. Reverse transcription can only occur inside an ordered retrovirus core particle and does not proceed to completion with the virus RNA free in solution. Eukaryotic viruses which replicate in the nucleus, such as members of the Herpesviridae, Adenoviridae, and Polyomaviridae, undergo structural changes following penetration, but overall remain largely intact. This is important because these capsids contain nuclear localization sequences responsible for attachment to the cytoskeleton and this interaction allows the transport of the entire capsid to the nucleus. At the nuclear pores, complete uncoating occurs and the nucleocapsid passes into the nucleus.
Transcription and Genome Replication
The replication strategy of a virus depends, in large part, on the structure and composition of its genome. For viruses with RNA genomes in particular, genome replication and transcription are often inextricably linked, and frequently carried out by the same enzymes. Therefore, it makes most sense to consider both of these aspects of virus replication together.
Group I: Double-Stranded DNA
This class can be further subdivided into two as follows:
-
1.
Replication is exclusively nuclear or associated with the nucleoid of prokaryotes. The replication of these viruses is relatively dependent on cellular factors. In some cases, no virus-encoded enzymes are packaged within these virus particles as this is not necessary, whereas in more complex viruses numerous enzymatic activities may be present within the particles.
-
2.
Replication occurs in cytoplasm. These viruses have evolved (or acquired from their hosts) all the necessary factors for transcription and replication of their genomes and are therefore largely independent of the cellular apparatus for DNA replication and transcription. Because of this independence from cellular functions, these viruses have some of the largest and most complex particles known, containing many different enzymes.
Group II: Single-Stranded DNA
The replication of these virus genomes occurs in the nucleus, involving the formation of a double-stranded intermediate which serves as a template for the synthesis of new single-stranded genomes. In general, no virus-encoded enzymes are packaged within the virus particle since most of the functions necessary for replication are provided by the host cell.
Group III: Double-Stranded RNA
These viruses all have segmented genomes, as each segment is transcribed separately to produce individual monocistronic messenger RNAs. Replication occurs in the cytoplasm and is largely independent of cellular machinery, as the particles contain many virus-encoded enzymes essential for RNA replication and transcription since these processes (involving copying RNA to make further RNA molecules) do not normally occur in cellular organisms.
Group IV: Single-Stranded (+)-Sense RNA
These viruses can be subdivided into two groups.
-
1.
Viruses with polycistronic mRNA such as flaviviruses and picornaviruses. As with all the viruses in this group, the genome RNA represents mRNA which is translated after infection, resulting in the synthesis of a polyprotein product, which is subsequently cleaved to form the mature proteins.
-
2.
Viruses with complex transcription such as coronaviruses and togaviruses. In this subgroup, two rounds of translation are required to produce subgenomic RNAs which serve as mRNAs in addition to the full-length RNA transcript which forms progeny virus genomes. Although the replication of these viruses involves copying RNA from an RNA template, no virus-encoded enzymes are packaged within the genome since the ability to express genetic information directly from the genome without prior transcription allows the virus replicase to be synthesized after infection has occurred.
Group V: Single-Stranded (–)-Sense RNA
The genomes of these viruses can also be divided into two types.
-
•
Segmented. The first step in the replication of these viruses (e.g., orthomyxoviruses) is transcription of the (–)-sense RNA genome by the virion RNA-dependent RNA polymerase packaged in virus particles to produce monocistronic mRNAs, which also serve as the template for subsequent genome replication.
-
•
Nonsegmented. Monocistronic mRNAs for each of the virus genes are produced by the virus transcriptase in the virus particle from the full-length virus genome. Subsequently, a full-length (+)-sense copy of the genome is synthesized which serves as a template for (–)-sense progeny virus genomes (e.g., paramyxoviruses and rhabdoviruses).
Group VI: Single-Stranded RNA with DNA Intermediate
Retrovirus genomes are composed of (+)-sense RNA but are unique in that they are diploid and do not serve directly as mRNA but as a template for reverse transcription into DNA. A complete replication cycle involves conversion of the RNA form of the virus genetic material into a DNA form, the provirus, which is integrated into the host cell chromatin. The enzyme reverse transcriptase needs to be packaged into virus particles to achieve this conversion, as virus genes are only expressed from the DNA provirus and not from the RNA genome found in retrovirus particles of retroviruses.
Group VII: Double-Stranded DNA with RNA Intermediate
This group of viruses also relies on reverse transcription, but unlike the retroviruses, this occurs inside the virus particle during maturation. On infection of a new cell, the first event to occur is repair of the gapped genome, followed by transcription. As with group VI viruses, a reverse transcriptase enzyme activity is present inside virus particles, but in this case, the enzyme carries out the conversion of virus RNA into the DNA genome of the virus inside the virus particle. This contrasts with retroviruses where reverse transcription occurs after the RNA genome has been released from the virus particle into the host cell.
Assembly
During assembly, the basic structure of the virus particle is formed as all the components necessary for the formation of the mature virion come together at a particular site in the cell. The site of assembly depends on the pattern of virus replication and the mechanism by which the virus is eventually released from the cell and so varies for different viruses. Although some DNA virus particles form in the nucleus, the cytoplasm is the most common site of particle assembly. In the majority of cases, cellular membranes are used to anchor virus proteins, and this initiates the process of assembly.
For enveloped viruses, the lipid covering is acquired through a process known as budding, where the virus particle is extruded through a cell membrane. Lipid rafts are membrane microdomains enriched in glycosphingolipids (or glycolipids), cholesterol and a specific set of associated proteins. Lipid rafts have been implicated in a variety of cellular functions, such as apical sorting of proteins and signal transduction, but they are also used by viruses as platforms for cell entry (e.g., for HIV-1, SV40, and the rotaviruses), and as sites for particle assembly, budding and release from the cell membrane (e.g., in influenza A virus, HIV, measles virus, and rotaviruses).
As with the earliest stages of replication, it is often not possible to identify the assembly, maturation, and release of virus particles as distinct and separate phases. The site of assembly has a profound influence on all these processes. In general terms, rising intracellular levels of virus proteins and genomes reach a critical concentration and this triggers assembly. Many viruses achieve high levels of newly synthesized structural components by concentrating these into subcellular compartments known as inclusion bodies. These are a common feature of the late stages of infection of cells by many different viruses. Alternatively, local concentrations of virus structural components can be boosted by lateral interactions between membrane-associated proteins. This mechanism is particularly important in enveloped viruses which are released from the cell by budding (see above).
Maturation
Maturation is the stage of the replication cycle at which virus particles become infectious. This often involves structural changes in the newly formed particle resulting from specific cleavages of virus proteins to form the mature products or from conformational changes in proteins which occur during assembly (e.g., hydrophobic interactions). Protein cleavage frequently leads to substantial structural changes in the capsid. Alternatively, internal structural alterations, for example, the condensation of nucleoproteins with the virus genome, often result in changes visible by electron microscopy.
Proteases are frequently involved in maturation, and virus-encoded enzymes, cellular proteases or a mixture of the two may be used. Virus-encoded proteases are usually highly specific for particular amino acid sequences and structures, only cutting a particular peptide bond in a particular protein. Moreover, they are often further controlled by being packaged into virus particles during assembly and only activated when brought into close contact with their target sequence by the conformation of the capsid, for example, by being placed in a local hydrophobic environment, or by changes of pH or cation cofactor concentrations inside the particle as it forms. Retrovirus proteases are good examples of enzymes involved in maturation which are under tight control. The retrovirus core particle is composed of proteins from the gag gene and the protease is packaged into the core before its release from the cell on budding. During the budding process, the protease cleaves the gag protein precursors into the mature products – the capsid, nucleocapsid, and matrix proteins of the mature virus particle. Other protease cleavage events involved in maturation are less closely controlled. Influenza A virus hemagglutinin must be cleaved into two fragments (HA1 and HA2) to be able to promote membrane fusion during infection. Cellular trypsin-like enzymes are responsible for this process, which occurs in secretory vesicles as the virus buds into them prior to release at the cell surface; however, this process is controlled by the virus M2 protein, which regulates the pH of intracellular compartments in influenza virus-infected cells.
Release
For lytic viruses (most nonenveloped viruses), release is a simple process – the infected cell breaks open and releases the virus. The reasons for lysis of infected cells are not always clear, but virus-infected cells often disintegrate because viral replication disrupts normal cellular function, for example, the expression of essential genes. Many viruses also encode proteins that stimulate (or in some cases suppress) apoptosis, which can also result in release of virus particles.
Enveloped viruses acquire their lipid membrane as the virus buds out of the cell through the cell membrane, or into an intracellular vesicle prior to subsequent release. Virion envelope proteins are picked up during this process as the virus particle is extruded. This process is known as budding. As mentioned earlier, assembly, maturation, and release are usually simultaneous processes for viruses which are released by budding. The release of mature virus particles from their host cells by budding presents a problem in that these particles are designed to enter, rather than leave, cells. Certain virus envelope proteins are involved in the release phase of replication as well as in receptor binding. The best-known example of this is the neuraminidase protein of influenza virus. In addition to being able to reverse the attachment of virus particles to cells via hemagglutinin, neuraminidase is also believed to be important in preventing the aggregation of influenza A virus particles and may well have a role in virus release. In addition to using specific proteins, viruses which bud have also solved the problem of release by the careful timing of the assembly-maturation-release pathway. Although it may not be possible to separate these stages by means of biochemical analysis, this does not mean that spatial separation of these processes has not evolved as a means to solve this problem.
Further Reading
- Cann A.J. 4th edn. Elsevier; Amsterdam: 2004. Principles of Molecular Virology. [Google Scholar]
- Freed E.O. HIV-1 and the host cell: An intimate association. Trends in Microbiology. 2004;12:170–177. doi: 10.1016/j.tim.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Kasamatsu H., Nakanishi A. How do animal DNA viruses get to the nucleus? Annual Review of Microbiology. 1998;52:627–686. doi: 10.1146/annurev.micro.52.1.627. [DOI] [PubMed] [Google Scholar]
- Lopez S., Arias C.F. Multistep entry of rotavirus into cells: A Versaillesque dance. Trends in Microbiology. 2004;12:271–278. doi: 10.1016/j.tim.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Moore J.P., Kitchen S.G., Pugach P., Zack J.A. The CCR5 and CXCR4 coreceptors – central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Research and Human Retroviruses. 2004;20:111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G., He Y., Kuhn R.J. Picornavirus–receptor interactions. Trends in Microbiology. 2002;10:324–331. doi: 10.1016/s0966-842x(02)02383-1. [DOI] [PubMed] [Google Scholar]
- Schneider-Schaulies J. Cellular receptors for viruses: Links to tropism and pathogenesis. Journal of General Virology. 2000;81:1413–1429. doi: 10.1099/0022-1317-81-6-1413. [DOI] [PubMed] [Google Scholar]
Glossary
- (+)-sense RNA (plus-sense RNA)
A virus with a single-stranded RNA genome of the same polarity (‘sense’) as mRNA.
- (−)-sense RNA (minus-sense RNA)
A virus with a single-stranded RNA genome of the opposite polarity (‘sense’) as mRNA.
- Assembly
The stage of replication during which all the structural components come together at one site in the cell and the basic structure of the virus particle is formed.
- Attachment
The binding of a virus particle to a specific receptor on the surface of a host cell.
- Capsid
A protein shell comprising the main structural unit of a virus particle.
- Envelope
A lipid membrane enveloping a virus particle.
- Fusion protein
The protein(s) on the surface of a virus particle responsible for fusion of the virus envelope with cellular membranes.
- Gene expression
An important stage of viral replication at which virus genetic information is expressed: one of the major control points in replication.
- Genome replication
The stage of viral replication at which the virus genome is copied to form new progeny genomes.
- Matrix protein
A structural protein of a virus particle which underlies the envelope and links it to the core.
- Maturation
The stage of viral replication at which a virus particle becomes infectious.
- Molecular epidemiology
The use of nucleotide sequence information to study the diversity and distribution of virus populations.
- mRNA
Messenger RNA, translated on ribosomes to produce proteins.
- Nucleocapsid
The core of a virus particle consisting of the genome plus a complex of proteins.
- Penetration
The stage of viral replication at which the virus genome enters the cell.
- Polyprotein
A long polypeptide encoding several mature proteins which are subsequently released by protease cleavage.
- Receptor
A specific molecule on the surface of a cell which is used by a virus for attachment.
- Release
The stage of viral replication at which virus particles escape the infected cell.
- Tropism
The ability of a virus to infect specific cell or tissue types.
- Uncoating
The stage of viral replication at which structural proteins are lost and the virus genome is exposed to the replication machinery.
- Virions
Structurally mature, extracellular virus particles.
- Virus attachment protein
The protein on the surface of a virus particle responsible for binding the receptor.


