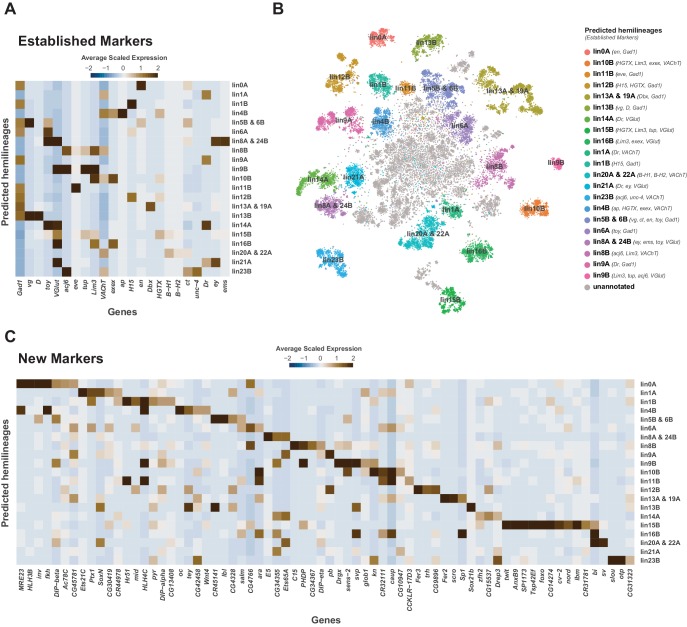
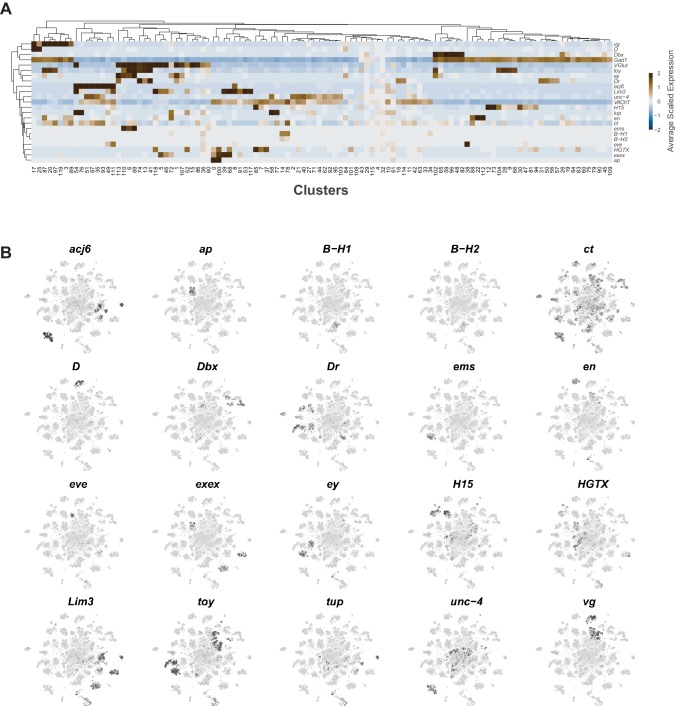
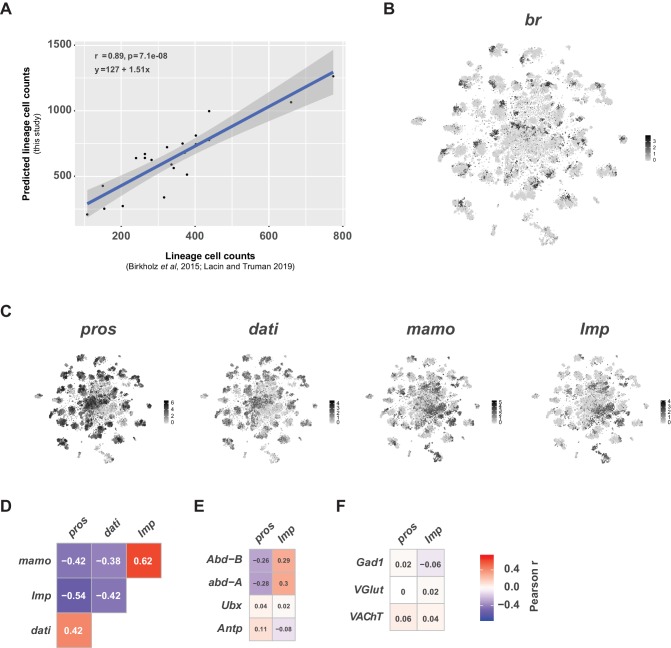
(
A) Scatter plot of hemilineage cell counts from thoracic neuromeres (calculated from
Birkholz et al., 2015 and
Lacin et al., 2019) compared to cell counts from predicted hemilineage identity in this single-cell data set (used
pros+,
abd-A-,
Abd-B- cells to approximate postembryonic, thoracic hemilineages). Each dot represents a hemilineage (or group of hemilineages). The regression line (blue line) and 95% confidence interval (grey bar) are shown. Pearson’s correlation coefficient (r), p-value (p) and equation of the regression line appear in the upper left corner. (
B) t-SNE plot of
br expression showing regional expression in clusters throughout the VNC. Expression shown in black, intensity is proportional to the log-normalized expression levels. (
C) t-SNE plots of
pros,
dati,
mamo, and
Imp in the VNC. Expression shown in black, intensity is proportional to the log-normalized expression levels. (
D) Heatmaps showing Pearson correlations coefficients (in text) of expression for
pros,
dati,
mamo, and
Imp in single cells. (
E) Heatmaps showing Pearson correlations coefficients (in text) of expression for
pros and
Imp with the Hox genes
Abd-B,
abd-A,
Ubx, and
Antp in single cells. (
F) Heatmaps showing Pearson correlations coefficients (in text) of expression for pros and Imp with fast-acting neurotransmitter markers
Gad1,
VGlut, and
VAChT in single cells. (
D–F) All heatmaps share the same scale and consistent patterns of correlation were seen at the cluster-level and between replicates (data not shown).