Abstract
During influenza infection, hemagglutinins (HAs) on the viral surface bind to sialic acids on the host cell’s surface. While all HAs bind sialic acids, human influenza targets terminal α2,6 sialic acids and avian influenza targets α2,3 sialic acids. For interspecies transmission (zoonosis), HA must mutate to adapt to these differences. Here, multivalent gold nanoparticles bearing either α2,6- or α2,3-sialyllactosamine have been developed to interrogate a panel of HAs from pathogenic human, low pathogenic avian, and other species’ influenza. This method exploits the benefits of multivalent glycan presentation compared to monovalent presentation to increase affinity and investigate how multivalency affects selectivity. Using a library-orientated approach, parameters including polymer coating and core diameter were optimized for maximal binding and specificity were probed using galactosylated particles and a panel of biophysical techniques [ultraviolet–visible spectroscopy, dynamic light scattering, and biolayer interferometry]. The optimized particles were then functionalized with sialyllactosamine and their binding analyzed against a panel of HAs derived from pathogenic influenza strains including low pathogenic avian strains. This showed significant specificity crossover, which is not observed in monovalent formats, with binding of avian HAs to human sialic acids and vice versa in agreement with alternate assay formats. These results demonstrate that precise multivalent presentation is essential to dissect the interactions of HAs and may aid the discovery of tools for disease and zoonosis transmission.
Introduction
Worldwide, annual epidemics of seasonal influenza are estimated to result in 3–5 million cases of severe illness, and up to 650,000 deaths annually are associated with respiratory diseases from seasonal influenza.1 While influenza A and B viruses cause seasonal epidemics, only A types have been evidenced to cause pandemics. Influenza A viruses are classified into subtypes according to the combination of proteins on their surface; hemagglutinins (HA) and neuraminidases (NA)—for example, the H1N1 strain pandemic in 2009.2 During infection, influenza virus particles display trimeric HAs that bind to sialosides on the surface of their host cells to facilitate internalization. While all influenza strains bind sialosides, human strains preferentially bind to terminal α2,6 sialic acids, whereas avian influenza viruses show preference for α2,3 sialic acids.3,4 Zoonosis (interspecies transmission) can occur due to mutations in the HA, enabling a switch from α2,3 to α2,6. This allows avian influenza to infect humans, and often occurs in porcine (pig) hosts, which present both α2,3 to α2,6 sialosides in their respiratory tracts.5−7 Humans can be infected with avian, porcine, and other zoonotic influenza viruses including avian influenza subtypes; H5N1, H7N9, and H9N2 and swine flu virus subtypes; H1N1 (swine flu pandemic 2009), H1N2, and H3N2.8 It has also emerged that N-glycolyl modifications (in addition to N-acetyl) play a significant role in species-dependent binding9 and that influenza can bind phosphorylated, non-sialylated glycans determined against an array of N-linked glycans from human lungs,10 suggesting that other glycans are also involved, or potential targets during infection.11 Therefore, the study of HA interactions with sialic acids is essential to increase our understanding of this global pathogen and to help guide the discovery of new diagnostics and treatments.
The binding affinity of a HA toward an individual sialoside is weak (Kd ≈ 2 mM),12 but because of the multivalent presentation of both the HA (∼200–1000 copies/virus) and the dense layer of sialic acids on the cell surface,13 the affinity is significantly increased by the cluster glycoside effect.14 Hence, in vivo, HAs engage with cell–surface sialic acids which are not presented as free molecules in solution, but rather multivalent ensembles as in the glycocalyx. Strategies to probe and dissect HA function should reproduce this multivalent environment to be truly predictive. There are many examples where multivalent materials, in particular polymers, have been used to engage with carbohydrate binding proteins, with most studies focusing on plant lectins, which are of limited biomedical relevance.15−19
Whitesides and co-workers synthesized sialic acid containing glycopolymeric inhibitors of influenza virus X-31 via a poly(N-acrylyloxy succinimide) reactive scaffold. Using hemagglutination and enzyme-linked immunosorbent assays, the inhibitory activity of these polymers were found to be between 103 and 106 times greater than the free glycan.20,21 However, the effect of the linkage was not examined. Miura and co-workers synthesized 2,3- and 2,6-sialyllactose-functional glycopolymers that were able to recognize two different influenza strains. They found that 2,6-sialyllactose-functional glycopolymers inhibited hemagglutination caused by H3N2 (A/Aichi/2/68) but not H1N1 (A/Puerto Rico/8/34); conversely, 2,3-sialyllactose-functional glycopolymers inhibited aggregation caused by the H1N1 strain but not the H3N2.22 The density and polymer length were found to affect the interaction with the virus. Trivalent presentation of 2,3 sialic acids was sufficient to give >400 fold increase in inhibitory activity against H3 influenzas.23 Godula and co-workers developed a microarray platform for the evaluation of virus binding based on 2,6- and 2,3-sialyllactose glycopolymers, which showed discrimination between different strains, with H3N2 (A/Aichi/2/68) binding exclusively to 2,6-siallylactose glycopolymers, whereas H1N1 (A/Puerto Rico/8/34) bound to both 2,3- and 2,6-sialyllactose polymers.24 In addition to the nature of the sialic acid, the 3-D presentation is a crucial determinant with bivalent ligands showing wide affinity trends based only on separation,25 for example, H3N2 strains have evolved specificity related to the length of the sialosides linker.26 Furthermore, linear sialic acid polymers were found to be more potent inhibitors than dendritic.27
Gibson and co-workers have previously developed multivalent gold/polymer hybrids to dissect glycan binding interactions.28−31 Reversible addition fragmentation chain transfer (RAFT) polymerization was used to produce telechelic polymers that have a gold immobilization site at one end (masked thiol) and a glycan installation site (an amine reactive pentafluorophenyl group) at the other end. The polymer linker was found to be important, ensuring a stable formulation and fast response readout.28,30 The use of gold nanoparticle cores, exploiting their unique aggregation-induced red-blue color shift, provides label-free readouts. This has been used for the discrimination of human (H3N2) and avian (H5N1) influenza using PEGylated gold particles presenting trivalent α2,6 sialic acid ligands.32 Fairbanks and co-workers investigated the interaction of HAs and whole viruses with gold nanoparticles decorated with biantennary sialyloligosaccharides (α2,6 linked) extracted from egg yolk. Selectivity was observed between two H1N1 strains, A/New Caledonia/20/1999 and A/Puerto Rico/8/34, which have specificity toward α2,6 sialic acid and α2,3 sialic acid, respectively.33 De Geest and co-workers immobilized α2,6 sialic acid pendant glycopolymers on AuNPs and observed an interaction with inactivated H1N1 (A/Puerto Rico/8/34) viruses as determined by DLS; however, the selectivity compared with other strains was not investigated.34 Furthermore, Papp et al. showed the importance of gold core size on the efficiency of virus inhibitions with 14 nm particles having higher binding affinity than 2 nm particles.35 The gold particle size enables the generation of large signal responses as well as multivalency; therefore, it is essential to optimize the core size, linker, and glycan.36 Lipid bilayers and dendrimer scaffolds have also been employed to probe or inhibit hemagglutinin binding.37,38
Considering the above, the aim of this work was to develop a multivalent nanoparticle platform to dissect the binding trends of influenza HAs toward multivalent sialic acid isomers. A library-oriented screen of 27 nanoparticle formulations was used to identify the optimum size and linker length, and then translated to enable 2,3- and 2,6-sialyllactosamine attachment to multivalent gold nanoparticles. The binding affinities of these particles were evaluated by colorimetric aggregation assays, dynamic light scattering (DLS) and biolayer interferometry (BLI) to ensure the whole binding landscape was probed. A panel of eight HAs, including human and avian, were interrogated with the particles, and it was demonstrated that the multivalent presentation enables significant cross-binding between human and avian strains.
Experimental Section
Materials
All chemicals were used as supplied unless otherwise stated. N-Hydroxyethyl acrylamide (HEA) (97%), 4,4′-azobis(4-cyanovaleric acid) (ACVA) (98%), mesitylene (reagent grade), triethylamine (>99%), sodium citrate tribasic dihydrate (>99%), gold(III) chloride trihydrate (99.9%), and ammonium carbonate (reagent grade) were all purchased from Sigma-Aldrich. d(+)-Galactosamine hydrochloride (99%) was purchased from Acros Organics. 3′-Sialyllactose and 6′-sialyllactose were purchased from Carbosynth. 2-(Dodecylthiocarbonothioylthio)-2-methylpropionic acid pentafluorophenyl ester was synthesized as previously outlined by Richards and Gibson.28 Soybean agglutinin (SBA) and wheat germ agglutinin (WGA) were purchased from Vector Laboratories. The following reagents were obtained through BEI resources, NIAID, NIH: H1 HA protein from influenza virus, A/California/07/2009 (H1N1)pdm09, recombinant from baculovirus, NR-44074, H2 HA protein from influenza virus, A/Singapore/1/1957 (H2N2), recombinant from baculovirus, NR-2668, H3 HA protein from influenza virus, A/Uruguay/716/2007 (H3N2), recombinant from baculovirus, NR-15168, H4 HA protein from influenza virus, A/mallard/Alberta/455/2015 (H4N6), recombinant from baculovirus, NR-51128, H5 HA protein from influenza virus, A/duck/Hunan/795/2002 (H5N1), recombinant from baculovirus, NR-43739, H7 HA protein from influenza virus, A/Canada/rv444/2004 (H7N3), recombinant from baculovirus, NR-43740, H9 HA protein from influenza virus, A/Hong Kong/33982/2009 (H9N2), recombinant from baculovirus, NR-41792 and H10 HA protein with C-terminal histidine tag from influenza virus, A/harbor seal/Germany/1/2014 (H10N7), recombinant from baculovirus, NR-50172. Clear and black half are 96-well plates that were purchased from Greiner Bio-One. Streptavidin (SA) biosensors were purchased from ForteBio. Lectins and HAs were biotinylated using EZ-Link sulfo-NHS-LC-biotin reagent from Thermo Fisher Scientific using standard procedure (20-fold molar excess of biotin reagent, conjugation performed in phosphate-buffered saline (PBS) buffer and isolated using Amicon Ultra-0.5 mL 3000 MWCO centrifugal filters from Merck Millipore).
Physical and Analytical Methods
1H NMR spectra were recorded on Bruker DPX-300 and DPX-400 spectrometers using deuterated solvents purchased from Sigma-Aldrich. Chemical shifts are reported relative to residual non-deuterated solvent. Size exclusion chromatography (SEC) was carried out in dimethylformamide (DMF) and run on an Agilent 1260 Infinity II-MDS instrument equipped with differential refractive index, dual wavelength UV detectors, viscometry and light scattering detectors. The system was equipped with 2× PLgel mixed D columns (300 × 7.5 mm) and a PLgel 5 μm guard column. The eluent is DMF with 5 mM NH4BF4 additive. Samples were run at 1 mL·min–1 at 50 °C. Poly(methyl methacrylate) (Agilent EasiVials) were used for calibration. Analyte samples were filtered through a nylon filter with 0.22 μm pore size before injection. Experimental molar mass (Mn,SEC) and dispersity (D̵) values of synthesized polymers were determined by conventional calibration using Agilent SEC/SEC software. The calibration range is 500–2,000,000 Da.
Lectin-Induced Aggregation Studies by Absorbance
A stock solution of lectin was made (0.1 mg·mL–1 for SBA and 1 mg·mL–1 for WGA) in 10 mM HEPES buffer with 0.15 M NaCl, 0.1 mM CaCl2, and 0.01 mM MnCl2. A serial dilution (25 μL) was made up in the same buffer in a clear, flat bottom, half area 96-well microtiter plate. GlycoAuNPs (250 μL) were added to each well and incubated at room temperature for 30 min. After 30 min, an absorbance spectrum was recorded from 450 to 700 nm with 10 nm intervals.
Biolayer Interferometry
BLI was carried out on ForteBio Octet Red96 (ForteBio, USA). Assays were performed in black 96-well half area plates. Assays were carried out at 30 °C and agitated at 1000 rpm. SA biosensor tips (ForteBio, USA) were hydrated in milliQ H2O water for at least 10 min prior to use. A stable baseline was established in milliQ water for 1 min. The biosensors were functionalized by loading with 10 μg/mL biotinylated lectin or HA in PBS for 5 min followed by a 1 min equilibration step in 10 mM HEPES with 0.15 M NaCl and 0.1 mM CaCl2 and MnCl2 to remove any unbound protein and to establish a stable baseline. Following protein immobilization, the binding association with galactosylated and sialylated AuNPs was carried out in 10 mM HEPES with 0.15 M NaCl and 0.1 mM CaCl2 and MnCl2 for 10 min followed by dissociation in 10 mM HEPES with 0.15 M NaCl and 0.1 mM CaCl2 and MnCl2 for 10 min.
Synthetic Section
Polymerization of Hydroxyethyl Acrylamide Using PFP-DMP
HEA (0.5 g, 4.34 mmol), pentafluorophenyl 2-(dodecylthiocarbonothioylthio)-2-methylpropionic acid (PFP-DMP) (0.092 g, 0.17 mmol), and ACVA (0.0097 g, 0.034 mmol) were dissolved in 50:50 toluene/methanol (4 mL). Mesitylene (150 μL) was added as an internal reference. An aliquot was taken for NMR analysis in CDCl3. The solution was degassed under N2 for 30 min. The reaction was stirred at 70 °C for 90 min. An aliquot was taken for NMR analysis in MeOD. The reaction was rapidly cooled in liquid nitrogen and precipitated into diethyl ether. The polymer was reprecipitated into diethyl ether from methanol twice to yield a yellow polymer product that was dried under vacuum. 96% conversion by NMR, Mn (theoretical) = 3400 g·mol–1Mn (SEC) = 5800 g·mol–1Mn/Mw (SEC) = 1.16.
Gold Nanoparticle Synthesis
Gold nanoparticles of differing sizes were synthesized by the method developed by Bastús et al.39 A solution of 2.2 mM sodium citrate in Milli-Q water (150 mL) was heated under reflux for 15 min under vigorous stirring. After boiling had commenced, 1 mL of HAuCl4 (25 mM) was injected. The color of the solution changed from yellow to bluish gray and then to soft pink in 10 min. Immediately after the synthesis of the Au seeds and in the same reaction vessel, the reaction was cooled until the temperature of the solution reached 90 °C. Then, 1 mL of HAuCl4 solution (25 mM) was injected. After 30 min, the reaction was finished. This process was repeated twice. After that, the sample was diluted by extracting 55 mL of the sample and adding 53 mL of MilliQ water and 2 mL of 60 mM sodium citrate. This solution was then used as a seed solution, and the process was repeated a further 7 times. Generations 2, 5, and 8 were used for this study (Table 2).
Table 2. Nanoparticles Synthesized and Characterization.
| diameter
(nm) |
|||||
|---|---|---|---|---|---|
| particle | generationa | λSPRb | ASPR/A450b | UV–visc | DLSd |
| Au30 | 2 | 523 | 1.84 | 30 | 28.2 ± 0.9 |
| Au50 | 5 | 531 | 2.00 | 50 | 48.8 ± 0.4 |
| Au70 | 7 | 540 | 2.05 | 68 | 72.5 ± 1.2 |
Generation of nanoparticles used from the seeding synthetic methodology (see Figure S1).
Maximum absorption wavelength from the surface plasmon resonance band of the particles and characteristic ratio were used for diameter estimation.
Estimated using the method of Haiss et al.(41) from UV–vis.
Diameter from DLS ± standard error from three measurements.
Results and Discussion
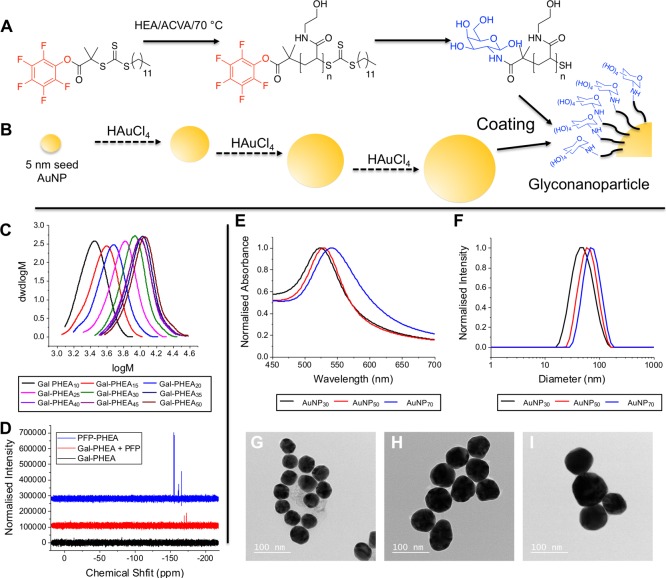
To access multivalent glycosylated gold nanoparticles, RAFT polymerization was employed to synthesize telechelic poly(N-hydroxyethyl acrylamide) (PHEA) ligands suitable for both nanoparticle immobilization, and capture of glycans via a terminal pentafluoroethyl ester (PFP),28,30,31,40 (Figure 1A). A range of degrees of polymerization (DP) was targeted from DP10–DP50 by varying the [M]/[CTA] ratio. The polymers were characterized by SEC confirming narrow dispersity (Figure 1C and Table 1). 2-Deoxy-2-amino galactosamine was conjugated to the ω-terminal PFP, which also removes the dithiocarbonate to reveal a thiol for subsequent AuNP conjugation. 19F NMR (Figure 1D) was used to confirm displacement of the PFP group. Note, GalNH2 was chosen as the glycan to conduct initial screening, before progressing to the sialic acid particles (vide infra).
Figure 1.
Synthetic strategy and characterization of polymers and particles. (A) Synthesis of galactose terminal PHEA; (B) step growth process for the synthesis of gold nanoparticles; (C) SEC of Gal-PHEAs; (D) 19F NMR showing displacement of the pentafluorophenyl unit; (E) UV–vis spectroscopy of uncoated AuNPs; (F) DLS of uncoated AuNPs; (G–I) TEM of uncoated AuNPs. Scale bar = 100 nm.
Table 1. Polymers Prepared in This Study.
| polymer | [M]/[CTA]a | conversion (%)b | Mn(NMR) (g·mol–1)b | Mn(SEC) (g·mol–1)c | Mw/Mnc |
| Gal-PHEA10 | 10 | 96 | 1600 | 2600 | 1.12 |
| Gal-PHEA15 | 15 | 99 | 2200 | 3300 | 1.16 |
| Gal-PHEA20 | 20 | 97 | 2800 | 4200 | 1.16 |
| Gal-PHEA25 | 25 | 98 | 3600 | 5800 | 1.16 |
| Gal-PHEA30 | 30 | 95 | 3800 | 7700 | 1.13 |
| Gal-PHEA35 | 35 | 97 | 4600 | 9500 | 1.14 |
| Gal-PHEA40 | 40 | 98 | 5000 | 8900 | 1.15 |
| Gal-PHEA45 | 45 | 94 | 5700 | 9200 | 1.15 |
| Gal-PHEA50 | 50 | 92 | 5800 | 10,300 | 1.15 |
Feed ratio of monomer to chain-transfer agent.
Determined by 1H NMR.
Determined by SEC in dimethylformamide using poly(methyl methacrylate) standards. Mw, weight average molecular weight; Mn, number average molecular weight.
To access a range of gold nanoparticles of defined size, the kinetically controlled seeded growth strategy based on the Turkevich/Frens method via the reduction of HAuCl4 by sodium citrate was used.39 This involved a growth process leading to the enlargement of presynthesized 10 nm gold seeds via the surface-catalyzed reduction of Au3+ by sodium citrate (Figure 1B). This was characterized by UV–vis spectroscopy (Figure 1E), DLS (Figure 1F), and transmission electron microscopy (TEM) (Figure 1G–I). The three different nanoparticle sizes were coated with the nine different polymers to generate a diverse library of 27 multivalent glyconanoparticles, this convergent strategy maximized structural diversity. DLS measurements showed an increase in hydrodynamic diameter, along with a red-shift in the SPR peak (λmax), both consistent with polymer coating. X-ray photoelectron spectroscopy (XPS) was used to confirm polymer coating on AuNPs by increased N 1s (from amide) as well as C 1s signals (Supporting Information, Tables S1 and S2, Figures S2–S5).
To enable protein binding studies, it was essential to first screen the library for colloidal stability in the buffer to prevent false positives in aggregation screens. Both particle size and polymer length impact this, and it is a crucial step in any binding assay. The galactose functional particles were screened for aggregation stability by observing the increase in Abs700. A NaCl gradient (0–1 M) as well as HEPES buffer were tested (full data set is in the Supporting Information, Figure S6). From this, it was clear that for all particle sizes, ligands with a DP above 25 were essential to obtain colloidally stable particles, in line with the previous reports.28,42 From this screen, it was decided to study protein binding using particles coated with polymers DP25–DP50 to avoid false positives due to buffer-induced aggregation. To screen for binding, the model lectin, SBA was used to validate the methods and to establish which particle structural requirements gave the strongest outputs, Figure 2.
Figure 2.
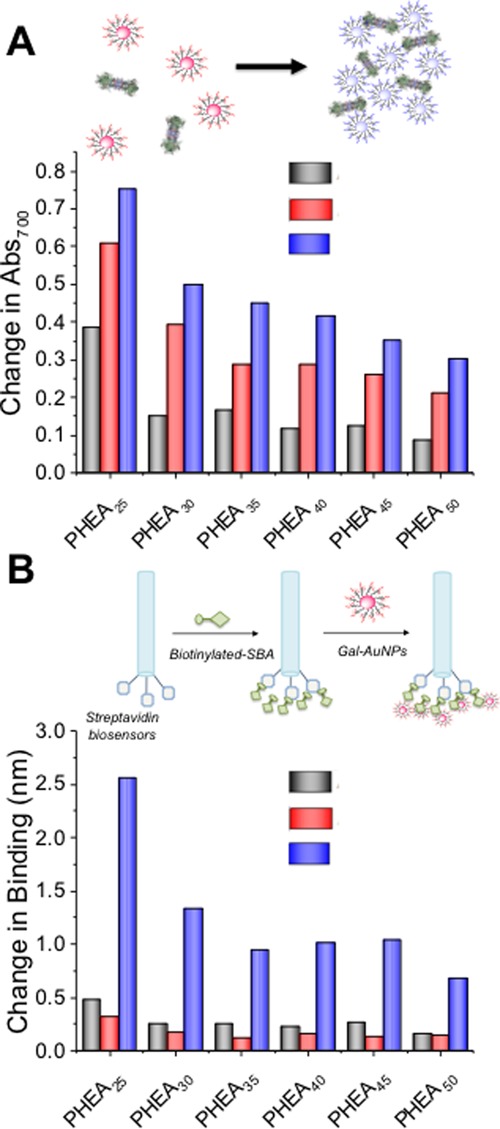
Galactosylated particles and SBA binding. (A) Aggregation of gold particles leading to red-blue color shift due to SPR band coupling. Change in Abs700 using 500 nM SBA as a function of coating and core size. Full binding curves from 500 to 0 nM are in the Supporting Information, Figure S7; (B) BLI schematic, showing capture of SBA and subsequent binding to AuNPs to generate signal. Total binding (Δmax) as a function of coating and core size. Full binding curves are in the Supporting Information, Figure S8.
Figure 2A shows the outputs of this aggregation screen as a function of polymer ligand length and nanoparticle size. Full dose-dependent curves are in the Supporting Information (Figure S6). For all polymer lengths, the largest changes were seen for the shortest polymer chain length (DP25) with a step-wise decrease as polymer length increases, and the biggest (70 nm) particles gave the largest outputs. Although the aggregation assay used above provides a convenient output of binding, it cannot rule out the case where lectins bind but fail to agglutinate. For example, the longer polymer linkers are likely able to bind to SBA, but also provide a steric block against agglutination, highlighting the need for tuning of these parameters. Therefore, a second assay using BLI was developed.43 In short, biotinylated SBA was immobilized onto SA functional BLI sensors (ForteBio) (Figure 2B) and the nanoparticles exposed to this. As BLI is mass sensitive, individual glycans do not give a strong signal, but when coupled in this nanoparticle format, larger signals are obtained. In addition, cluster glycoside enhancement is achieved and is a key benefit of this approach. The total BLI signal as a function of particle parameters is shown in Figure 2D. The shorter polymers at all particle sizes gave the largest outputs; however, there was little binding observed to the SBA when DP50 polymers were used. This suggests that the lower density of longer polymers is reducing overall affinity and multivalent enhancement. Brewer and co-workers,44,45 and Godula and Bertozzi46 observed valency-dependent binding of SBA to GalNAc-functionalized mucins/mucin-like materials. Furthermore, Gibson and co-workers have previously shown density-dependent binding of SBA to heterogenous AuNPs.31 Larger particles gave more signal, but it should be noted that BLI is a mass-weighted analysis method and hence only qualitative trends can be compared for nanoparticle size.
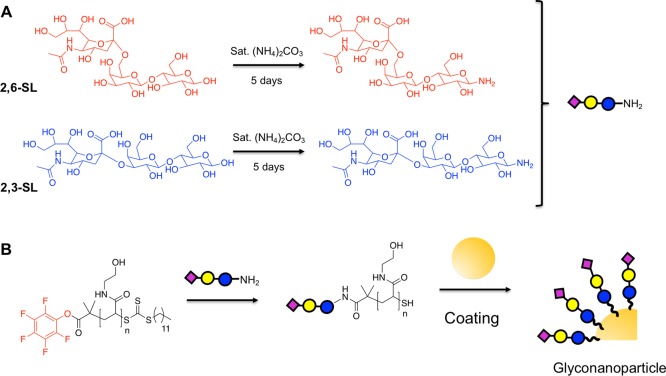
Guided by the above data, using galactose functional nanoparticles, the primary aim of studying multivalent sialic acid particles could be investigated on the optimized scaffold. 1-Deoxy, 1-amino, 2,3- and 2,6-sialyllactose were synthesized by the single-step Kochetkov amination method47 (Figure 3A, with the amination confirmed by mass spectrometry). Using the same procedure as GalNH2, the sialyllactosamines (SLs) were conjugated to DP25 PHEA and subsequently used to coat 70 nm AuNPs. This single polymer/particle combination was chosen based on the above screening (Figure 2), which ensured that particles that generate the largest signals with consistent grafting densities were used.
Figure 3.
Synthesis of sialylated gold nanoparticles. (A) Amination of 2,3- and 2,6- sialyllactoses using Kochetkov method; (B) reaction of DP25 PFP-PHEA with sialyllactoamines and subsequent immobilization of the polymers onto 70 nm gold nanoparticles.
These glycoparticles were first tested against WGA, a lectin known to have some affinity to sialic acids.48 Using UV–vis to monitor aggregation, it appeared that both 2,3- and 2,6-sialyllactosamines had similar affinities toward WGA (Figure 4A), which was not expected. However, parallel DLS investigations (Figure 4B) showed 2,6-sialyllactosamine to cluster faster and to give larger cluster sizes, which is consistent with it having a stronger interaction than 2,3-sialyllactosamine. BLI analysis against WGA immobilized on a surface showed greater specificity for the 2,6-sialyllactosamine compared to 2,3-sialyllactosamine, which is in line with the previous reports. This set of analyses shows that the colorimetric changes of glyconanoparticles are useful for comparisons of affinity, but more detailed methods (such as the BLI) are required, especially if unknown/new interactions are being evaluated.
Figure 4.
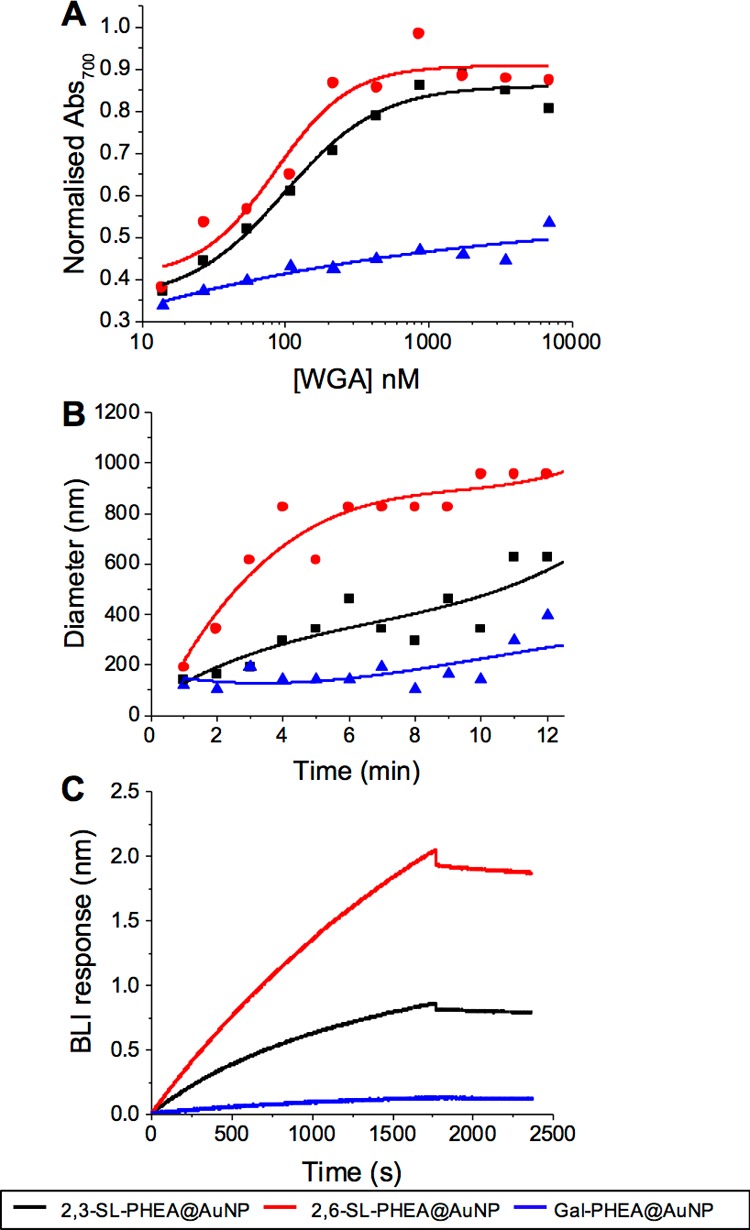
Interaction of SL-functionalized particles: 2,3-SL-PHEA@AuNP (black ■), 2,6-SL-PHEA@AuNP (red ●), and Gal-PHEA@AuNP (blue ▲) with WGA by (A) UV–vis; (B) DLS; and (C) BLI.
The above data shows that DP25 PHEA ligands on 70 nm particles give strong binding, and that the amidation/conjugation methodology enables installation of sialic acid isomers with retention of their biological binding specificities and affinities. The particles were then taken forward into a screen against a panel of HAs. A single nanoparticle/polymer length combination was used to ensure that grafting density differences did not affect the observed outcomes, where glycan density display is likely to play a significant role.
Influenza A can be categorized by the different subtypes of the proteins present of which there are at least 18 different HA subtypes named H1 to H18.2 The panel of HAs used were from the five strains with human hosts: H1 from A/California/07/2009 (H1N1)pdm09, H2 from A/Singapore/1/1957 (H2N2), H3 from A/Uruguay/716/2007 (H3N2), H7 from, A/Canada/rv444/2004 (H7N3), and H9 from A/Hong Kong/33982/2009 (H9N2); and two from avian hosts: H4 from A/mallard/Alberta/455/2015 (H4N6) and H5 from A/duck/Hunan/795/2002 (H5N1); and another zoonotic strain, H10 from A/harbor seal/Germany/1/2014 (H10N7) (seal host), and it enables the study of pathogenic processes without the need for intact viral particles. HAs were immobilized onto the BLI sensors using biotin/SA (as above). BLI has been widely used to evaluate hemagglutinin/neuraminidase function using isolated proteins and the whole virus.49−51Figure 5A shows example BLI curves of 2,6-sialyllactosamine nanoparticles against the HA panel and Figure 5B shows BLI curves of 2,3-sialyllactosamine nanoparticles. There was a difference in response in terms of total mass captured and the rate of binding, reiterating the range of binding affinities shown by HAs from different pathogenic strains (low pathogenic avian strains) when presented in a multivalent format. To enable comparison, Figure 5C shows the maximal binding of the particles to the HAs for both 2,3- and 2,6-sialyllactosamine-functionalized AuNPs. A key feature here is that although many of the avian HAs are characterized as being α2,3-selective based on previous studies using monovalent compounds, there is clearly cross-affinity in most cases using our multivalent system. This shows the importance of this study, as it shows that glycan specificity can change when in multivalent format because of the enhancement of weak interactions, as well as stronger ones. H9 in particular showed significant binding to both 2,3-sialyllactosamine and 2,6-sialyllactosamine AuNPs, indicating that it can bind to both human and avian sialic acids, which is consistent with the virus being of avian origins infecting a human host. This crossover in binding has also been observed by Godula and co-workers,24 where significant binding of H1N1 (A/Puerto Rico/8/34 which transmitted into humans) was noted toward both α2,6 and α2,3 sialylated polymer microarrays. In contrast, more discrete binding was observed with H3N2 (A/Aichi/2/68) which bound only to α2,6 but not to α2,3 sialylated polymer arrays (note that the specific strains were not the same as used in this study). Furthermore, McCauley and co-workers have investigated the mammalian transmissibility of many avian and other zoonotic influenza strains including H7N9,49 H9N2,50 and H10N852 by looking at the binding of sialylated polymers to viruses. In particular, a panel of H9N2 viruses from different origins were assessed for their binding to α2,6 and α2,3 sialylated polymer-functionalized BLI sensors.50 Wide-ranging binding variability was noted to different H9N2 viruses, with the H9N2 (A/Hong Kong/33982/2009) virus (the same H9 used in this study) showing greater binding toward α2,6 sialylated polymers over α2,3 sialylated polymers, which is in agreement with our results here, supporting the approach taken (dose-dependent BLI binding curves for H1 from A/California/07/2009 (H1N1)pdm09, H7 from A/Canada/rv444/2004 (H7N3), and H9 from H9N2 (A/Hong Kong/33982/2009) in the Supporting Information, Figures S12–S14). The H7N9 strain has been reported to retain its avian affinity preference even with mutations to target human receptors, which we also observed using this nanoparticle system.53 To visualize these differences, Figure 5D plots the relative binding preferences, defined as the maximal binding of 2,6-sialyllactosamine nanoparticles, minus the maximal binding of 2,3-sialyllactosamine to each hemagglutinin. As the same coating and same nanoparticle core was used in each case, this enabled visualization of the differences in total particle captured to the haemagglutinin. These results show that our nanoparticle system generates sufficient signal amplification to enable the evaluation of glycans for binding haemagglutinins and to unravel the cross-affinity due to the multivalent presentation. It is important to note that these results may be specific to the sialyllactose ligands being employed here, which are readily available compared to, for example, complex branched glycans. It does show that, however, in multivalent format both the extent of binding and relative binding between the isomers varies between the hemagglutinins and may offer routes to multiplexed sensing and discrimination.
Figure 5.
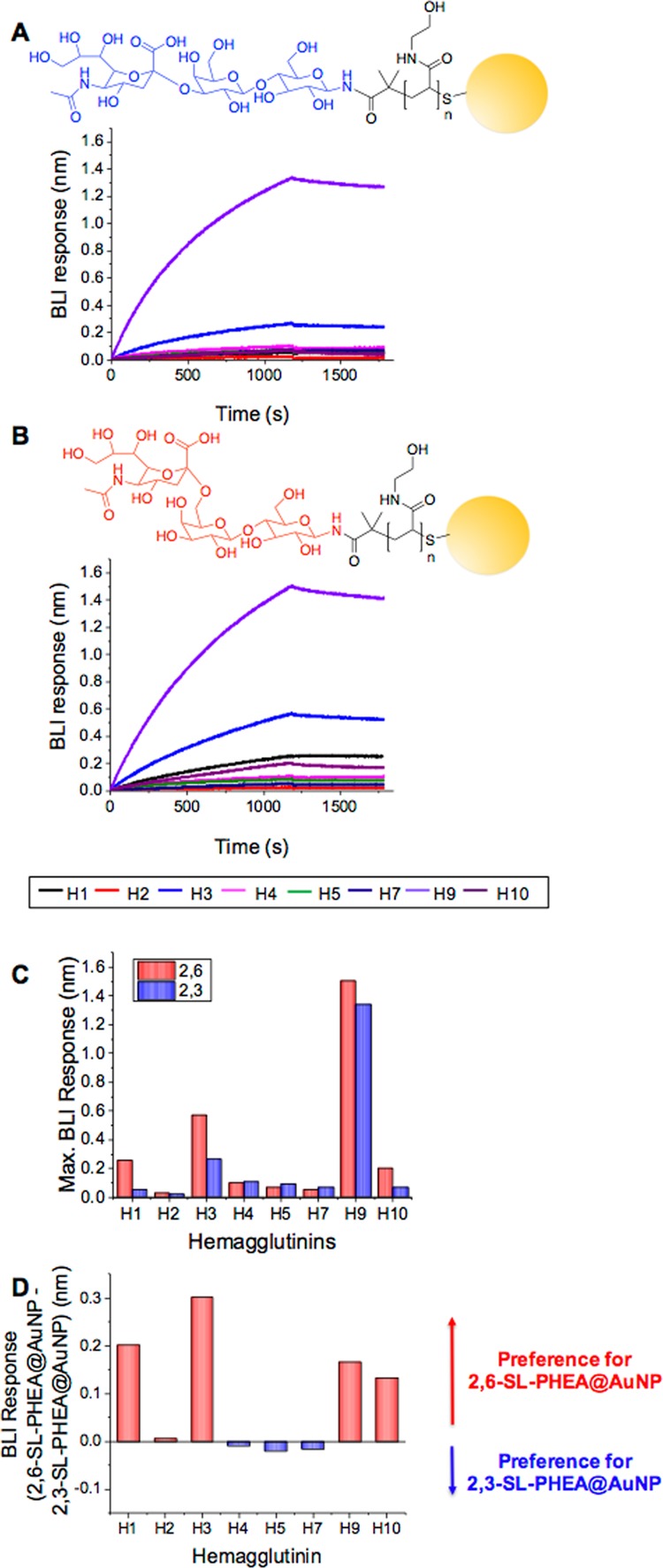
Interaction of SL-functionalized nanoparticles with a panel of HAs using BLI. (A) 2,3-SL-PHEA@AuNP and (B) 2,6-SL-PHEA@AuNP BLI binding curves to HA panel (H1 from A/California/07/2009 (H1N1)pdm09, H2 from A/Singapore/1/1957 (H2N2), H3 from A/Uruguay/716/2007 (H3N2), H4 from A/mallard/Alberta/455/2015 (H4N6), H5 from A/duck/Hunan/795/2002 (H5N1), H7 from, A/Canada/rv444/2004 (H7N3), NR-43740, H9 from A/Hong Kong/33982/2009 (H9N2), and H10 from A/harbor seal/Germany/1/2014 (H10N7)); (C) comparison of the maximal binding of 2,3-SL-PHEA@AuNP and 2,6-SL-PHEA@AuNP to the HA panel; (D) difference (subtraction) between maximal binding to 2,6-SL-PHEA@AuNP and 2,3-SL-PHEA@AuNP to visualize relative binding preferences.
Conclusions
Here, we have developed a multivalent nanoparticle approach to investigate the affinity of sialic acid isomers against a panel of influenza A HAs to map their affinity. This allows for the probing of specificity and affinity trends while benefitting from signal amplification due to particle size. The multivalent nanoparticles were synthesized using polymeric ligands bearing a glycan capture (pentafluorophenyl ester) and a gold immobilization (thiol) termini. Initial screening of a library of 27 nanoparticles, varying in ligand length and core diameter, enabled identification of the optimum multivalent probe, using a model lectin and complementary UV–vis, DLS, and BLI analysis. 2,3- and 2,6-sialyllactosamine were conjugated to these particles via a Kochetkov amination step, which were then interrogated for their binding against a panel of influenza A HAs including three mammalian strains, four avian strains, and a zoonotic influenza strain. As expected, 2,3-sialyllactosamine had strong affinity toward avian strains and 2,6-sialyllactosamine against mammalian. However, the analysis also revealed that in multivalent format, there was significant crossover in affinities. All avian influenzas showed affinity toward the multivalent 2,6-sialyllactosamine particles suggesting the potential for crossover due to multivalency. It is important to note that this study does not predict zoonosis potential, but rather highlights that multivalent nanoparticle presentation affects the affinity/selectivity trends compared to monovalent compounds. The identification of off-specific binding profiles is crucial to help develop nanomaterials for biosensing and monitoring of emerging infectious diseases.
Acknowledgments
M.I.G. holds an ERC starting grant (CRYOMAT 638661). The Royal Society are also thanked for funding the cryo-microscopes used in this study. BBSRC/Innovate are thanked for funding the Specialty Glycans project BB/M02878X/1. The BBSRC-funded MIBTP program (BB/M01116X/1) and Iceni Diagnostics ltd are thanked for a studentship for A.N.B. The Warwick Polymer Research Technology Platform is acknowledged for the SEC analysis. Laura Wilkins (UoW) is thanked for the TEM analysis. The following reagents were obtained through BEI Resources, NIAID, NIH: H1 HA protein from influenza virus, A/California/07/2009 (H1N1)pdm09, recombinant from baculovirus, NR-44074, H2 HA protein from influenza virus, A/Singapore/1/1957 (H2N2), recombinant from baculovirus, NR-2668, H3 HA protein from influenza virus, A/Uruguay/716/2007 (H3N2), recombinant from baculovirus, NR-15168, H4 HA protein from influenza virus, A/mallard/Alberta/455/2015 (H4N6), recombinant from baculovirus, NR-51128, H5 HA protein from influenza virus, A/duck/Hunan/795/2002 (H5N1), recombinant from baculovirus, NR-43739, H7 HA protein from influenza virus, A/Canada/rv444/2004 (H7N3), recombinant from baculovirus, NR-43740, H9 HA protein from influenza virus, A/Hong Kong/33982/2009 (H9N2), recombinant from baculovirus, NR-41792 and H10 HA protein with C-terminal histidine tag from influenza virus, A/harbor seal/Germany/1/2014 (H10N7), recombinant from baculovirus, NR-50172.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biomac.0c00179.
Additional nanoparticle characterization (XPS and DLS), full UV–visible spectroscopy curves for the lectin binding assays, and underpinning BLI binding curves (PDF)
The authors declare no competing financial interest.
Notes
The research data supporting this publication can be found at http://wrap.warwick.ac.uk.
Supplementary Material
References
- Iuliano A. D.; Roguski K. M.; Chang H. H.; Muscatello D. J.; Palekar R.; Tempia S.; Cohen C.; Gran J. M.; Schanzer D.; Cowling B. J.; Wu P.; Kyncl J.; Ang L. W.; Park M.; Redlberger-Fritz M.; Yu H.; Espenhain L.; Krishnan A.; Emukule G.; van Asten L.; Pereira da Silva S.; Aungkulanon S.; Buchholz U.; Widdowson M.-A.; Bresee J. S.; Azziz-Baumgartner E.; Cheng P.-Y.; Dawood F.; Foppa I.; Olsen S.; Haber M.; Jeffers C.; MacIntyre C. R.; Newall A. T.; Wood J. G.; Kundi M.; Popow-Kraupp T.; Ahmed M.; Rahman M.; Marinho F.; Sotomayor Proschle C. V.; Vergara Mallegas N.; Luzhao F.; Sa L.; Barbosa-Ramírez J.; Sanchez D. M.; Gomez L. A.; Vargas X. B.; Acosta Herrera a.; Llanés M. J.; Fischer T. K.; Krause T. G.; Mølbak K.; Nielsen J.; Trebbien R.; Bruno A.; Ojeda J.; Ramos H.; an der Heiden M.; del Carmen Castillo Signor L.; Serrano C. E.; Bhardwaj R.; Chadha M.; Narayan V.; Kosen S.; Bromberg M.; Glatman-Freedman A.; Kaufman Z.; Arima Y.; Oishi K.; Chaves S.; Nyawanda B.; Al-Jarallah R. A.; Kuri-Morales P. A.; Matus C. R.; Corona M. E. J.; Burmaa A.; Darmaa O.; Obtel M.; Cherkaoui I.; van den Wijngaard C. C.; van der Hoek W.; Baker M.; Bandaranayake D.; Bissielo A.; Huang S.; Lopez L.; Newbern C.; Flem E.; Grøneng G. M.; Hauge S.; de Cosío F. G.; de Moltó Y.; Castillo L. M.; Cabello M. A.; von Horoch M.; Medina Osis J.; Machado A.; Nunes B.; Rodrigues A. P.; Rodrigues E.; Calomfirescu C.; Lupulescu E.; Popescu R.; Popovici O.; Bogdanovic D.; Kostic M.; Lazarevic K.; Milosevic Z.; Tiodorovic B.; Chen M.; Cutter J.; Lee V.; Lin R.; Ma S.; Cohen A. L.; Treurnicht F.; Kim W. J.; Delgado-Sanz C.; de mateo Ontañón S.; Larrauri A.; León I. L.; Vallejo F.; Born R.; Junker C.; Koch D.; Chuang J.-H.; Huang W.-T.; Kuo H.-W.; Tsai Y.-C.; Bundhamcharoen K.; Chittaganpitch M.; Green H. K.; Pebody R.; Goñi N.; Chiparelli H.; Brammer L.; Mustaquim D. Estimates of Global Seasonal Influenza-Associated Respiratory Mortality: A Modelling Study. Lancet 2018, 391, 1285–1300. 10.1016/s0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Heath Organisation . Fact Sheet—Influenza (Seasonal). https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed March 10, 2020).
- Rogers G. N.; D’Souza B. L. Receptor Binding Properties of Human and Animal H1 Influenza Virus Isolates. Virology 1989, 173, 317–322. 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- Connor R. J.; Kawaoka Y.; Webster R. G.; Paulson J. C. Receptor Specificity in Human, Avian, and Equine H2 and H3 Influenza Virus Isolates. Virology 1994, 205, 17–23. 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- Webster R. G.; Laver W. G.; Air G. M.; Schild G. C. Molecular Mechanisms of Variation in Influenza Viruses. Nature 1982, 296, 115–121. 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- Gamblin S. J.; Haire L. F.; Russell R. J.; Stevens D. J.; Xiao B.; Ha Y.; Vasisht N.; Steinhauer D. A.; Daniels R. S.; Elliot A.; Wiley D. C.; Skehel J. J. The Structure and Receptor Binding Properties of the 1918 Influenza Hemagglutinin. Science 2004, 303, 1838–1842. 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- Rogers G. N.; Paulson J. C.; Daniels R. S.; Skehel J. J.; Wilson I. A.; Wiley D. C. Single Amino Acid Substitutions in Influenza Haemagglutinin Change Receptor Binding Specificity. Nature 1983, 304, 76–78. 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- World Health Organisation . Fact Sheet—Influenza (Avian and other zoonotic). https://www.who.int/news-room/fact-sheets/detail/influenza-(avian-and-other-zoonotic) (accessed March 10, 2020).
- Broszeit F.; Tzarum N.; Zhu X.; Nemanichvili N.; Eggink D.; Leenders T.; Li Z.; Liu L.; Wolfert M. A.; Papanikolaou A.; Martínez-Romero C.; Gagarinov I. A.; Yu W.; García-Sastre A.; Wennekes T.; Okamatsu M.; Verheije M. H.; Wilson I. A.; Boons G.-J.; de Vries R. P. N-Glycolylneuraminic Acid as a Receptor for Influenza A Viruses. Cell Rep. 2019, 27, 3284–3294.e6. 10.1016/j.celrep.2019.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd-Leotis L.; Jia N.; Dutta S.; Trost J. F.; Gao C.; Cummings S. F.; Braulke T.; Müller-Loennies S.; Heimburg-Molinaro J.; Steinhauer D. A.; Cummings R. D. Influenza Binds Phosphorylated Glycans from Human Lung. Sci. Adv. 2019, 5, eaav2554 10.1126/sciadv.aav2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S.; Lasanajak Y.; Smith D. F.; Cummings R. D.; Air G. M. Glycan Array Analysis of Influenza H1N1 Binding and Release. Cancer Biomarkers 2014, 14, 43–53. 10.3233/cbm-130376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter N. K.; Bednarski M. D.; Wurzburg B. A.; Hanson J. E.; Whitesides G. M.; Skehel J. J.; Wiley D. C. Hemagglutinins from Two Influenza Virus Variants Bind to Sialic Acid Derivatives with Millimolar Dissociation Constants: A 500-MHz Proton Nuclear Magnetic Resonance Study. Biochemistry 1989, 28, 8388–8396. 10.1021/bi00447a018. [DOI] [PubMed] [Google Scholar]
- Weis W.; Brown J. H.; Cusack S.; Paulson J. C.; Skehel J. J.; Wiley D. C. Structure of the Influenza Virus Haemagglutinin Complexed with Its Receptor, Sialic Acid. Nature 1988, 333, 426–431. 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- Lundquist J. J.; Toone E. J. The Cluster Glycoside Effect. Chem. Rev. 2002, 102, 555–578. 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]
- Gou Y.; Richards S.-J.; Haddleton D. M.; Gibson M. I. Investigation of Glycopolymer-Lectin Interactions Using QCM-d: Comparison of Surface Binding with Inhibitory Activity. Polym. Chem. 2012, 3, 1634–1640. 10.1039/c2py20140b. [DOI] [Google Scholar]
- Gou Y.; Geng J.; Richards S.-J.; Burns J.; Remzi Becer C.; Haddleton D. M. A Detailed Study on Understanding Glycopolymer Library and Con A Interactions. J. Polym. Sci., Part A: Polym. Chem. 2013, 51, 2588–2597. 10.1002/pola.26646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Chen G.; Stenzel M. H. Synthesis and Lectin Recognition of Glyco Star Polymers Prepared by “Clicking” Thiocarbohydrates onto a Reactive Scaffold. Macromolecules 2010, 43, 8109–8114. 10.1021/ma100919x. [DOI] [Google Scholar]
- Ponader D.; Wojcik F.; Beceren-Braun F.; Dernedde J.; Hartmann L. Sequence-Defined Glycopolymer Segments Presenting Mannose: Synthesis and Lectin Binding Affinity. Biomacromolecules 2012, 13, 1845–1852. 10.1021/bm300331z. [DOI] [PubMed] [Google Scholar]
- Baradel N.; Fort S.; Halila S.; Badi N.; Lutz J.-F. Synthesis of Single-Chain Sugar Arrays. Angew. Chem., Int. Ed. 2013, 52, 2335–2339. 10.1002/anie.201209052. [DOI] [PubMed] [Google Scholar]
- Mammen M.; Dahmann G.; Whitesides G. M. Effective Inhibitors of Hemagglutination by Influenza Virus Synthesized from Polymers Having Active Ester Groups. Insight into Mechanism of Inhibition. J. Med. Chem. 1995, 38, 4179–4190. 10.1021/jm00021a007. [DOI] [PubMed] [Google Scholar]
- Sigal G. B.; Mammen M.; Dahmann G.; Whitesides G. M. Polyacrylamides Bearing Pendant α-Sialoside Groups Strongly Inhibit Agglutination of Erythrocytes by Influenza Virus: The Strong Inhibition Reflects Enhanced Binding through Cooperative Polyvalent Interactions. J. Am. Chem. Soc. 1996, 118, 3789–3800. 10.1021/ja953729u. [DOI] [Google Scholar]
- Nagao M.; Fujiwara Y.; Matsubara T.; Hoshino Y.; Sato T.; Miura Y. Design of Glycopolymers Carrying Sialyl Oligosaccharides for Controlling the Interaction with the Influenza Virus. Biomacromolecules 2017, 18, 4385–4392. 10.1021/acs.biomac.7b01426. [DOI] [PubMed] [Google Scholar]
- Lu W.; Du W.; Somovilla V. J.; Yu G.; Haksar D.; de Vries E.; Boons G.-J.; de Vries R. P.; de Haan C. A. M.; Pieters R. J. Enhanced Inhibition of Influenza A Virus Adhesion by Di- and Trivalent Hemagglutinin Inhibitors. J. Med. Chem. 2019, 62, 6398–6404. 10.1021/acs.jmedchem.9b00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. L.; Cohen M.; Fisher C. J.; Schooley R. T.; Gagneux P.; Godula K. Determination of Receptor Specificities for Whole Influenza Viruses Using Multivalent Glycan Arrays. Chem. Commun. 2015, 51, 5326–5329. 10.1039/c4cc08613a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandlow V.; Liese S.; Lauster D.; Ludwig K.; Netz R. R.; Herrmann A.; Seitz O. Spatial Screening of Hemagglutinin on Influenza A Virus Particles: Sialyl-LacNAc Displays on DNA and PEG Scaffolds Reveal the Requirements for Bivalency Enhanced Interactions with Weak Monovalent Binders. J. Am. Chem. Soc. 2017, 139, 16389–16397. 10.1021/jacs.7b09967. [DOI] [PubMed] [Google Scholar]
- Peng W.; de Vries R. P.; Grant O. C.; Thompson A. J.; McBride R.; Tsogtbaatar B.; Lee P. S.; Razi N.; Wilson I. A.; Woods R. J.; Paulson J. C. Recent H3N2 Viruses Have Evolved Specificity for Extended, Branched Human-Type Receptors, Conferring Potential for Increased Avidity. Cell Host Microbe 2017, 21, 23–34. 10.1016/j.chom.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S.; Lauster D.; Bardua M.; Ludwig K.; Angioletti-Uberti S.; Popp N.; Hoffmann U.; Paulus F.; Budt M.; Stadtmüller M.; Wolff T.; Hamann A.; Böttcher C.; Herrmann A.; Haag R. Linear Polysialoside Outperforms Dendritic Analogs for Inhibition of Influenza Virus Infection in Vitro and in Vivo. Biomaterials 2017, 138, 22–34. 10.1016/j.biomaterials.2017.05.028. [DOI] [PubMed] [Google Scholar]
- Richards S.-J.; Gibson M. I. Optimization of the Polymer Coating for Glycosylated Gold Nanoparticle Biosensors to Ensure Stability and Rapid Optical Readouts. ACS Macro Lett. 2014, 3, 1004–1008. 10.1021/mz5004882. [DOI] [PubMed] [Google Scholar]
- Richards S.-J.; Otten L.; Gibson M. I.; Besra G. S.; Gibson M. I. Glycosylated Gold Nanoparticle Libraries for Label-Free Multiplexed Lectin Biosensing. J. Mater. Chem. B 2016, 4, 3046–3053. 10.1039/c5tb01994j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S.-J.; Biggs C. I.; Gibson M. I.. Multivalent Glycopolymer-Coated Gold Nanoparticles. Macro-Glycoligands; Methods in Molecular Biology; Springer, 2016; Vol. 1367, pp 169–179. [DOI] [PubMed] [Google Scholar]
- Otten L.; Vlachou D.; Richards S.-J.; Gibson M. I. Glycan Heterogeneity on Gold Nanoparticles Increases Lectin Discrimination Capacity in Label-Free Multiplexed Bioassays. Analyst 2016, 141, 4305–4312. 10.1039/c6an00549g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín M. J.; Rashid A.; Rejzek M.; Fairhurst S. A.; Wharton S. A.; Martin S. R.; McCauley J. W.; Wileman T.; Field R. A.; Russell D. A. Glyconanoparticles for the Plasmonic Detection and Discrimination between Human and Avian Influenza Virus. Org. Biomol. Chem. 2013, 11, 7101. 10.1039/c3ob41703d. [DOI] [PubMed] [Google Scholar]
- Poonthiyil V.; Nagesh P. T.; Husain M.; Golovko V. B.; Fairbanks A. J. Gold Nanoparticles Decorated with Sialic Acid Terminated Bi-Antennary N-Glycans for the Detection of Influenza Virus at Nanomolar Concentrations. ChemistryOpen 2015, 4, 708–716. 10.1002/open.201500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.; Schepens B.; Nuhn L.; Saelens X.; Schotsaert M.; Callewaert N.; De Rycke R.; Zhang Q.; Moins S.; Benali S.; Mespouille L.; Hoogenboom R.; De Geest B. G. Influenza-Binding Sialylated Polymer Coated Gold Nanoparticles Prepared via RAFT Polymerization and Reductive Amination. Chem. Commun. 2016, 52, 3352–3355. 10.1039/c6cc00501b. [DOI] [PubMed] [Google Scholar]
- Papp I.; Sieben C.; Ludwig K.; Roskamp M.; Böttcher C.; Schlecht S.; Herrmann A.; Haag R. Inhibition of Influenza Virus Infection by Multivalent Sialic-Acid- Functionalized Gold Nanoparticles. Small 2010, 6, 2900–2906. 10.1002/smll.201001349. [DOI] [PubMed] [Google Scholar]
- Richards S.-J.; Fullam E.; Besra G. S.; Gibson M. I. Discrimination between Bacterial Phenotypes Using Glyco-Nanoparticles and the Impact of Polymer Coating on Detection Readouts. J. Mater. Chem. B 2014, 2, 1490–1498. 10.1039/c3tb21821j. [DOI] [PubMed] [Google Scholar]
- Di Iorio D.; Verheijden M. L.; Van Der Vries E.; Jonkheijm P.; Huskens J. Weak Multivalent Binding of Influenza Hemagglutinin Nanoparticles at a Sialoglycan-Functionalized Supported Lipid Bilayer. ACS Nano 2019, 13, 3413–3423. 10.1021/acsnano.8b09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S.-J.; Na D. H.; Kwak J. H.; Douaisi M.; Zhang F.; Park E. J.; Park J.-H.; Youn H.; Song C.-S.; Kane R. S.; Dordick J. S.; Lee K. B.; Linhardt R. J. Nanostructured Glycan Architecture Is Important in the Inhibition of Influenza A Virus Infection. Nat. Nanotechnol. 2017, 12, 48–54. 10.1038/nnano.2016.181. [DOI] [PubMed] [Google Scholar]
- Bastús N. G.; Comenge J.; Puntes V. Kinetically Controlled Seeded Growth Synthesis of Citrate-Stabilized Gold Nanoparticles of up to 200 Nm: Size Focusing versus Ostwald Ripening. Langmuir 2011, 27, 11098–11105. 10.1021/la201938u. [DOI] [PubMed] [Google Scholar]
- Richards S.-J.; Otten L.; Gibson M. I. Glycosylated Gold Nanoparticle Libraries for Label-Free Multiplexed Lectin Biosensing. J. Mater. Chem. B 2016, 4, 3046–3053. 10.1039/c5tb01994j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiss W.; Thanh N. T. K.; Aveyard J.; Fernig D. G. Determination of Size and Concentration of Gold Nanoparticles from UV–Vis Spectra. Anal. Chem. 2007, 79, 4215–4221. 10.1021/ac0702084. [DOI] [PubMed] [Google Scholar]
- Won S.; Richards S.-J.; Walker M.; Gibson M. I. Externally Controllable Glycan Presentation on Nanoparticle Surfaces to Modulate Lectin Recognition. Nanoscale Horiz. 2017, 2, 106–109. 10.1039/c6nh00202a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen R. L. Strategies Using Bio-Layer Interferometry Biosensor Technology for Vaccine Research and Development. Biosensors 2017, 7, 49. 10.3390/bios7040049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam T. K.; Gerken T. A.; Brewer C. F. Thermodynamics of Multivalent Carbohydrate-Lectin Cross-Linking Interactions: Importance of Entropy in the Bind and Jump Mechanism. Biochemistry 2009, 48, 3822–3827. 10.1021/bi9002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam T. K.; Gerken T. A.; Cavada B. S.; Nascimento K. S.; Moura T. R.; Brewer C. F. Binding Studies of Alpha-GalNAc-Specific Lectins to the Alpha-GalNAc (Tn-Antigen) Form of Porcine Submaxillary Mucin and Its Smaller Fragments. J. Biol. Chem. 2007, 282, 28256–28263. 10.1074/jbc.m704677200. [DOI] [PubMed] [Google Scholar]
- Godula K.; Bertozzi C. R. Density Variant Glycan Microarray for Evaluating Cross-Linking of Mucin-like Glycoconjugates by Lectins. J. Am. Chem. Soc. 2012, 134, 15732–15742. 10.1021/ja302193u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson G. T.Functional Targets. Bioconjugate Techniques, 3rd ed.; Academic Press: London, 2013; Chapter 1. [Google Scholar]
- Peters B. P.; Ebisu S.; Goldstein I. J.; Flashner M. Interaction of Wheat Germ Agglutinin with Sialic Acid. Biochemistry 1979, 18, 5505–5511. 10.1021/bi00591a038. [DOI] [PubMed] [Google Scholar]
- Benton D. J.; Martin S. R.; Wharton S. A.; McCauley J. W. Biophysical Measurement of the Balance of Influenza A Hemagglutinin and Neuraminidase Activities. J. Biol. Chem. 2015, 290, 6516–6521. 10.1074/jbc.m114.622308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T. P.; Benton D. J.; Sadeyen J. R.; Chang P.; Sealy J. E.; Bryant J. E.; Martin S. R.; Shelton H.; McCauley J. W.; Barclay W. S.; Iqbal M. Variability in H9N2 Haemagglutinin Receptor-Binding Preference and the PH of Fusion. Emerging Microbes Infect. 2017, 6, 1–7. 10.1038/emi.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W.; Guo H.; Nijman V. S.; Doedt J.; van der Vries E.; van der Lee J.; Li Z.; Boons G.-J.; van Kuppeveld F. J. M.; de Vries E.; Matrosovich M.; de Haan C. A. M. The 2nd Sialic Acid-Binding Site of Influenza A Virus Neuraminidase Is an Important Determinant of the Hemagglutinin-Neuraminidase-Receptor Balance. PLoS Pathog. 2019, 15, e1007860 10.1371/journal.ppat.1007860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachieri S. G.; Xiong X.; Collins P. J.; Walker P. A.; Martin S. R.; Haire L. F.; Zhang Y.; McCauley J. W.; Gamblin S. J.; Skehel J. J. Receptor Binding by H10 Influenza Viruses. Nature 2014, 511, 475–477. 10.1038/nature13443. [DOI] [PubMed] [Google Scholar]
- Xiong X.; Martin S. R.; Haire L. F.; Wharton S. A.; Daniels R. S.; Bennett M. S.; McCauley J. W.; Collins P. J.; Walker P. A.; Skehel J. J.; Gamblin S. J. Receptor Binding by an H7N9 Influenza Virus from Humans. Nature 2013, 499, 496–499. 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.