Abstract
Atrial septal defect (ASD) is one of the most common congenital heart defects diagnosed in children. Sarcomeric genes has been attributed to ASD and knockdown of MYH3 functionally homologues gene in chick models indicated abnormal atrial septal development. Here, we report for the first time, a case-control study investigating the role of MYH3 among non-syndromic ASD patients in contributing to septal development. Four amplicons which will amplifies the 40 kb MYH3 were designed and amplified using long range-PCR. The amplicons were then sequenced using indexed paired-end libraries on the MiSeq platform. The STREGA guidelines were applied for planning and reporting. The non-synonymous c. 3574G>A (p.Ala1192Thr) [p = 0.001, OR = 2.30 (1.36–3.87)] located within the tail domain indicated a highly conserved protein region. The mutant model of c. 3574G>A (p.Ala1192Thr) showed high root mean square deviation (RMSD) values compared to the wild model. To our knowledge, this is the first study to provide compelling evidence on the pathogenesis of MYH3 variants towards ASD hence, suggesting the crucial role of non-synonymous variants in the tail domain of MYH3 towards atrial septal development. It is hoped that this gene can be used as panel for diagnosis of ASD in future.
Introduction
Atrial septal defect (ASD) is one of the most common forms of congenital heart defects (CHD), with an estimated incidence of an approximately 100 per 100 000 live births [1] and accounting for 10–15% of CHD worldwide [2–4]. Based on the location relative to the fosa ovalis, ASD refers to a communication between the right and left atria, anatomically classified into the deficient of atrial septum structure [5]. ASD is categorized into 1) ostium secundum 2) ostium primum 3) sinus venosus and [6]. An uncorrected ASD can cause a pulmonary over-circulation thus generating a right heart volume overload. The increased pulmonary resistance and pressure due to histopathological pulmonary arteriolar changes can contribute to a reduced functional capacity, leading to cyanosis, arrhythmia, stroke and a premature death [7]. Factors such as environment, maternal factors, specialised medications during pregnancy and genetic variants may disturb the regulatory factors of heart, leading to aberrant structure and function of heart [8].
The incidence of CHD among Asians has been reported to be 50% higher compared to the Caucasians and Blacks, especially for severe and complex CHD that have a high infant mortality[9]. In Asian countries, the incidence of ASD is high in Malaysia (9.6%) [10], Korea (44.2%) [11] and Thailand (43%) [12]. Studies have described that approximately 5% of children with serious CHD dies without undergoing any procedures [13]. The influence of genetic factors in contributing to a high incidence of CHD among Asians has not been clearly described despite reports indicating a potentially higher exposure due to genetic factors [14]. This underpins the importance of investigating the genetic causative factor of ASD among Asian, especially in Malaysia.
The molecular genetics of ASD have been elucidated with mutations in transcription factor genes; Nkx2.5, Tbx5 and Gata4 and structural proteins; MYH6, MYH7 and α-cardiac action [15]. Although these mutations have provided important insights into cardiac morphogenesis and ASD formation, the causative gene remains unknown in many families and individuals [16]. A study by Ruthland and colleagues (2011)on chick model showed that knockdown of embryonic myosin heavy chain (eMYH) causes abnormal atrial septal development.According to the study, the atrial septum either failed to form or only a small outgrowth of the dorsocranial wall was observed, or a small septum is formed in comparison with controls. These lead to an interest in comprehending the possible homologous relationships between the human and the chick’ MYH clusters. Thus, an expression profile of human skeletal myosin heavy chain genes carried out suggested that the human myosin heavy chain 3 (MYH3) is the functional homolog of the chick’ eMYH gene [17].
MYH3 (OMIM#160720) is a sarcomeric muscle gene mapped to chromosome 17, consisting of 40 coding exons spanning to a total of 40 kb [18]. The MYH3 belongs to the myosin II family, which is composed of globular motor domain, a short neck and hinge region connected to a long coiled-coil rod domain [19]. MYH3 which is expressed during fetal development and muscle regeneration has previously been associated with Freeman-Sheldon and Sheldon-Hall syndromes [20], although defects on the heart is yet to be described [18].
Studies have reported the contributing factors of MYH3 towards distal arthrogryposis type I, 2A (Freeman-Sheldon syndrome) and 2B (Sheldon-Hall syndrome), contractures, pterygia, and variable skeletal fusions syndrome 1A (OMIM 178110) and 1B (OMIM 618469). However, to the best of our knowledge, the association of MYH3 towards heart abnormalities in humans has not been reported. Therefore, this study for the first time reports genetic association of MYH3 towards atrial septal formation among non-syndromic ASD patients.
Materials and methods
Study population and sample collection
This was a case-control study approved by the Research and Ethics Committee, School of Medical Sciences, USM Health Campus (USMKK/PPP/JEPeM [252.3(13)]) and Ministry of Health Malaysia (MOH) (KKM/NIHSEC/BOO-2/2/2/P13-147) which complies with the Declaration of Helsinki. Written informed consents to participate in this study were obtained from either the parents of patients below 18 years old or directly from the patients who are 18 years old and above. Additionally, all patients and/or parent’ of patients has signed written informed consents to allow publication of their medical and/or genetic information.
A total of 69 non-syndromic ASD patients (cases) confirmed with ASD were recruited within almost one year. Baseline assessment of individual and family histories, review of the medical records and two-dimensional echocardiography with colour flow Doppler by an experienced paediatric cardiologist was recorded. For each case recruited, a gender-ethnicity matched control was recruited, this is to avoid analysis stratification. Echocardiography was also carried-out on the controls to eliminate potential bias. The recruitment of patients and controls cohort was conducted at Echocardiography Unit, Hospital Universiti Sains Malaysia (HUSM), Kubang Kerian, Kelantan, which is the main tertiary cardiac referral centre in the northeast region of Peninsular Malaysia. Following signing of written informed consents, peripheral blood (3 ml) was collected in EDTA tubes and stored at 4°C until further analysis.
Long-range PCR development and amplification
Genomic DNA of the recruited samples were extracted from peripheral blood from patients (n = 44) using a commercially available kit GeneAll® ExgeneTM Blood SV mini, GeneAll, Korea, at the Human Genome Center, Universiti Sains Malaysia (HGC-USM). For patients who refused to give their blood (n = 25), saliva samples were collected instead using PSP® SalivaGene DNA kit, STRATEC. Findings from a high-throughput genotyping study reported that DNA isolates from blood draws and DNA isolates form saliva showed no comparable differences in term of results [21]. The concentration and purity of the extracted DNA was measured using NanoQuant (Tecan, USA).
MYH3 sequence from Ensembl Genome Browser (ID: ENSG00000109063, GRCh38) was used to design the LR-PCR primers. The entire promoter, 5’ and 3’ MYH3 untranslated and coding regions were amplified in four distinct LR-PCR reactions. LR-PCR was performed using a Max Taq Polymerase (Vivantis, USA), according to the manufacturer's protocol at the HGC-USM.
DNA thermal cycling was performed using a SureCycler 8800 (Agilent Technologies, UK); 94°C for 2 min, 94°C 12 sec, annealing temperature for 30 secs, 68°C for 10 mins, 68°C for 7 mins for a total of 35 cycles. The size of the amplified PCR products was determined by gel electrophoresis using SYBR® Green I nucleic acid gel stain (Life Technologies, USA) under ultraviolet light. The amplified products were then purified by using an Illustra Exo-ProStar (GE Healthcare, UK). Subsequently, the LR-PCR fragments were quantified using Qubit® 1.0 Fluorometer (Invitrogen, USA) with Qubit dsDNA BR Assay Kits (Invitrogen, USA) and were pooled together at equal molar ratios.
NGS library preparation and sequencing
For each sample, 5 μl (1 ng in total) at 0.2 ng/ul of pooled LR-PCR products was used to generate indexed paired-end libraries with Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA) according to the manufacturer's protocol. A fragment length of the libraries was ascertained using the High Sensitivity DNA kit (Agilent Technologies, Santa Clara, CA) on the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Normalized libraries were subjected to a 300-cycle sequencing run with MiSeq Reagent Nano Kit v2 (Illumina, San Diego, CA) on the MiSeq (Illumina, San Diego, CA), were carried out the Genetic Laboratory, faculty of Science, University of Malaya.
Data enrichment and variant calling
The sequencing data was evaluated with FastQC (version 1.0, Babraham Bioinformatics, UK) for quality control on the raw data. Subsequently, the reads were aligned to a reference genome (hg19) by BWA-MEM (version 1.0) algorithm based on the default settings. The SNPs were filtered by a set of filters. A variant Studio v2.1 software (Illumina, San Diego, CA) was used to identify and annotate exonic and intronic variants and to determine if the variants have been reported in public databases. An integrative Genomics Viewer (IGV version v1.0.0) was used to examine the read counts of the target amplicons and confirm the detected variants. All variants identified by NGS were resequenced with Sanger sequencing.
Genotyping in controls cohort
The identified variants were genotyped in the controls cohort by made-to-order TaqMan SNP Genotyping Assay (ThermoFisher, USA). Variants that are not readily-available were custom designed and genotyped using the TaqMan SNP Genotyping Assay (ThermoFisher, USA). Variants that were not readily available as pre-designed Made-to-order TaqMan® SNP Genotyping Assay and cannot be custom designed were then genotyped using Sanger sequencing method.
Statistical and bioinformatics analyses
The study protocol and reporting were developed according to the STREGA guidelines for case-control studies [22]. The allelic and genotypic frequencies of the patient and control groups were determined using a Fisher’s exact test with odds ratio and 95% confidence interval (CI). A x2 P-value of < 0.05 was considered as statistically significant. Association was evaluated for every variant and False Discovery Rate and Bonferroni adjustments were used for multiple-testing corrections. Haploview version 4.2 (Broad Institute of MIT and Harvard, USA) was used for analysis of linkage disequilibrium (LD) and haplotype block [23]. LD blocks were defined according to the haplotype block definition of Gabriel and colleagues [24]. The method defines pairs to be in strong LD if the one-sided upper 95% confidence bound on D′ is >0.98 and the lower bound is above 0.7.
In-silico analysis of pathogenic potential of identified non-synonymous variants were predicted using Sorting Intolerant From Tolerant (SIFT) and Polymorphism Phenotyping v2 (PolyPhen-2). SIFT scores which range between 0 and 1 as well as scores below 0.05 suggest that the amino acid change is not tolerated whereas, PolyPhen-2 scores >0.85 are interpreted as “probably damaging” and scores 0.15–0.85 as “possibly damaging”. The evolutionary conservation of the identified non-synonymous variants was conducted using a Clustal Omega programme (EMBL-EBI, European Bioinformatics Institute). The splicing effects were predicted by the Human Splicing Finder (HSF) version 3.0 (http://www.umd.be/HSF/) and ESEfinder 3.0 (http://rulai.cshl.edu/).
Modelling of the mutant protein structure
Prior to the template selection, both wild and mutant types of complete sequences of human myosin MYH3 were aligned against the sequences from Protein Data Bank (PDB), (www.rcsb.org/pdb). Our results showed that protein structures with the PDB ID: 5TBY and 5WJ7 with 72% and 83% sequence identity, respectively are suitable templates for homology modelling. Multiple templates approach homology modelling was adopted in this study. Both the selected templates indicated to belong to the family protein from Homo sapiens myosin 7, which is closely related to the modeled sequences.
A total of 20 models for wild and mutant type sequence were built using Modeller 9v20 (www.salilab.org/modeller) and the best models with the lowest discrete optimize potential energy (DOPE) scoring was selected for each wild and mutant types. The structural differences between the mutant against the wild type built model was calculated using a root mean square deviation (RMSD) method.
Nomenclature and classification for detected variants
All sequence variants are named according to the nomenclature guidelines published by the Human Genome Variation Society (HGVS) [25]. Nucleotide and amino acid numbering were based on GenBank reference sequences NM_002470.3 and NP_002461.2 respectively.
Results
In the present study, a total of 69 non-syndromic ASD patients were recruited. However, only 51 samples were further proceeded as 18 samples were excluded due to low genomic DNA concentration (<50 ng/ul) and low sequencing libraries of <10 nM.
The entire 40 kb of MYH3 was successfully sequenced in 51 non-syndromic ASD patients using LRPCR-NGS approach. The validity of the results was confirmed with Sanger sequencing.
Demographic and clinical data
Although a total of 69 non-syndromic ASD patients where recruited (blood sample, n = 44 and saliva sample, n = 25), only 51 samples were further proceeded for sequencing. The demographic data of these 51 samples were; aged between 3–62 years old with mean age of 4.72 ± 1.77 years. Based on ethnicity, 47 (92.16%) were Malays, two (3.92%) were Chinese and two (3.92%) were Siamese. In terms of gender, 4 (28.6%) were male and 47 (71.4%) were female patients. The cardiac phenotypes of the recruited patients were categorized into four groups 1) ASD secundum (82.2%) 2) ASD primum (6.5%) and 3) fenestrated ASD (11.3%). The recruited controls cohort consisting of 73 healthy individuals were matched for age, ethnicity and gender with the cases. The age of controls ranged from 3–62 years old with mean age of 4.72 ± 1.77 years. In term of ethnicity and gender; 69 Malays, two Chinese and two Siamese, with 59 of them being females and 14 males.
Coverage and sequencing depth of targets of LRPCR-NGS
Four pairs of LR-PCR primers were designed to amplify the entire 40 kb of MYH3. These primers were optimized at annealing temperatures of 63.1°C, 62.4°C, 64.7°C and 64.7°C respectively, employing LR-PCR cycling method within a short period of time. Targeted sequencing using Illumina MiSeq paired-end sequence resulted in 11,947,785 reads (98.05%) of which 99.8% may be aligned with the human genome (S1 Fig).
This provided 99.8% coverage of the sequence at a minimum sequencing depth of 6×. The run had a cluster density of 615 +/- 6 K/mm2 with approximately 92.63 +/- 2.05% of the clusters passing the QC filter. The observed read depth in all the samples was > 100 X per base with Q ≥ 30 (99.9% base call accuracy).
MYH3 variants analysis
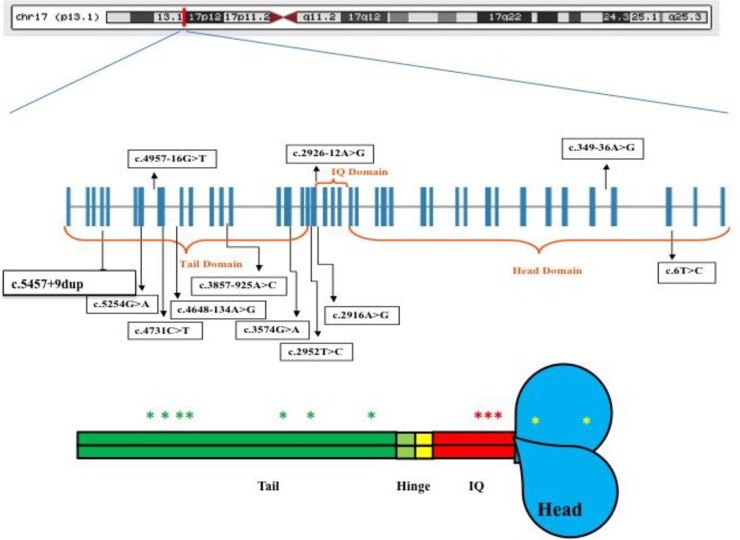
Variants with minor allele frequency (MAF) <5% and phred scale of <30 were excluded, resulting in 11 variants and one indel. Fig 1B and 1C show the diagrammatic position of the MYH3 variants (Fig 1) detected while Table 1 summarizes the characteristic of the identified variants. To evaluate the performance and validity of the LRPCR-NGS, resequencing of the identified variants was carried out using a Sanger sequencing.
Fig 1. Genomic model of MYH3 and diagrammatic illustration of the spectrum of variants.
(A) Position of MYH3 at Chromosome 17. (B) Boxes represent exons and adjoining lines represent introns. (C) Schematic representation of MYH3 domains with the approximate location of MYH3 variants indicated with coloured asterisk mark.
Table 1. MYH3 variants detected using LRPCR-NGS approach.
| Variants | HGVS | rs ID | Location | Amico Acid Change | Type of variant | PolyPhen-2 | SIFT | Number of patients | Type of ASD |
|---|---|---|---|---|---|---|---|---|---|
| (n = number of patients) | |||||||||
| c.6T>C | NC_000017.11:g.10655059A>G | rs17817203 | Exonic | - | Synonymous | - | - | 2 | • Fenestrated ASD (n = 1) |
| • ASD secundum (n = 1) | |||||||||
| c.2952T>C | NC_000017.11:g.10639448A>G | rs2285474 | Exonic | - | Synonymous | - | - | 22 | • ASD secundum (n = 19) |
| • ASD primum (n = 2) | |||||||||
| • Fenestrated ASD (n = 1) | |||||||||
| c.4648-134A>G | NC_000017.11:g.10632918T>C | rs1981514 | Intronic | - | - | - | - | 28 | • ASD secundum (n = 23) |
| • ASD primum (n = 3) | |||||||||
| • Fenestrated ASD (n = 2) | |||||||||
| c.5254G>A | NC_000017.11:g.10631643C>A | rs34393601 | Exonic | p.Ala1752Thr | Missense | 0.144 | 0.01 | 2 | • ASD secundum (n = 1) |
| (Benign) | (Deleterious) | • Fenestrated ASD (n = 1) | |||||||
| c.4957-16G>T | ENST00000583535.6:c.4957-16G>T | rs2239936 | Intronic | - | - | - | - | 36 | • ASD secundum (n = 30) |
| • ASD primum (n = 3) | |||||||||
| • Fenestrated ASD (n = 3) | |||||||||
| c.2916A>G | ENST00000583535.6:c.2916A>G | rs2285472 | Exonic | - | Synonymous | - | - | 25 | • ASD secundum (n = 22) |
| • ASD primum (n = 2) | |||||||||
| • Fenestrated ASD (n = 1) | |||||||||
| c.2926-12A>G | ENST00000583535.6:c.2926-12A>G | rs2285473 | Intronic | - | - | - | - | 22 | • ASD secundum (n = 20) |
| • ASD primum (n = 2) | |||||||||
| c.3574G>A | ENST00000583535.6:c.3574G>A | rs2285477 | Exonic | p.Ala1192Thr | Missense | 0.003 | 0.44 | 27 | • ASD secundum (n = 21) |
| (Benign) | (Tolerated) | • ASD primum (n = 3) | |||||||
| • Fenestrated ASD (n = 3) | |||||||||
| c.4731C>T | ENST00000583535.6:c.4731C>T | rs2285479 | Exonic | - | Synonymous | - | - | 32 | • ASD secundum (n = 30) |
| • ASD primum (n = 1) | |||||||||
| • Fenestrated ASD (n = 1) | |||||||||
| c.3857-925A>C | ENST00000583535.6:c.3857-925A>C | rs55980976 | Intronic | - | - | - | - | 31 | • ASD secundum (n = 29) |
| • ASD primum (n = 2) | |||||||||
| c.5457+9dup | NC_000017.11:g.10630279dup | rs397750512 INDEL | Intronic | - | Insertion | - | - | 33 | • ASD secundum (n = 30) |
| • ASD primum (n = 2) | |||||||||
| • Fenestrated ASD (n = 1) | |||||||||
| c.349-36A>G | ENST00000583535.6:c.349-36A>G | rs2285467 | Exonic | - | Synonymous | - | - | 12 | • ASD secundum (n = 12) |
Statistical and bioinformatics analysis
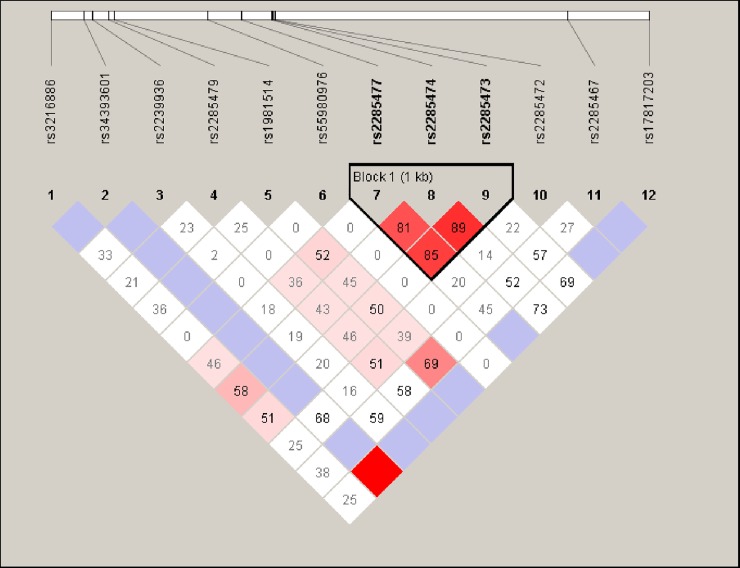
Fisher exact x2 analysis showed c.4648-134A>G (p = 0.007, OR = 2.05 [CI: 1.18–3.57]), c.2952T>C (p = 0.0003, OR = 2.62 [CI: 1.54–4.45]), c.4957-16G>T (p = 0.044, OR = 0.62 [CI:0.371.03]), c.2916A>G (p = 0.0003, OR = 2.55 [CI: 1.50–4.32]), c.2926-12A>G (p = 0.00006, OR = 2.90 [CI: 1.69–4099]), c.3574G>A (p = 0.001, OR = 2.30 [CI: 1.36–3.87]), c. 349–36 A>G (p = 0.033, OR = 0.37[CI: 0.14–0.97]) and c.3857-925A>C (p = 0.000, OR = 0.00 [CI:0. 00–0.06]) to be significantly correlated with ASD (Table 2). These variants were further subjected to haplotype analysis. The LD block of rs2285477 (c.3574G>A), rs2285474 (c.2952T>C) and rs2285473 (c.2926-12A>G) showed a strong correlation with ASD (p-value; GTA: 0.0003, ACG: 2.878e-5) (Fig 2), haplotypes: GTA, ACG and ATG also showed statistical significant (p<0.05) when compared to the control group.
Table 2. Genotype and allelic frequencies distribution of the variants in a cohort of ASD patients and healthy controls.
| Variant | Variables | ASD patients (n = 51) | Control population (n = 73) | p-value | Odds Ratio (95% CI) |
|---|---|---|---|---|---|
| c.6T>C | Genotype | ||||
| T/T | 49 | 70 | |||
| C/C | - | 3 | |||
| T/C | 2 | - | 0.288 | 2.14 (0.42–10.83) | |
| Allele Frequency | |||||
| T | 0.980 | 0.958 | |||
| C | 0.019 | 0.042 | |||
| c.4648-134A>G | Genotype | ||||
| T/T | 23 | 11 | |||
| C/C | 8 | 13 | |||
| T/C | 20 | 32 | 0.011 | 1.97 (1.13–3.41) | |
| Allele Frequency | |||||
| T | 0.647 | 0.369 | |||
| C | 0.353 | 0.397 | |||
| c.2952T>C | Genotype | ||||
| T/T | 27 | 15 | |||
| C/C | 7 | 17 | |||
| T/C | 17 | 41 | P = 0.0002 | 2.62 (1.54–4.45) | |
| Allele Frequency | |||||
| T | 0.696 | 0.486 | |||
| C | 0.303 | 0.514 | |||
| c.5254G>A | Genotype | ||||
| G/G | 49 | 73 | |||
| A/A | - | 0 | |||
| G/A | 2 | - | |||
| Allele Frequency | 0.172 | 0.00 (0.00–0.00 | |||
| G | 0.980 | 1.000 | |||
| A | 0.019 | 0 | |||
| c.4957-16G>T | Genotype | ||||
| G/G | 16 | 28 | |||
| T/T | 14 | 6 | |||
| G/T | 21 | 38 | 0.044 | 0.62 (0.37–1.03) | |
| Allele Frequency | |||||
| G | 0.519 | 0.644 | |||
| T | 0.480 | 0.342 | |||
| c.2916A>G | Genotype | ||||
| A/A | 28 | 15 | |||
| G/G | 4 | 18 | |||
| A/G | 19 | 38 | p = 0.0003 | 2.55 (1.50–4.32) | |
| Allele Frequency | |||||
| A | 0.735 | 0.466 | |||
| G | 0.265 | 0.507 | |||
| c.2926-12A>G | Genotype | ||||
| A/A | 29 | 15 | |||
| G/G | 3 | 17 | |||
| A/G | 19 | 40 | p = 00006 | 2.90 (1.69–4.99) | |
| Allele Frequency | |||||
| A | 0.754 | 0.479 | |||
| G | 0.245 | 0.507 | |||
| c.3574G>A | Genotype | ||||
| G/G | 27 | 14 | |||
| A/A | 4 | 16 | |||
| G/A | 20 | 42 | 0.001 | 2.30 (1.36–3.87) | |
| Allele Frequency | |||||
| G | 0.725 | 0.479 | |||
| A | 0.274 | 0.507 | |||
| c.4731C>T | Genotype | ||||
| C/C | 17 | 12 | |||
| T/T | 10 | 14 | |||
| C/T | 24 | 37 | 0.164 | 1.34 (0.79–2.26) | |
| Allele Frequency | |||||
| C | 0.568 | 0.418 | |||
| T | 0.431 | 0.445 | |||
| c.3857-925A>C | Genotype | ||||
| AA | 21 | 73 | |||
| CC | 9 | 0 | |||
| AC | 21 | 0 | p = 0000 | 0.00 (0.00–0.06) | |
| Allele Frequency | |||||
| A | 0.617 | 1.000 | |||
| C | 0.382 | 0.000 | |||
| c.5457+9dup | Genotype | ||||
| G/GA | 38 | 7 | p≤0.050* | 0.01 | |
| GA/GA | 13 | 66 | (0.01–0.09) | ||
| c.349-36A>G | Genotype | ||||
| AA | 38 | 60 | |||
| GG | 1 | - | |||
| AG | 12 | 7 | 0.033 | 0.37 (0.14–0.97) | |
| Allele Frequency | |||||
| A | 0.862 | 0.869 | |||
| G | 0.137 | 0.048 |
* p≤0.050 is considered significant
Fig 2. Haploview generated graphical representation of LD in MYH3 variants identified among ASD patients.
Twelve SNPs spanning from position 17:10655059 to 17:10631643 were grouped into one block consisting of rs2285477 (c.3574G>A), rs2285474 (c.2952T>C) and rs2285473 (c.2926-12A>G) according to the LD structure. Length of the block is provided in kilobases (kb), and pairwise LD (D′) is given for each SNP combination (showed in box). The color code in the haploview plot follows the standard color scheme for haploview: white, |D′|<1, LOD<2; shades of pink, |D′|<1, LOD≥2; blue, | D′| = 1, LOD<2; red, |D′| = 1, LOD≥2.
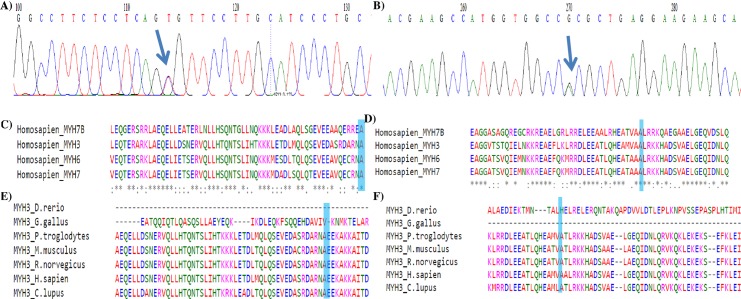
The non-synonymous c.5254G>A (p. Ala1752Thr) was predicted to be deleterious by SIFT tool (0.01) whereas PolyPhen-2 predicts it to be benign (0.144). The non-synonymous c.3574G>A (p. Ala1192Thr) was predicted to be tolerated and benign based on SIFT and PolyPhen-2 tool with scores of 0.44 and 0.003 respectively (Table 1). The Ala1751 (p. Ala1752Thr) and Ala1191 (p. Ala1192Thr) residues were identified to be highly conserved in human MYH7B, MYH3, MYH6 and MYH7 whereas in orthologous sequence it is conserved among chimp, mouse, rat, human and dog (Fig 3).
Fig 3.
Evolutionary conservation of the non-synonymous (a) c.5254G>A (p.Ala1752Thr, rs34393601) (b) c.3574G>A (p.Ala1192Thr, rs2285477) variant in MYH3. The Ensembl accession codes of the paralogous sequences; MYH7B: ENSG00000078814, MYH3: ENSG00000109063, MYH6: ENSG00000197616, MYH7, ENSG00000092054; The Ensembl accession codes of the orthologous sequences; zebra fish: C7MYH3.1, chicken: ENSGALG00000002678, chimp: ENSPTRG00000008773, mouse: ENSMUSG00000057003; rat: ENSRNOG00000046276; dog: ENSCAFG0000002309.
The putative 5’ and 3’ splice sites as well as branch sites were predicted to determine if the identified mutations would disrupt these sites. The results were interpreted by comparing the percentage of consensus values (CV%) between the wild-type and the mutant variant [26]. CV of higher than 80 indicates strong sites, and a negative CV variation percentage between the wild-type and mutant variant indicates that the variant affects splicing mechanism. The c.2916A>G mutation that occurs at late exonic position was predicted to affect the splicing mechanisms. The HSF analysis for c.2916A>G generated consensus values of 80.66 and 51.71 for the wild and mutant types respectively with predicted consensus deviation value of -35.89%. The A to G synonymous substitution (exon 23, 324th nucleotide) affects the SC35 binding motif TGCCACAG, reducing the score from 2.90 to 0.0 (below the default threshold) and creating a sequence containing a potential SRp40 binding motif (TGCCACG score: 2.80) (S2 Fig)
Modelling of the mutant protein structure
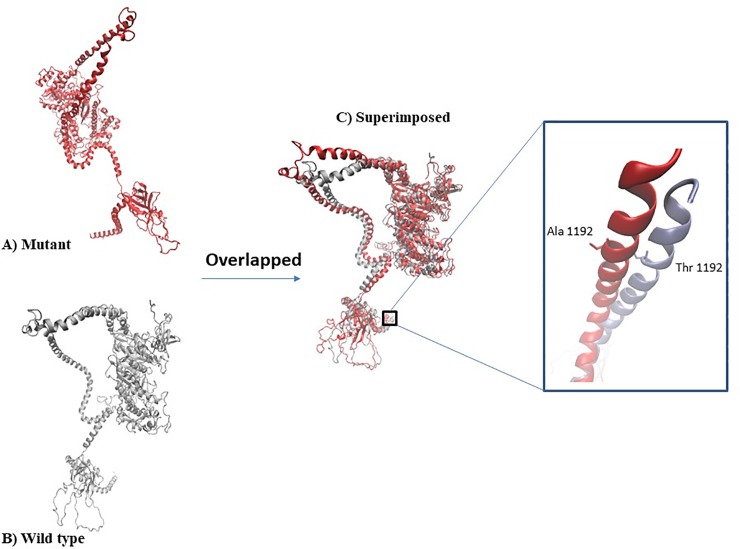
Fig 4 shows the 3D structures of the predicted models. The homology modelling of the c. 3574G>A (p.Ala1192Thr) was performed using Visual Molecular Dynamic 1.9.3. (Urbana, IL). The native and wild types models were superimposed, generating RMSD value of 5.4Å. To assess the stereochemical properties of the built model, Ramachandran plot analysis was conducted. Ramachandran plot analysis showed that out of 1121 total residues in both the built models, the wild and mutant type models were observed with 86.5% and 86.1%, residues fall in most favoured region, respectively.
Fig 4.
Structural comparison via RMSD analysis (a) Wild type built model (b) Mutant type built model (c) Superimposition of wild and mutant type model with RMSD value of 5.4Å.
Discussion
To our knowledge, this is the first study to elucidate the role of MYH3 towards ASD in non-syndromic patients employing LRPCR-NGS approach. A total of 12 genetic variants were detected of which seven (58%) were located within the tail domain of MYH3. The non-synonymous variant c.5254G>A (p. Ala1752Thr) was identified in two female ASD patients having ASD secundum with defect size of 0.5 cm and 0.8 cm respectively but was absent in the 73 unrelated controls. Nevertheless, the non-synonymous c.3574G>A (p. Ala1192Thr) variant was identified in six patients and was associated with ASD [p = 0.001 (OR = 2.30 [1.36–3.87]). Evolutionary conservation analysis of MYH3 amino acid sequence showed that these variants are highly conserved in the sarcomeric MYH genes expressed in the heart, and orthologous sequence alignment showed it to be conserved in chimp, mouse, rat, human and dog. Haplotype analysis showed that; the c.3574G>A (rs2285477) located at the tail domain, c.2952T>C (rs2285474) and c.2926-12A>G (rs2285473) located at the IQ domain showed strong correlation with ASD (p-value; GTA: 0.0003, ACG: 2.878e-5, ATG: 0.0293), the same LD block also showed association in 10,000 permutations [S3 Fig].
The amino acid substitution of hydrophobic alanine to hydrophilic theornine in p. Ala1752Thr and p. Ala1192Thr, places a hydrophobic side chain into a hydrophilic environment in the tail domain, which suggest that the mutant complex might be destabilized (ΔΔG of binding) relative to the wild-type complex. This action may pertubate the ability of the long coiled-coil tails of the individual myosin molecules to form thick filaments of the sarcomere. Thus further differing the ability of C-terminal tail domain’s ability for the heavy chains to self-associate and/or bind the myosin to its cargo [27]. A less stable thick filament could also affect the coupling between ATP hydrolysis and force generation by deficient transmission of energy from the motor domain to the sarcomeric structure [28].
Studies have also reported that mutations in the tail domain could lead towards disruption of either the head or the rod domain of embryonic myosin [29]. This result is similar to that conducted by Tajsharghi and colleagues [30], indicating that the tail domain is a functionally important residue for myosin heavy change (MHC) association, which binds thick filament in the cardiac muscle. A study by Ching and colleagues [31] identified I820N mutation that causes hydrophobic isoleucine to change to hydrophilic asparagine, thus predicting that the change may affect the binding of MHC to its regulatory light chain thus suggesting that these mutations may affect the normal formation of the myofibrils, which is in-hand with the findings of current study. However, histological analysis of the myocardium is substantially important to justify the said postulation.
The present study also identified c.2916A>G which is located at the late exonic position of exon 23 of the tail domain to affect splicing mechanism. In silico analysis suggested that this mutation will result in alterations of the exonic splicing enhancer (ESE) site, that would cause either skipping of the respective exon, activation of cryptic site or intron retention. Homology modelling of c. 3574G>A (p.Ala1192Thr) mutant against the wild type protein, showed some degree of difference with RMSD value of 5.4Å. However, these predictions were identified by the combined use of different algorithms where it is not yet possible to consistently predict the effects of nucleotide substitutions on splicing [32]. Therefore, in vitro analysis can be employed to further validate these predictions thus allowing a more accurate determination of the type of nucleotide changes which may affect the splicing mechanism.
MYH3 plays important role in skeletal muscle development [33]. Other genes have additionally been reported to play important roles in skeletal muscle developments such as MYH2, MYH7 and MYH8 where myosin myopathies involving these genes have been widely reported. However, the phenotypes associated with these genes vary from one skeletal muscle gene to another, ranging from prenatal non-progressive arthrogryposis syndromes to adult-onset progressive muscle weakness [33]. Additionally, mutations in MYH2 have been associated with dominant myopathy characterized by ophthalmoplegia, congenital joint contractures, and rimmed vacuoles in muscle fibers [34, 35]. Mutations in MYH7 cause skeletal myopathies such as myosin storage myopathy and Laing early-onset distal myopathy [33, 34, 36, 37].
Although studies have reported the contributing factors of MYH3 towards various syndromes including distal arthrogryposis type I, 2A (Freeman-Sheldon syndrome) and 2B (Sheldon-Hall syndrome), the association of MYH3 towards heart abnormalities in humans has not been reported [29, 38]. The data presented in this study provided novel insights into the contribution of molecular genetics of MYH3 towards the development of atrial septa.
Nevertheless, our study has some limitations, only MYH3 gene was focussed, thus novel variants or other genomic loci beyond MYH3, was not identified. The identified variants were also detected among controls, therefore further study on the mendelian segregation among familial ASD patient for mutation in MYH3 is important. However, despite its limitations, targeted NGS is able to obtain high-quality data, within a hypothesis-driven approach, while remaining less expensive than its whole genome sequencing and whole exome sequencing counterparts. This methodology is appropriate for efficient and directed research, in elucidating molecular pathways of various diseases.
Conclusion
In conclusion, this study proposes non-synonymous mutations in the tail domain of MYH3 as a potential autosomal cause of ASD among non-syndromic patients. Hence, if a number of genes with contributions of similar magnitude as MYH3 to the risk of ASD could be identified, these genes can be used as panel for diagnosis of ASD.
Supporting information
A) Schematic representation of MYH3 long-range amplicons. B) Gel electrophoresis picture showing optimised amplicons; Lane M = 1 kb ladder, Lane A1 = Amplicon 1, Lane A2 = Amplicon 2, Lane 3 = Amplicon 3, Lane A4 = Amplicon 4, Lane–ve = Negative control; 0.7% agarose gel at 85V for 60 mins. C): Miseq reads aligned with MYH3 loci of human genome assembly hg19 using BWA program and viewed using IGV; Red areas: reads from the plus DNA strands; blue area:reads from the minus strands.
(PDF)
ESE finder analysis of (A) wild-type and (B) mutated MYH3 for c.2916A>G variant. The default threshold values for SF2/ASF (SRSF1), SC35 (SRSF2), SRp40 (SRSF5) and SRp55 (SRSF6) were 1.956, 2.383, 2.67 and 2.676 respectively. The width of each bar reflects the length of the motif, the placement of each bar along the X-axis represents the position of a motif along the DNA sequence, the height of the bar represents the numerical score on the Y-axis.
(PDF)
The X-axis corresponds to chi-square value of permutated SNPs and the Y-axis corresponds to number of permutation. Permutation progress bar indicates the tallest bar with highest permutated χ2 at >900 permutation.
(PDF)
Data Availability
The original uncropped and unadjusted images of gel blot and minimal anonymized data set from this study has been deposited at DRYAD (doi: 10.5061/dryad.pvmcvdnh3). The study protocol has been deposited at protocols.io (https://dx.doi.org/10.17504/protocols.io.bbjtiknn).
Funding Statement
This work was funded by the Research University Grant (RU), Universiti Sains Malaysia [1001/PPSP/813066] and Fundamental Research Grant Scheme, Ministry of Education Malaysia [FRGS/1/2018/SKK11/PERDANA/02/1]. THL and SHEL were the respective Principle investigators. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Geva T, Martins JD, Wald RM. Atrial septal defects. The Lancet. 2014;383(9932):1921–32. [DOI] [PubMed] [Google Scholar]
- 2.Schwedler G, Lindinger A, Lange PE, Sax U, Olchvary J, Peters B, et al. Frequency and spectrum of congenital heart defects among live births in Germany: a study of the Competence Network for Congenital Heart Defects. Clinical research in cardiology: official journal of the German Cardiac Society. 2011;100(12):1111–7. Epub 2011/09/13. 10.1007/s00392-011-0355-7 . [DOI] [PubMed] [Google Scholar]
- 3.Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, Correa A. Prevalence of congenital heart defects in metropolitan Atlanta, 1998–2005. The Journal of pediatrics. 2008;153(6):807–13. Epub 2008/07/29. 10.1016/j.jpeds.2008.05.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu MH, Chen HC, Lu CW, Wang JK, Huang SC, Huang SK. Prevalence of congenital heart disease at live birth in Taiwan. The Journal of pediatrics. 2010;156(5):782–5. Epub 2010/02/09. 10.1016/j.jpeds.2009.11.062 . [DOI] [PubMed] [Google Scholar]
- 5.Cory N, Mark L. Left to Right Shunts In: Elzouki AY, Harfi H.A., Nazer H., Oh W., Stapleton F.B., Whitley R.J., editor. Textbook of Clinical Pediatrics. 1. 2 ed New York: Springer; 2012. [Google Scholar]
- 6.Celermajer DS. Atrial septal defects: even simple congenital heart diseases can be complicated. European heart journal. 2018;39(12):999–1001. Epub 2017/12/02. 10.1093/eurheartj/ehx633 . [DOI] [PubMed] [Google Scholar]
- 7.van der Linde D, Konings EE, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJ, et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. Journal of the American College of Cardiology. 2011;58(21):2241–7. Epub 2011/11/15. 10.1016/j.jacc.2011.08.025 . [DOI] [PubMed] [Google Scholar]
- 8.Wang H, Liu Y, Li Y, Wang W, Li L, Meng M, et al. Analysis of NKX2-5 in 439 Chinese Patients with Sporadic Atrial Septal Defect. Med Sci Monit. 2019;25:2756–63. 10.12659/MSM.916052 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knowles RL, Ridout D, Crowe S, Bull C, Wray J, Tregay J, et al. Ethnic and socioeconomic variation in incidence of congenital heart defects. Archives of Disease in Childhood. 2017;102(6):496 10.1136/archdischild-2016-311143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung LC, Mohan AJ, Soo TL, Ng HP. Pattern of congenital heart disease and access to tertiary cardiac care in Malaysia. The Medical journal of Malaysia. 2000;55(4):424–32. Epub 2001/02/28. . [PubMed] [Google Scholar]
- 11.Lamichhane DK, Leem J-H, Park M, Kim JA, Kim HC, Kim JH, et al. Increased prevalence of some birth defects in Korea, 2009–2010. BMC Pregnancy and Childbirth. 2016;16(1):61 10.1186/s12884-016-0841-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratanasit N, Karaketklang K, Jakrapanichakul D, Kittipovanonth M, Punlee K, Rochanasiri W, et al. Prevalence and echocardiographic characteristics of common congenital heart disease in adult patients at Siriraj Hospital: 10-year study. Journal of the Medical Association of Thailand = Chotmaihet thangphaet. 2015;98(1):7–13. Epub 2015/03/18. . [PubMed] [Google Scholar]
- 13.Knowles RL, Bull C, Wren C, Wade A, Goldstein H, Dezateux C. Modelling survival and mortality risk to 15 years of age for a national cohort of children with serious congenital heart defects diagnosed in infancy. PloS one. 2014;9(8):e106806 Epub 2014/09/11. 10.1371/journal.pone.0106806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Chen S, Zühlke L, Black GC, Choy M-k, Li N, et al. Global birth prevalence of congenital heart defects 1970–2017: updated systematic review and meta-analysis of 260 studies. International Journal of Epidemiology. 2019;48(2):455–63. 10.1093/ije/dyz009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wessels MW, Willems PJ. Genetic factors in non-syndromic congenital heart malformations. Clinical genetics. 2010;78(2):103–23. Epub 2010/05/26. 10.1111/j.1399-0004.2010.01435.x . [DOI] [PubMed] [Google Scholar]
- 16.Zaidi S, Brueckner M. Genetics and Genomics of Congenital Heart Disease. Circulation Research. 2017;120(6):923–40. 10.1161/CIRCRESAHA.116.309140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rutland CS, Polo-Parada L, Ehler E, Alibhai A, Thorpe A, Suren S, et al. Knockdown of embryonic myosin heavy chain reveals an essential role in the morphology and function of the developing heart. Development (Cambridge, England). 2011;138(18):3955–66. 10.1242/dev.059063 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.England J, Loughna S. Heavy and light roles: myosin in the morphogenesis of the heart. Cellular and molecular life sciences: CMLS. 2013;70(7):1221–39. Epub 2012/09/06. 10.1007/s00018-012-1131-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chong JX, Burrage LC, Beck AE, Marvin CT, McMillin MJ, Shively KM, et al. Autosomal-Dominant Multiple Pterygium Syndrome Is Caused by Mutations in MYH3. American journal of human genetics. 2015;96(5):841–9. Epub 2015/05/11. 10.1016/j.ajhg.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tajsharghi H, Oldfors A. Myosinopathies: pathology and mechanisms. Acta neuropathologica. 2013;125(1):3–18. Epub 2012/08/05. 10.1007/s00401-012-1024-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poehls UG, Hack CC, Ekici AB, Beckmann MW, Fasching PA, Ruebner M, et al. Saliva samples as a source of DNA for high throughput genotyping: an acceptable and sufficient means in improvement of risk estimation throughout mammographic diagnostics. Eur J Med Res. 2018;23(1):20–. 10.1186/s40001-018-0318-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, Von Elm E, et al. STrengthening the REporting of Genetic Association Studies (STREGA)—an extension of the STROBE statement. Genetic Epidemiology: The Official Publication of the International Genetic Epidemiology Society. 2009;33(7):581–98. [DOI] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. Epub 2004/08/07. 10.1093/bioinformatics/bth457 . [DOI] [PubMed] [Google Scholar]
- 24.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9. Epub 2002/05/25. 10.1126/science.1069424 . [DOI] [PubMed] [Google Scholar]
- 25.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Human mutation. 2000;15(1):7–12. Epub 1999/12/29. . [DOI] [PubMed] [Google Scholar]
- 26.Desmet F-O, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37(9):e67–e. Epub 2009/04/01. 10.1093/nar/gkp215 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korn ED. Coevolution of head, neck, and tail domains of myosin heavy chains. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(23):12559–64. Epub 2000/11/01. 10.1073/pnas.230441597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granados-Riveron JT, Ghosh TK, Pope M, Bu'Lock F, Thornborough C, Eason J, et al. Alpha-cardiac myosin heavy chain (MYH6) mutations affecting myofibril formation are associated with congenital heart defects. Human molecular genetics. 2010;19(20):4007–16. Epub 2010/07/27. 10.1093/hmg/ddq315 . [DOI] [PubMed] [Google Scholar]
- 29.Toydemir RM, Rutherford A, Whitby FG, Jorde LB, Carey JC, Bamshad MJ. Mutations in embryonic myosin heavy chain (MYH3) cause Freeman-Sheldon syndrome and Sheldon-Hall syndrome. Nature genetics. 2006;38(5):561–5. Epub 2006/04/28. 10.1038/ng1775 . [DOI] [PubMed] [Google Scholar]
- 30.Tajsharghi H, Thornell LE, Lindberg C, Lindvall B, Henriksson KG, Oldfors A. Myosin storage myopathy associated with a heterozygous missense mutation in MYH7. Annals of neurology. 2003;54(4):494–500. Epub 2003/10/02. 10.1002/ana.10693 . [DOI] [PubMed] [Google Scholar]
- 31.Ching YH, Ghosh TK, Cross SJ, Packham EA, Honeyman L, Loughna S, et al. Mutation in myosin heavy chain 6 causes atrial septal defect. Nature genetics. 2005;37(4):423–8. Epub 2005/03/01. 10.1038/ng1526 . [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14(5):802–13. Epub 2008/03/29. 10.1261/rna.876308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajsharghi H, Kimber E, Kroksmark A-K, Jerre R, Tulinius M, Oldfors A. Embryonic Myosin Heavy-Chain Mutations Cause Distal Arthrogryposis and Developmental Myosin Myopathy That Persists Postnatally. Archives of Neurology. 2008;65(8):1083–90. 10.1001/archneur.65.8.1083 [DOI] [PubMed] [Google Scholar]
- 34.Darin N, Kyllerman M, Wahlstrom J, Martinsson T, Oldfors A. Autosomal dominant myopathy with congenital joint contractures, ophthalmoplegia, and rimmed vacuoles. Annals of neurology. 1998;44(2):242–8. Epub 1998/08/26. 10.1002/ana.410440215 . [DOI] [PubMed] [Google Scholar]
- 35.Tajsharghi H, Thornell LE, Darin N, Martinsson T, Kyllerman M, Wahlstrom J, et al. Myosin heavy chain IIa gene mutation E706K is pathogenic and its expression increases with age. Neurology. 2002;58(5):780–6. Epub 2002/03/13. 10.1212/wnl.58.5.780 . [DOI] [PubMed] [Google Scholar]
- 36.Laing NG, Ceuterick-de Groote C, Dye DE, Liyanage K, Duff RM, Dubois B, et al. Myosin storage myopathy: slow skeletal myosin (MYH7) mutation in two isolated cases. Neurology. 2005;64(3):527–9. Epub 2005/02/09. 10.1212/01.WNL.0000150581.37514.30 . [DOI] [PubMed] [Google Scholar]
- 37.Meredith C, Herrmann R, Parry C, Liyanage K, Dye DE, Durling HJ, et al. Mutations in the slow skeletal muscle fiber myosin heavy chain gene (MYH7) cause laing early-onset distal myopathy (MPD1). American journal of human genetics. 2004;75(4):703–8. Epub 2004/08/24. 10.1086/424760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarado DM, Buchan JG, Gurnett CA, Dobbs MB. Exome sequencing identifies an MYH3 mutation in a family with distal arthrogryposis type 1. The Journal of bone and joint surgery American volume. 2011;93(11):1045–50. Epub 2011/05/03. 10.2106/JBJS.J.02004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Schematic representation of MYH3 long-range amplicons. B) Gel electrophoresis picture showing optimised amplicons; Lane M = 1 kb ladder, Lane A1 = Amplicon 1, Lane A2 = Amplicon 2, Lane 3 = Amplicon 3, Lane A4 = Amplicon 4, Lane–ve = Negative control; 0.7% agarose gel at 85V for 60 mins. C): Miseq reads aligned with MYH3 loci of human genome assembly hg19 using BWA program and viewed using IGV; Red areas: reads from the plus DNA strands; blue area:reads from the minus strands.
(PDF)
ESE finder analysis of (A) wild-type and (B) mutated MYH3 for c.2916A>G variant. The default threshold values for SF2/ASF (SRSF1), SC35 (SRSF2), SRp40 (SRSF5) and SRp55 (SRSF6) were 1.956, 2.383, 2.67 and 2.676 respectively. The width of each bar reflects the length of the motif, the placement of each bar along the X-axis represents the position of a motif along the DNA sequence, the height of the bar represents the numerical score on the Y-axis.
(PDF)
The X-axis corresponds to chi-square value of permutated SNPs and the Y-axis corresponds to number of permutation. Permutation progress bar indicates the tallest bar with highest permutated χ2 at >900 permutation.
(PDF)
Data Availability Statement
The original uncropped and unadjusted images of gel blot and minimal anonymized data set from this study has been deposited at DRYAD (doi: 10.5061/dryad.pvmcvdnh3). The study protocol has been deposited at protocols.io (https://dx.doi.org/10.17504/protocols.io.bbjtiknn).