Abstract
Introduction
Prediction models for gestational hypertension and preeclampsia have been developed with data and assumptions from developed countries. Their suitability and application for low resource settings have not been tested. This review aimed to identify and assess the methodological quality of prediction models for gestational hypertension and pre-eclampsia with reference to their application in low resource settings.
Methods
Using combinations of keywords for gestational hypertension, preeclampsia and prediction models seven databases were searched to identify prediction models developed with maternal data obtained before 20 weeks of pregnancy and including at least three predictors (Prospero registration CRD 42017078786). Prediction model characteristics and performance measures were extracted using the CHARMS, STROBE and TRIPOD checklists. The National Institute of Health quality assessment tools for observational cohort and cross-sectional studies were used for study quality appraisal.
Results
We retrieved 8,309 articles out of which 40 articles were eligible for review. Seventy-seven percent of all the prediction models combined biomarkers with maternal clinical characteristics. Biomarkers used as predictors in most models were pregnancy associated plasma protein-A (PAPP-A) and placental growth factor (PlGF). Only five studies were conducted in a low-and middle income country.
Conclusions
Most of the studies evaluated did not completely follow the CHARMS, TRIPOD and STROBE guidelines in prediction model development and reporting. Adherence to these guidelines will improve prediction modelling studies and subsequent application of prediction models in clinical practice. Prediction models using maternal characteristics, with good discrimination and calibration, should be externally validated for use in low and middle income countries where biomarker assays are not routinely available.
Introduction
Hypertensive disorders of pregnancy (HDPs) are important causes of maternal morbidity and mortality globally but the burden is greatest in low- and middle-income countries (LMIC) [1–3]. These disorders of pregnancy include gestational hypertension, preeclampsia and eclampsia and are characterized by an increase in blood pressure and multi-organ derangements which range from mild to severe [4]. There is no known cure but daily administration of low dose aspirin early in the first trimester has been shown to reduce the incidence and the severity of preeclampsia [5–8]. Preeclampsia is a major indication for preterm delivery, accounting for about 15% of all preterm deliveries [9–13] and is a cause of increased healthcare costs through the prolonged stay of the mother or newborn in intensive care units [14].
Prediction models provide estimates of the probability or risk of the future occurrence of a particular outcome or event in individuals at risk of such an event [15]. Prediction models have also been used to identify women at high risk of developing HDPs later in pregnancy so as to provide for closer monitoring from early pregnancy onwards, including low dose aspirin prophylaxis [5–8] which has been shown to reduce the risk of developing preeclampsia.
The aim of this systematic review was to evaluate the performance of multivariate prediction models to address the question of the effectiveness of prediction models in identifying pregnant women at risk of gestational hypertension and preeclampsia. The objectives were to identify prediction models for gestational hypertension and preeclampsia; assess the methodological quality of the studies to develop and externally validate the prediction models using the CHARMS [16] checklist; and to identify prediction models that can be applied in low and middle income country settings.
Methods
This study was conducted using the critical appraisal and data extraction for systematic reviews of prediction modelling studies (CHARMS) [16], strengthening the reporting of observational studies in epidemiology (STROBE) [17] and the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) [18] checklists. The Population, Intervention, Comparator and Outcome (PICO) format for the review was as follows: P (pregnant women), I (prediction models), C (none) and O (gestational hypertension or preeclampsia). The study protocol was registered with the Prospero International Prospective Register of Systematic Reviews (CRD 42017078786).
Search strategy
A comprehensive systematic literature search with was conducted in PubMed/Medline, Embase, Cochrane Library, Web of Science and CINAHL databases from their inception through 18 September 2017. The search was updated to 15 October 2019 (DLV,EA). The MeSH database, EMTREE subject headings and CINAHL subject headings were used to construct the search strategy along with author keywords and general keywords. In addition, an electronic hand search was conducted in a number of journals from 10th September through 25th September, 2017 and from October 1 to October 15, 2019. Finally, grey literature was searched using the New York Academy of Medicine Grey Literature, OCLC’s OAISTER, and Open Grey databases.
The search strategy is provided as a supplementary file (S1 Data).
Eligibility/Inclusion criteria
Cohort studies, nested-case control studies and randomized controlled trials were eligible for inclusion in the study. Case-control, cross-sectional, animal studies, bio-molecular studies, letters, reviews and case reports were excluded because for prediction modeling studies we require absolute risks whereas case-control or cross-sectional studies only give relative risks. The primary outcomes for the included studies were gestational hypertension and preeclampsia.
Definition of terms
Gestational hypertension was defined as elevated systolic blood pressure equal to or greater than 140mmHg and/or diastolic blood pressure equal or greater than 90mmHg on at least two occasions four hours apart and appearing for the first time after 20 weeks of gestation without proteinuria [4]. Pre-eclampsia was defined as gestational hypertension with proteinuria of 300mg or more in a 24-hour urine sample or spot urine protein/creatinine ratio of 30mg/mmol [4]. Pre-eclampsia was further divided into early-onset preeclampsia (requiring preterm delivery before 34 weeks gestation) and late-onset preeclampsia (with delivery at or after 34 weeks gestation or later) as an outcome by some studies [19–24].
A prediction model [25] was defined as a logistic regression formula or a survival model with three or more predictors that could be used to estimate risk probabilities for individual patients or to distinguish between groups of patients of different risks.
Screening methods for study identification
Two reviewers (EA, MAC) independently assessed the titles and abstracts of the search results to select relevant papers for further screening. After removal of duplicates, the articles were obtained for screening/reading of the full text after which eligible papers were selected for inclusion in the systematic review. Discrepancies between the reviewers were resolved through consensus.
Data extraction and management
Data extraction of the identified studies was done by using the CHARMS checklists (EA). Extracted data were checked (MAC) and disagreements were resolved by consensus (EA, MAC). In case of disagreement a third reviewer (KKG) was consulted. Studies were analysed qualitatively given the large variability of the studies included.
The following categories were extracted: authors, journal, year of publication, region or place where study was conducted, period of data collection, study design, inclusion and exclusion criteria, the sample size of the derivation cohort and/or the validation cohort, the gestational age at which women were enrolled into the study and the number of outcomes. Other information extracted were the number and types of predictors, the target population for whom the prediction model is intended for, the handling of missing data, the modeling method used, the model selection method, the handling of continuous data, the method used for internal validation and whether or not an external validation was done.
Quality assessment
Quality of the studies was assessed using the CHARMS, STROBE and TRIPOD checklists and the National Institute of Health (NIH) [26] quality assessment tools for observational cohort and cross-sectional studies was independently assessed by two authors (EA, MAC). The NIH quality assessment tools focus on concepts that are key for critical appraisal of the internal validity of a study. The tool uses a 14-item checklist to assess the study design, inclusion criteria, outcome and variable description and collection and loss to follow up among others. Each item is scored as yes, no or other (not reported, not applicable or cannot determine). The tool also provides guidance on grading the studies as good, fair or poor. The studies were finally graded for risk of bias as”low” if risk of bias was unlikely, “moderate” if there were no essential flaws, but not all criteria had been satisfied and “high” if there were flaws in one or more important items. We adapted the tool and used 13 out of the 14 items, because one item, “for exposures that can vary in amount or level, did the study examine different levels of the exposure as related to the outcome (e.g., categories of exposure, or exposure measured as continuous variable)?” was not relevant to our review.
Meta-analysis
We performed a meta-analysis on 22 of the studies with preeclampsia as outcome, using the MedCalc Statistical Software version 19.1.7 (MedCalc Software Ltd, Ostend, Belgium; https://www.medcalc.org; 2020). These 22 studies had fully reported the area under the curve with 95% confidence intervals. We used the random effects model.
Results
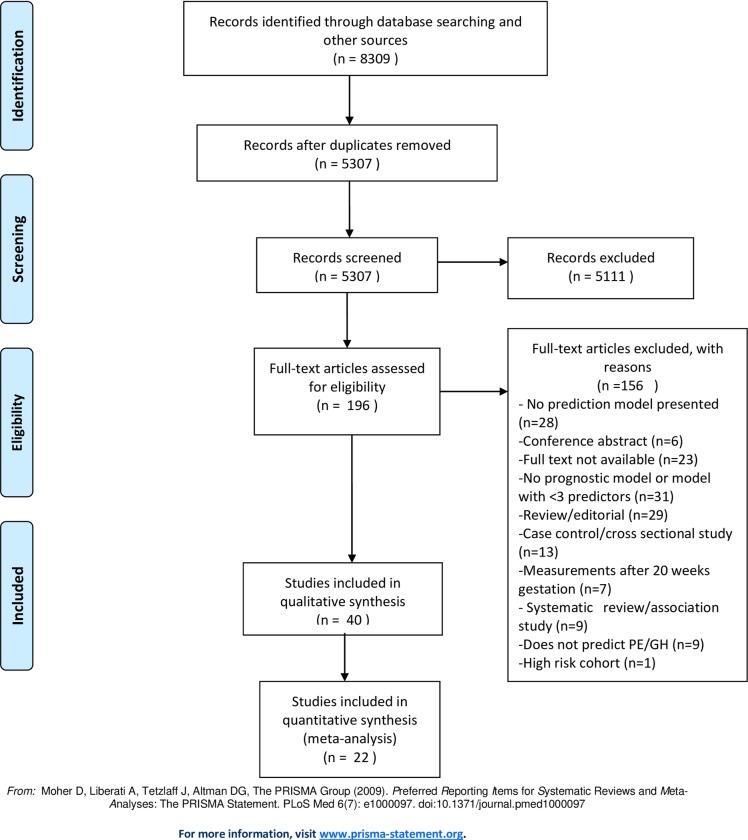
Fig 1 shows the flow diagram for inclusion and exclusion of relevant articles. The search yielded 8,309 papers. After removing 3,002 duplicates, 5307 papers were screened further for relevance and 196 papers selected for full text assessment. 156 articles were excluded based on reasons such as not presenting a prediction model, measurement of predictors done after 20 weeks of gestation and the prediction outcome not being preeclampsia or gestational hypertension. Finally 40 papers, published between 2000 and 2019, were selected for the review.
Fig 1. Flow diagram for inclusion and exclusion of relevant articles.
Prediction models for gestational hypertension and pre-eclampsia
All forty studies included in this review were conducted between 2000 and 2019. Table 1 gives an overview of important parameters of the selected studies. The studies have been grouped in the following order: maternal characteristics only, maternal characteristics and uterine artery Doppler, maternal characteristics with biomarkers and maternal characteristics with biomarkers and uterine artery Doppler.
Table 1. Overview of prediction models.
| Study | Study design | Centre | Study population | Outcome | Women, n (outcome events; predictors) | Number of events per variable |
| Mello et al, 2002 [14] | Prospective cohort | Single | Italian (Caucasian) | Preeclampsia | 187 (47; 8)) | 5.9 |
| Poon, et al, 2010 [34] | Prospective cohort | Single | United Kingdom (multi racial) | Early Preeclampsia, late preeclampsia, gestational hypertension. | 8366 (165; 8) | 20.6 |
| Muto et al, 2016 [42] | Prospective cohort | Single | Japanese | Preeclampsia, gestational hypertension | 1986 (50; 6) | 8.3 |
| Kuijk et al, 2014 [32] | Combined prospective and retrospective cohort | Multi centre | Dutch (multi racial) | Early onset preeclampsia | 229(15; 5) | 3 |
| Poon et al, 2008 [35] | Prospective cohort | Single | United Kingdom (multi racial) | Preeclampsia, gestational hypertension | 5193 (104; 5) | 5 |
| Benko et al, 2019 [53] | Prospective cohort | Multicentre | United Kingdom, Bulgaria, Spain (Multi racial) | Preeclampsia in twin pregnancies. | 2219 (171;11) | 15.5 |
| Boutin et al, 2018 [58] | Prospective cohort | Single | Canadian (multi ethnic) | Preterm preeclampsia, all preeclampsia. | 4612 (232;6) | 38.7 |
| Antwi et al, 2017 [47] | Prospective cohort | Multi centre | Ghanaian | Gestational hypertension | 2529 (261; 6) | 43.5 |
| Becker Rolf, 2011 [49] | Retrospective cohort | Single | German (Caucasian) | Preeclampsia, preterm delivery, intrauterine fetal growth restriction, placental abruption, intrauterine fetal death, early neonatal fetal death (within first week of postnatal life) | 15,855(172; 6) | 28.7 |
| North et al, 2011 [48] | Prospective cohort | Multi centre | United Kingdom, New Zealand, Ireland, Australia (multi racial) | Preeclampsia | 3529(186; 13) | 14.3 |
| Sepulvelda-Martinez et al, 2019 [56] | Nested case control (Prospective cohort) | Single | Chilean | Preterm preeclampsia, term preeclampsia. | 1756 (49; 7) | 7 |
| Myatt L. et al, 2012 [50] | Prospective cohort | Multi centre | American (multi racial) | Preeclampsia | 2,394 (176; 7) | 25.1 |
| Goetzinger et al,2010 [51] | Retrospective cohort | Single | American (multi racial) | Preeclampsia | 3716 (293; 5) | 58.6 |
| Odibo et al, 2011 [52] | Retrospective cohort | Single | American (multi racial) | Preeclampsia | 452(42;6) | 7 |
| Kuijk et al. 2011 [19] | Prospective cohort | Multi centre | Dutch (multi racial) | Early onset preeclampsia | 407 (28; 5) | 5.6 |
| Stamilio et al, 2000 [31] | Retrospective cohort | Single | American (multi racial) | Preeclampsia, Severe preeclampsia | 1998 (49; 4) | 12.2 |
| Gabbay-Benziv et al, [23] | Prospective cohort | Multi centre | American (multi racial) | Preeclampsia | 2433 (108; 5) | 21.6 |
| Allen et al, 2017 [44] | Prospective cohort | Single | United Kingdom (multi racial) | Preeclampsia, gestational hypertension, small-for-gestational age | 1045 (56; 5) | 11.2 |
| Mello et al, 2001 [45] | Prospective cohort | Single | Italian (Caucasian) | Pregnancy induced hypertension | 303 (76; 9) | 8.4 |
| Antwi et al, 2018 [60] | Prospective cohort | Multi centre | Ghananian | Gestational hypertension | 373 (25;6) | 4.1 |
| Zhang et al, 2019 [57] | Prospective cohort | Single | Chinese | Early preeclampsia, late preeclampsi, small-for-gestational age baby. | 3270 (43;8) | 5.3 |
| O’Gorman et al, 2016 [27] | Prospective cohort | Single | United Kingdom (multi racial) | Preterm Preeclampsia, term preeclampsia. | 35,948 (1058; 15) | 70.5 |
| Paré et al, 2014 [28] | Prospective cohort | Multi centre | American (multi racial) | Preeclampsia, gestational hypertension, HELLP* syndrome, eclampsia | 2,637 (431; 8) | 29.6 |
| Moon et al, 2015 [29] | Prospective cohort | Single | United Kingdom (multi racial) | Preeclampsia | 1177(102;11) | 9.3 |
| Park et al, 2013 [30] | Prospective cohort | Multi centre | Australian (multi racial) | Early Preeclampsia, late preeclampsia, gestational hypertension. | 3066 (83; 7) | 11.9 |
| Kenny et al, 2014 [33] | Prospective cohort | Multi center | New Zealand, Australia, United Kingdom, Ireland (multi racial) | Early onset preeclampsia, Preeclampsia | 3529 (278; 5) | 55.6 |
| Poon et al, 2009 [21] | Prospective cohort | Single | United Kingdom (multi racial) | Early Preeclampsia, Late preeclampsia, gestational hypertension. | 7797 (157; 8) | 19.6 |
| Herraiz et al, 2009 [36] | Prospective cohort | Single | Spanish (multi racial) | Early Preeclampsia, late preeclampsia | 152 (20;4) | 5 |
| Di Lorenzo et al, 2012 [37] | Prospective cohort | Single | Italian (multi racial) | Early onset preeclampsia, late onset preeclampsia, overall Preeclampsia, gestational hypertension | 2118 (preeclampsia(25), gestational hypertension (46); 8) | 3.1 |
| Goetzinger et al, 2014 [38] | Prospective cohort | Single | American (multi racial) | Preeclampsia | 578(49; 6) | 8.1 |
| Crovetto et al, 2014 [39] | Nested case-control (Prospective cohort) | Single | Spanish (multi racial) | Early Preeclampsia, late preeclampsi | 5759 (112; 10) | 11.2 |
| Gallo et al, 2016 [40] | Prospective cohort | Multi centre | United Kingdom (multi racial) | Preterm Preeclampsia, term preeclampsia. | 7748 (268; 11) | 24.4 |
| Skrastad et al, 2015 [41] | Prospective cohort | Single | Norway | Preeclampsia, gestational hypertension | 541 (21; 11) | 1.9 |
| Antonio et al, 2017 [43] | Prospective cohort | Single | Brazilian (multi racial) | Preeclampsia, gestational hypertension | 617 (34; 4) | 8.5 |
| Parra-Cordero et al, 2013 [24] | Nested case-control (Prospective cohort) | Single | Chilean | Early onset Preeclampsia, late onset preeclampsia. | 2619 (83; 4) | 20.7 |
| Myers et al, 2013 [20] | Prospective cohort | Multi centre | United Kingdom, New Zealand, Australia (multi racial) | Preterm preeclampsia | 3529 (55; 7) | 7.9 |
| Baschat et al, 2014 [46] | Prospective cohort | Multi centre | American (multi racial) | Early onset preeclampsia, Preeclampsia | 2441 (108; 5) | 21.6 |
| Scazzocchio, et al, 2017 [54] | Prospective cohort | Single | Spain | Early onset preeclampsia, late onset preeclampsia. | 4203 (169; 7) | 24.1 |
| Wright et al, 2019 [55]. | Prospective cohort | Multicentre | United Kingdom, Spain, Belgium, Italy, Greece | Early preeclampsia, pre-term preeclampsia. All preeclampsia. | 61,174 (1770; 11) | 160.9 |
| Lobo et al, 2019 [59] | Prospective cohort | Single | Brazil (multi ethnic) | Preterm Preeclampsia, term preeclampsia | 617 (34;8) | 4.2 |
| Study | Predictors | Type of model | Internal validation | External validation | Calibration (p-value Hosmer-Lemeshow test or calibration plot) | Model performance: PPV, NPV, Sensitivity, Specificity, |
| Mello et al, 2002 [14] | Maternal characteristics | Logistic regression | Yes | No | No | Yes |
| Poon, et al, 2010 [34] | Maternal characteristics | Logistic regression | Not stated | No | No | Yes |
| Muto et al, 2016 [42] | Maternal characteristics | Logistic regression | Not stated | No | No | Yes |
| Kuijk et al, 2014 [32] | Maternal characteristics | Logistic regression | Not applicable | Yes. Study externally validated a previously developed prediction model | Yes. Calibration plot and Hosmer-Lemeshow goodnesss -of-fit test. | Yes |
| Poon et al, 2008 [35] | Maternal characteristics | Logistic regression | Not stated | No | No | Yes |
| Benko et al, 2019 | Maternal characteristics | Parametric survival model | Not stated | Yes | Yes | Yes |
| Boutin et al, 2018 | maternal age, BMI, hypertension, chronic inflammatory disease, ovulation induction, in vitro fertilization | Proportional hazard model | Not stated | No | No | Yes |
| Antwi et al, 2017 | Maternal weight, height, parity, diastolic blood pressure, history of gestational hypertension, family history of hypertension | Logistic regression | Bootstrapping | Yes | No | No |
| Becker Rolf, 2011 [49] | Maternal characteristics, uterine artery pulsatility index | Logistic regression | Not stated | Yes | No | No |
| North et al, 2011 [48] | Maternal characteristics, uterine artery pulsatility index | Logistic regression | Cross validation | No | Yes. Calibration plot | Yes |
| Sepulveda-Martinez et al, 2019 | maternal characteristics, uterine artery pulsatility index | Logistic regression | Not stated | No | No | Yes |
| Myatt L. et al, 2012 [50] | Maternal characteristics, serum biomarkers | Logistic regression | Not stated | No | No | Yes |
| Goetzinger et al,2010 [51] | Maternal characteristics, serum biomarkers | Logistic regression | Not stated | No | No | Yes |
| Odibo et al, 2011 [52] | Maternal characteristics, serum biomarkers | Logistic regression | Not stated | No | No | Yes |
| Kuijk et al. 2011 [19] | Maternal characteristics, fasting blood glucose. | Logistic regression | Bootstrapping | No | Yes. Hosmer-Lemeshow goodnesss-of-fit test. | Yes |
| Stamilio et al, 2000 [31] | Maternal characteristics, serum biomarkers. | Logistic regression | Not stated | No | No | Yes |
| Gabbay-Benziv et al, [23] | Maternal characteristics, biomarkers. | Logistic regression | Not stated | No | No | Yes |
| Allen et al, 2017 [44] | Maternal characteristics, biomarkers. | Logistic regression | Not stated | No | No | Yes |
| Mello et al, 2001 [45] | Maternal characteristics, hematological and biochemical indices. | Logistic regression | Cross validation | No | No | Yes |
| Antwi et al, 2018 [47] | Maternal characteristics, serum biomarkers. | Logistic regression | Bootstrapping | Yes | Yes. Calibration plot | Yes |
| Zhang et al, 2019 | BMI, ethicity, parity, history of preeclampsia, chronic hypertension, PAPP-A, PlGF | Not stated | No | No | Yes | |
| O’Gorman et al, 2016 [27] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Not stated | No | No | Yes |
| Paré et al, 2014 [28] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Not stated | No | No | No |
| Moon et al, 2015 [29] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Not stated | No | No | Yes |
| Park et al, 2013 [30] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Not applicable because this study is an external validation of a previously developed prediction model | No | No | Yes |
| Kenny et al, 2014 [33] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Yes | No | No | Yes |
| Poon et al, 2009 [21] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Not stated | No | No | Yes |
| Herraiz et al, 2009 [36] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Not stated | Yes. Study externally validated a previously developed prediction model | No | Yes |
| Di Lorenzo et al, 2012 [37] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Not stated | No | No | Yes |
| Goetzinger et al, 2014 [38] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Not stated | Yes | Yes | Yes |
| Crovetto et al, 2014 [39] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Not stated | No | No | Yes |
| Gallo et al, 2016 [40] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Cross validation | No | No | Yes |
| Skrastad et al, 2015 [41] | Maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Not stated | Yes. Study externally validated a previously developed prediction model | No | Yes |
| Antonio et al, 2017 [43] | Maternal characteristics, biomarkers, Uterine artery pulsatility index. | Logistic regression | Not stated | No | No | Yes |
| Parra-Cordero et al, 2013 [24] | Maternal characteristics, biomarkers, Uterine artery pulsatility index. | Logistic regression | Not stated | No | No | Yes |
| Myers et al, 2013 [20] | Maternal characteristics, biomarkers, Uterine artery pulsatility index. | Logistic regression | Cross validation | No | No | Yes |
| Baschat et al, 2014 [46] | Maternal characteristics, biomarkers, Uterine artery pulsatility index. | Logistic regression | Cross validation | No | No | Yes |
| Scazzocchio et al 2017 | maternal characteristics, serum biomarkers, uterine artery pulsatility index | Logistic regression | Bootstrapping | Yes | Yes | Yes |
| Wright et al, 2019 | maternal characteristics, MAP, Uterine artery pulsatility index, PlGF | Logistic regression | Not stated | Yes | Yes | Yes |
| Lobo et al, 2019 | Maternal age, ethnicity, smoking status, MAP, Urerine artery pulsatility index, PlGF, PAPP-A | Fetal Medicine Foundation Algorithm | Not stated | Yes | No | Yes |
| Study | Discrimination (AUC) | Prediction rule/score chart/nomogram | Handling of missing values | Model selection: Stepwise selection, Univariate p-values, No selection | Handling of continuous data: Kept linear, categorized, dichotomized | |
| Mello et al, 2002 [14] | Yes; AUC (development) = 0.984; AUC (after external validation) = 0.892. | No | Not stated | Stepwise selection | Categorized | |
| Poon, et al, 2010 [34] | Yes; PE < 34 weeks: AUC = 0.794 (0.720 to 0.869); | Model formula with regression coefficients | Complete case analysis | Not stated | Kept linear | |
| PE ≥ 34 weeks: AUC = 0.796 (0.761 to 0.830). | ||||||
| Muto et al, 2016 [42] | No | Model formula with regression coefficients | Complete case analysis | Not stated | Categorized | |
| Kuijk et al, 2014 [32] | Yes; PE< 37 weeks: AUC = 62.4 (51.0 to 73.7). All PE:AUC = 61.4 (51.9 to 70.9) | Model formula with regression coefficients, score chart. | Regression imputation | Not stated | Categorized | |
| Poon et al, 2008 [35] | Yes; AUC = 0.852. | Model formula with regression coefficients | Complete case analysis | Not stated | Kept linear | |
| Benko et al, 2019 | Yes; development cohort: AUC = 0.65 (0.60 to 0.69); validation cohort: AUC not stated. | Regression coefficients | Not stated | survival analysis | Not stated | |
| Boutin et al, 2018 | AUC: 0.62 (0.58–0.66) | No | Complete case analysis | Univariate p-value | Not stated | |
| Antwi et al, 2017 [47] | Yes; development cohort: AUC = 0.70 (0.67 to 0.74); validation cohort: AUC = 0.68 (0.60 to 0.77). | Model formula with regression coefficients, score chart. | Multiple imputation | Stepwise backward selection | Kept linear | |
| Becker Rolf, 2011 [49] | No | Model formula with regression coefficients, algorithm. | Not stated | Not stated | Categorized | |
| North et al, 2011 [48] | Yes; AUC = 0.710 (0.706 to 0.714) | Model formula with regression coefficients | Imputation by expectation maximization method. | Stepwise backward selection | Kept linear, BMI categorized. | |
| Sepulveda-Martinez et al 2019 | AUC: 0.890 (0.837–0.955) | Algorithm | Not stated | Stepwise backward selection | Not stated | |
| Myatt L. et al, 2012 [50] | Yes; AUC = 0.73 (0.69 to 0.77). | No | Complete case analysis | Stepwise backward selection | Kept linear | |
| Goetzinger et al,2010 [51] | Yes; AUC = 0.70 (0.65 to 0.72). | Model formula with regression coefficients | Complete case analysis | Stepwise backward selection | Categorized | |
| Odibo et al, 2011 [52] | Yes; AUC = 0.77 (0.63 to 0.81). | Model formula with regression coefficients | Complete case analysis | Stepwise backward selection | Kept linear | |
| Kuijk et al. 2011 [19] | Yes; AUC = 0.65 (0.56 to 0.74). | Model formula with regression coefficients | Single regression imputation | Not stated | Kept linear | |
| Stamilio et al, 2000 [31] | Yes; AUC = 0.75. | Model formula with regression coefficients | Complete case analysis | Stepwise backward selection | Categorized | |
| Gabbay-Benziv et al, [23] | Yes; 0.78 (0.72 to 0.85) | Prediction rule | Complete case analysis | Not stated | Categorized | |
| Allen et al, 2017 [44] | Yes; AUC = 0.81 (0.69 to 0.93) | Model formula with regression coefficients | Complete case analysis | Stepwise selection | Kept linear | |
| Mello et al, 2001 [45] | Yes; prediction at 16 weeks: AUC = 0.952 (0.895 to 1.000); prediction at 20 weeks: AUC = 0.851 (0.739 to 0.941) | Model formula with regression coefficients | Complete case analysis | Not stated | Categorized | |
| Antwi et al, 2018 | AUC: 0.82 (0.74–0.89) | Model formula with regression coefficients | Complete case analysis | Stepwise backward selection | Kept linear | |
| Zhang et al, 2019 | AUC for early PE: 0.90 (0.89–0.91); AUC for late PE: 0.82 (0.81–0.84) | PREDICTOR Algorithm | Complete case analysis | Not stated | Not stated | |
| O’Gorman et al, 2016 [27] | Yes; PE< 37 weeks: AUC = 0.907; PE ≥37 weeks: AUC = 0.796. | Model formula with regression coefficients | Complete case analysis | Stepwise backward selection | Kept linear | |
| Paré et al, 2014 [28] | No | Model formula with regression coefficients | Not stated | Stepwise backward selection | Kept linear | |
| Moon et al, 2015 [29] | Yes; Model nulliparous: AUC = 0.88 (0.80 to 0.94); Model multiparous: AUC = 0.84 (0.75 to 0.91). | Model formula with regression coefficients | Complete case analysis | Stepwise backward selection | Not stated | |
| Park et al, 2013 [30] | Yes; AUC = 0.926 (0.916–0.936). | Model formula with regression coefficients | Complete case analysis | Not stated | Kept linear | |
| Kenny et al, 2014 [33] | Yes; development cohort: AUC = 0.73(0.70 to 0.77); validation cohort: AUC = 0.68(0.63 to 0.74). | Model formula with regression coefficients | Imputation by expextation maximization method, complete case analysis for uterine artery pulsatility index | Stepwise backward selection | Kept linear | |
| Poon et al, 2009 [21] | No | model formula with regression coefficients | Complete case analysis | Not stated | Kept linear | |
| Herraiz et al, 2009 [36] | Yes; PE< 34 weeks: AUC = 0.779 (0.641 to 0.917); PE 34 weeks: AUC = 0.641 (0.481 to 0.801). | Model formula with regression coefficients | Not stated | Not applicable | Kept linear | |
| Di Lorenzo et al, 2012 [37] | Yes; AUC = 0.895 | Model formula with regression coefficients | Complete case analysis | Step down procedure | Kept linear | |
| Goetzinger et al, 2014 [38] | Yes; development cohort: AUC = 0.80 (0.73 to 0.86); validation cohort: AUC = 0.78 (0.69 to 0.86). | Model formula with regression coefficients | Complete case analysis | Stepwise backward selection | Categorized | |
| Crovetto et al, 2014 [39] | Yes; AUC = 0.960 (0.919 to 0.999). | Model formula with regression coefficients | Not stated | Stepwise forward selection | Kept linear | |
| Gallo et al, 2016 [40] | Yes; PE<32 weeks: AUC = 0.995 (0.990 to 0.999); PE< 32 weeks: AUC = 0.930 (0.892 to 0.968); PE ≥ 37 weeks:AUC = 0.773 (0.771 to 0.805). | Model formula with regression coefficients | Complete case analysis | Not stated | Kept linear | |
| Skrastad et al, 2015 [41] | Yes; AUC (FMF*) = 0.77(0.67 to 0.87), AUC (PREDICTOR¥) = 0.74 (0.63–0.84) | Fetal Medicine Foundation algorithm | Complete case analysis | Not stated | Kept linear | |
| Antonio et al, 2017 [43] | Yes; PE <34 weeks: AUC = 0.946 (0.919 to 0.973); PE< 37 weeks: AUC = 0.870 (0.798 to 0.942); PE< 42 weeks: AUC = 0.857 (0.807 to0.907) | Model formula with regression coefficients | Complete case analysis | Not stated | Kept linear | |
| Parra-Cordero et al, 2013 [24] | ROC curve presented but AUC values not provided. | Model formula with regression coefficients | Complete case analysis | Not stated | Kept linear | |
| Myers et al, 2013 [20] | Yes; AUC = 0.84 (0.77 to 0.91) | No | Complete case analysis | Stepwise selection (forward selection followed by series of backward selection) | Age and blood pressure kept linear, BMI categorized | |
| Baschat et al, 2014 [46] | Yes; PE < 34 weeks: AUC = 0.83 (0.74 to 0.91); all PE: AUC = 0.82 (0.78 to 0.86). | Model formula with regression coefficients | Complete case analysis | Lasso logistic regression | Categorized | |
| Scazzocchio et al, 2017 | Early onset PE AUC = 0.94 (95% CI, 0.88–0.99), late onset PE AUC = 0.72 (95% CI, 0.66–0.77) | Regression coefficients | Not stated | Not stated | Not stated | |
| Wright et al, 2019 | Early PE:AUC = 0.95 (0.93–0.97); Pretem PE = 0.91 (0.89–0.91); All PE = 0.83 (0.81–0.84) | Algorithm | Not stated | Not stated | Not stated | |
| Lobo et al, 2019 | Preterm PE AUC:0.94 (0.92–0.97); Term PE AUC: 0.87 (0.79–094) | FMF Algorithm | Complete case analysis | Not stated | Not stated | |
Twelve studies were conducted in the United Kingdom, eight in the United States of America, four each in Australia, Spain and Italy and three in New Zealand. Two studies were done in the Netherlands, Ireland, Brazil, Chile and Ghana with one each in Japan, China, Germany, Norway, Bulgaria, Greece, Belgium and Canada.
Most of the studies were prospective cohort studies (33/40 = 82.5%), four were retrospective cohort studies (10%), three were nested-case control studies (7.5%) and one study combined a retrospective and prospective cohort design for data collection. The prediction models were derived through logistic regression or parametric survival modeling.
The gestational age at inclusion into the studies ranged between eight and twenty weeks. All the gestational ages were confirmed by ultrasound. The sample size for the studies ranged between 173 and 35,948. The events per variable in the studies ranged between 2.1 and 88.2.
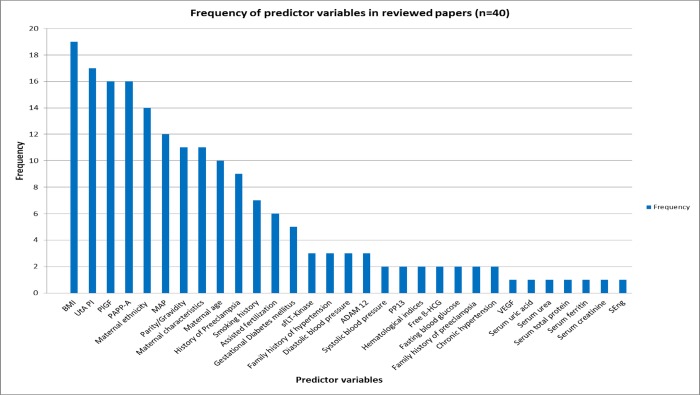
Seventy seven percent of all the prediction models combined biomarkers with maternal clinical characteristics. Body mass index (BMI) was the most frequently used predictor (19/40). Other maternal clinical predictors used in the models were first trimester systolic blood pressure and diastolic blood pressure, mean arterial pressure, maternal ethnicity, parity, previous history of preeclampsia, family history of hypertension, family history of preeclampsia, history of smoking and history of gestational diabetes mellitus. The following biomarkers were included: uterine artery pulsatility index (UtA PI, 17/40), pregnancy associated plasma protein-A (PAPP-A) (16/40) and placental growth factor (PlGF) (16/40). The following predictors were used less than ten times in the studies under review: free beta human chorionic gonadotropin (fß-HCG), alpha feto protein (AFP), soluble fms-like tyrosine kinase-1 (sFlt‐1), placental protein 13 (PP13), A disintegrin and metalloproteinase 12 (ADAM12), soluble endoglin (sEng) and vascular endothelial growth factor (VEGF). Fig 2 shows the frequency of predictor variables in the prediction models.
Fig 2. Frequency of predictor variables in the prediction models.
Methodological quality of the studies to develop or validate prediction models using the CHARMS, STROBE and TRIPOD checklists
Source of data
All the studies indicated the type of study design used to obtain data for the prediction modeling. 37 were cohort studies whilst three were nested case-control studies.
Participants
All the studies indicated the participant eligibility and recruitment criteria, including the study location, number of centres and the inclusion and exclusion criteria.
Outcomes to be predicted
All the studies gave a standard definition for the outcome(s) to be predicted. Most of the studies had a single outcome while eleven studies had two or more outcomes.
Candidate predictors
All the studies defined and described the candidate predictors and the methods for their measurement. The timing of predictor measurements was also provided in all studies. Handling of predictors in the modeling process was described by 31 out of the 40 studies. Nine of the studies categorized continuous variables whilst 21 studies kept continuous variables linear.
Sample size
All studies provided the number of participants and the number of outcomes. Only nine of the studies explicitly estimated the sample size before the onset of the study. The number of outcomes in relation to the number of candidate predictors (events per variable) were deduced from the data and ranged between 2.1 and 88.2.
Missing data
The number of participants with any missing value for each predictor was not provided by the studies. Nine of the studies did not indicate how missing data were handled. Complete case analysis was used by 26 out of the 40 studies whilst five studies imputed missing data using the single regression imputation method [19,32], expectation maximization method [33,48] and multiple imputation [47].
Model development
All the studies selected candidate predictors for inclusion in the model through univariate analysis using a pre-determined p-value. Logistic regression and survival modelling were used to derive the prediction models. For selection of predictors during multivariable modeling, one study used the stepwise forward selection method, 14 studies used the stepwise backward selection method and two studies used stepwise selection without further specification. One study [46] applied the Lasso regression approach and another survival analysis whilst 21 studies did not state the method used for deriving the model.
Model performance
Discrimination of the prediction models, depicted by the c-statistic or the area under the receiver operating characteristic (ROC) curve was reported by 34 (85%) of the studies while calibration was reported by five (12.5%) studies. Classification measures were reported by 37 (92.5%) of the studies (Table 1).
Model evaluation
Internal and external validation
Internal validation was reported by eleven out of 40 studies, using bootstrapping [19,47,54,60], cross validation [14,20,40,46,48], split sample [61] and back propagation of error method for artificial neural networks [45]. Nine out of the 40 prediction models were externally validated.
Risk of bias assessment
Risk of bias refers to the extent that flaws in the design, conduct, and analysis of the primary prediction modelling study lead to biased, often overly optimistic, estimates of predictive performance measures such as model calibration, discrimination, or (re)classification (usually due to overfitted models).
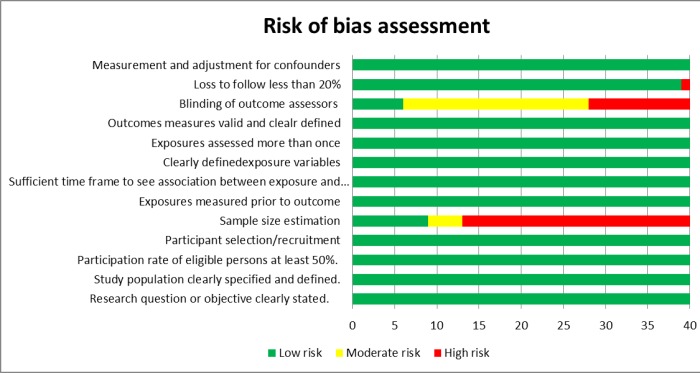
Fig 3 shows the risk of bias assessment of the studies. Most of the studies had a low risk of bias. The major source of bias related to sample size estimations, only stated in detail by nine out of 40 studies.
Fig 3. Risk of bias assessment of the prediction studies.
Details of the risk of bias assessment are presented in Table 2.
Table 2. Quality assessment of prediction model studies using the National Institute of Health criteria.
| Study | Research question or objective in this paper clearly stated? | Study population clearly specified and defined? | Participation rate of eligible persons at least 50%? | Study subjects recruited from the same or similar populations (including the same time period)? Inclusion and exclusion criteria prespecified and applied uniformly to all participants? | Sample size justification, power description, or variance and effect estimates provided? | Exposure(s) of interest measured prior to the outcome(s) being measured? | Sufficient time frame to reasonably expect to see an association between exposure and outcome if it existed? | Exposure measures (independent variables) clearly defined, valid, reliable, and implemented consistently across all study participants? | Exposure(s) assessed more than once over time? | Outcome measures clearly defined, valid, reliable, and implemented consistently across all study participants? | Outcome assessors blinded to the exposure status of participants? | Loss to follow-up after baseline 20% or less? | Key potential confounding variables measured and adjusted statistically for their impact on the relationship between exposure(s) and outcome(s)? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G.Mello et al 2002 | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes (0) | Yes |
| Becker Rolf. | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes (0) | Yes |
| Myatt L. et al. | Yes | Yes | Yes (100%) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes (1.9%) | Yes |
| Goetzinger et al | Yes | Yes | Yes (100%) | Yes | NR | Yes | Yes | Yes | Yes | Yes | Yes | Yes (7%) | Yes |
| Odibo et al. | Yes | Yes | Yes (94.8%) | Yes | NR | Yes | Yes | Yes | Yes | Yes | Cd | Yes (5.2%) | Yes |
| O’Gorman et al. | Yes | Yes | Yes (100%) | Yes | NR | Yes | Yes | Yes | Yes | Yes | CD | Yes (0) | Yes |
| Paré et al. | Yes | Yes | Yes (100%) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | CD | No | Yes |
| Moon et al | Yes | Yes | Yes (100%) | Yes | CD | Yes | Yes | Yes | Yes | Yes | CD | Yes (1.9%) | Yes |
| Park et al. | Yes | Yes | Yes (98.1%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (1.9%) | Yes |
| Van Kuijk et al. | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes (0) | Yes |
| Stamilio et al. | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (0) | Yes |
| Kenny et al. | Yes | Yes | Yes (99%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (1%) | Yes |
| Poon, et al. | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (0) | Yes |
| Poon et al | Yes | Yes (91.9%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (8.1%) | Yes | |
| Herraiz et al. | Yes | Yes | Yes (87.9%) | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes (12.1%) | Yes |
| Di Lorenzo et al. | Yes | Yes | Yes (98%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (2.4%) | Yes |
| Goetzinger et al. | Yes | Yes | Yes (98%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (2%) | Yes |
| Crovetto et al. | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (0) | Yes |
| Gallo et al. | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (0) | Yes |
| Skrastad et al | Yes | Yes | Yes (96.6%) | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes (3.4%) | Yes |
| Muto et al | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (0) | Yes |
| Antonio et al. | Yes | Yes | 87.6% | Yes | Yes | Yes | Yes | Yes | Yes | Yes | CD | Yes (12.4%) | Yes |
| Van Kuijk et al. | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (0) | Yes |
| Gabbay-Benziv et al. | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (0) | Yes |
| Poon et al. | Yes | Yes | Yes (92.9%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (7.1%) | Yes |
| Allen et al. | Yes | Yes | Yes (83.6%) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | CD | Yes (16.4%) | Yes |
| Parra-Cordero et al. | Yes | Yes | Yes (100%) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | CD | Yes (0) | Yes |
| Myers et al. | Yes | Yes | Yes (99%) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | CD | Yes (1%) | Yes |
| Mello et al. | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes (0) | Yes |
| Baschat et al. | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | CD | Yes (0) | Yes |
| Antwi et al. | Yes | Yes | Yes (100%) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes (0) | Yes |
| North et al. | Yes | Yes | Yes (94.8%) | Yes | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes (5.2%) | Yes |
| Antwi et al, 2018 | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Benko et al | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Scazzocchio et al | Yes | Yes | Yes (100%) | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Sepulvelda-Martinez | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Wright et al | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Zhang et al | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Boutin et al | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Lobo et al | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes |
CD- Could not be determined; NR- Not reported.
Prediction models applicable in low and middle income settings
Apart from two models each from Brazil and Chile, both Upper middle income countries in Latin America, and two models from Ghana, all the other models in the literature that met our inclusion criteria were developed in high income countries of Europe, Japan, Australia, New Zealand, China, Canada and the United States of America.
Meta-analysis
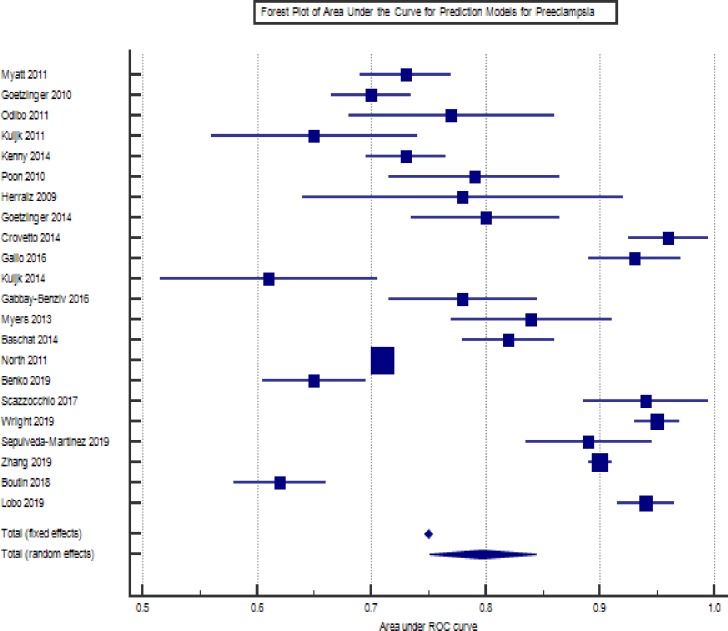
The forest plot of the meta-analysis of the prediction models for preeclampsia is presented in Fig 4. The I2 was 99%. Overall area under the curve was 0.79 (0.75–0.84).
Fig 4. Forest plot of prediction models for preeclampsia.
Discussion
We set out to review the evidence in the published literature on the performance of multivariate prediction models for gestational hypertension and preeclampsia to assess the effectiveness of prediction models in identifying pregnant women at risk for gestational hypertension and preeclampsia. The specific objectives of this study were to identify prediction models for gestational hypertension and preeclampsia in the literature, assess the methodological quality of the prediction modeling studies by applying the CHARMS checklist and identify prediction models that can be applied in low and middle income country settings.
Prediction models for gestational hypertension and preeclampsia
Our study identified 40 prediction models for gestational hypertension and preeclampsia, most of which had been developed and validated in high-income countries in Europe, Australia and the USA. Only two of such studies had been conducted in a low and middle income country setting. Most of the prediction models were developed in single centres but a few had been developed using data from multiple centres in one or more countries.
Methodological quality of prediction modeling studies
The STROBE (Strengthening the reporting of observational studies in epidemiology), TRIPOD (Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis) and the CHARMS checklists have outlined steps for developing and validating prediction models. The CHARMS checklist in particular provides guidance as to the items to extract when conducting a systematic review of prediction studies. An assessment of the methods used in model development in the studies evaluated in this review showed gaps in application of recommendations in the CHARMS, TRIPOD and STROBE checklists. The following domains of the CHARMS checklist were not adequately addressed in most of the studies: the source of data, study participants, outcome(s) to be predicted, candidate predictors, sample size, missing data, model development, model performance, model evaluation, results, interpretation and discussion. For example continuous predictors were dichotomized in some of the studies despite evidence and recommendations to the contrary [62–65]. Bias in predictor selection is known to occur when continuous predictors are categorized. Again, categorizing continuous variables assumes that there is a stepwise change in risk from one cut-off point to another. Bodnar et al [66] have demonstrated a dose-dependent relationship between pre-pregnancy BMI and the risk of preeclampsia. As BMI increases, so does the risk of preeclampsia. Therefore categorizing the predictor variable makes the functional relationship between the continuous variable (predictor) and the outcome variable linear, hence nonlinear transformations such as restricted cubic splines or fractional polynomials cannot be applied [62,67,68].
To prevent overestimation of risks by prediction models, it is recommended that the number of outcomes in relation to the number of predictors (events-per-variable) should be at least ten to one [69,70]. This requires an adequate sample size that ensures that there are enough outcomes in the study. Hence sample size estimation is an important methodological consideration so that at the onset of the study an adequate events-per-variable can be assured and thereby prevent overestimation of the predictive performance of the models (overfitting). Unfortunately, most of the studies under review did not report on sample size estimation. An adequate sample size also minimizes predictor selection bias. Predictor selection bias tends to be greater in smaller datasets when the events-per-variable ratio is small, especially when there are weak predictors in the dataset [16].
Information on missing data should be reported as part of the results of the studies. This includes the number of participants with any missing value (including values for both predictors and outcomes), number of participants with missing data for each predictor and how the missing data were handled, for example by complete case analysis, imputation or other methods. Information about missing data gives an idea as to the extent of bias, dependent on the reasons for the missing data. Where data were not missing completely at random, the prediction estimates are likely to be biased [64,71–75]. Missing data are seldom missing completely at random and may often be related to other observed participant data. Consequently, participants with completely observed data are likely to be different from those with missing data. Complete-case analysis which was the commonest method used to handle missing data in most studies deletes participants with a missing value from the analysis, thereby resulting in loss of information from a subset of the study population. This may result in over or under estimation of the predictive effect and reduced performance in an external population.
Prediction model performance is one of the important domains to be in the reported on [71]. Model performance indicators include calibration, discrimination and classification. It is recommended that discrimination and calibration should always be reported for prediction models. Discrimination indicates how well the prediction model distinguishes between two outcomes such as disease or non-disease and is assessed using the c-statistic or the area-under-the-curve (AUC) of a receiver operating characteristic curve [76–78]. The AUC ranges from 0.5 to 1 and represents the prediction model’s ability to correctly classify a randomly selected individual as being from one of two hypothetical populations [78–81]. An AUC value of 1.0 is considered perfect, 0.9–0.99 excellent, 0.8–0.89 good, 0.7–0.79 fair and 0.51–0.69 poor. An AUC of 0.5 is considered non-informative. The AUC in the studies under review ranged between 0.65 and 0.98. Apart from the study by Kuijk et al [19] which had an AUC of 0.65, all the other studies reported AUC greater than or equal to 0.70, indicating good to excellent discrimination. Calibration refers to how well the predicted risks compare to the observed outcomes. Usually this is evaluated in a calibration plot by graphically plotting observed against predicted event rates [16,67,82]. Calibration plots may be supplemented by the Hosmer-Lemeshow test, which is a formal statistical test to determine whether calibration is adequate. Unfortunately most of the studies under review did not report the calibration plot. This shortcoming leaves room for uncertainty in applying the model in clinical practice because one cannot determine the probability range within which the model works well. Both discrimination and calibration are essential in determining model performance.
Prediction model evaluation can be undertaken by internal validation (using the same dataset as that used to develop the model) and external validation (using a different dataset to that used in developing the model). The external dataset should be collected using the same predictor and outcome definitions and measurements. Again most of the studies did not report whether or not internal validation had been performed thus breaching an important methodological consideration. Most of the studies did not follow the guidelines in the TRIPOD, STROBE and CHARMS checklists. A possible explanation may be that some of studies were conducted prior to the development of these guidelines so the investigators may not have had the benefit of these methodological guidelines.
Prediction models applicable in low and middle income settings
Only five of the studies had been conducted in a low-and-middle income country setting. Given contextual differences between high and low-and-middle income countries, many of the prediction models under review which have been developed in high income countries at present may not be applicable in most low-and-middle income countries. This is because these prediction models included biomarkers and uterine artery pulsatility index as predictors in addition to maternal clinical characteristics [20,21,23,24,27,28,30,36–41,44,46,48–52,61,83]. At present uterine Doppler measurement and serum biomarker assays are not widely available in many low-and-middle income countries. Therefore prediction models using biomarkers and uterine artery pulsatility index may not be routinely applied in these settings.
Generally, prediction models developed in one setting have to be externally validated in new populations to assess their performance before applying them in clinical decision-making. The model intercept and the regression coefficients often have to be updated to fit the new context or population to which the prediction model is being applied to. Thus prediction models developed elsewhere may be updated for use in other settings provided the predictors and outcome are the same. In situations where a prediction model includes variables which cannot be measured in the setting where the model is to be applied, that model cannot be used in that population. Consequently most prediction models developed in high income countries and including variables like serum biomarkers and uterine artery pulsatility index are at present not applicable in most low-and-middle income countries where the burden of hypertensive disorders of pregnancy is greater. Presently prediction models using maternal clinical characteristics, and which give optimum predictions can be externally validated and applied in low resource settings.
Conclusion
Most of the studies evaluated did not completely follow the CHARMS, TRIPOD and STROBE guidelines in prediction model development and reporting. Adherence to these guidelines will improve prediction modelling studies and subsequent application of prediction models in clinical practice. Prediction models using maternal characteristics, with good discrimination and calibration, should be externally validated for use in low and middle income countries where biomarker assays are not routinely available.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
We acknowledge the assistance of Dr. Daniel Boateng (UMC Utrecht) with the risk of bias assessment.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Say L, Chou D, Gemmill A, Tunçalp Ö, Moller A-B, Daniels J, et al. Global causes of maternal death: a WHO systematic analysis. Lancet Glob Health 2014;2:e323–33. 10.1016/S2214-109X(14)70227-X [DOI] [PubMed] [Google Scholar]
- 2.Steegers Eric A P, Dadelszen Peter von, Duvekot RP, Johnnes J. Pre-eclampsia. Lancet. 2010;376(9741):631–44. 10.1016/S0140-6736(10)60279-6 [DOI] [PubMed] [Google Scholar]
- 3.WHO, UNICEF, UNFPA, The World Bank and the United Nations Population Division. Trends in maternal mortality: 1990 to 2013. Estimates by WHO, UNICEF, UNFPA, The World bank and the United Nations Population Division. 2014 Available from: www.who.int
- 4.Tranquilli A, Dekker G, Magee L, Roberts J, Sibai B, Steyn W, et al. The classification, diagnosis and management of the hypertensive disorders of pregnancy: A revised statement from the ISSHP. Pregnancy Hypertens. 2014;4:97–104. 10.1016/j.preghy.2014.02.001 [DOI] [PubMed] [Google Scholar]
- 5.Bujold E, Roberge S, Lacasse Y, Bureau M, Audibert F, Marcoux S, et al. Prevention of Preeclampsia and Intrauterine Growth Restriction With Aspirin Started in Early Pregnancy. Obstet Gynecol. 2010. August;116(2, Part 1):402–14. [DOI] [PubMed] [Google Scholar]
- 6.Duley L, Henderson-Smart DJ, Meher S, King JF. Antiplatelet agents for preventing pre-eclampsia and its complications. Cochrane Database Syst Rev. 2007. April 18;(2):1–146. [DOI] [PubMed] [Google Scholar]
- 7.Roberge S, Villa P, Nicolaides K, Giguère Y, Vainio M, Bakthi A, et al. Early Administration of Low-Dose Aspirin for the Prevention of Preterm and Term Preeclampsia: A Systematic Review and Meta-Analysis. Fetal Diagn Ther. 2012;31:141–6. 10.1159/000336662 [DOI] [PubMed] [Google Scholar]
- 8.Rolnik DL, Wright D, Poon LCY, Syngelaki A, O’gorman N, De C, et al. ASPRE trial: performance of screening for preterm pre-eclampsia. Ultrasound Obs Gynecol. 2017;50:492–5. [DOI] [PubMed] [Google Scholar]
- 9.Caughey AB, Stotland NE, Washington AE, Escobar GJ. Maternal Ethnicity, Paternal Ethnicity, and Parental Ethnic Discordance. Obstet Gynecol. 2005. July;106(1):156–61. 10.1097/01.AOG.0000164478.91731.06 [DOI] [PubMed] [Google Scholar]
- 10.Jeyabalan A. Epidemiology of preeclampsia: impact of obesity. Nutr Rev. 2013;71(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mol BWJ, Roberts CT, Thangaratinam S, Magee LA, De Groot JM, Justus Hofmeyr G. Pre-eclampsia. The Lancet. 2016;387:999–1011. [DOI] [PubMed] [Google Scholar]
- 12.Rao AK, Cheng YW, Caughey AB. Perinatal complications among different Asian-American subgroups. Am J Obstet Gynecol. 2006;194(5):39–41. [DOI] [PubMed] [Google Scholar]
- 13.Saleem S, McClure EM, Goudar SS, Patel A, Esamai F, Garces A, et al. A prospective study of maternal, fetal and neonatal deaths in low- and middle-income countries. Bull World Health Organ. 2014. August 1;92(8):605–12. 10.2471/BLT.13.127464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mello G, Parretti E, Cioni R, Lagazio C, Mealli F, Pratesi M. Individual longitudinal patterns in biochemical and hematological markers for the early prediction of pre-eclampsia. J Matern Neonatal Med. 2002. January 7;11(2):93–9. [DOI] [PubMed] [Google Scholar]
- 15.Moons KGM, Donders RART, Stijnen T, Harrell FE. Using the outcome for imputation of missing predictor values was preferred. J Clin Epidemiol. 2006;59:1092–101. 10.1016/j.jclinepi.2006.01.009 [DOI] [PubMed] [Google Scholar]
- 16.Moons KGM, De Groot JAH, Bouwmeester W, Vergouwe Y, Mallett S, Altman DG, et al. Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies: The CHARMS Checklist. PLOS Med. 2014;11(10):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandenbroucke JP, Von Elm E, Altman DG, Gøtzsche PC, Mulrow CD, Pocock SJ, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): Explanation and Elaboration. PLoS Med. 2007;4(10):e297 10.1371/journal.pmed.0040297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. BMC Med. 2015;13(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Kuijk SMJ, Nijdam M-E, Janssen KJM, Sep SJS, Peeters LL, Delahaije DHJ, et al. A Model for Preconceptional Prediction of Recurrent Early-Onset Preeclampsia: Derivation and Internal Validation. Reprod Sci. 2011;18(11):1154–9. 10.1177/1933719111410708 [DOI] [PubMed] [Google Scholar]
- 20.Myers J, Kenny L, McCowan L, Chan E, Dekker G, Poston L, et al. Angiogenic factors combined with clinical risk factors to predict preterm pre-eclampsia in nulliparous women: a predictive test accuracy study. BJOG An Int J Obstet Gynaecol. 2013. September 1;120(10):1215–23. [DOI] [PubMed] [Google Scholar]
- 21.Poon LCY, Kametas NA, Maiz N, Akolekar R, Nicolaides KH. First-Trimester Prediction of Hypertensive Disorders in Pregnancy. Hypertension. 2009;53(5):812–8. 10.1161/HYPERTENSIONAHA.108.127977 [DOI] [PubMed] [Google Scholar]
- 22.Crovetto F, Figueras F, Triunfo S, Crispi F, Rodriguez-Sureda V, Dominguez C, et al. First trimester screening for early and late preeclampsia based on maternal characteristics, biophysical parameters, and angiogenic factors. Prenat Diagn. 2015;35(2):183–91. 10.1002/pd.4519 [DOI] [PubMed] [Google Scholar]
- 23.Gabbay-Benziv R, Oliveira N, Baschat AA. Optimal first trimester preeclampsia prediction: a comparison of multimarker algorithm, risk profiles and their sequential application. Prenat Diagn. 2016. January 1;36(1):34–9. 10.1002/pd.4707 [DOI] [PubMed] [Google Scholar]
- 24.Parra-Cordero M, Rodrigo R, Barja P, Bosco C, Rencoret G, Sepúlveda-Martinez A, et al. Prediction of early and late pre-eclampsia from maternal characteristics, uterine artery Doppler and markers of vasculogenesis during first trimester of pregnancy. Ultrasound Obstet Gynecol. 2013. May 1;41(5):538–44. 10.1002/uog.12264 [DOI] [PubMed] [Google Scholar]
- 25.Kleinrouweler CE, Cheong-See FM, Collins GS, Kwee A, Thangaratinam S, Khalid et al. Prognostic models in obstetrics: available, but far from applicable. Am J Obstet Gynecol. 2016;214(1):79–90. 10.1016/j.ajog.2015.06.013 [DOI] [PubMed] [Google Scholar]
- 26.National Heart Lung and Blood Institute. Study Quality Assessment Tools. National Heart, Lung, and Blood Institute. [cited 2018 Oct 31]. Available from: https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools
- 27.O’Gorman N, Wright D, Syngelaki A, Ranjit Akolekar R;, Wright A, Poon LC, et al. Competing risks model in screening for preeclampsia by maternal factors and biomarkers at 11–13 weeks gestation. Am J Epidemiol. 2016;214:103.e1-103.e12. [DOI] [PubMed] [Google Scholar]
- 28.Paré E, Parry S, McElrath TF, Pucci D, Newton A, Lim K-H. Clinical Risk Factors for Preeclampsia in the 21st Century. Obstet Gynecol. 2014. October;124(4):763–70. 10.1097/AOG.0000000000000451 [DOI] [PubMed] [Google Scholar]
- 29.Moon M, Odibo A. First-trimester screening for preeclampsia: impact of maternal parity on modeling and screening effectiveness. J Matern Neonatal Med. 2015. November 22;28(17):2028–33. [DOI] [PubMed] [Google Scholar]
- 30.Park FJ, Leung CHY, Poon LCY, Williams PF, Rothwell SJ, Hyett JA. Clinical evaluation of a first trimester algorithm predicting the risk of hypertensive disease of pregnancy. Aust New Zeal J Obstet Gynaecol. 2013. December 1;53(6):532–9. [DOI] [PubMed] [Google Scholar]
- 31.Stamilio DM, Sehdev HM, Morgan MA, Propert K, Macones GA, Philadelphia C, et al. Can antenatal clinical and biochemical markers predict the development of severe preeclampsia? Am J Obstet Gynecol. 2000;182(3):589–94. 10.1067/mob.2000.103890 [DOI] [PubMed] [Google Scholar]
- 32.van Kuijk SM, Delahaije DH, Dirksen CD, Scheepers HC, Spaanderman ME, Ganzevoort W, et al. External validation of a model for periconceptional prediction of recurrent early-onset preeclampsia. Hypertens Pregnancy. 2014. August 6;33(3):265–76. 10.3109/10641955.2013.872253 [DOI] [PubMed] [Google Scholar]
- 33.Kenny LC, Black MA, Poston L, Taylor R, Myers JE, Baker PN, et al. Early Pregnancy Prediction of Preeclampsia in Nulliparous Women, Combining Clinical Risk and Biomarkers The Screening for Pregnancy Endpoints (SCOPE) International Cohort Study Preeclampsia. Hypertension. 2014;64(3):644–52. 10.1161/HYPERTENSIONAHA.114.03578 [DOI] [PubMed] [Google Scholar]
- 34.Poon L, Kametas NA, Chelemen T, Leal A, Nicolaides KH. Maternal risk factors for hypertensive disorders in pregnancy: a multivariate approach. J Hum Hypertens. 2010;24(2):104–10. 10.1038/jhh.2009.45 [DOI] [PubMed] [Google Scholar]
- 35.Poon LCY, Kametas NA, Pandeva I, Valencia C, Nicolaides KH. Mean arterial pressure at 11+0 to 13+6 weeks in the prediction of preeclampsia. Hypertension. 2008;51(4):1027–33. 10.1161/HYPERTENSIONAHA.107.104646 [DOI] [PubMed] [Google Scholar]
- 36.Herraiz I, Arbues J, Camano I, Gomez-Montes E, Graneras A, Galindo A. Application of a first-trimester prediction model for pre-eclampsia based on uterine arteries and maternal history in high-risk pregnancies. Prenat Diagn. 2009. December 1;29(12):1123–9. 10.1002/pd.2383 [DOI] [PubMed] [Google Scholar]
- 37.Lorenzo G Di, Ceccarello M, Cecotti V, Ronfani L, Monasta L, Brumatti LV, et al. First trimester maternal serum PIGF, free β-hCG, PAPP-A, PP-13, uterine artery Doppler and maternal history for the prediction of preeclampsia. Placenta. 2012;33:495–501. 10.1016/j.placenta.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 38.Goetzinger KR, Tuuli MG, Cahill AG, Macones GA, Odibo ACEO. Development and Validation of a Risk Factor Scoring System for First-Trimester Prediction of Pre-Eclampsia. Am J Perinatol. 2014;31(12):1049–56. 10.1055/s-0034-1371705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crovetto F, Figueras F, Triunfo S, Crispi F, Rodriguez-Sureda V, Peguero A, et al. Added Value of Angiogenic Factors for the Prediction of Early and Late Preeclampsia in the First Trimester of Pregnancy. 2014;35(4):258–66. [DOI] [PubMed] [Google Scholar]
- 40.Gallo DM, Wright D, Casanova C, Campanero M, Nicolaides KH. Competing risks model in screening for preeclampsia by maternal factors and biomarkers at 19–24 weeks’ gestation. Am J Obstet Gynecol. 2016;214(5):619.e1-619.e17. [DOI] [PubMed] [Google Scholar]
- 41.Skråstad R, Hov G, Blaas H-G, Romundstad P, Salvesen K. Risk assessment for preeclampsia in nulliparous women at 11–13 weeks gestational age: prospective evaluation of two algorithms. BJOG An Int J Obstet Gynaecol. 2015. December 1;122(13):1781–8. [DOI] [PubMed] [Google Scholar]
- 42.Muto H, Yamamoto R, Ishii K, Kakubari R, Takaoka S, Mabuchi A, et al. Risk assessment of hypertensive disorders in pregnancy with maternal characteristics in early gestation: A single-center cohort study. Taiwan J Obstet Gynecol. 2016;55:341–5. 10.1016/j.tjog.2016.04.009 [DOI] [PubMed] [Google Scholar]
- 43.Antonio G, Lobo R, Nowak PM, Panigassi AP, Iara A, Lima F, et al. Validation of Fetal Medicine Foundation algorithm for prediction of pre-eclampsia in the first trimester in an unselected Brazilian population. J Matern Neonatal Med 2017;1–8. [DOI] [PubMed] [Google Scholar]
- 44.Allen R, Aquilina J. Prospective observational study to determine the accuracy of first-trimester serum biomarkers and uterine artery Dopplers in combination with maternal characteristics and arteriography for the prediction of women at risk of preeclampsia and other adverse. J Matern Neonatal Med. 2017;31(21):2789–806. [DOI] [PubMed] [Google Scholar]
- 45.Mello G, Parretti E, Ognibene A, Mecacci F, Cioni R, Scarselli G, et al. Prediction of the Development of Pregnancy-Induced Hypertensive Disorders in High-Risk Pregnant Women by Artificial Neural Networks. Vol. 39, Clin Chem Lab Med. 2001. [DOI] [PubMed] [Google Scholar]
- 46.Baschat AA, Magder LS, Doyle LE. Prediction of preeclampsia utilizing the first trimester screening examination. Am J Obs Gynecol. 2014;211(5):514–5. [DOI] [PubMed] [Google Scholar]
- 47.Antwi E, Groenwold RHH, Browne JL, Franx A, Agyepong IA, Koram KA, et al. Development and validation of a prediction model for gestational hypertension in a Ghanaian cohort. BMJ Open. 2017;7(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.North RA, McCowan LME, Dekker GA, Poston L, Chan EHY, Stewart AW, et al. Clinical risk prediction for pre-eclampsia in nulliparous women: Development of model in international prospective cohort. BMJ. 2011;342(7803). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Becker R. Identification of high-risk patients for adverse pregnancy outcome based on multivariate logistic regression analysis at 20–23 gestational weeks. J Perinat Med. 2011. January 1;39(6):667–72. 10.1515/JPM.2011.079 [DOI] [PubMed] [Google Scholar]
- 50.Myatt L, Clifton RG, Roberts JM, Spong CY, Hauth JC, Varner MW, et al. First-Trimester Prediction of Preeclampsia in Low-Risk Nulliparous Women NIH Public Access Author Manuscript. Obs Gynecol. 2012;119(6):1234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goetzinger KR, Singla A, Gerkowicz S, Dicke JM, Gray DL, Odibo AO. Predicting the risk of pre-eclampsia between 11 and 13 weeks’ gestation by combining maternal characteristics and serum analytes, PAPP-A and free β-hCG. Prenat Diagn. 2010. December 1;30(12–13):1138–42. 10.1002/pd.2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Odibo AO, Zhong Y, Goetzinger KR, Odibo L, Bick JL, Bower CR, et al. First-trimester placental protein 13, PAPP-A, uterine artery Doppler and maternal characteristics in the prediction of pre-eclampsia. Placenta. 2011;32:598–602. 10.1016/j.placenta.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benkő Z, Chaveeva P, Paco Matallana C, Zingler E, Wright D, Nicolaides KH. Revised competing‐risks model in screening for pre‐eclampsia in twin pregnancy by maternal characteristics and medical history. Ultrasound Obstet Gynecol. 2019. November 14;54(5):617–24. 10.1002/uog.20411 [DOI] [PubMed] [Google Scholar]
- 54.Scazzocchio E, Crovetto F, Triunfo S, Gratacós E, Figueras F. Validation of a first-trimester screening model for pre-eclampsia in an unselected population. Ultrasound Obstet Gynecol. 2017. February 1;49(2):188–93. 10.1002/uog.15982 [DOI] [PubMed] [Google Scholar]
- 55.Wright D, Tan MY, O’Gorman N, Poon LC, Syngelaki A, Wright A, et al. Predictive performance of the competing risk model in screening for preeclampsia. Am J Obstet Gynecol. 2019. February 1;220(2):199.e1-199.e13. [DOI] [PubMed] [Google Scholar]
- 56.Sepúlveda-Martínez A, Rencoret G, Silva MC, Ahumada P, Pedraza D, Muñoz H, et al. First trimester screening for preterm and term pre-eclampsia by maternal characteristics and biophysical markers in a low-risk population. J Obstet Gynaecol Res. 2019. January 1;45(1):104–12. 10.1111/jog.13809 [DOI] [PubMed] [Google Scholar]
- 57.Zhang J, Han L, Li W, Chen Q, Lei J, Long M, et al. Early prediction of preeclampsia and small-for-gestational-age via multi-marker model in Chinese pregnancies: a prospective screening study. BMC Pregnancy Childbirth. 2019. December;19(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boutin A, Gasse C, Demers S, Giguère Y, Tétu A, Bujold E. Maternal Characteristics for the Prediction of Preeclampsia in Nulliparous Women: The Great Obstetrical Syndromes (GOS) Study. J Obstet Gynaecol Canada. 2018;40(5):572–8. [DOI] [PubMed] [Google Scholar]
- 59.Lobo GAR, Nowak PM, Panigassi AP, Lima AIF, Araujo Júnior E, Nardozza LMM, et al. Validation of Fetal Medicine Foundation algorithm for prediction of pre-eclampsia in the first trimester in an unselected Brazilian population. J Matern Neonatal Med. 2019;32(2):286–92. [DOI] [PubMed] [Google Scholar]
- 60.Antwi E, Klipstein-Grobusch K, Browne JL, Schielen PC, Koram KA, Agyepong IA, et al. Improved prediction of gestational hypertension by inclusion of placental growth factor and pregnancy associated plasma protein-a in a sample of Ghanaian women. Reprod Health. 2018;15(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kenny LC, Black MA, Poston L, Taylor R, Myers JE, Baker PN, et al. Early Pregnancy Prediction of Preeclampsia in Nulliparous Women, Combining Clinical Risk and Biomarkers. Hypertension. 2014. September;64(3):644–52. 10.1161/HYPERTENSIONAHA.114.03578 [DOI] [PubMed] [Google Scholar]
- 62.Altman DG, Royston P. Statistics Notes The cost of dichotomising continuous variables. BMJ. 2006;332:1 10.1136/bmj.332.7532.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bennette C, Vickers A. Against quantiles: categorization of continuous variables in epidemiologic research, and its discontents. BMC Med Res Methodol. 2012;12(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bouwmeester W, A Zuithoff NP, Mallett S, Geerlings MI, Vergouwe Y, Steyerberg EW, et al. Reporting and Methods in Clinical Prediction Research: A Systematic Review. PLoS Med. 2012;9(5):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mallett S, Royston P, Dutton S, Waters R, Altman DG. Reporting methods in studies developing prognostic models in cancer: a review. BMC Med. 2010;8(20):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bodnar LM, Ness RB, Markovic N, Roberts JM. The Risk of Preeclampsia Rises with Increasing Prepregnancy Body Mass Index. Ann Epidemiol. 2005;15:475–82. 10.1016/j.annepidem.2004.12.008 [DOI] [PubMed] [Google Scholar]
- 67.Harrell FE. Multivariable Modeling Strategies Regression modelling strategies. In Springer, Cham; 2015. p. 63–102. Available from: http://link.springer.com/10.1007/978-3-319-19425-7_4 [Google Scholar]
- 68.Shariat SF, Karakiewicz PI, Margulis V, Kattan MW. Inventory of prostate cancer predictive tools. Curr Opin Urol. 2008. May;18(3):279–96. 10.1097/MOU.0b013e3282f9b3e5 [DOI] [PubMed] [Google Scholar]
- 69.Peduzzi P, Concato J, Feinstein AR, Holford’ TR. Importance of events per independent variable in proportional hazards regression analysis. Accuracy and precision of regression estimates. J Clin Epidemiol. 1995;48(12):1503–10. 10.1016/0895-4356(95)00048-8 [DOI] [PubMed] [Google Scholar]
- 70.Vittinghoff E, McCulloch CE. Relaxing the rule of ten events per variable in logistic and Cox regression. Am J Epidemiol. 2007. March 15;165(6):710–8. 10.1093/aje/kwk052 [DOI] [PubMed] [Google Scholar]
- 71.Burton A, Altman DG. Missing covariate data within cancer prognostic studies: a review of current reporting and proposed guidelines. Br J Cancer. 2004;91:4–8. 10.1038/sj.bjc.6601907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collins GS, Mallett S, Omar O, Yu L-M. Developing risk prediction models for type 2 diabetes: a systematic review of methodology and reporting. BMC Med. 2011;9(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Collins GS, Omar O, Shanyinde M, Yu L-M. A systematic review finds prediction models for chronic kidney disease were poorly reported and often developed using inappropriate methods. J Clin Epidemiol. 2013;66(3):268–77. 10.1016/j.jclinepi.2012.06.020 [DOI] [PubMed] [Google Scholar]
- 74.Little RJA. Regression with Missing X ‘s: A Review. J Am Stat Assoc. 1992. December;87(420):1227–37. [Google Scholar]
- 75.Mackinnon A. The use and reporting of multiple imputation in medical research—a review. J Intern Med. 2010. December 1;268(6):586–93. 10.1111/j.1365-2796.2010.02274.x [DOI] [PubMed] [Google Scholar]
- 76.Cook NR, Romanens M. Use and Misuse of the Receiver Operating Characteristic Curve in Risk Prediction. Circulation. 2007;115(7):928–35. 10.1161/CIRCULATIONAHA.106.672402 [DOI] [PubMed] [Google Scholar]
- 77.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982. April;143(1):29–36. 10.1148/radiology.143.1.7063747 [DOI] [PubMed] [Google Scholar]
- 78.Hosmer DW, Lemeshow S, Sturdivant RX. Applied logistic regression. 3rd ed Wiley; 2013. 528 p. [Google Scholar]
- 79.Bamber D, Service P. The Area above the Ordinal Dominance Graph and the Area below the Receiver Operating Characteristic Graph. Vol. 12, Journal of Mathematical Psychology. 1975. [Google Scholar]
- 80.Simpson AJ, Fitter MJ. What is the best index of detectability? Psychol Bull. 1973;80(6):481–8. [Google Scholar]
- 81.Zhang DD, Zhou X-H, Freeman DH, Freeman JL. A non-parametric method for the comparison of partial areas under ROC curves and its application to large health care data sets. Stat Med. 2002. March 15;21(5):701–15. 10.1002/sim.1011 [DOI] [PubMed] [Google Scholar]
- 82.Ewout W. Steyerberg Clinical Prediction Models: A Practical Approach to Development, Validation. Springer; 2009. 497 p. [Google Scholar]
- 83.Moon M, Odibo A. First-trimester screening for preeclampsia: impact of maternal parity on modeling and screening effectiveness. J Matern Neonatal Med. 2015;28(17):2028–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.