Abstract
This chapter focuses on the mechanisms by which cells express the information stored in genes, and examines the genome coding struggles of different virus groups. Control of gene expression is a vital element of virus replication. Coordinate expression of groups of virus genes results in successive phases of gene expression. Immediate–early genes encode activator proteins, early genes encode further regulatory proteins, and late genes encode virus structural proteins. Viruses make use of the biochemical apparatus of their host cells to express their genetic information as proteins and, consequently, utilize the appropriate biochemical language recognized by the cell. Thus viruses of prokaryotes produce polycistronic mRNAs, while viruses with eukaryotic hosts produce more monocistronic mRNAs. Some viruses of eukaryotes do produce polycistronic mRNA to assist with the coordinate regulation of multiple genes. Viruses rely on specific cis- and trans-acting mechanisms to manipulate the biology of their host cells, and to enhance and coordinate the expression of their genetic information. The chapter discusses control of prokaryote gene expression, expression in bacteriophage λ, eukaryote gene expression, and also discusses genome coding strategies. The chapter ends with a discussion of the transcription and post-transcriptional methods of control of gene expression.
What’s in this chapter?
-
▪
We start by looking at the mechanisms by which cells express the information stored in genes.
-
▪
After that we examine the genome coding struggles of different virus groups.
-
▪
Then we look at how viruses control gene expression via transcription and by using posttranscriptional methods.
Expression of Genetic Information
As described in Chapter 1, no virus yet discovered has the genetic information that encodes the tools necessary for the generation of metabolic energy or for protein synthesis (ribosomes). So all viruses are dependent on their host cells for these functions, but the way in which viruses persuade their hosts to express their genetic information for them varies considerably. Patterns of virus replication are determined by tight controls on virus gene expression. There are fundamental differences in the control mechanisms of these processes in prokaryotic and eukaryotic cells, and these differences inevitably affect the viruses that use them as hosts. In addition, the relative simplicity and compact size of most virus genomes (compared with those of cells) creates further limits. Cells have evolved varied and complex mechanisms for controlling gene expression by using their extensive genetic capacity. Viruses have had to achieve highly specific quantitative, temporal, and spatial control of expression with much more limited genetic resources. Viruses have counteracted their genetic limitations by the evolving of a range of solutions to these problems. These mechanisms include:
-
▪
Powerful positive and negative signals that promote or repress gene expression
-
▪
Highly compressed genomes in which overlapping reading frames are common
-
▪
Control signals that are frequently nested within other genes
-
▪
Strategies that allow multiple polypeptides to be created from a single messenger RNA
Gene expression involves regulatory loops mediated by signals that act either in cis (affecting the activity of neighboring genetic regions) or in trans (giving rise to diffusible products that act on regulatory sites anywhere in the genome). For example, transcription promoters are cis-acting sequences that are located adjacent to the genes whose transcription they control, while proteins such as transcription factors, which bind to specific sequences present on any stretch of nucleic acid present in the cell, are examples of trans-acting factors. The relative simplicity of virus genomes and the elegance of their control mechanisms are models that form the basis of our current understanding of genetic regulation. This chapter assumes that you are familiar with the mechanisms involved in cellular control of gene expression. However, before we get into the details of virus gene expression, we’ll start with a brief reminder of some important aspects.
Box 5.1. It’s all about the gene.
Even before Richard Dawkins wrote The Selfish Gene in the 1970s, the molecular biology revolution in the 1960s had ensured that the gene became the biological object around which our thinking revolved. In Dawkins’s view, genes are only concerned with their own survival. When we look at how malleable virus genomes are, and how easily genes flow from host to virus and from one virus to another, it’s easy to believe Dawkins was telling the truth. So in many ways, this chapter on gene expression is at the very heart of this book. And understanding the mechanisms of transmission and expression that act on genes is central to understanding modern biology.
Control of Prokaryote Gene Expression
Bacterial cells are second only to viruses in the specificity and economy of their genetic control mechanisms. In bacteria, genetic control operates both at the level of transcription and at subsequent (posttranscriptional) stages of gene expression.
The initiation of transcription is regulated primarily in a negative way by the synthesis of trans-acting repressor proteins, which bind to operator sequences upstream of protein coding sequences. Collections of metabolically related genes are grouped together and coordinately controlled as operons. Transcription of these operons typically produces a polycistronic mRNA that encodes several different proteins. During subsequent stages of expression, transcription is also regulated by a number of mechanisms that act, in Mark Ptashne’s famous phrase, as “genetic switches,” turning on or off the transcription of different genes. Such mechanisms include antitermination, which is controlled by trans-acting factors that promote the synthesis of longer transcripts encoding additional genetic information, and by various modifications of RNA polymerase. Bacterial σ (sigma) factors are apoproteins that affect the specificity of the RNA polymerase holoenzyme (active form) for different promoters. Several bacteriophages (e.g., phage SP01 of Bacillus subtilis) encode proteins that function as alternative σ factors, sequestering RNA polymerase and altering the rate at which phage genes are transcribed. Phage T4 of Escherichia coli encodes an enzyme that carries out a covalent modification (adenosine diphosphate [ADP]-ribosylation) of the host-cell RNA polymerase. This is believed to eliminate the requirement of the polymerase holoenzyme for σ factor and to achieve an effect similar to the production of modified σ factors by other bacteriophages.
At a posttranscriptional level, gene expression in bacteria is also regulated by control of translation. The best known virus examples of this phenomenon come from the study of bacteriophages of the family Leviviridae, such as R17, MS2, and Qβ. In these phages, the secondary structure of the single-stranded RNA phage genome not only regulates the quantities of different phage proteins that are translated but also operates temporal (timed) control of a switch in the ratios between the different proteins produced in infected cells.
Control of Expression in Bacteriophage λ
The genome of phage λ has been studied in great detail and illustrates several of the mechanisms just described, including the action of repressor proteins in regulating lysogeny versus lytic replication and antitermination of transcription by phage-encoded trans-acting factors. Such has been the impact of these discoveries that no discussion of the control of virus gene expression is complete without detailed examination of this phage.
Phage λ was discovered by Esther Lederberg in 1949. Experiments at the Pasteur Institute by André Lwoff in 1950 showed that some strains of Bacillus megaterium, when irradiated with ultraviolet light, stopped growing and subsequently lysed, releasing a crop of bacteriophage particles. Together with Francois Jacob and Jacques Monod, Lwoff subsequently showed that the cells of some bacterial strains carried a bacteriophage in a dormant form, known as a prophage, and that the phage could be made to alternate between the lysogenic (nonproductive) and lytic (productive) growth cycles. After many years of study, our understanding of λ has been refined into a picture that represents one of the best understood and most elegant genetic control systems yet to be investigated. A simplified genetic map of λ is shown in Figure 5.1 .
Figure 5.1.

Simplified genetic map of bacteriophage λ.
The top part of this figure shows the main genetic regions of the phage genome and the bottom part is an expanded view of the main control elements described in the text.
For regulation of the growth cycle of the phage, the structural genes encoding the head and tail components of the virus capsid can be ignored. The components involved in genetic control are as follows:
-
▪
PL is the promoter responsible for transcription of the left-hand side of the λ genome, including N and cIII.
-
▪
OL is a short noncoding region of the phage genome (approximately 50 bp), which lies between the cI and N genes next to PL.
-
▪
PR is the promoter responsible for transcription of the right-hand side of the λ genome, including cro, cII, and the genes encoding the structural proteins.
-
▪
OR is a short noncoding region of the phage genome (approximately 50 bp), which lies between the cI and cro genes next to PR.
-
▪
cI is transcribed from its own promoter and encodes a repressor protein of 236 amino acids that binds to OR, preventing transcription of cro but allowing transcription of cI, and to OL, preventing transcription of N and the other genes at the left-hand end of the genome.
-
▪
cII and cIII encode activator proteins that bind to the genome, enhancing the transcription of the cI gene.
-
▪
cro encodes a 66-amino-acid protein that binds to OR, blocking binding of the repressor to this site.
-
▪
N encodes an antiterminator protein that acts as an alternative ρ (rho) factor for host-cell RNA polymerase, modifying its activity and permitting extensive transcription from PL and PR.
-
▪
Q is an antiterminator similar to N, but it only permits extended transcription from PR.
In a newly infected cell, N and cro are transcribed from PL and PR, respectively (Figure 5.2 ). The N protein allows RNA polymerase to transcribe a number of phage genes, including those responsible for DNA recombination and integration of the prophage, as well as cII and cIII. The N protein acts as a positive transcription regulator. In the absence of the N protein, the RNA polymerase holoenzyme stops at certain sequences located at the end of the N and Q genes, known as the nut and qut sites, respectively. However, RNA polymerase-N protein complexes are able to overcome this restriction and permit full transcription from PL and PR. The RNA polymerase-Q protein complex results in extended transcription from PR only. As levels of the cII and cIII proteins in the cell build up, transcription of the cI repressor gene from its own promoter is turned on.
Figure 5.2.
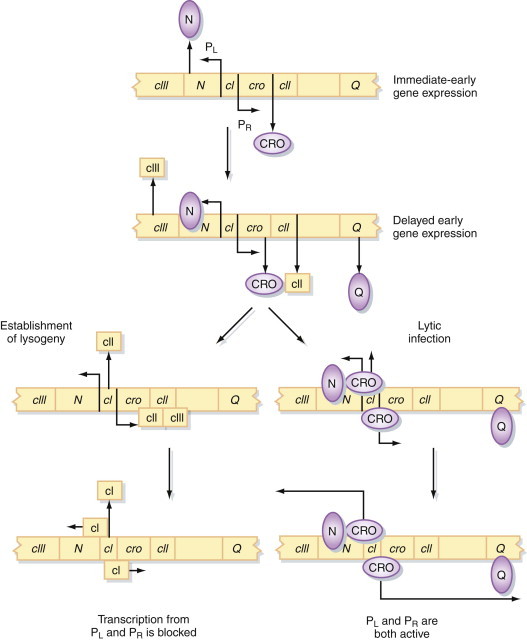
Control of expression of the bacteriophage λ genome.
See text for a detailed description of the events that occur in a newly infected cell and during lytic infection or lysogeny.
At this point, the critical event that determines the outcome of the infection occurs. The cII protein is constantly degraded by host cell proteases. If levels of cII remain below a critical level, transcription from PR and PL continues, and the phage undergoes a productive replication cycle that culminates in lysis of the cell and the release of phage particles. This is the sequence of events that occurs in the vast majority of infected cells. However, in a few rare instances, the concentration of cII protein builds up, transcription of cI is enhanced, and intracellular levels of the cI repressor protein rise. The repressor binds to OR and OL, which prevents transcription of all phage genes (particularly cro; see later) except itself. The level of cI protein is maintained automatically by a negative feedback mechanism, since at high concentrations the repressor also binds to the left-hand end of OR and prevents transcription of cI (Figure 5.3 ). This autoregulation of cI synthesis keeps the cell in a stable state of lysogeny.
Figure 5.3.

Control of lysogeny in bacteriophage λ.
See text for a detailed description of the events that occur in the establishment and maintenance of lysogeny.
If this is the case, how do such cells ever leave this state and enter a productive, lytic replication cycle? Physiological stress and particularly ultraviolet irradiation of cells result in the induction of a host-cell protein, RecA. This protein, the normal function of which is to induce the expression of cellular genes that permit the cell to adapt to and survive in altered environmental conditions, cleaves the cI repressor protein. In itself, this would not be sufficient to prevent the cell from reentering the lysogenic state; however, when repressor protein is not bound to OR, cro is transcribed from PR. Cro also binds to OR but, unlike cI, which preferentially binds the right-hand end of OR, the Cro protein binds preferentially to the left-hand end of OR, preventing the transcription of cI and enhancing its own transcription in a positive-feedback loop. The phage is then locked into a lytic cycle and cannot return to the lysogenic state.
This description is a highly simplified version of the genetic control of expression in phage λ. A great deal of detail is known about the molecular mechanisms by which these systems work, but because this topic could easily fill an entire book on its own, there is not enough space to describe all of it in detail here.
The molecular details of the λ gene have also contributed to our understanding of genetic regulation in prokaryotic and eukaryotic cells. Determination of the structures of the proteins involved in this scheme has allowed us to identify the fundamental principles behind the observation that many proteins from unrelated organisms can recognize and bind to specific sequences in DNA molecules. The concepts of proteins with independent DNA-binding and dimerization domains, protein cooperativity in DNA binding, and DNA looping allowing proteins bound at distant sites to interact with one another have all risen from the study of λ. The references given at the end of this chapter explain more fully the nuances of gene expression in this complex bacteriophage.
Box 5.2. Bacteriophages are, like, so last century.
No they’re not. Apart from the contribution of bacteriophages to understanding viruses as a whole—and there’s no better example of that than λ—some of the most exciting work in virology over the last decade has been about phages. When we finally raised our sights from the glassware in our laboratories and went out hunting for viruses in the natural environment, we were staggered by what we found. Phages in particular are everywhere, and in staggering quantities and variety. It has been estimated that every second on Earth, 1×1025 bacteriophage infections occur. That means that phages control the turnover of such large quantities of organic material that this has a major impact on nutrient cycling and the global climate. Last century? Wrong.
Control of Eukaryote Gene Expression
Control of gene expression in eukaryotic cells is much more complex than in prokaryotic cells and involves a multilayered approach in which diverse control mechanisms exert their effects at different levels. The first level of control occurs prior to transcription and depends on the local configuration of the DNA. DNA in eukaryotic cells has an elaborate structure, forming complicated and dynamic but far from random complexes with numerous proteins to form chromatin. Although the contents of eukaryotic cell nuclei appear amorphous in electron micrographs (at least in interphase), they are actually highly ordered. Chromatin interacts with the structural backbone of the nucleus—the nuclear matrix—and these interactions are thought to be important in controlling gene expression.
Locally, nucleosome configuration and DNA conformation, particularly the formation of left-handed helical Z-DNA, are also important. DNAse I digestion of chromatin does not give an even, uniform digestion pattern but reveals a pattern of DNAse hypersensitive sites believed to indicate differences in the function of various regions of the chromatin. It is probable, for example, that retroviruses are more likely to integrate into the host-cell genome at these sites than elsewhere. Transcriptionally active DNA is also hypomethylated; that is, there is a relative scarcity of nucleotides modified by the covalent attachment of methyl groups in these regions compared with the frequency of methylation in transcriptionally quiescent regions of the genome. The methylation of Moloney murine leukemia virus sequences in preimplantation mouse embryos has been shown to suppress the transcription of the provirus genome.
The second level of control rests in the process of transcription itself, which again is much more complex than in prokaryotes. There are three forms of RNA polymerase in eukaryotic cells that can be distinguished by their relative sensitivity to the drug α-amanitin and that are involved in the expression of different classes of genes (Table 5.1 ). The rate at which transcription is initiated is a key control point in eukaryotic gene expression. Initiation is influenced dramatically by sequences upstream of the transcription start site, which function by acting as recognition sites for families of highly specific DNA-binding proteins known as transcription factors. Immediately upstream of the transcription start site is a relatively short region known as the promoter. It is at this site that transcription complexes, consisting of RNA polymerase plus accessory proteins, bind to the DNA and transcription begins.
Table 5.1.
Forms of RNA Polymerase in Eukaryotic Cells
| RNA Polymerase | Sensitivity to α-amanitin |
Cellular Genes Transcribed | Virus Genes Transcribed |
|---|---|---|---|
| I | Unaffected | Ribosomal RNAs | — |
| II | Highly sensitive | Most single-copy genes | Most DNA virus genomes |
| III | Moderately sensitive | 5S rRNA, tRNAs | Adenovirus VA RNAs |
However, sequences further upstream from the promoter also influence the efficiency with which transcription complexes form. The rate of initiation depends on the combination of transcription factors bound to these transcription enhancers. The properties of these enhancer sequences are remarkable in that they can be inverted and moved around relative to the position of the transcription start site without losing their activity and can exert their influence even from a distance of several kilobases away. This emphasizes the flexibility of DNA, which allows proteins bound at distant sites to interact with one another, as also shown by the protein–protein interactions seen in regulation of phage λ gene expression (earlier). Transcription of eukaryotic genes results in the production of monocistronic mRNAs, each of which is transcribed from its own individual promoter.
At the next stage, gene expression is influenced by the structure of the mRNA produced. The stability of eukaryotic mRNAs varies considerably, some having comparatively long half-lives in the cell (e.g., many hours). The half-lives of others, typically those that encode regulatory proteins, may be very short (e.g., a few minutes). The stability of eukaryotic mRNAs depends on the speed with which they are degraded. This is determined by such factors as its terminal sequences, which consist of a methylated cap structure at the 5′ end and polyadenylic acid at the 3′ end, as well as on the overall secondary structure of the message. However, gene expression is also regulated by differential splicing of heterogeneous (heavy) nuclear RNA (hnRNA) precursors in the nucleus, which can alter the genetic meaning of different mRNAs transcribed from the same gene. In eukaryotic cells, control is also exercised during export of RNA from the nucleus to the cytoplasm.
Finally, the process of translation offers further opportunities for control of expression. The efficiency with which different mRNAs are translated varies greatly. These differences result largely from the efficiency with which ribosomes bind to different mRNAs and recognize AUG translation initiation codons in different sequence contexts, as well as the speed at which different sequences are converted into protein. Certain sequences act as translation enhancers, performing a function analogous to that of transcription enhancers.
The point of this extensive list of eukaryotic gene expression mechanisms is that they are all utilized by viruses to control gene expression. Examples of each type are given in the following sections. If this seems remarkable, remember that the control of gene expression in eukaryotic cells was unraveled largely by using viruses as model systems, therefore finding examples of these mechanisms in viruses is really only a self-fulfilling prophecy.
Genome Coding Strategies
Box 5.3. So many viruses, how am I going to remember them all?
Good question. You don’t need to remember all the details about every virus—even people who write virology textbooks can’t do that. What you do need to do is to have a framework that allows you to think, “Yes, I’ve seen something like this before, so I can guess what’s likely to happen.” And that’s where the seven classes of virus genomes described in the previous chapter come in. Add to that an understanding of how gene expression works for each type and you’re pretty much there. There’s one small catch. Even for viruses with very similar genome structures, there are often surprising differences in mechanisms of gene expression. Hey, this is biology, it’s all about variation. If you wanted everything to be predictable, you should have signed up for the physics class.
In Chapter 4, genome structure was one element of a classification scheme used to divide viruses into seven groups. The other part of this scheme is the way in which the genetic information of each class of virus genomes is expressed. The replication and expression of virus genomes are inseparably linked, and this is particularly true in the case of RNA viruses. Here, the seven classes of virus genomes described in Chapter 4 and Appendix 1 are reviewed again, this time examining the way in which the genetic information of each class is expressed.
Class I: Double-stranded DNA
Chapter 4 said that this class of virus genomes can be subdivided into two further groups: those in which genome replication is exclusively nuclear (e.g., Adenoviridae, Polyomaviridae, Herpesviridae) and those in which replication occurs in the cytoplasm (Poxviridae). In one sense, all of these viruses can be considered to be similar; because their genomes all resemble double-stranded cellular DNA, they are essentially transcribed by the same mechanisms as cellular genes. However, there are profound differences between them relating to the degree to which each family is reliant on the host-cell machinery.
Polyomaviruses and papillomaviruses
Polyomaviruses are heavily dependent on cellular machinery for both replication and gene expression. Polyomaviruses encode trans-acting factors (T-antigens) that stimulate transcription (and genome replication). Papillomaviruses in particular are dependent on the cell for replication, which occurs only in terminally differentiated keratinocytes and not in other cell types, although they do encode several trans-regulatory proteins (Chapter 7).
Adenoviruses
Adenoviruses are also heavily dependent on the cellular apparatus for transcription, but they possess various mechanisms that specifically regulate virus gene expression. These include trans-acting transcriptional activators such as the E1A protein, and posttranscriptional regulation of expression, which is achieved by alternative splicing of mRNAs and the virus-encoded VA RNAs (Chapter 7). Adenovirus infection of cells is divided into two stages, early and late, with the latter phase commencing at the time when genome replication occurs; however, in adenoviruses, these phases are less distinct than in herpesviruses (next).
Herpesviruses
These viruses are less reliant on cellular enzymes than the previous groups. They encode many enzymes involved in DNA metabolism (e.g., thymidine kinase) and a number of trans-acting factors that regulate the temporal expression of virus genes, controlling the phases of infection. Transcription of the large, complex genome is sequentially regulated in a cascade fashion (Figure 5.4 ). At least 50 virus-encoded proteins are produced after transcription of the genome by host-cell RNA polymerase II. Three distinct classes of mRNAs are made:
-
▪
α: Immediate-early (IE) mRNAs encode trans-acting regulators of virus transcription.
-
▪
β: (Delayed) early mRNAs encode further nonstructural regulatory proteins and some minor structural proteins.
-
▪
γ: Late mRNAs encode the major structural proteins.
Figure 5.4.

Control of expression of the herpes simplex virus genome.
HSV particles contain a protein called α-gene transcription initiation factor (α-TIF), which turns on α-gene expression in newly infected cells, beginning a cascade of closely regulated events that control the expression of the entire complement of the 70 or so genes in the virus genome.
Gene expression in herpesviruses is tightly and coordinately regulated, as indicated by the following observations (see Figure 5.4). If translation is blocked shortly after infection (e.g., by treating cells with cycloheximide), the production of late mRNAs is blocked. Synthesis of the early gene product turns off the immediate-early products and initiates genome replication. Some of the late structural proteins (γ1) are produced independently of genome replication; others (γ2) are only produced after replication. Both the immediate-early and early proteins are required to initiate genome replication. A virus-encoded DNA-dependent DNA polymerase and a DNA-binding protein are involved in genome replication, together with a number of enzymes (e.g., thymidine kinase) that alter cellular biochemistry. The production of all of these proteins is closely controlled.
Poxviruses
Genome replication and gene expression in poxviruses are almost independent of cellular mechanisms (except for the requirement for host-cell ribosomes). Poxvirus genomes encode numerous enzymes involved in DNA metabolism, virus gene transcription, and posttranscriptional modification of mRNAs. Many of these enzymes are packaged within the virus particle (which contains >100 proteins), enabling transcription and replication of the genome to occur in the cytoplasm (rather than in the nucleus, like all the families just described) almost totally under the control of the virus. Gene expression is carried out by virus enzymes associated with the core of the particle and is divided into two rather indistinct phases:
-
▪
Early genes: These comprise about 50% of the poxvirus genome and are expressed before genome replication inside a partially uncoated core particle (Chapter 2), resulting in the production of 5′ capped, 3′ polyadenylated but unspliced mRNAs.
-
▪
Late genes: These are expressed after genome replication in the cytoplasm, but their expression is also dependent on virus-encoded rather than on cellular transcription proteins (which are located in the nucleus). Like herpesviruses, late gene promoters are dependent on prior DNA replication for activity.
More detailed consideration of some of these mechanisms is given later in this chapter (see “Transcriptional Control of Expression” and “Posttranscriptional Control of Expression,” later).
Class II: Single-stranded DNA
Both the autonomous and the helper virus-dependent parvoviruses are highly reliant on host cell assistance for gene expression and genome replication. This is presumably because the very small size of their genomes does not permit them to encode the necessary biochemical apparatus. There viruses show an extreme form of parasitism, utilizing the normal functions present in the nucleus of their host cells for both expression and replication (Figure 5.5 ). The members of the replication-defective Dependovirus genus of the Parvoviridae are entirely dependent on adenovirus or herpesvirus superinfection for the provision of further helper functions essential for their replication beyond those present in normal cells. The adenovirus genes required as helpers are the early, transcriptional regulatory genes such as E1A rather than late structural genes, but it has been shown that treatment of cells with ultraviolet light, cycloheximide, or some carcinogens can replace the requirement for helper viruses. Therefore, the help required appears to be for a modification of the cellular environment (probably affecting transcription of the defective parvovirus genome) rather than for a specific virus protein.
Figure 5.5.
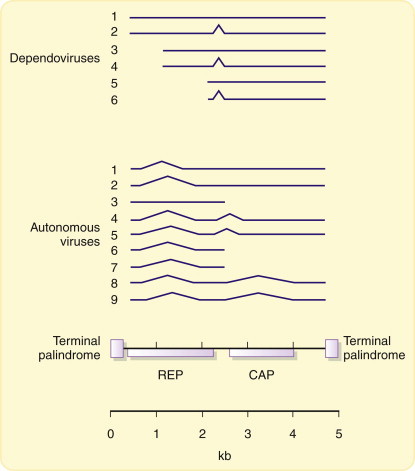
Transcription of parvovirus genomes.
Transcription of parvovirus genomes is heavily dependent on host-cell factors and results in the synthesis of a series of spliced, subgenomic mRNAs that encode two proteins: Rep, which is involved in genome replication, and Cap, the capsid protein (see text).
The Geminiviridae also fall into this class of genome structures (Figure 3.16). The expression of their genomes is quite different from that of parvoviruses but nevertheless still relies heavily on host-cell functions. There are open reading frames in both orientations in the virus DNA, which means that both (+) and (–)sense strands are transcribed during infection. The mechanisms involved in control of gene expression have not been fully investigated, but at least some geminiviruses (subgroup I) may use splicing.
Class III: Double-stranded RNA
All viruses with RNA genomes differ fundamentally from their host cells, which of course possess double-stranded DNA genomes. Therefore, although each virus must be biochemically compatible with its host cell, there are fundamental differences in the mechanisms of virus gene expression from those of the host cell. Reoviruses have multipartite genomes (see Chapter 3) and replicate in the cytoplasm of the host cell. Characteristically for viruses with segmented RNA genomes, a separate monocistronic mRNA is produced from each segment (Figure 5.6 ). Early in infection, transcription of the dsRNA genome segments by virus-specific transcriptase activity occurs inside partially uncoated subvirus particles. At least five enzymatic activities are present in reovirus particles to carry out this process, although these are not necessarily all separate peptides (Table 5.2 ).
Figure 5.6.
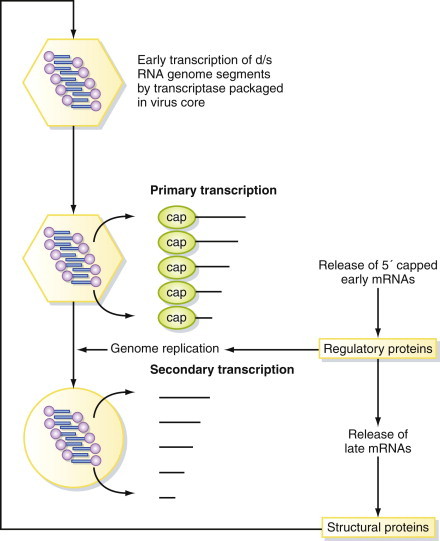
Expression of reovirus genomes.
Expression of reovirus genomes is initiated by a transcriptase enzyme packaged inside every virus particle. Subsequent events occur in a tightly regulated pattern, with the expression of late mRNAs encoding the structural proteins being dependent on prior genome replication.
Table 5.2.
Enzymes in Reovirus Particles
| Activity | Virus Protein | Encoded by Genome Segment |
|---|---|---|
| d/s RNA-dependent RNA polymerase (Pol) | λ3 | L3 |
| RNA triphosphatase | μ2 | M1 |
| Guanyltransferase (Cap) | λ2 | L2 |
| Methyltransferase | λ2 | L2 |
| Helicase (Hel) | λ1 | L1 |
This primary transcription results in capped transcripts that are not polyadenylated and that leave the virus core to be translated in the cytoplasm. The various genome segments are transcribed/translated at different frequencies, which is perhaps the main advantage of a segmented genome. RNA is transcribed conservatively; that is, only (–)sense strands are used, resulting in synthesis of (+)sense mRNAs, which are capped inside the core (all this occurs without de novo protein synthesis). Secondary transcription occurs later in infection inside new particles produced in infected cells and results in uncapped, nonpolyadenylated transcripts. The genome is replicated in a conservative fashion (cf. semiconservative DNA replication). Excess (+)sense strands are produced, which serve as late mRNAs and as templates for (–)sense strand synthesis (i.e., each strand leads to many (+) strands, not one-for-one as in semiconservative replication).
Class IV: Single-stranded (+)sense RNA
This type of genome occurs in many animal viruses and plant viruses (Appendix 2
![]() ). In terms of both the number of different families and the number of individual viruses, this is the largest single class of virus genomes. Essentially, these virus genomes act as messenger RNAs and are themselves translated immediately after infection of the host cell (Chapter 3). Not surprisingly with so many representatives, this class of genomes displays a very diverse range of strategies for controlling gene expression and genome replication. However, in very broad terms, the viruses in this class can be subdivided into two groups:
). In terms of both the number of different families and the number of individual viruses, this is the largest single class of virus genomes. Essentially, these virus genomes act as messenger RNAs and are themselves translated immediately after infection of the host cell (Chapter 3). Not surprisingly with so many representatives, this class of genomes displays a very diverse range of strategies for controlling gene expression and genome replication. However, in very broad terms, the viruses in this class can be subdivided into two groups:
-
▪
Production of a polyprotein encompassing the whole of the virus genetic information, which is subsequently cleaved by proteases to produce precursor and mature polypeptides. These cleavages can be a subtle way of regulating the expression of genetic information. Alternative cleavages result in the production of various proteins with distinct properties from a single precursor (e.g., in picornaviruses and potyviruses; Figure 5.7 ). Certain plant viruses with multipartite genomes use a very similar strategy for controlling gene expression, although a separate polyprotein is produced from each of the genome segments. The best studied example of this is the comoviruses, whose genome organization is very similar to that of the picornaviruses and may represent another member of this superfamily (Figure 5.7).
-
▪
Production of subgenomic mRNAs, resulting from two or more rounds of translation of the genome. This strategy is used to achieve temporal separation of what are essentially early and late phases of replication, in which nonstructural proteins, including a virus replicase, are produced during the early phase followed by structural proteins in the late phase (Figure 5.8 ). The proteins produced in each of these phases may result from proteolytic processing of a polyprotein precursor, although this encompasses only part of the virus genome rather than the entire genome, as before. Proteolytic processing offers further opportunities for regulation of the ratio of different polypeptides produced in each phase of replication (e.g., in togaviruses and tymoviruses). In addition to proteolysis, some viruses employ another strategy to produce alternative polypeptides from a subgenomic mRNA, either by read-through of a leaky translation stop codon (e.g., tobamoviruses such as TMV; see Figure 3.13), or by deliberate ribosomal frame-shifting at a particular site (see “Posttranscriptional Control of Expression,” later).
Figure 5.7.
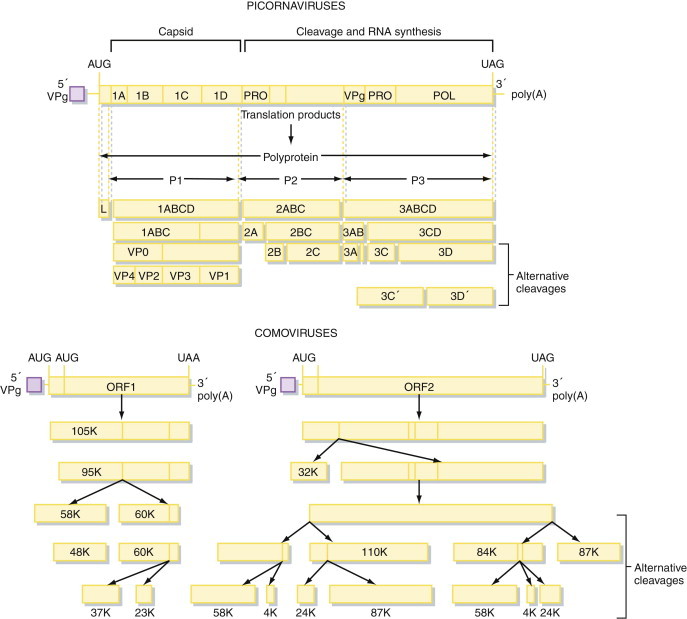
Gene expression in positive-sense RNA viruses.
Many positive-sense RNA virus genomes are frequently translated to form a long polyprotein, which is subsequently cleaved by a highly specific virus-encoded protease to form the mature polypeptides. Picornaviruses and comoviruses are examples of this mechanism of gene expression.
Figure 5.8.
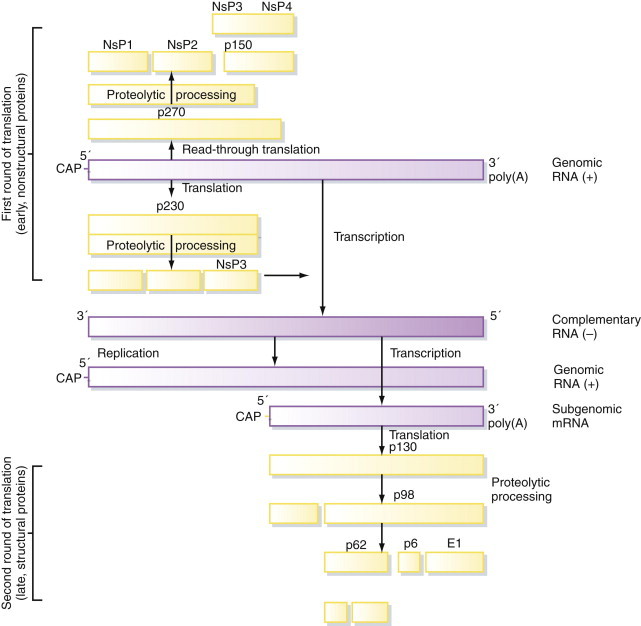
Subgenomic RNAs in positive-sense RNA viruses.
Some positive-sense RNA virus genomes (e.g., togaviruses) are expressed by two separate rounds of translation, involving the production of a subgenomic mRNA at a later stage of replication.
All the viruses in this class have evolved mechanisms that allow them to regulate their gene expression in terms of both the ratios of different virus-encoded proteins and the stage of the replication cycle when they are produced. Compared with the two classes of DNA virus genomes described earlier, these mechanisms operate largely independently of those of the host cell. The power and flexibility of these strategies are reflected very clearly in the overall success of the viruses in this class, as determined by the number of different representatives known and the number of different hosts they infect.
Class V: Single-stranded (–)sense RNA
As discussed in Chapter 3, the genomes of these viruses may be either segmented or nonsegmented. The first step in the replication of segmented orthomyxovirus genomes is transcription of the (–)sense vRNA by the virion-associated RNA-dependent RNA polymerase to produce (predominantly) monocistronic mRNAs, which also serve as the template for subsequent genome replication (Figure 5.9 ). As with all (–)sense RNA viruses, packaging of this virus-specific transcriptase/replicase within the virus nucleocapsid is essential because no host cell contains any enzyme capable of decoding and copying the RNA genome.
Figure 5.9.
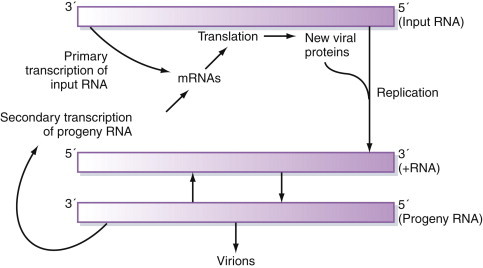
General scheme for the expression of negative-sense RNA virus genomes.
All negative-sense RNA viruses face the problem that the information stored in their genome cannot be interpreted directly by the host cell. They must include mechanisms for converting the genome into mRNAs within the virus particle.
In the other families that have nonsegmented genomes, monocistronic mRNAs are also produced. Here, however, these messages must be produced from a single, long (–)sense RNA molecule. Exactly how this is achieved is not clear. It is possible that a single, genome-length transcript is cleaved after transcription to form the separate mRNAs, but it is more likely that these are produced individually by a stop-and-start mechanism of transcription regulated by the conserved intergenic sequences present between each of the virus genes (Chapter 3). Splicing mechanisms cannot be used because these viruses replicate in the cytoplasm.
On the surface, such a scheme of gene expression might appear to offer few opportunities for regulation of the relative amounts of different virus proteins. If this were true, it would be a major disadvantage, since all viruses require far more copies of the structural proteins (e.g., nucleocapsid protein) than of the nonstructural proteins (e.g., polymerase) for each virion produced. In practice, the ratio of different proteins is regulated both during transcription and afterward. In paramyxoviruses, for example, there is a clear polarity of transcription from the 3′ end of the virus genome to the 5′ end that results in the synthesis of far more mRNAs for the structural proteins encoded in the 3′ end of the genome than for the nonstructural proteins located at the 5′ end (Figure 5.10 ). Similarly, the advantage of producing monocistronic mRNAs is that the translational efficiency of each message can be varied with respect to the others (see “Posttranscriptional Control of Expression,” later).
Figure 5.10.

Expression of paramyxovirus genomes.
Paramyxovirus genomes exhibit transcriptional polarity. Transcripts of genes at the 3′ end of the virus genome are more abundant than those of genes at the 5′ end of the genome, permitting regulation of the relative amounts of structural (3′ genes) and nonstructural (5′ genes) proteins produced.
Viruses with ambisense genome organization (where genetic information is encoded in both the positive [i.e., virus-sense] and negative [i.e., complementary] orientations on the same strand of RNA; see Chapter 4) must express their genes in two rounds of expression so that both are turned into decodable mRNA at some point (Figure 5.11 ).
Figure 5.11.
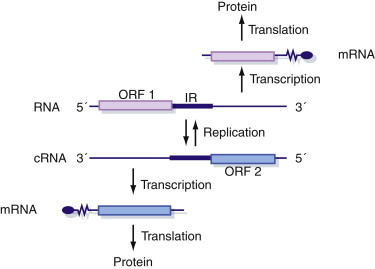
Expression of ambisense virus genomes.
Ambisense virus genomes (which have both positive and negative sense information in the same strand of RNA) require two rounds of gene expression so that information encoded in both strands of the genome is turned into decodable mRNA at some point in the replication cycle.
Class VI: Single-stranded (+)sense RNA with DNA intermediate
The retroviruses are the ultimate case of reliance on the host-cell transcription machinery. The RNA genome forms a template for reverse transcription to DNA—these are the only (+)sense RNA viruses whose genome does not serve as mRNA on entering the host cell (Chapter 3). Once integrated into the host-cell genome, the DNA provirus is under the control of the host cell and is transcribed exactly as are other cellular genes. Some retroviruses, however, have evolved a number of transcriptional and posttranscriptional mechanisms that allow them to control the expression of their genetic information, and these are discussed in detail later in this chapter.
Class VII: Double-stranded DNA with RNA intermediate
Expression of the genomes of these viruses is complex and relatively poorly understood. The hepadnaviruses contain a number of overlapping reading frames clearly designed to squeeze as much coding information as possible into a compact genome. The hepatitis B virus X gene encodes a transcriptional trans-activator believed to be analogous to the human T-cell leukemia virus (HTLV) tax protein (see later). At least two mRNAs are produced from independent promoters, each of which encodes several proteins and the larger of which is also the template for reverse transcription during the formation of the virus particle (Chapter 3). Expression of caulimovirus genomes is similarly complex, although there are similarities with hepadnaviruses in that two major transcripts are produced, 35S and 19S. Each of these encodes several polypeptides, and the 35S transcript is the template for reverse transcription during the formation of the virus genome.
Transcriptional Control of Expression
Having looked at general strategies used by different groups of viruses to regulate gene expression, the rest of this chapter concentrates on more detailed explanations of specific examples from some of the viruses mentioned earlier, beginning with control of transcription in SV40, a member of the Polyomaviridae. Few other genomes, virus or cellular, have been studied in as much detail as SV40, which has been a model for the study of eukaryotic transcription mechanisms (particularly DNA replication; see Chapter 6) for many years. In this sense, SV40 provides a eukaryotic parallel with the bacteriophage λ genome. In vitro systems exist for both transcription and replication of the SV40 genome, and it is believed that all the virus and cellular DNA-binding proteins involved in both of these processes are known. The SV40 genome encodes two T-antigens (tumor antigens) known as large T-antigen and small T-antigen after the sizes of the proteins (Figure 5.12 ). Replication of the double-stranded DNA genome of SV40 occurs in the nucleus of the host cell. Transcription of the genome is carried out by host-cell RNA polymerase II, and large T-antigen plays a vital role in regulating transcription of the virus genome. Small T-antigen is not essential for virus replication but does allow virus DNA to accumulate in the nucleus. Both proteins contain nuclear localization signals that result in their accumulation in the nucleus, where they migrate after being synthesized in the cytoplasm.
Figure 5.12.
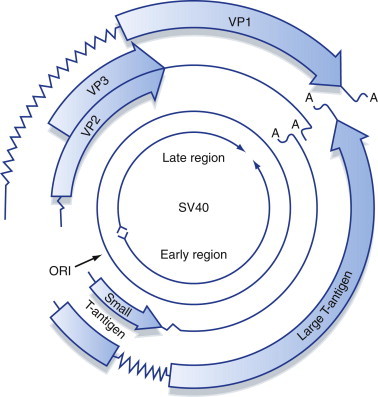
Organization and protein-coding potential of the SV40 genome.
The highly compact SV40 genome includes genetic information encoded in overlapping reading frames on both strands of the DNA genome and mRNA splicing to produce alternative polypetides from one open reading frame.
Soon after infection of permissive cells, early mRNAs are expressed from the early promoter, which contains a strong transcription enhancer element (the 72-bp sequence repeats), allowing it to be active in newly infected cells (Figure 5.13 ). The early proteins made are the two T-antigens. As the concentration of large T-antigen builds up in the nucleus, transcription of the early genes is repressed by direct binding of the protein to the origin region of the virus genome, preventing transcription from the early promoter and causing the switch to the late phase of infection. Large T-antigen is also required for replication of the genome, and this is discussed further in Chapter 7. After DNA replication has occurred, transcription of the late genes occurs from the late promoter and results in the synthesis of the structural proteins VP1, VP2, and VP3. This process illustrates two classic features of control of virus gene expression. First, the definition of the early and late phases of replication, when different sets of genes tend to be expressed, is before and after genome replication. Second, there is usually a crucial protein, in this case T-antigen, whose function is comparable that of a switch—you should compare the pattern of replication of SV40 with the description of bacteriophage λ gene expression control given earlier in this chapter.
Figure 5.13.

Control of transcription of the SV40 genome.
Multiple virus-encoded (T-antigen) and cellular proteins bind to the ori region of the SV40 genome to control gene expression during different phases of replication (see text for details).
Another area where control of virus transcription has received much attention is in the human retroviruses, human T-cell leukemia virus (HTLV) and human immunodeficiency virus (HIV). Integrated DNA proviruses are formed by reverse transcription of the RNA retrovirus genome, as described in Chapter 3. The presence of numerous binding sites for cellular transcription factors in the long terminal repeats (LTRs) of these viruses have been analyzed by DNAse I footprinting and gel-shift assays (Figure 5.14 ). Together, the distal elements (such as NF-kB and SP1 binding sites) and proximal elements (such as the TATA box) make up a transcription promoter in the U3 region of the LTR (Chapter 3). However, the basal activity of this promoter on its own is relatively weak, and results in only limited transcription of the provirus genome by RNA polymerase II. Both HTLV and HIV encode proteins that are trans-acting positive regulators of transcription: the Tax protein of HTLV and the HIV Tat protein (Figure 5.15 ). These proteins act to increase transcription from the virus LTR by a factor of at least 50 to 100 times that of the basal rate from the unaided promoter.
Figure 5.14.
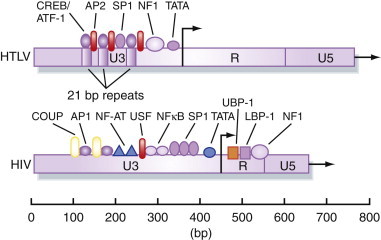
Cellular transcription factors that interact with retrovirus LTRs.
Many cellular DNA-binding proteins are involved in regulating both the basal and trans-activated levels of transcription from the promoter in the U3 region of the retrovirus LTR.
Figure 5.15.

Expression of the HTLV and HIV genomes.
These complex retroviruses contain additional genes to the usual retrovirus pattern of gag, pol, and env, and these are expressed via a complicated mixture of spliced mRNAs.
Unlike T-antigen and the early promoter of SV40, neither the Tax nor the Tat protein (which have no structural similarity to one another) bind directly to its respective LTR. Instead, these proteins function indirectly by interacting with cellular transcription factors, which in turn bind to the promoter region of the virus LTR. So the HTLV Tax and HIV Tat proteins are positive regulators of the basal promoter in the provirus LTR and are under the control of the virus, since synthesis of these proteins is dependent on the promoters that they themselves activate (Figure 5.16 ). On its own, this would be an unsustainable system because it would result in unregulated positive feedback, which might be acceptable in a lytic replication cycle but would not be appropriate for a retrovirus integrated into the genome of the host cell. Therefore, each of these viruses encodes an additional protein (the Rex and Rev proteins in HTLV and HIV, respectively), which further regulates gene expression at a posttranscriptional level (see “Posttranscriptional Control of Expression,” next).
Figure 5.16.

Trans-acting regulation of HTLV and HIV gene expression by virus-encoded proteins.
The Tax (HTLV) and Tat (HIV) proteins act at a transcriptional level and stimulate the expression of all virus genes. The Rex (HTLV) and Rev (HIV) proteins act posttranscriptionally and regulate the balance of expression between virion proteins and regulatory proteins.
Control of transcription is a critical step in virus replication and in all cases is closely regulated. Even some of the simplest virus genomes, such as SV40, encode proteins that regulate their transcription. Many virus genomes encode trans-acting factors that modify and/or direct the cellular transcription apparatus. Examples of this include HTLV and HIV, as described earlier, but also the X protein of hepadnaviruses, Rep protein of parvoviruses, E1A protein of adenoviruses (see later), and the immediate-early proteins of herpesviruses. The expression of RNA virus genomes is similarly tightly controlled, but this process is carried out by virus-encoded transcriptases and has been less intensively studied and is generally much less well understood than transcription of DNA genomes.
Posttranscriptional Control of Expression
In addition to control of the process of transcription, the expression of virus genetic information is also governed at a number of additional stages between the formation of the primary RNA transcript and completion of the finished polypeptide. Many generalized subtle controls, such as the differential stability of various mRNAs, are employed by viruses to regulate the flow of genetic information from their genomes into proteins. This section describes only a few well-researched, specific examples of posttranscriptional regulation.
Many DNA viruses that replicate in the nucleus encode mRNAs that must be spliced by cellular mechanisms to remove intervening sequences (introns) before being translated. This type of modification applies only to viruses that replicate in the nucleus (and not, for example, to poxviruses) because it requires the processing of mRNAs by nuclear apparatus before they are transported into the cytoplasm for translation. However, several virus families have taken advantage of this capacity of their host cells to compress more genetic information into their genomes. A good example of such a reliance on splicing are the parvoviruses, transcription of which results in multiple spliced, polyadenylated transcripts in the cytoplasm of infected cells, enabling them to produce multiple proteins from their 5 kb genomes (Figure 5.5), and similarly, polyomaviruses such as SV40 (Figure 5.12). In contrast, the large genetic capacity of herpesviruses makes it possible for these viruses to produce mostly unspliced monocistronic mRNAs, each of which is expressed from its own promoter, thereby rendering unnecessary extensive splicing to produce the required range of proteins.
One of the best-studied examples of the splicing of virus mRNAs is the expression of the adenovirus genome (Figure 5.17 ). Several families of adenovirus genes are expressed via differential splicing of precursor hnRNA transcripts. This is particularly true for the early genes that encode trans-acting regulatory proteins expressed immediately after infection. The first proteins to be expressed, E1A and E1B, are encoded by a transcriptional unit on the r-strand at the extreme left-hand end of the adenovirus genome (Figure 5.17). These proteins are primarily transcriptional trans-regulatory proteins comparable to the Tax and Tat proteins described earlier, but are also involved in transformation of adenovirus-infected cells (Chapter 6). Five polyadenylated, spliced mRNAs are produced (13S, 12S, 11S, 10S, 9S) that encode five related E1A polypeptides (containing 289, 243, 217, 171, and 55 amino acids, respectively; Figure 5.18 ). All of these proteins are translated from the same reading frame and have the same amino and carboxy termini. The differences between them are a consequence of differential splicing of the E1A transcriptional unit and result in major differences in their functions.
Figure 5.17.

Transcription of the adenovirus genome.
The arrows in this figure show the positions of exons in the virus genome that are joined by splicing to produce families of related but unique virus proteins.
Figure 5.18.

Expression of the adenovirus E1A proteins.
The number shown above each box is the number of amino acids encoded by each exon.
The 289 and 243 amino acid peptides are transcriptional activators. Although these proteins activate transcription from all the early adenovirus promoters, it has been discovered that they also seem to be “promiscuous,” activating most RNA polymerase II-responsive promoters that contain a TATA box. There are no obvious common sequences present in all of these promoters, and there is no evidence that the E1A proteins bind directly to DNA. E1A proteins from different adenovirus serotypes contain three conserved domains: CR1, CR2, and CR3. The E1A proteins interact with many other cellular proteins, primarily through binding to the three conserved domains. By binding to components of the basal transcription machinery, activating proteins that bind to upstream promoter and enhancer sequences and regulatory proteins that control the activity of DNA-binding factors, E1A can both activate and repress transcription.
Synthesis of the adenovirus E1A starts a cascade of transcriptional activation by turning on transcription of the other adenovirus early genes: E1B, E2, E3, and E4 (Figure 5.17). After the virus genome has been replicated, this cascade eventually results in transcription of the late genes encoding the structural proteins. The transcription of the E1A itself is a balanced, self-regulating system. The immediate-early genes of DNA viruses typically have strong enhancer elements upstream of their promoters. This is because in a newly infected cell there are no virus proteins present and the enhancer is required to kick-start expression of the virus genome. The immediate-early proteins synthesized are transcriptional activators that turn on expression of other virus genes, and E1A functions in exactly this way. However, although E1A trans-activates its own promoter, the protein represses the function of the upstream enhancer element so, at high concentrations, it also downregulates its own expression (Figure 5.19 ).
Figure 5.19.
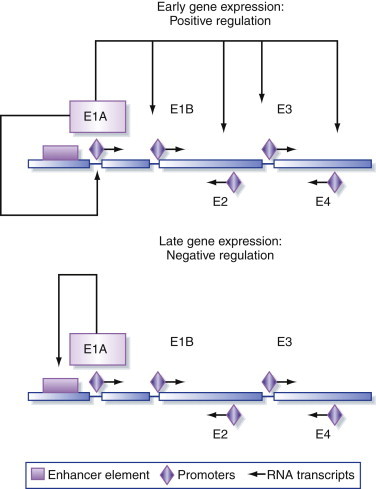
Regulation of adenovirus gene expression.
Adenovirus early proteins are involved in complex positive and negative regulation of gene expression.
The next stage at which expression can be regulated is during export of mRNA from the nucleus and preferential translation in the cytoplasm. Again, the best studied example of this phenomenon comes from the Adenoviridae. The virus-associated (VA) genes encode two small (~160 nt) RNAs transcribed from the r-strand of the genome by RNA polymerase III (whose normal function is to transcribe similar small RNAs such as 5S ribosomal RNA and tRNAs) during the late phase of virus replication (Figure 5.17). Both VA RNA I and VA RNA II have a high degree of secondary structure, and neither molecule encodes any polypeptide (in these two respects they are similar to tRNAs), and they accumulate to high levels in the cytoplasm of adenovirus-infected cells. The way in which these two RNAs act is not completely understood, but their net effect is to boost the synthesis of adenovirus late proteins. The VA RNAs are processed by the host cell to form virus-encoded miRNAs. These operate through RNA interference (see Chapter 6) to downregulate a large number of cellular genes involved in RNA binding, splicing, and translation. In addition, virus infection of cells stimulates the production of interferons (Chapter 6). One of the actions of interferons is to activate a cellular protein kinase known as PKR that inhibits the initiation of translation. VA RNA I binds to this kinase, preventing its activity and relieving inhibition on translation. The effects of interferons on the cell are generalized (discussed in Chapter 6) and result in inhibition of the translation of both cellular and virus mRNAs. The effect of the VA RNAs is to promote selectively the translation of adenovirus mRNAs at the expense of cellular mRNAs whose translation is inhibited.
The HTLV Rex and HIV Rev proteins mentioned earlier also act to promote the selective translation of specific virus mRNAs. These proteins regulate the differential expression of the virus genome but do not substantially alter the expression of cellular mRNAs. Both of these proteins appear to function in a similar way, and, although not related to one another in terms of their amino acid sequences, the HTLV Rex protein can substitute functionally for the HIV Rev protein. Negative-regulatory sequences in the HIV and HTLV genomes cause the retention of virus mRNAs in the nucleus of the infected cell. These sequences are located in the intron regions that are removed from spliced mRNAs encoding the Tax/Tat and Rex/Rev proteins (Figure 5.15), therefore, these proteins are expressed immediately after infection. Tax and Tat stimulate enhanced transcription from the virus LTR (Figure 5.16). However, unspliced or singly spliced mRNAs encoding the gag, pol, and env gene products are expressed only when sufficient Rex/Rev protein is present in the cell. Both proteins bind to a region of secondary structure formed by a particular sequence in the mRNA and shuttle between the nucleus and the cytoplasm as they contain both a nuclear localization signal and a nuclear export signal, increasing the export of unspliced virus mRNA to the cytoplasm where it is translated and acts as the virus genome during particle formation.
The efficiency with which different mRNAs are translated varies considerably and is determined by a number of factors, including the stability and secondary structure of the RNA, but the main one appears to be the particular nucleotide sequence surrounding the AUG translation initiation codon that is recognized by ribosomes. The most favorable sequence for initiation is GCC(A/G)CCAUGGG, although there can be considerable variation within this sequence. A number of viruses use variations of this sequence to regulate the amounts of protein synthesized from a single mRNA. Examples include the Tax and Rex proteins of HTLV, which are encoded by overlapping reading frames in the same doubly spliced 2.1 kb mRNA (Figure 5.15). The AUG initiation codon for the Rex protein is upstream of that for Tax but provides a less favorable context for initiation of translation than the sequence surrounding the Tax AUG codon. This is known as the leaky scanning mechanism because it is believed that the ribosomes scan along the mRNA before initiating translation. Therefore, the relative abundance of Rex protein in HTLV-infected cells is considerably less than that of the Tax protein, even though both are encoded by the same mRNA.
Picornavirus genomes illustrate an alternative mechanism for controlling the initiation of translation. Although these genomes are genetically economical (i.e., have discarded most cis-acting control elements and express their entire coding capacity as a single polyprotein), they have retained long noncoding regions (NCRs) at their 5′ ends, comprising approximately 10% of the entire genome. These sequences are involved in the replication and possibly packaging of the virus genome. Translation of most cellular mRNAs is initiated when ribosomes recognize the 5′ end of the mRNA and scan along the nucleotide sequence until they reach an AUG initiation codon. Picornavirus genomes are not translated in this way. The 5′ end of the RNA is not capped and thus is not recognized by ribosomes in the same way as other mRNAs, but it is modified by the addition of the VPg protein (see Chapters 3 and 6Chapter 3Chapter 6). There are also multiple AUG codons in the 5′ NCR upstream of the start of the polyprotein coding sequences that are not recognized by ribosomes. In picornavirus-infected cells, a virus protease cleaves the 220-kDa cap-binding complex (CBC) involved in binding the m7 G cap structure at the 5′ end of the mRNA during initiation of translation. Translation of artificially mutated picornavirus mRNAs in vitro and the construction of bicistronic picornavirus genomes bearing additional 5′ NCR signals in the middle of the polyprotein have resulted in the concept of the ribosome landing pad, or internal ribosomal entry site (IRES). Rather than scanning along the RNA from the 5′ end, ribosomes bind to the RNA via the IRES and begin translation internally. This is a precise method for controlling the translation of virus proteins. Very few cellular mRNAs use this mechanism but it has been shown to be used by a variety of viruses, including picornaviruses, hepatitis C virus, coronaviruses, and flaviviruses.
Many viruses belonging to different families compress their genetic information by encoding different polypeptides in overlapping reading frames. The problem with this strategy lies in decoding the information. If each polypeptide is expressed from a monocistronic mRNA transcribed from its own promoter, the additional cis-acting sequences required to control and coordinate expression might cancel out any genetic advantage gained. More importantly, there is the problem of coordinately regulating the transcription and translation of multiple different messages. Therefore, it is highly desirable to express several polypeptides from a single RNA transcript, and the examples described earlier illustrate several mechanisms by which this can be achieved (e.g., differential splicing and control of RNA export from the nucleus, or initiation of translation).
Box 5.4. Viruses: Making more from less.
If viruses were as wasteful with their genomes as cells are, they would struggle to exist. For most cellular genes, it’s one gene, one protein. And while some of the big DNA viruses do have that genetic arrangement, most viruses have to work harder and get more than one protein out of a gene. There are lots of ways they do this. Splicing, alternative start codons, ribosomal frameshifting, alternate protease cleavages—all these are used by different viruses to squeeze the maximum amount of information into the minimum space. Of course the trick is to ensure that you can get the information, in the form of proteins, out again. The host cell is tricked into doing this, so in addition to the range of virus genomes, the range of host organisms also means that many different gene expression mechanisms are used.
An additional mechanism known as ribosomal frameshifting is used by several groups of viruses to achieve the same effect. The best studied examples of this phenomenon come from retrovirus genomes, but many viruses use a similar mechanism. Such frameshifting was first discovered in viruses but is now known to occur also in prokaryotic and eukaryotic cells. Retrovirus genomes are transcribed to produce at least two 5′ capped, 3′ polyadenylated mRNAs. Spliced mRNAs encode the envelope proteins, as well as, in more complex retroviruses such as HTLV and HIV, additional proteins such as the Tax/Tat and Rex/Rev proteins (Figure 5.15). A long, unspliced transcript encodes the gag, pro, and pol genes and also forms the genomic RNA packaged into virions. The problem faced by retroviruses is how to express three different proteins from one long transcript. The arrangement of the three genes varies in different viruses. In some cases (e.g., HTLV) they occupy three different reading frames, while in others (e.g., HIV) the protease (pro) gene forms an extension at the 5′ end of the pol gene (Figure 5.20 ). In the latter case, the protease and polymerase (i.e., reverse transcriptase) are expressed as a polyprotein that is autocatalytically cleaved into the mature proteins in a process that is similar to the cleavage of picornavirus polyproteins.
Figure 5.20.

Ribosomal frameshifting and termination suppression in retroviruses.
Ribosomal frameshifting and termination suppression are posttranscriptional methods used to extend the range of proteins produced by retrovirus genomes.
At the boundary between each of the three genes is a particular sequence that usually consists of a tract of reiterated nucleotides, such as UUUAAAC (Figure 5.21 ). This sequence is rarely found in protein-coding sequences and therefore appears to be specifically used for this type of regulation. Most ribosomes encountering this sequence will translate it without difficulty and continue on along the transcript until a translation stop codon is reached. However, a proportion of the ribosomes that attempt to translate this sequence will slip back by one nucleotide before continuing to translate the message, but now in a different (i.e., –1) reading frame. Because of this, the UUUAAAC sequence has been termed the slippery sequence, and the result of this frameshifting is the translation of a polyprotein containing alternative information from a different reading frame. This mechanism also allows the virus to control the ratios of the proteins produced. Because only a proportion of ribosomes undergoes frameshifting at each slippery sequence, there is a gradient of translation from the reading frames at the 5′ end of the mRNA to those at the 3′ end.
Figure 5.21.

RNA pseudoknot formation.
RNA pseudoknot formation is the mechanism by which ribosomal frameshifting occurs in a number of different viruses and a few cellular genes (see text for details).
The slippery sequence alone results in only a low frequency of frameshifting, which appears to be inadequate to produce the amount of protease and reverse transcriptase protein required by the virus. So there are additional sequences that further regulate this system and increase the frequency of frameshift events. A short distance downstream of the slippery sequence is an inverted repeat that allows the formation of a stem–loop structure in the mRNA (Figure 5.21). A little further on is an additional sequence complementary to the nucleotides in the loop that allows base-pairing between these two regions of the RNA. The net result of this combination of sequences is the formation of what is known as an RNA pseudoknot. This secondary structure in the mRNA causes ribosomes translating the message to pause at the position of the slippery sequence upstream, and this slowing or pausing of the ribosome during translation increases the frequency at which frameshifting occurs, thus boosting the relative amounts of the proteins encoded by the downstream reading frames. It is easy to imagine how this system can be fine-tuned by subtle mutations that alter the stability of the pseudoknot structure and thus the relative expression of the different genes.
Yet another method of translational control is termination suppression. This is a mechanism similar in many respects to frameshifting that permits multiple polypeptides to be expressed from individual reading frames in a single mRNA. In some retroviruses, such as murine leukemia virus (MLV), the pro gene is separated from the gag gene by a UAG termination codon rather than a slippery sequence and pseudoknot (Figure 5.20). In the majority of cases, translation of MLV mRNA terminates at this sequence, giving rise to the Gag proteins. However in a few instances, the UAG stop codon is suppressed and translation continues, producing a Gag–Pro–Pol polyprotein, which subsequently cleaves itself to produce the mature proteins. The overall effect of this system is much the same as ribosomal frameshifting, with the relative ratios of Gag and Pro/Pol proteins being controlled by the frequency with which ribosomes traverse or terminate at the UAG stop codon.
Summary
Control of gene expression is a vital element of virus replication. Coordinate expression of groups of virus genes results in successive phases of gene expression. Typically, immediate-early genes encode activator proteins, early genes encode further regulatory proteins, and late genes encode virus structural proteins. Viruses make use of the biochemical apparatus of their host cells to express their genetic information as proteins and, consequently, utilize the appropriate biochemical language recognized by the cell. Thus viruses of prokaryotes produce polycistronic mRNAs, while viruses with eukaryotic hosts produce more monocistronic mRNAs. Some viruses of eukaryotes do produce polycistronic mRNA to assist with the coordinate regulation of multiple genes. In addition, viruses rely on specific cis- and trans-acting mechanisms to manipulate the biology of their host cells and to enhance and coordinate the expression of their own genetic information.
Further Reading
- Alberts B., editor. Molecular Biology of the Cell. Fifth ed. Garland Science; New York: 2007. Freed, E.O., Martin, M. (2001). Human immunodeficiency viruses and their replication. In: Fields Virology, Fourth ed. Fields, B.N., Knipe, D.M., Howley, P.M. (Eds.), Lippincott Williams & Wilkins, Philadelphia, PA. pp. 1971–2042. [Google Scholar]
- Giedroc D.P. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J Mol Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannian P., Green P.L. Human T lymphotropic virus type 1 (HTLV-1): molecular biology and oncogenesis. Viruses. 2010;2(9):2037–2077. doi: 10.3390/v2092037. doi:10.3390/v2092037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman D. BIOS Scientific; Oxford: 2002. Gene Regulation: A Eukaryotic Perspective. [Google Scholar]
- López-Lastra M. Translation initiation of viral mRNAs. Rev Med Virol. 2010;20(3):177–195. doi: 10.1002/rmv.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2004. A Genetic Switch: Phage Lambda Revisited. [Google Scholar]
- Skalsky R.L., Cullen B.R. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Marsh J.W. Gene transcription in HIV infection. Microbes Infec. 2003;5:1023–1027. doi: 10.1016/s1286-4579(03)00187-4. [DOI] [PubMed] [Google Scholar]


