Abstract
Microalga-based biomaterial production has attracted attention as a new source of drugs, foods, and biofuels. For enhancing the production efficiency, it is essential to understand its differences between heterogeneous microalgal subpopulations. However, existing techniques are not adequate to address the need due to the lack of single-cell resolution or the inability to perform large-scale analysis and detect small molecules. Here we demonstrated large-scale single-cell analysis of Euglena gracilis (a unicellular microalgal species that produces paramylon as a potential drug for HIV and colon cancer) with our recently developed high-throughput broadband Raman flow cytometer at a throughput of >1,000 cells/s. Specifically, we characterized the intracellular content of paramylon from single-cell Raman spectra of 10,000 E. gracilis cells cultured under five different conditions and found that paramylon contents in E. gracilis cells cultured in an identical condition is given by a log-normal distribution, which is a good model for describing the number of chemicals in a reaction network. The capability of characterizing distribution functions in a label-free manner is an important basis for isolating specific cell populations for synthetic biology via directed evolution based on the intracellular content of metabolites.
1. Introduction
Natural products of microalgae have great potential for use in industry and medicine [1,2]. Compared to the conventional chemical engineering scheme, the production of various molecules based on microalgae is advantageous in carbon neutrality, ease and rapidity of production under outdoor atmospheric conditions, and ability to synthesize complicated and valuable materials. For this reason, significant efforts have been made to increase bioproduction efficiency through genetic engineering [3–5] and modification of culturing conditions [6–8]. To further enhance the efficiency of microalga-based bioproduction, it is important to comprehend differences in productivity between heterogeneous subpopulations of microalgal cells [9,10]. Specifically, it has been demonstrated that minor subpopulations can dominate under stressful conditions such as those used to induce the production of specific biomolecules [11]. Also, identifying rare but highly productive cells is crucial for both selection and evaluation in bioproduction-focused directed evolution. To this end, a technique to evaluate natural products of cellular ensembles without sacrificing single-cell resolution is demanded.
However, conventional methods such as dry-mass measurements, chromatograms, and mass spectrometry of cellular extracts are inadequate for meeting the need as they only quantify ensemble-averaged bioproducts and are incapable of performing large-scale analysis of single live cells. On the other hand, fluorescence-based flow cytometry [12] is an attractive approach to overcoming these limitations as it can analyze more than 1,000 single live cells per second without sacrificing single-cell resolution. By staining target molecules with fluorescent labels, it can characterize the heterogeneity of a large population of microbial cells [13]. Recent advances in imaging flow cytometry enable us to perform even more detailed phenotyping in combination with artificial-intelligence-based image analysis [14,15]. Although fluorescence-based flow cytometry is a powerful method for quantifying relatively larger intracellular biomolecules such as proteins and nucleic acids, its application to small metabolites is limited by the low binding specificity of fluorescent tags for smaller target molecules. To circumvent this problem, Raman microscopy has been exploited as a tool for evaluating various metabolites due to the availability of information about small molecules found in Raman spectra. Its label-free nature is suitable for use in selective breeding applications because the measurement process does not introduce probes that may affect cellular functions. However, the number of cells evaluated in Raman microscopy is typically limited to ∼1 cell per second or less, which is not sufficient for large-scale screening of cells [16–18].
In this paper, we demonstrate large-scale single-cell analysis of Euglena gracilis with our recently developed high-throughput broadband Raman flow cytometer at a throughput of >1,000 cells/s. [19]. E. gracilis is a unicellular microalgal species that has attracted a great deal of attention from researchers because of its high productivity of lipids and carbohydrates [20]. Among the latter, paramylon, a β-1,3-glucan, has high potential as a drug for HIV and colon cancer [21,22]. Paramylon is also important as an intermediate in the synthesis of wax ester, which can be converted to a biofuel [23]. By using a rapid-scan Fourier-transform coherent anti-Stokes Raman scattering (FT-CARS) flow cytometer as an analyzer, we acquired 10,000 single-cell Raman spectra of E. gracilis cultured under different conditions at a throughput of up to 80 cells/s and quantified their heterogeneity in terms of intracellular paramylon content. We found that the intracellular paramylon content was well fitted by a log-normal distribution, which is a good model for describing the amount of chemicals in a reaction network [24]. Although the log-normal distribution is widely observed in nature [25], it has not been discussed in the context of Raman spectroscopic data mainly due to the small number of sampled cells with conventional Raman spectroscopy techniques. We overcome this with the ability to interrogate numerous cells and hence characterize their distribution. As cell populations generally obey log-normal functions, distribution analysis is important for finding exceptional cells which potentially have high impact on their productivity of specific chemicals. Our findings pave the way towards a rapid and easy approach to assessing the biomaterial productivity of microalgae without the need for pretreatment of cells such as staining or target molecule extraction.
2. Results and discussion
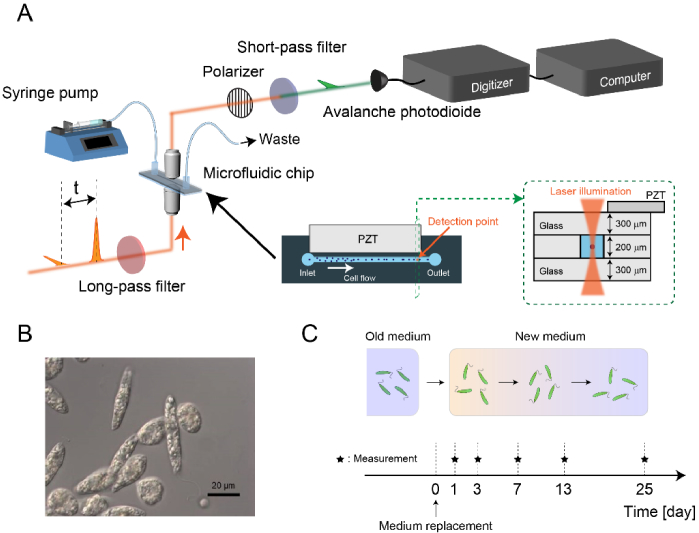
Figure 1(A) shows the schematic of the high-throughput FT-CARS flow cytometer. The detail of the setup is described in our previous report [19]. Briefly, a Ti:Sapphire femtosecond mode-locked laser (Coherent, Vitara-T-HP) is used as a light source. The output pulse of the laser is divided into two pulses, pump and probe, by a Michelson-interferometer-based pulse pair generator. The interval between the pulses τ is rapidly scanned at a rate of 24,000 scans/s, which realizes a high spectral acquisition rate of 24,000 spectra/s. The pulse pair is focused on the cells flowing in a microchannel on a microfluidic chip (a custom-made flow cell with a channel cross-section of 200 µm x 200 µm, Translume), equipped with a 3.66-MHz piezoelectric transducer (PZT) for acoustic cell focusing. The blue-shifted component of the probe pulse is isolated using an optical short-pass filter and detected by an avalanche photodiode (APD). The detected signals are digitized by a digitizer (AlazarTech, ATS9440) at a sampling rate of 100 MHz to rapidly transfer the data to a storage medium. The acquired data is Fourier-transformed to generate Raman spectra of flowing cells. For accurate Fourier transformation, the pump-probe interval is calibrated by measuring interferograms of a narrowband continuous-wave (CW) laser propagating collinearly with the Ti:Sapphire laser beam. Each signal acquisition is triggered by a forward scattering signal of the pump pulse.
Fig. 1.
Large-scale analysis of numerous single live E. gracilis cells. A: Setup of the high-throughput broadband FT-CARS flow cytometer. B: Image of E. gracilis cells. C: Procedure of preparing E. gracilis cells.
In order to investigate paramylon production in E. gracilis cells without being disturbed by the strong Raman signal of chlorophyll, we used SMZK strain of E. gracilis, a chloroplastless mutant of wild type Z strain [26] (Fig. 1(B)). The cells were cultured in 100 mL Erlenmeyer flasks using 50 mL of KH medium [27] with rotary shaking at 100 rpm at room temperature (26°C) without illumination. FT-CARS flow cytometric measurements were performed at 1, 3, 7, 13, and 25 days after the replacement of the culturing medium (Fig. 1(C)). 10,000 cells were measured under each condition at throughputs ranging from 20 to 80 cells/s at a cell flowing speed of 2 cm/s. All the measurements were performed sequentially on the same day to minimize fluctuations of the data originating from a long-term drift of the sensitivity of the FT-CARS flow cytometer. For each interrogated cell, 24 spectra were acquired and their average was used in our large-scale single-cell analysis described below.
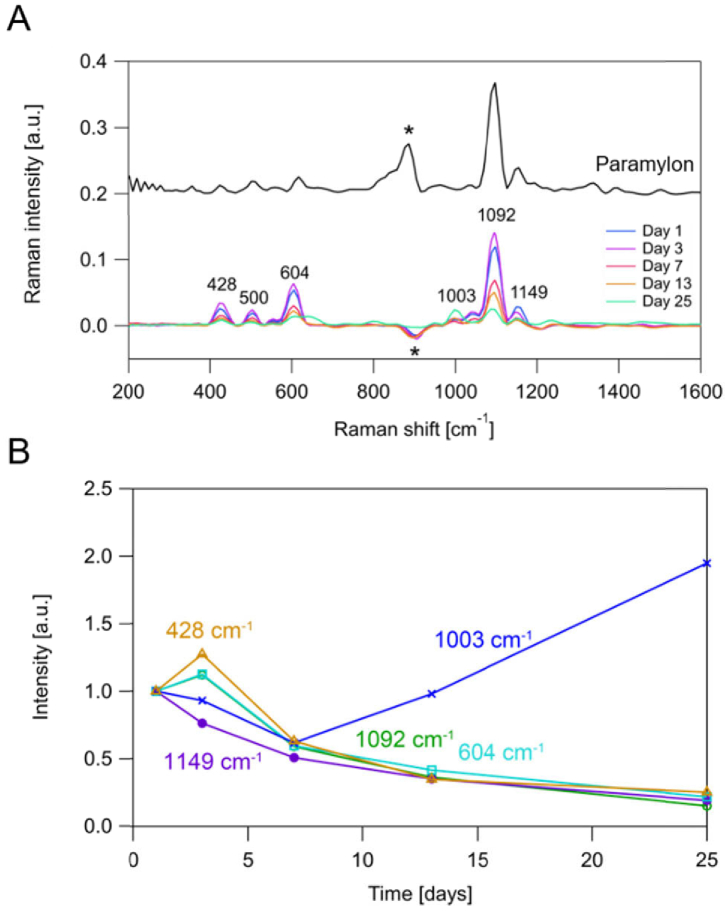
Figure 2(A) shows the averaged single-cell Raman spectra (N = 10,000) obtained from the E. gracilis samples as well as spectra of purified paramylon (paramylon particles) produced by our same spectrometer. Comparing these spectra indicates that the peaks at 428, 500, 604, and 1092 cm−1 are assignable to paramylon while the peak at 1003 cm−1 is assignable to proteins. The negative peak at around 900 cm−1 originated from the vibration of the PZT on the microfluidic chip, which is not related to active Raman modes within the sample. The peak at 1149 cm−1 seems assignable to paramylon based on the comparison of the spectra obtained from the cells and paramylon particles, but its time dependence is different from those of the other paramylon peaks (Fig. 2(B)). One possible origin of the difference is that paramylon particles in E. gracilis cells have different chemical characteristics, such as different degrees of polymerization and different crystallinity, depending on the culture conditions, which can result in different spectral profiles. Further investigation with cell sorting, size-exclusion chromatography, and X-ray crystallography can elucidate the difference. Our averaged data shows good agreement with a previous report [28] that the intracellular paramylon content of E. gracilis cells increases in the first few days following the introduction of the fresh growth medium and then decreases as the medium remains unchanged.
Fig. 2.
Time-dependent change of the averaged Raman spectral profiles (N = 10,000 for each condition). A: Raman spectrum of paramylon particles (top). Averaged Raman spectra of E. gracilis cells 1-25 days after the replacement of the culture (bottom). The asterisk (*) indicates the artificial peaks originating from the acoustic wave. B: Time-dependent changes of Raman intensities at 1092, 604 and 428 cm−1 (assignable to paramylon), 1149 cm−1 (unknown), and 1003 cm−1 (assignable to proteins).
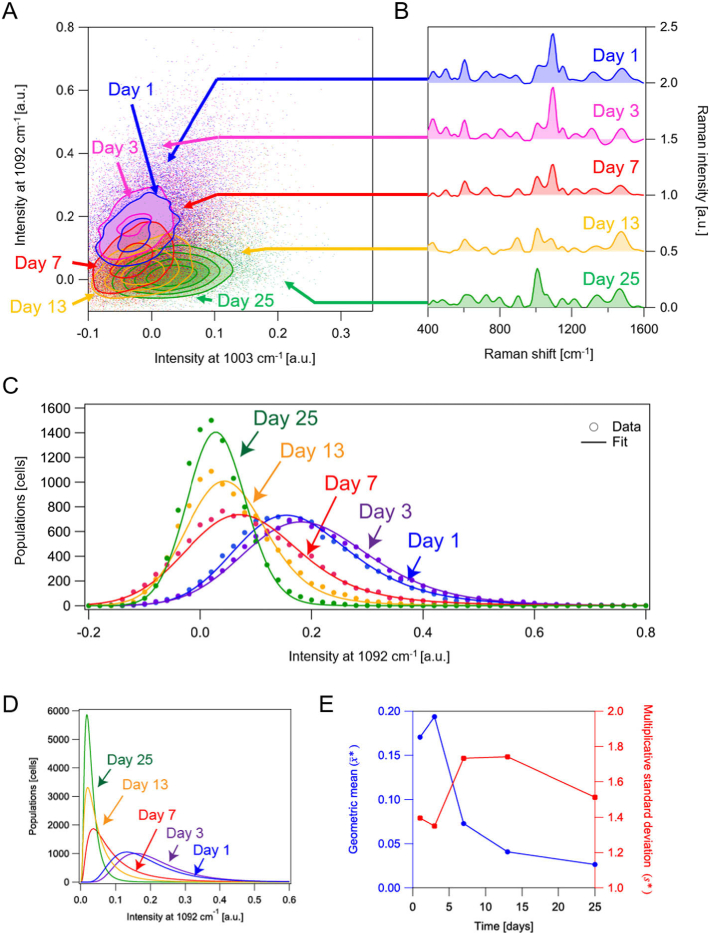
The FT-CARS flow cytometer revealed not only the population-averaged but also cell-to-cell heterogeneity of intracellular paramylon content. Figure 3(A) shows the contour plot of Raman intensities at 1092 and 998 cm−1 for the 10,000 E. gracilis cells under each condition, where minor populations outside the contours are shown by scatter plots. The time-dependent change of the position of each contour is consistent with that of the averaged spectra shown in Fig. 2(B). Figure 3(B) shows single-cell Raman spectra of E. gracilis cells whose positions in the scatter plot are indicated by the corresponding arrows. The spectra show that the characteristic Raman peaks of paramylon and proteins are detectable at a single-cell level. In Fig. 3(C), histograms of the intensities at 1092 cm−1 are shown for more detailed analysis of the heterogeneity of the intracellular paramylon content. All the histograms show non-symmetric probability distributions, which are often seen in conventional fluorescence flow cytometry data and fitted by lognormal functions [24]. Also in our data, each histogram is well fitted by a lognormal function convoluted by a Gaussian function as shown in Fig. 3(C), with the Gaussian function accounting for instrumental fluctuations, fluctuations of the cell position in the microfluidic device, and intracellular spatial heterogeneity of the paramylon distribution.
Fig. 3.
Statistical analysis of bioproducts in E. gracilis cells. A: Scatter plots of E. gracilis cells 1-25 days after the replacement of the culture in 998- and 1092-cm−1 Raman intensities. B: Raman spectra of single E. gracilis cells under different conditions. Corresponding plots in the scatter plots are indicated by the arrows. C: Histograms of single E. gracilis cells in paramylon content at different points in time. The solid curves are probability density functions modeled by lognormal functions convoluted by a Gaussian function. D: Extracted log-normal distributions by the fitting analysis in C. E: Time-dependent changes of the geometric mean and multiplicative standard deviation .
The extracted log-normal distributions (Fig. 3(D)) show that the paramylon content has considerable cell-to-cell variations despite the condition the E. gracilis cells used in this study are monoclonal. As a log-normal distribution is asymmetric, geometric mean and multiplicative standard deviation are good estimators such that the random variable is in the range of with 68.3% confidence, where is a times-divide operator [25]. Time-dependent changes of the and values are shown in Fig. 3(E). Although the decreasing trend in can be understood by the consumption of paramylon by the cells, the time-dependent change of is not straightforward to comprehend. The maxima of on days 7 and 13 are presumably due to the results of multiplicative propagation of inhomogeneity by time while the inhomogeneity is lowered on day 25 because most of the cells consumed almost all of the paramylon. The log-normal distribution implies that the cells were under steady growth, which is theoretically rationalized by considering a catalytic reaction network model with temporal fluctuations of reaction rates [24]. In general, the distribution deviates from a log-normal function when the population consists of multiple subpopulations having different metabolic pathways or the cells undergo a change of state [29]. Therefore, the present method is useful for identifying subpopulations as a deviation from a lognormal distribution in a label-free manner. Specifically, finding subpopulations with high productivity of valuable metabolites is highly effective for improving breeding based on metabolic engineering.
3. Summary
In conclusion, we demonstrated a rapid and easy way to screen the microalgal productivity of metabolites in a label-free manner. Specifically, we quantified intracellular paramylon content of E. gracilis cells at a single-cell level using the recently developed high-throughput broadband FT-CARS flow cytometer. The paramylon content of numerous single E. gracilis cells at 1-25 days after the replacement of the culturing medium was evaluated by the intensities of the characteristic Raman peaks of paramylon. The distributions of the intracellular paramylon content were well reproduced by a lognormal function convoluted by a Gaussian function, which indicates that the population consists of a single steady-state population. As the distribution of intracellular metabolite content is a good measure of cell state and the existence of subpopulations, our strategy is useful for label-free tracking of the change of cellular state and identification of different subpopulations. In the future, in combination with a cell sorting function [14,15], we can isolate specific cell populations for selective breeding based on the intracellular content of metabolites.
Funding
Cabinet Office, Government of Japan10.13039/501100002770; New Energy and Industrial Technology Development Organization10.13039/501100001863; Japan Science and Technology Agency10.13039/501100002241 (JPMJPR1878); Japan Society for the Promotion of Science10.13039/501100001691 (17K19099, 19H05633); Japan Society for the Promotion of Science10.13039/501100001691 (Core-to-core program); White-Rock Foundation; Kanagawa Institute of Industrial Science and Technology; Nakatani Foundation for Advancement of Measuring Technologies in Biomedical Engineering10.13039/501100008884; Ogasawara Foundation for the Promotion of Science and Engineering10.13039/501100008663.
Disclosures
K.G. is one of the inventors on a pending patent related to this work filed by the Japan Patent Office (nos. PCT/JP2016/089069 and WO2017119389A1). K.G. is a shareholder of a cell analysis startup. The authors declare that they have no other competing interests.
References
- 1.Bajhaiya A. K., Ziehe Moreira J., Pittman J. K., “Transcriptional Engineering of Microalgae: Prospects for High-Value Chemicals,” Trends Biotechnol. 35(2), 95–99 (2017). 10.1016/j.tibtech.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 2.Kottuparambil S., Thankamony R. L., Agusti S., “Euglena as a potential natural source of value-added metabolites. A review,” Algal Res. 37, 154–159 (2019). 10.1016/j.algal.2018.11.024 [DOI] [Google Scholar]
- 3.Joseph A., Aikawa S., Sasaki K., Matsuda F., Hasunuma T., Kondo A., “Increased biomass production and glycogen accumulation in apcE gene deleted Synechocystis sp. PCC 6803,” AMB Express 4(1), 17 (2014). 10.1186/s13568-014-0017-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumaraswamy G. K., Guerra T., Qian X., Zhang S., Bryant D. A., Dismukes G. C., “Reprogramming the glycolytic pathway for increased hydrogen production in cyanobacteria: metabolic engineering of NAD+-dependent GAPDH,” Energy Environ. Sci. 6(12), 3722–3731 (2013). 10.1039/c3ee42206b [DOI] [Google Scholar]
- 5.Chow T.-J., Su H.-Y., Tsai T.-Y., Chou H.-H., Lee T.-M., Chang J.-S., “Using recombinant cyanobacterium (Synechococcus elongatus) with increased carbohydrate productivity as feedstock for bioethanol production via separate hydrolysis and fermentation process,” Bioresour. Technol. 184, 33–41 (2015). 10.1016/j.biortech.2014.10.065 [DOI] [PubMed] [Google Scholar]
- 6.Aikawa S., Ho S.-H., Nakanishi A., Chang J.-S., Hasunuma T., Kondo A., “Improving polyglucan production in cyanobacteria and microalgae via cultivation design and metabolic engineering,” Biotechnol. J. 10(6), 886–898 (2015). 10.1002/biot.201400344 [DOI] [PubMed] [Google Scholar]
- 7.González-Fernández C., Ballesteros M., “Linking microalgae and cyanobacteria culture conditions and key-enzymes for carbohydrate accumulation,” Biotechnol. Adv. 30(6), 1655–1661 (2012). 10.1016/j.biotechadv.2012.07.003 [DOI] [PubMed] [Google Scholar]
- 8.Chen C.-Y., Zhao X.-Q., Yen H.-W., Ho S.-H., Cheng C.-L., Lee D.-J., Bai F.-W., Chang J.-S., “Microalgae-based carbohydrates for biofuel production,” Biochem. Eng. J. 78, 1–10 (2013). 10.1016/j.bej.2013.03.006 [DOI] [Google Scholar]
- 9.Lidstrom M. E., Konopka M. C., “The role of physiological heterogeneity in microbial population behavior,” Nat. Chem. Biol. 6(10), 705–712 (2010). 10.1038/nchembio.436 [DOI] [PubMed] [Google Scholar]
- 10.Paszek P., Ryan S., Ashall L., Sillitoe K., Harper C. V., Spiller D. G., Rand D. A., White M. R. H., “Population robustness arising from cellular heterogeneity,” Proc. Natl. Acad. Sci. 107(25), 11644–11649 (2010). 10.1073/pnas.0913798107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang N., Guan B., Kong Q., Sun H., Geng Z., Duan L., “Enhancement of astaxanthin production from Haematococcus pluvialis mutants by three-stage mutagenesis breeding,” J. Biotechnol. 236, 71–77 (2016). 10.1016/j.jbiotec.2016.08.009 [DOI] [PubMed] [Google Scholar]
- 12.Shapiro H. M., Practical Flow Cytometry (John Wiley & Sons, Inc., 2003). [Google Scholar]
- 13.Álvarez-Barrientos A., Arroyo J., Cantón R., Nombela C., Sánchez-Pérez M., “Applications of flow cytometry to clinical microbiology,” Clin. Microbiol. Rev. 13(2), 167–195 (2000). 10.1128/CMR.13.2.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nitta N., Sugimura T., Isozaki A., Mikami H., Hiraki K., Sakuma S., Iino T., Arai F., Endo T., Fujiwaki Y., Fukuzawa H., Hase M., Hayakawa T., Hiramatsu K., Hoshino Y., Inaba M., Ito T., Karakawa H., Kasai Y., Koizumi K., Lee S., Lei C., Li M., Maeno T., Matsusaka S., Murakami D., Nakagawa A., Oguchi Y., Oikawa M., Ota T., Shiba K., Shintaku H., Shirasaki Y., Suga K., Suzuki Y., Suzuki N., Tanaka Y., Tezuka H., Toyokawa C., Yalikun Y., Yamada M., Yamagishi M., Yamano T., Yasumoto A., Yatomi Y., Yazawa M., Di Carlo D., Hosokawa Y., Uemura S., Ozeki Y., Goda K., “Intelligent Image-Activated Cell Sorting,” Cell 175(1), 266–276.e13 (2018). 10.1016/j.cell.2018.08.028 [DOI] [PubMed] [Google Scholar]
- 15.Isozaki A., Mikami H., Hiramatsu K., Sakuma S., Kasai Y., Iino T., Yamano T., Yasumoto A., Oguchi Y., Suzuki N., Shirasaki Y., Endo T., Ito T., Hiraki K., Yamada M., Matsusaka S., Hayakawa T., Fukuzawa H., Yatomi Y., Arai F., Di Carlo D., Nakagawa A., Hoshino Y., Hosokawa Y., Uemura S., Sugimura T., Ozeki Y., Nitta N., Goda K., “A practical guide to intelligent image-activated cell sorting,” Nat. Protoc. 14(8), 2370–2415 (2019). 10.1038/s41596-019-0183-1 [DOI] [PubMed] [Google Scholar]
- 16.Song Y., Yin H., Huang W. E., “Raman activated cell sorting,” Curr. Opin. Chem. Biol. 33, 1–8 (2016). 10.1016/j.cbpa.2016.04.002 [DOI] [PubMed] [Google Scholar]
- 17.Zhang P., Ren L., Zhang X., Shan Y., Wang Y., Ji Y., Yin H., Huang W. E., Xu J., Ma B., “Raman-Activated Cell Sorting Based on Dielectrophoretic Single-Cell Trap and Release,” Anal. Chem. 87(4), 2282–2289 (2015). 10.1021/ac503974e [DOI] [PubMed] [Google Scholar]
- 18.Jing X., Gou H., Gong Y., Su X., Xu L., Ji Y., Song Y., Thompson I. P., Xu J., Huang W. E., “Raman-activated cell sorting and metagenomic sequencing revealing carbon-fixing bacteria in the ocean,” Environ. Microbiol. 20(6), 2241–2255 (2018). 10.1111/1462-2920.14268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiramatsu K., Ideguchi T., Yonamine Y., Lee S., Luo Y., Hashimoto K., Ito T., Hase M., Park J.-W., Kasai Y., Sakuma S., Hayakawa T., Arai F., Hoshino Y., Goda K., “High-throughput label-free molecular fingerprinting flow cytometry,” Sci. Adv. 5(1), eaau0241 (2019). 10.1126/sciadv.aau0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wakisaka Y., Suzuki Y., Iwata O., Nakashima A., Ito T., Hirose M., Domon R., Sugawara M., Tsumura N., Watarai H., Shimobaba T., Suzuki K., Goda K., Ozeki Y., “Probing the metabolic heterogeneity of live Euglena gracilis with stimulated Raman scattering microscopy,” Nat. Microbiol. 1(10), 16124 (2016). 10.1038/nmicrobiol.2016.124 [DOI] [PubMed] [Google Scholar]
- 21.Feldman S. C., Reynaldi S., Stortz C. A., Cerezo A. S., Damonte E. B., “Antiviral properties of fucoidan fractions from Leathesia difformis,” Phytomedicine 6(5), 335–340 (1999). 10.1016/S0944-7113(99)80055-5 [DOI] [PubMed] [Google Scholar]
- 22.Watanabe T., Shimada R., Matsuyama A., Yuasa M., Sawamura H., Yoshida E., Suzuki K., “Antitumor activity of the β-glucan paramylon from Euglena against preneoplastic colonic aberrant crypt foci in mice,” Food Funct. 4(11), 1685–1690 (2013). 10.1039/c3fo60256g [DOI] [PubMed] [Google Scholar]
- 23.Demirbas A., Fatih Demirbas M., “Importance of algae oil as a source of biodiesel,” Energy Convers. Manage. 52(1), 163–170 (2011). 10.1016/j.enconman.2010.06.055 [DOI] [Google Scholar]
- 24.Furusawa C., Suzuki T., Kashiwagi A., Yomo T., Kaneko K., “Ubiquity of log-normal distributions in intra-cellular reaction dynamics,” Biophysics (Biophys. Soc. Jpn.) 1, 25–31 (2005). 10.2142/biophysics.1.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Limpert E., Stahel W. A., Abbt M., “Log-normal Distributions across the Sciences: Keys and Clues,” BioScience 51(5), 341–352 (2001). 10.1641/0006-3568(2001)051[0341:LNDATS]2.0.CO;2 [DOI] [Google Scholar]
- 26.Oda Y., Nakano Y., Kitaoka S., “Utilization and Toxicity of Exogenous Amino Acids in Euglena gracilis,” Microbiology (London, U. K.) 128(4), 853–858 (1982). 10.1099/00221287-128-4-853 [DOI] [Google Scholar]
- 27.Koren L. E., Hutner S. H., “High-yield media for photosynthesizing Euglena gracilis Z,” J. Protozool. 14, 17 (1967). [Google Scholar]
- 28.Barsanti L., Vismara R., Passarelli V., Gualtieri P., “Paramylon (β-1,3-glucan) content in wild type and WZSL mutant of Euglena gracilis. Effects of growth conditions,” J. Appl. Phycol. 13(1), 59–65 (2001). 10.1023/A:1008105416065 [DOI] [Google Scholar]
- 29.Yamada K., Suzuki H., Takeuchi T., Kazama Y., Mitra S., Abe T., Goda K., Suzuki K., Iwata O., “Efficient selective breeding of live oil-rich Euglena gracilis with fluorescence-activated cell sorting,” Sci. Rep. 6(1), 26327 (2016). 10.1038/srep26327 [DOI] [PMC free article] [PubMed] [Google Scholar]