ABSTRACT
Little is known about the influence of host genotype and phytohormones on the composition of fungal endophytic communities. We investigated the influence of host genotype and phytohormones on the structure of the fungal endophytic communities of tomato roots by amplicon sequencing of the ITS1 region and combined this approach with isolation and functional characterization of the isolates. A significant effect of the host genotype on the dominant fungal species was found by comparing the cultivars Castlemart and UC82B and, surprisingly, root pathogens were among the most abundant taxa. In contrast, smaller changes in the relative abundance of the dominant species were found in mutants impaired in jasmonic acid biosynthesis (def1) and ethylene biosynthesis (8338) compared to the respective wild types. However, def1 showed significantly higher species richness compared to the wild type. Analysis of the phytohormone profiles of these genotypes indicates that changes in the phytohormone balance may contribute to this difference in species richness. Assessing the lifestyle of isolated fungi on tomato seedlings revealed the presence of both beneficial endophytes and latent pathogens in roots of asymptomatic plants, suggesting that the interactions between members of the microbiome maintain the equilibrium in the community preventing pathogens from causing disease.
Keywords: fungal endophytes, microbiome, amplicon sequencing, phytohormones, host genotype, fungal lifestyle
This article provides novel insights into the role of plant hormones and host genotype in the structure of the fungal communities inhabiting tomato roots, while also exploring the fungal lifestyle.
INTRODUCTION
Endophytes are microorganisms capable of colonizing the inner part of living plant tissues (endosphere) without causing diseases (Hardoim et al. 2015; Collinge et al. 2019). Specific fungal and bacterial endophytes are known to be important as biocontrol agents against plant pathogens (Porras-Alfaro and Bayman 2011; Busby, Ridout and Newcombe 2016; Card et al. 2016) and as biostimulants alleviating plant abiotic stresses, such as salt and drought stress (du Jardin 2015; López-Bucio, Pelagio-Flores and Herrera-Estrella 2015). Nevertheless, the endophytic microbiome is complex and asymptomatic plant tissues harbour both beneficial (mutualistic), neutral (commensal) and potentially harmful (pathogenic) microorganisms (Hardoim et al. 2015; Brader et al. 2017; Collinge et al. 2019). Therefore, interactions between the members of the microbiome could influence the virulence of pathogenic microbes, resulting in a ‘natural biocontrol’ of the pathogen (Busby, Peay and Newcombe 2016; Ritpitakphong et al. 2016; Durán et al. 2018). The symptomless infection of the host tissues by a pathogenic species can be also explained by a lack of compatibility with the host plant or the lack of pathogenicity genes in certain strains belonging to a pathogenic species (Kogel, Franken and Hückelhoven 2006; Brader et al. 2017). In order to get more insights into the role of microbe–microbe and host–microbe interactions in shaping the mycobiome and plant health, it is necessary to perform the isolation and functional characterization of endophytic fungi. However, the majority of the community studies rely only on literature to assign a potential function to the taxa detected by cultivation-independent techniques.
Despite the recognized importance of endophytes in plant health and agriculture, little is known about the general mechanisms that guide plant–endophyte interactions.
Several studies have reported the influence of the host genotype on the structure of endophytic communities using sequencing-based, cultivation-independent approaches. For example, the composition of the bacterial endophytic community was influenced by the host genotype in roots of Arabidopsis thaliana (Bulgarelli et al. 2012; Lundberg et al. 2012). Whereas the bacterial communities in the root endosphere have been extensively studied (Yu and Hochholdinger 2018), little is known about the role of host genotype in the fungal endophytic root microbiome (endophytic root mycobiome). However, it was recently shown by ITS-amplicon sequencing that the fungal endophytic communities of A. thaliana are significantly influenced by the host genotype (Urbina et al. 2018). Nonetheless, more integrative studies combining isolation and sequencing-based, cultivation-independent approaches are necessary to expand our knowledge of the fungal endophytic root communities in relation to the host genotype. Moreover, we need to identify the factors driving these shifts in endophytic communities in order to deepen our knowledge on how to shape the community and attract beneficial endophytes for future applications in agriculture.
In this context, phytohormones influence the ability of endophytes to colonize plant tissues. Several studies have been conducted on the model endophytic fungus Serendipita indica (formerly Piriformospora indica) in order to understand the role of different phytohormones in the interaction with A. thaliana and barley (Xu et al. 2018). However, the role of phytohormones in the structure of fungal endophytic communities has, to the best of our knowledge, not been studied yet. Few studies on the bacterial endophytic microbiome suggest that salicylic acid (SA) has a pronounced effect on the composition of the endophytic microbiome in A. thaliana (Kniskern, Traw and Bergelson 2007; Lebeis et al. 2015). Moreover, ethylene (ET) was also shown to play a role in sculpting the bacterial endophytic community of Nicotiana attenuata (Long et al. 2010). In contrast, jasmonic acid (JA) did not result in major changes in the endophytic bacterial communities of either leaves or roots of N. attenuata (Santhanam et al. 2014), although MeJA treatment was shown to decrease significantly the diversity of the bacterial endophytic communities of wheat roots (Liu et al. 2017). Therefore, we hypothesized that phytohormones can play a role in structuring the endophytic mycobiome of roots based on the observed effects on bacterial communities.
This study combines cultivation-independent and cultivation-dependent approaches in order to shed light on the role of host genotype and phytohormones in the composition of the endophytic mycobiome of tomato roots and to functionally characterize the mycobiome. In detail, we hypothesize that (i) host genotype significantly affects the composition of fungal endophytic communities of tomato roots, (ii) that phytohormones modulate the diversity and structure of the root endophytic mycobiome and (iii) that interactions between members of the microbiome maintain the equilibrium in the community, preventing pathogens from causing disease. We performed amplicon sequencing of the internal transcribed spacer 1 (ITS1) region in order to reconstruct the root fungal endophytic communities of the tomato lines def1 (impaired in JA biosynthesis) and 8338 (impaired in ET biosynthesis) and their respective wild types Castlemart and UC82B. This setup allowed investigation of whether reduced JA and ET biosynthesis affects the composition of the endophytic mycobiome as well as the role of the host genotype on the fungal endophytic community structure (Castlemart vs UC82B). Moreover, the phytohormone profiles of these tomato genotypes were analysed by UHPLC/TQ-MS to investigate the effect of hormone patterns on the endophytic community structure. Isolation of endophytic fungi from roots enabled us to identify the isolates and to determine their lifestyle by inoculating them in tomato seedlings. Additionally, we confirmed the endophytic capability of the isolates (capability of colonizing the inner part of plant tissues) by re-isolation of the fungi from surface sterilized roots of the inoculated seedlings. Moreover, we characterized the ability of the isolates to cope with abiotic stress in vitro (temperature and osmotic stress) in order to evaluate the functional diversity of the mycobiome using physiologic parameters.
MATERIALS AND METHODS
Plant growth for amplicon sequencing, endophyte isolation and phytohormone measurement
Four different genotypes of tomato (Solanum lycopersicum) were used for fungal endophyte isolation, Illumina amplicon sequencing analysis and phytohormone measurements: Castlemart, defenceless1 (def1), UC82B and 8338. The mutant line def1 was produced with the cv. Castlemart as background and characterized as a JA-biosynthesis mutant (Howe et al. 1996). The transgenic line 8338 was produced by inserting the bacterial ACC-deaminase gene in cv. UC82B, which is therefore impaired in ET biosynthesis (Klee et al. 1991). The tomato seeds were surface sterilized in 70% ethanol for 1 min followed by 1% NaClO (v v−1) for 10 min and five times rinsing with sterile MilliQ water. The sterilized seeds were germinated for 8 days in a growth chamber (200 µEm−2 s−1, Philips Master IL-D 36 w/865, France/22°C/60% RH and 12 h darkness/18°C/80% RH) in Petri dishes on sterile filter paper soaked in sterile water. The fully germinated seedlings were transferred to a soil mixture made of organic soil (Ekologisk plantjord, Fagerhults Torv AB) that was previously used for tomato plant cultivation in a greenhouse (for ∼5 months), mixed 1:1 (v v−1) with peat soil (Substrate no. 2, Pindstrup Mosebrug A/S) that was autoclave-sterilized (three times at 120°C for 1 h, with a 24 h interval between sterilizations). Each seedling was transferred to a pot and grown in the greenhouse (25°C day and 20°C night).
Root harvest for amplicon sequencing and endophyte isolation
The whole root system was harvested from 6 weeks-old plants and processed separately for each plant. The soil was carefully removed and the roots were washed under tap water. Roots with a diameter of ≥2 mm were selected and surface sterilized with 2% NaClO (v v−1) by shaking the root system for 4 min. After treatment with NaClO, the roots were rinsed five times in sterile MilliQ water in order to remove residues of NaClO that could interfere with the further steps. A volume of 200 µl of the last rinsing water was plated on potato dextrose agar (PDA, BD Difco™) in order to check for microbial growth. Plates were examined after 1 week of incubation at room temperature and no fungal growth was detected.
DNA extraction, amplicon library preparation and Illumina sequencing
The roots from five plants for each genotype were surface sterilized as described above and pooled together in one biological replication. A total of four biological replications per genotype were used for the DNA extractions. The sterilized roots were freeze-dried and ground in liquid nitrogen using a mortar and pestle. DNA extraction was performed using the FastDNA™ SPIN Kit for Soil (MP Biomedicals, USA) following the manufacturer's instructions.
The amplicon libraries for Illumina HiSeq were prepared by amplifying the ITS1 region using the primers ITS1-F_KYO1 and ITS2_KYO1 (Toju et al. 2012) with a ‘pad sequence’ added to the 5′ end in order to attach the barcode before sequencing. PCR was performed with the following thermocycler programme (Toju et al. 2012): initial denaturation at 95°C for 10 min, 40 cycles of denaturation at 94°C for 20 s, annealing at 49°C for 30 s and extension at 72°C for 20 s, with a final extension step at 72°C for 7 min. The Taq-&GO™ Ready-to-use PCR Mix (MP Biochemicals, USA) was used for the amplification, adding MgCl2 (0.75 mM final conc.), each primer (0.2 µM final conc.) and 50 ng of DNA template. The presence of the expected amplification product was checked on agarose gels (1% in TAE buffer). Barcodes were attached by PCR using the following thermocycler programme: initial denaturation at 95°C for 5 min, 15 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s and extension at 72°C for 30 s, with a final extension step at 72°C for 5 min. The barcodes used in this study are suggested by the Earth Microbiome Project (http://www.earthmicrobiome.org/). The amplicons were purified using the Wizard® SV Gel and PCR Clean-Up System (Promega, USA). The barcoded libraries were sent for Illumina HiSeq sequencing (GATC Biotech, Germany). The raw data obtained were deposited in the NCBI database under the BioProject ID: PRJNA622681.
Determination of fungal community structure based on ITS1 amplicon sequencing
The raw reads went through an initial quality check. The whole dataset was demultiplexed and quality filtered with QIIME (version 1.9.0, Caporaso et al. 2010) default parameters (Bokulich et al. 2013). Obtained high quality reads were dereplicated and clustered with a similarity threshold of 97% using vsearch (version 2.4.3). A set of representative sequences was created and chimaeras were filtered via both de novo and reference-based approaches while mapping high quality sequences (vsearch, Rognes et al. 2016). Taxonomical assignment was obtained using the BLAST algorithm (Altschul et al. 1990) in the QIIME environment in combination with the UNITE ITS database (version 7.2–dynamic; Kõljalg et al. 2013). OTUs that were unassigned or assigned to non-fungal contaminants were filtered from the resulting OTU table.
Analysis of alpha and beta diversity was performed using R (R Core Team, 2018). Alpha diversity was estimated on the rarefied OTU counts. The raw OTU counts were rarefied to 100 000 reads per sample using the R package ‘phyloseq’ (McMurdie and Holmes 2013) and the alpha diversity indices (Observed species, Chao1, Shannon and Inverse Simpson) were computed using the same package. Rarefaction curves were estimated for each replication individually using the R package ‘vegan’ (Dixon 2003).
Before analysis of the beta diversity, the OTU table was summarized at genus level (in the QIIME environment) and low-abundant taxa (relative abundance <0.5%) were filtered out. Principal coordinate analysis (PCoA) was used to investigate differences between the samples based on relative abundance (RA) values (non-rarefied). The Bray–Curtis dissimilarity matrix used for the PCoA was computed using the R package ‘vegan’ and PCoA was computed using the R package ‘ape’ (Paradis and Schliep 2019).
The single taxa RA (phylum, class and genus level) was SQRT-transformed (Hellinger transformation) in order to fit the data to the normal distribution (Borcard, Gillet and Legendre 2011). Only the taxa whose average RA was ≥0.5% in at least one of the genotypes were included in the plots.
Hormone extraction and quantification by UHPLC/TQ-MS
Phytohormone extraction and quantification was performed on leaf and root samples harvested from the same plants processed for amplicon sequencing analysis, but without surface sterilization of the tissue. Leaf disks and part of the root system were harvested and processed separately. A total of four biological replications per genotype were used for the extractions and each replication was made of a pool of five plants. The samples were ground in liquid nitrogen using a mortar and pestle and ∼200 mg of fresh material was used for the hormone extraction in ethylacetate:formic acid, 99:1 (v v−1) following the protocol from Stingl et al. (2013). For all samples, the UHPLC/TQ-MS procedure was as follows: phytohormones were analysed by UHPLC/TQ-MS on an AdvanceTM-UHPLC/EVOQTM Elite-TQ-MS instrument (Bruker) equipped with a C-18 reversed-phase column (Kinetex 1.7 u XB-C18, 10 cm [1] 2.1 mm, 1.7 mm particle size, Phenomenex) by using a 0.05% formic acid in water (v v−1), pH 4.0 (solvent A)–methanol (solvent B) gradient at a flow rate of 0.4 ml min−1 at 40°C. The gradient applied was as follows: 10–50% B (15 min), 50% B (2 min), 50–100% B (0.1 min), 100% B (2.9 min), 100–10% B (0.1 min) and 10% B (5 min). Compounds were ionized by ESI with a spray voltage of +4500 and −4000 V in positive and negative mode, respectively. Heated probe temperature was 350°C and cone temperature was 300°C. Quantification was based on response factors relative to (2H6)abscisic acid (ABA) (negative ionization mode) and (2H5)tZ (positive ionization mode). The individual hormones were monitored based on the following MRM transitions: (2H6)ABA, (−) 269 > 159 [7 V]; (2H5)trans-zeatine (tZ), (+) 225 > 137 [15 V]; ABA, (−)263 > 153[7 V]; 1-aminocyclopropane-1-carboxylic acid (ACC), (+)102 > 56[15 V]; JA, (−)209 > 59[11 V]; JA-Ile, (−)322 > 130[17 V]; SA, (−)137 > 93[20 V]; indole-3-acetic acid (IAA), (+)176 > 130[10 V]; 3-indoleacetamide, (+)175 > 130[15 V]; indole-3-carboxylic acid, (+)162 > 118[15 V]; indole-3-aldehyde (IAld), (+)146 > 118[15 V]; N6-(Δ2-isopentenyl)adenine (iP), (+)204 > 136[10 V]; N6-(Δ2-isopentenyl)adenine riboside (iPR), (+)336 > 204[15 V]; dihydrozeatin (DHZ), (+)222 > 136[15 V]; dihydrozeatin riboside (DHZR), (+)354 > 222[15 V]; trans‐zeatin‐7‐glucoside (tZ7G)/trans‐zeatin‐9‐glucoside (tZ9G)/trans-zeatin-O-glucoside (tZOG), (+)382 > 220[17 V]; trans- zeatine riboside (tZR), (+)352 > 220[15 V]; cis-zeatin (cZ)/trans-zeatin (tZ), (+)220 > 136[15 V]. tZ7G, tZ9G and tZOG, as well as tZ and cZ were distinguished based on retention times in comparison to those of known standards.
The quantity (in pmol) of each hormone was normalized to the exact amount of fresh tissue used for the extraction for each sample. Both the box plots and the principal component analysis (PCA) were computed in the R environment. The data were SQRT-transformed before proceeding with the PCA in order to reduce the relative weight of the high values of specific hormones in the dataset (pseudo scaling) (van den Berg et al. 2006).
Isolation and identification of endophytic fungi
The roots from five plants for Castlemart and def1, seven plants for 8338 and eight plants for UC82B were harvested and sterilized as described above. Each plant was treated separately and kept as a single biological replication. The surface-sterilized roots were cut into pieces of 0.5–1 cm length using a sterile scalpel and incubated on one-third strength PDA. The root pieces were distributed on five plates for each plant (five root pieces per plate). As soon as fungal mycelium was growing from the tip of the root pieces, it was transferred to a new PDA plate under sterile conditions.
Identification of the isolates was carried out both by morphology and marker gene sequencing. The isolates were grouped based on morphology (morphotypes) and a subset of fungi from each morphotype was identified by marker gene sequencing in order to identify the species. The isolates were assigned to the different morphotypes by observation of the mycelium morphology on PDA and microscope observations of conidiophores and conidia (size, colour and shape) using stereo and light microscopes. The subset of fungi selected for marker gene sequencing were grown in potato dextrose broth liquid cultures at room temperature (∼23°C) for 3–10 days, depending on the isolate. The mycelium was harvested from the liquid culture and washed with sterile MilliQ water. DNA extraction was performed with the DNeasy® Plant Mini kit (Qiagen, Hilden, Germany) after grinding the mycelium (60–90 mg) in buffer AP1 using glass beads and a Tissuelyser II (Qiagen, Hilden, Germany). After the grinding step, DNA extraction was performed following the manufacturer's instructions. The ITS regions were used as molecular markers for the identification of the isolates. For the isolate belonging to the genus Penicillium, the ribosomal RNA large subunit (LSU) was used as a molecular marker since it was not possible to obtain a specific PCR product targeting the ITS regions. The primers used for the PCR amplification of the ITS regions were ITS1F (Gardes and Bruns 1993) and ITS4 (White et al. 1990). For the amplification of the LSU region, the primers LR0R (Cubeta et al. 1991) and LR5 (Vilgalys and Hester 1990) were used. Phusion High-Fidelity DNA Polymerase (Thermo Fisher Scientific, USA) was used to amplify the target regions. The thermocycler settings used in the PCR for both primer pairs were as follows: initial denaturation at 98°C for 30 s, 30 cycles of denaturation at 98°C for 8 s, annealing at 55°C for 30 s and extension at 72°C for 20 s, and final extension at 72°C for 8 min. The presence of the expected amplification product was checked on agarose gel (1% in TAE buffer). The PCR products were purified using the Wizard® SV Gel and PCR Clean-Up System (Promega, USA) and the purified amplicon sent for sequencing (GATC Biotech, Germany). The full-sequenced ITS and LSU regions were blasted in the UNITE database (https://unite.ut.ee/index.php) and NCBI database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) in order to identify the isolate. The sequences obtained were deposited in the GenBank database of NCBI with accession numbers from MT221562 to MT221594.
In planta assay to assess fungal lifestyle and endophytic colonization
The tomato seeds were germinated under sterile conditions as described above. Fully germinated seedlings were inoculated with spore suspensions of the fungi of interest. One isolate for each morphotype was tested in the in planta assay in order to assess the effect of the fungus on the plant (fungal lifestyle). Isolates obtained from Castlemart/def1 and isolates from UC82B/8338 were tested on Castlemart and UC82B, respectively. Spores were harvested in sterile MilliQ water from pure cultures of the fungus cultured on PDA at room temperature or 28°C (depending on the isolate) for at least 15 days and the spore concentration determined using a haemocytometer. Eight day-old seedlings were dipped overnight (16–18 h) by shaking at 150 rpm in a spore suspension diluted to 3 × 105 spores ml−1 and then transferred to sterile plastic boxes (70 × 94 × 96 mm, Duchefa Biochemie BV, The Netherlands) containing Murashige and Skoog (MS) Basal Salt Medium (Sigma-Aldrich, USA) supplemented with 1.5% (w w−1) agar (six seedlings in each box). The spore suspensions of the isolates belonging to Colletotrichum coccodes and Chaetomium globosum were diluted to 3 × 104 spores ml−1. The control plants were treated with sterile MilliQ water and then transferred to the MS medium. The plants were grown for 12 days in a growth chamber under the same conditions described above for seed germination. The lifestyle of the isolate in planta was assessed by plant phenotype observation at day 12 by comparing the inoculated plants with the controls (Supplementary Fig. 4, see online supplementary material).
After the isolate effect in planta was assessed, the roots from non-symptomatic plants were harvested and surface sterilized with NaClO 0.8% (v v−1), rinsed five times in sterile MilliQ water and cut into 0.5–1 cm pieces with a sterile scalpel. The root pieces were incubated on one-third strength PDA in order to re-isolate the fungus and assess the endophytic colonization of the root tissue by the isolate. The roots inoculated with isolates belonging to Trichoderma spp. were sterilized in NaClO 1.5% (v v−1) and the root pieces incubated on one-fifth strength PDA due to the fast-growing mycelium of Trichoderma spp. Mycelial growth from the root tip was interpreted as successful colonization of the inner part of the root by the fungus. Three endophytic isolates were tested for growth-promoting effect on tomato plants (cv. Castlemart): isolates RFE 177, RFE 84 and RFE 166, identified as Trichoderma viride, Penicillium sp. and Pseudeurotium sp., respectively. A mixture of these three endophytes in a ratio 1:1:1 was also tested. Treatment with Thielaviopsis basicola (isolate RFE 190) was used in order to validate the results of the experiment by using a fungus that would decrease significantly the plant fresh weight due to its pathogenic capability. Each fungus was inoculated using a spore suspension of 3 × 104 spores ml−1, both in the case of a single-fungus inoculation with each of the four isolates mentioned above and in the three-endophyte mixture. The plants were grown for 8 days under the same conditions used for the in planta assays described above. After 8 days the fresh weight of the whole seedling was measured. The experiment was conducted using three biological replications for each treatment, pooling eight plants in each replication. The experiment was performed twice to validate the results.
Functional characterization of the isolates in vitro: salt tolerance and growth at 36°C
The fungal endophytes were cultured on PDA and the spores harvested as described above for the in planta assay. PDA plates were inoculated with 5 × 103 spores in the centre of the plate. Four different NaCl concentrations were tested (0, 100, 200 and 300 mM) for each isolate by supplementing the medium with NaCl, and the assay was conducted in triplicate. The radial growth of the mycelium was measured at 3, 5, 7 and 10 days after inoculation. The growth temperature tests were performed following the same procedure, but incubating the plates (in triplicate) at 36°C and measuring the radial growth after 10 days.
Statistical analysis
The alpha diversity data were subjected to a t-test in the R environment in order to assess the significance of the differences between the four genotypes. The test was conducted for each of the four diversity indices and the different genotypes were compared pairwise (mutant–wild type and wild type–wild type).
Permutational multivariate analysis of variance (PERMANOVA) was used to investigate whether there was a statistically significant shift of the mycobiome between the different genotypes. The comparison was performed pairwise (as described above for the alpha diversity) and it was based on the Bray–Curtis dissimilarity matrices calculated pairwise on each comparison. The function ‘adonis’ of the R package ‘vegan’ was used for the PERMANOVA, testing the significance of the F-value with 10 000 permutations. The differences between the genotypes in the RA of each fungal taxon (phylum, class or genus) were assessed by t-test (pairwise as described above) on the SQRT-transformed RA.
A t-test was performed on the phytohormones data in R environment in order to compare the genotypes pairwise (mutant-wild type and wild type-wild type) for each hormone detected.
The resulting P-values from the t-test on alpha diversity indices, single taxa RA and phytohormone measurements were adjusted for multiple hypotheses testing using the false discovery rate (FDR) procedure Benjamini–Hochberg on each pairwise comparison.
Statistical analyses of the measurements of the mycelial radial growth in the in vitro assays were performed for each isolate separately. Both the data for radial growth measured on plate and plant fresh weight measurements of the growth promotion assay represent continuous variables and were analysed by analysis of variance (one-way ANOVA), assuming a normal distribution. The data from the two repetitions of the plant growth promotion assay were merged since no interaction between treatment (inoculations with different isolates) and experiment (the two experiment repetitions) was detected using two-way ANOVA. ANOVA was performed by SAS (release 9.4, SAS Institute, Cary, NC). For all the statistical tests performed in this study, the hypotheses were rejected at P < 0.05.
RESULTS
Higher species richness in phytohormone mutants compared to the respective wild types
ITS1 amplicon sequencing on the roots of the four genotypes resulted in an average of 337 961 high-quality reads (Supplementary Data 1, see online supplementary material), after removing chimaeras, singletons, doubletons and plant ITS sequences. A total of 647 fungal OTUs were detected in the amplicon libraries obtained from the four tomato genotypes.
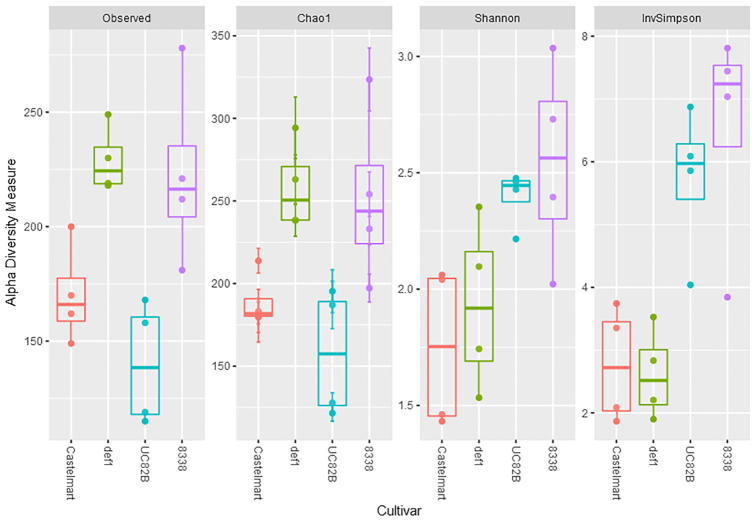
The alpha diversity for each genotype was estimated using four different diversity indices (Fig. 1). Based on the rarefaction analysis (Supplementary Fig. 1, see online supplementary material), the OTU counts were rarefied to 100 000 reads per sample for the alpha diversity analysis. Based on the observed species number and Chao1 indices, the species richness in the endophytic communities was higher in def1 and 8338 compared to the wild types Castlemart and UC82B, respectively, but statistically significant only in def1 compared to Castlemart (Table 1). On the other hand, the Inverse Simpson index showed a statistically significant lower diversity in Castlemart compared to UC82B based on the dominant fungal species present in the mycobiome, while no difference was detected between the mutant lines and the respective wild types using this index (Table 1). The results of the Shannon index did not show statistically significant differences between the four genotypes tested pairwise.
Figure 1.
Estimation of the alpha diversity of the root endophytic mycobiome of the four tomato genotypes based on amplicon sequencing data. The observed species number, Chao1, Shannon and Inverse Simpson indices were used in the analysis of the alpha diversity.
Table 1.
Statistical analysis on the alpha diversity of the fungal endophytic communities of the different tomato genotypes. The table shows the P-values resulting from the pairwise comparison of the genotypes using the t-test for each diversity index. The P-values were adjusted using the FDR correction rate (Benjamini–Hochberg procedure). An asterisk indicates statistically significant differences between the two genotypes (P-value < 0.05).
| Index | Castlea/def1b | UC82Bc/8338d | Castle/UC82B |
|---|---|---|---|
| Observed | 0.013558* | 0.06352 | 0.1769333 |
| Chao1 | 0.013558* | 0.06352 | 0.2087000 |
| Shannon | 0.662400 | 0.54820 | 0.0561800 |
| Inverse Simpson | 0.811900 | 0.54820 | 0.0361560* |
aCastlemart: wild type cultivar;bdef1 (defenceless1): impaired in JA biosynthesis (Castlemart background); cUC82B: wild type cultivar; d8338: impaired in ET biosynthesis (UC82B background).
Host genotype significantly shifts the composition of the endophytic mycobiome
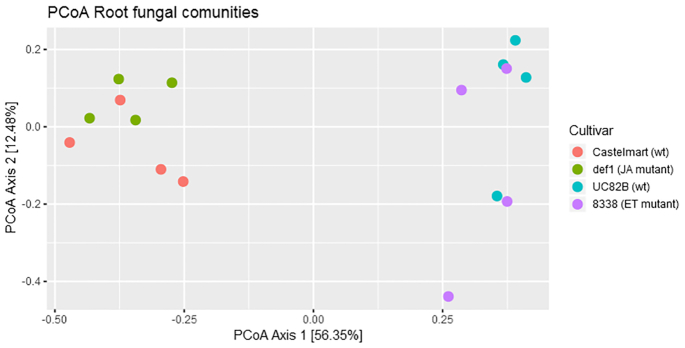
To investigate the differences between the mutant lines and the respective wild types as well as the influence of the host genotype (wild types) on the structure (beta diversity) of the fungal endophytic communities of tomato roots, Bray–Curtis dissimilarities were visualized using PCoA (Fig. 2). PCoA showed a cluster for each genotype and the differences between the clusters were tested for significance using PERMANOVA. PERMANOVA showed a statistically significant effect of the plant genotype on the composition of the mycobiome when the model was run on the four cultivars together (R2 = 73.92%, F value = 11.337, P-value < 0.001), therefore the pairwise comparisons were assessed. A statistically significant shift in the whole endophytic mycobiome composition was found between the wild type cultivars Castlemart and UC82B (R2 = 76.57%, F value = 19.61, P-value < 0.05). On the other hand, no statistically significant change in the mycobiome structure between Castlemart and def1 (R2 = 20.76%, F value = 1.5716, P-value > 0.05) and between UC82B and 8338 (R2 = 17.01%, F value = 1.2295, P-value > 0.05) was found.
Figure 2.
PCoA of root endophytic mycobiome based on amplicon sequencing data. The sample clustering was based on the Bray–Curtis dissimilarity matrix. Each dot in the plot corresponds to a single sample (biological replication). Only genera with average relative abundance >0.5% in at least one of the genotypes were included in the analysis.
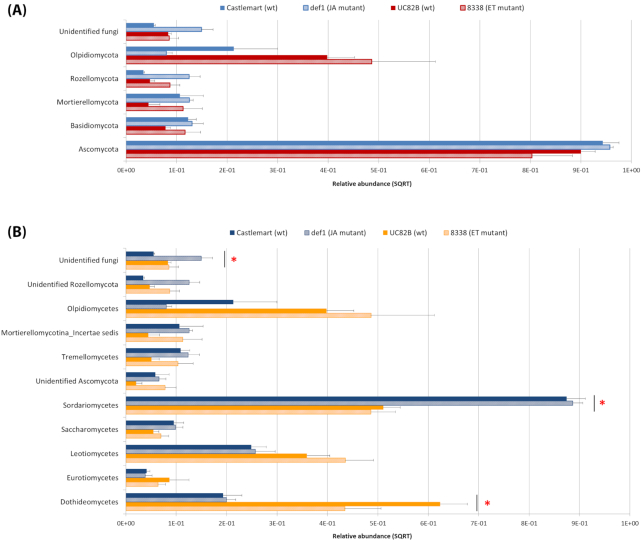
Analysis of the fungal endophytic communities at the phylum level showed the Ascomycota as the dominant phylum in the four genotypes, followed by Olpidiomycota (Fig. 3A). However, there was no statistically significant effect of the host genotype (Castlemart/UC82B comparison) or the mutant lines (Castlemart/def1 and UC82B/8338, comparisons) on the relative abundance of each phylum.
Figure 3.
Composition of the endophytic mycobiome in tomato roots at (A) phylum and (B) class level based on amplicon sequencing data. Relative abundance (SQRT-transformed) is shown as the average of the four biological replications for each of the genotypes (Castlemart, def1, UC82B and 8338). Error bars represent standard error of the mean. Only taxa with average relative abundance >0.5% in at least one of the genotypes were included. The red asterisks indicate statistically significant differences (P-value < 0.05) in the pairwise comparison between Castlemart and UC82B. P-values were adjusted using the FDR correction rate (Benjamini–Hochberg procedure).
Based on the class level taxonomy, the endophytic mycobiome was dominated by Sordariomycetes. Additionally, Dothideomycetes, Leotiomycetes and Olpidiomycetes were also among the most abundant classes detected (Fig. 3B). Interestingly, the relative abundance of Sordariomycetes was significantly higher in Castlemart compared to UC82B, whereas the class Dothideomycetes was significantly more abundant in UC82B compared to Castlemart. On the other hand, comparison between the two mutant lines and the respective wildtypes did not show any statistically significant difference at this taxonomic level.
At the genus level, several genera, such as Apiotrichum, Plectosphaerella, Fusarium, Ilyonectria, Pyrenochaeta and Thielaviopsis, differed significantly in relative abundance between the cultivars Castlemart and UC82B (Fig. 4). On the other hand, comparison between the tomato line 8338 (impaired in ET biosynthesis) and the wild type UC82B showed a statistically significant difference only in the relative abundance of the genus Pseudogymnoascus. In contrast, no difference in the relative abundance of any of the fungal genera was found when comparing def1 (impaired in JA biosynthesis) with its wild type Castlemart.
Figure 4.
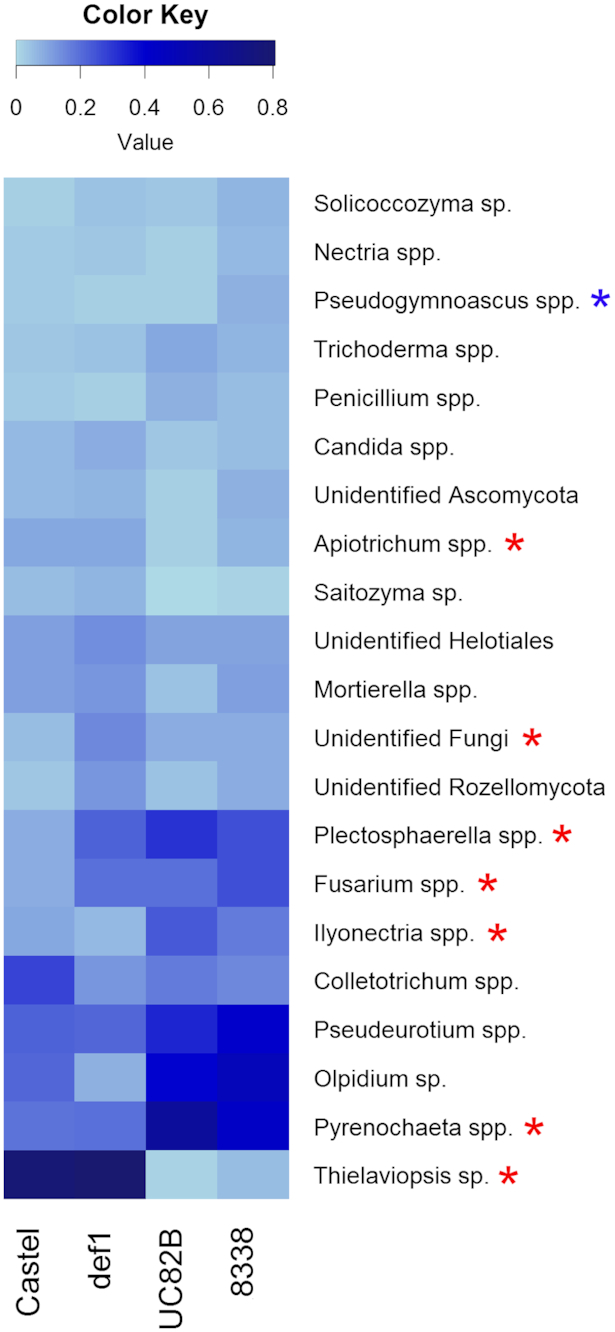
Heatmap showing the composition of the endophytic mycobiome of tomato roots at the genus level based on amplicon sequencing data. The average relative abundance (SQRT-transformed) of the most abundant fungal genera (average relative abundance >0.5% in at least one plant genotype) is shown for the four genotypes tested (Castlemart wild-type, def1 impaired in JA biosynthesis, UC82B wild-type and 8338 impaired in ET biosynthesis). The red and the blue asterisks indicate statistically significant differences (P-value < 0.05) in the pairwise comparison between Castlemart/UC82B and UC82B/8338, respectively. The P-values were adjusted using the FDR correction rate (Benjamini–Hochberg procedure).
Composition of the root mycobiome of tomato at the OTU level
Eight dominant genera were recognized among the most abundant genera detected by amplicon sequencing in the four different tomato genotypes: Plectosphaerella, Fusarium, Ilyonectria, Colletotrichum, Pseudeurotium, Olpidium, Pyrenochaeta and Thielaviopsis (Fig. 4). The tomato genotypes Castlemart and def1 were mostly colonized by Thielaviopsis sp. (average relative abundance >60% in both cultivars), whereas the mycobiome of the genotypes UC82B and 8338 was dominated by the genera Plectosphaerella (10 and 7%, respectively), Pseudeurotium (12 and 18%, respectively), Olpidium (17 and 28%, respectively) and Pyrenochaeta (39 and 20%, respectively).
Several OTUs were detected for each of the genera showed in Fig. 4 (Supplementary Data 3, see online supplementary material). However, for the genera Solicoccozyma, Saitozyma, Olpidium and Thielaviopsis the OTUs were assigned to only one species each (S. terricola, S. podzolica, O. brassicae and T. basicola, respectively). The genus Nectria was mostly represented by N. ramularie while in the yeast genera Pseudogymnoascus, Candida and Apiotrichum the most abundant species detected were P. verrucosus, C. subhashii and A. xylopini, respectively. Most of the OTUs belonging to Plectosphaerella spp. were assigned to P. cucumerina and in the genus Fusarium the most abundant species detected was F. oxysporum. The genera Ilyonectria, Colletotrichum and Pyrenochaeta were mainly represented by I. mors-panacis, C. coccodes and P. lycopersici, respectively. Finally, Pseudeurotium bakeri was the most abundant species in the genus Pseudeurotium. The genera Trichoderma, Penicillium and Mortierella, showed higher species diversity compared to the genera mentioned above. The most represented species in the genus Trichoderma was T. neokoningii, while Penicillium spp. was dominated by P. astrolabium and P. canescens among the 10 different species detected. The OTUs belonging to the genus Mortierella were assigned to 7 different species where M. hyalina and M. indohii were the most abundant.
Phytohormone levels differ between tomato genotypes
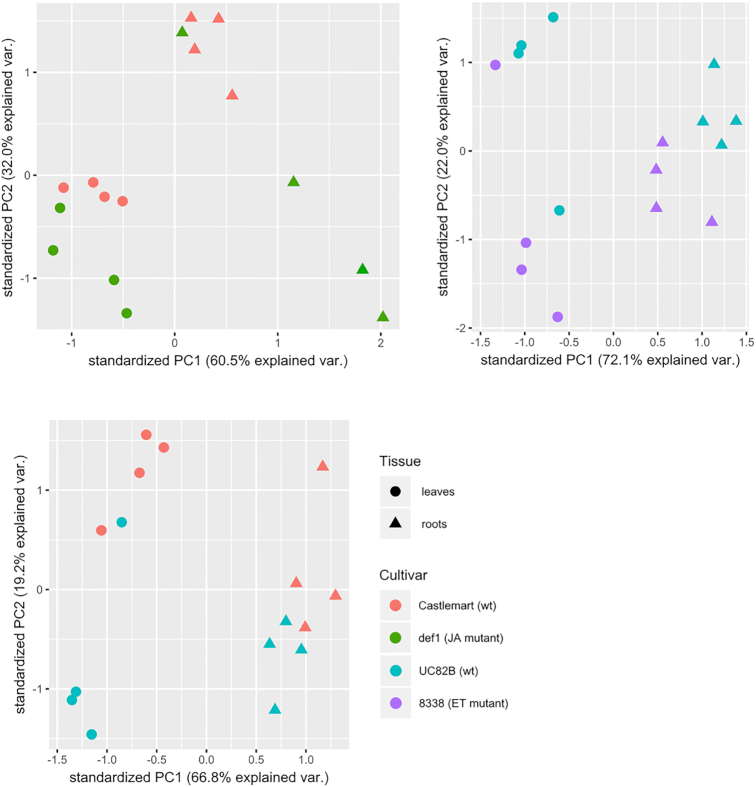
The phytohormone profiles for the four genotypes were analysed by UHPLC/TQ-MS for both leaves and roots. A total of seven hormones, such as ABA, SA, IAld, IAA, iPR, iP and tZR, were detected in the roots and only four (ABA, SA, IAld and IAA) in the leaves. PCA was used to show the differences in the entire phytohormone profiles between the genotypes (pairwise comparisons: Castlemart/def1, UC82B/8338 and Castlemart/UC82B) (Fig. 5).
Figure 5.
PCA on the phytohormone profiles of tomato leaves and roots. PCA was computed on the SQRT-tranformed measurements (pmol mg−1) of the phytorhormones extracted from leaves and roots of the four tomato genotypes used in this study. The analysis was computed pair wise (Castlemart/def1, UC82B/8338 and Castlemart/UC82B) and in each plot the hormone measurements for both leaves and roots were included. Each dot/triangle in the plot corresponds to a single sample (biological replication).
In each comparison there was a clear separation of the hormone profiles between leaves and roots. Moreover, in both tissues, it was possible to distinguish separate clusters for the two genotypes compared. However, when the levels of the single hormones were analysed, only few statistically significant changes were found (Supplementary Figs 2 and 3, see online supplementary material). In the leaves, the ABA levels in both def1 (impaired in JA biosynthesis) and 8338 (impaired in ET biosynthesis) were significantly higher compared to the respective wild types, whereas the level of SA was significantly lower in UC82B compared to Castlemart (Supplementary Fig. 2). In the roots, none of the seven hormones detected showed statistically significant differences between the genotypes, despite the clear separation of the clusters in the PCA plot.
Endophytic taxa isolated from tomato roots were also detected by amplicon sequencing
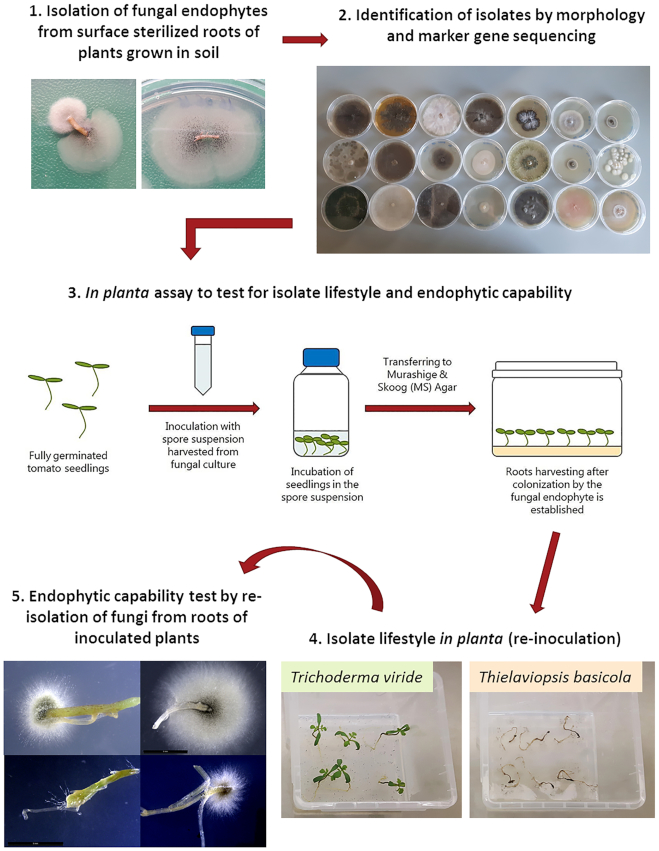
The isolation of fungal endophytes from surface-sterilized roots was performed on the same genotypes used for the amplicon sequencing analysis in order to perform a functional analysis of the mycobiome by evaluating the effect of the isolate when reintroduced in the plant roots. The procedure used for the isolations strongly decreased the risk of isolating non-endophytic fungi, since only mycelium growing from the tips of the root pieces was transferred to new PDA plates (Fig. 6).
Figure 6.
Scheme of the cultivation-dependent approaches used for the functional characterization of the fungal microbial communities of tomato roots. 1. Roots from plants grown in soil were harvested, surface sterilized and incubated on PDA medium in order to isolate endophytic fungi. 2. The isolates were identified at the species or genus level by morphology and marker gene sequencing. 3. In planta assays were performed by inoculating tomato seedlings with the spore suspension of the isolated fungi in order to assess their effect on the plant and the endophytic capability. 4. Symptoms assessment was performed 12 days after the inoculated plants were transferred to the medium. 5. The roots from symptomless plants were harvested, surface sterilized and plated on PDA medium in order to assess the endophytic colonization of the roots by the endophytic (non-pathogenic) isolates.
A total of 107 isolates were isolated from the roots (Supplementary Data 4, see online supplementary material). The taxonomical characterization resulted in a total of 18 species within 15 different genera. Pyrenochaeta lycopersici and Colletotrichum coccodes were the most abundant species isolated, constituting 25% and 18% of the total isolates, respectively. All the genera isolated were also detected by amplicon sequencing (Supplementary Data 2, see online supplementary material). However, the relative abundance of Blastobotrys spp., Chaetomium spp., Cladosporium spp., Mucor spp., Phialemonium spp., Alternaria spp. and Umbelopsis spp. was lower than the 0.5% threshold value set for the amplicon sequencing analysis and therefore they were not included in the mycobiome data analysis presented in Fig. 4.
Healthy tomato roots harbour both beneficial endophytes and latent pathogens
A subset of 27 isolates representing all the 18 identified species were selected in order to study their lifestyle in planta (Table 2). The in planta assays were conducted by growing tomato seedlings inoculated with the fungal spores on MS medium in sterile plastic boxes in order to test the interaction between the fungus and the plant, reducing the interference from external factors (Fig. 6).
Table 2.
In planta and in vitro assays performed on selected fungal endophytes isolated from tomato roots. The effect of inoculation with the isolate on the two different genotypes was assessed 12 days after inoculation based on the index shown in Supplementary Figure 4. In vitro tests for growth at different NaCl concentrations (0, 100, 200 and 300 mM NaCl) and at 36°C were performed on the isolates. The salt effect in vitro and the growth test at 36°C reported in the table are based on data shown in Supplementary Figs 6 and 7, respectively.
| Salt effect in vitroc (NaCl) | |||||||
|---|---|---|---|---|---|---|---|
| Isolatea | Species | Cultivar tested | In planta effectb | 100 mM | 200 mM | 300 mM | Growth at 36°Cd |
| RFE 192A* | Blastobotrys sp. | Castlemart | E | − | − | − | + |
| RFE 174 | Chaetomium globosum | E | + | − | − | + | |
| RFE 186A | Cladosporium sphaerospermum | E | + | + | + | − | |
| RFE 175 | Colletotrichum coccodes | P+ | + | + | − | − | |
| RFE 173A | Mucor sp. | E | 0 | − | − | − | |
| RFE 204* | Phialemonium atrogriseum | E | − | − | − | − | |
| RFE 202 | Phialemonium inflatum | E | + | + | + | + | |
| RFE 176 | Plectosphaerella sp. | P++ | + | + | + | − | |
| RFE 191 | Pyrenochaeta lycopersici | P+ | 0 | − | − | − | |
| RFE 190 | Thielaviopsis basicola | P+++ | − | − | − | − | |
| RFE 171 | Trichoderma asperellum | E | − | − | − | + | |
| RFE 183 | Trichoderma harzianum | E | − | − | − | + | |
| RFE 177 | Trichoderma viride | B | − | − | − | − | |
| RFE 339* | Alternaria infectoria | UC82B | P+ | 0 | − | − | + |
| RFE 21 | Chaetomium globosum | E | + | + | 0 | + | |
| RFE 314 | Colletotrichum coccodes | P+ | + | + | − | − | |
| RFE 315 | Fusarium sp. | P+ | + | + | + | + | |
| RFE 84 | Penicillium sp. | B | − | − | − | + | |
| RFE 343 | Phialemonium atrogriseum | E | + | + | 0 | − | |
| RFE 337 | Plectosphaerella sp. | P++ | + | + | − | − | |
| RFE 166 | Pseudeurotium sp. | B | − | − | − | − | |
| RFE 300 | Pyrenochaeta lycopersici | P++ | − | − | − | − | |
| RFE 50 | Thielaviopsis basicola | P+++ | − | − | − | − | |
| RFE 312* | Trichoderma asperellum | E | − | − | − | + | |
| RFE 310* | Trichoderma harzianum | E | − | − | − | + | |
| RFE 306 | Trichoderma viride | E | − | − | − | − | |
| RFE 66 | Umbelopsis isabellina | E | − | − | − | − | |
a‘*’ Isolates found only in the mutants (def1 or 8338) and tested in the respective wild type cultivars; b‘E’ endophytic (non-pathogenic), ‘B’ endophytic and beneficial, ‘P+’ weak pathogen, ‘P++’ pathogen, ‘P+++’ strong pathogen; c‘0’ no difference with growth at 0 mM, ‘+’ higher growth compared to 0 mM, ‘−’ lower growth compared to 0 mM; d‘+’ growth, ‘−’ no growth.
The isolates belonging to Colletotrichum coccodes, Plectosphaerella sp., Pyrenochaeta lycopersici, Thielaviopsis basicola, Alternaria infectoria and Fusarium sp. showed a pathogenic lifestyle in the in planta assay (Table 2). The pathogenic species isolated, except for A. infectoria, were among the most abundant species detected in the amplicon sequencing analysis (Fig. 4) despite the fact that the soil-grown plants analysed did not show disease symptoms. For example, T. basicola was the most abundant pathogen found in the mycobiome study in the genotypes Castlemart and def1, and the isolate tested (RFE 190) showed a strong pathogenic lifestyle in the plants re-inoculated with the spores of this isolate (Table 2 and Supplementary Fig. 4).
In contrast, isolates of the remaining 12 species caused no symptoms when applied to tomato seedlings. In order to verify that these isolates had actually colonized the roots endophytically, re-isolation of the fungi from surface sterilized roots of the inoculated seedlings was performed (Fig. 6). From all symptomless roots, it was possible to re-isolate the originally applied fungus, confirming the endophytic capability of all 17 isolates tested. Interestingly, among these endophytic (non-pathogenic) genera, Penicillium spp., Trichoderma spp. and Pseudeurotium spp. were part of the most abundant genera detected by amplicon sequencing (Fig. 4). Therefore, one isolate for each of these genera was further tested for plant growth promotion effects after the inoculation of tomato seedlings using the same method described above for the fungal lifestyle evaluation in planta (Supplementary Fig. 5, see online supplementary material). The inoculation with Penicillium sp. (RFE 84), T. viride (RFE 177) and Pseudeurotium sp. (RFE 166) as well as the inoculation with the mixture of the three endophytes resulted in a statistically significant increase in the plant fresh weight compared to the control plants. Moreover, the plant growth promotion effect of Penicillium sp. (RFE 84) and T. viride (RFE 177) was significantly higher compared to the effect of Pseudeurotium sp. and the mixture of the three endophytes. T. basicola (RFE 190) significantly decreased the fresh weight of the plants compared to the control as expected from the strong pathogenic lifestyle shown in the in planta assay (Table 2 and Supplementary Fig. 4).
In vitro functional characterization of fungal isolates for salt and temperature tolerance
The in vitro assays were performed on PDA medium in order to further characterize the fungal isolates, testing the effect of increasing salt concentrations (0, 100, 200 or 300 mM NaCl) on the fungi and investigating their ability to grow at 23 and 36°C (Table 2 and Supplementary Figs 6 and 7). The isolates belonging to the species Cladosporium sphaerospermum, Phialemonium inflatum, Fusarium sp. and C. coccodes showed an overall higher mycelial radial growth at increasing NaCl concentrations compared to 0 mM NaCl, except for C. coccodes whose radial growth at 300 mM was significantly reduced compared to that at 0 mM. Moreover, C. globosum, Phialemonium atrogriseum and Plectosphaerella sp. showed different mycelial radial growth in response to different NaCl concentrations depending on the isolate tested for each species. Mucor sp. (RFE 173A), P. lycopersici (RFE 191) and A. infectoria (RFE 339) showed decreased mycelial radial growth at 200 and 300 mM NaCl, but 100 mM NaCl had no effect compared to 0 mM NaCl. The rest of the isolates tested showed decreased mycelial radial growth at all the three salt concentrations compared to 0 mM NaCl.
Eleven out of the 27 isolates tested had detectable mycelial growth at 36°C (Table 2 and Supplementary Fig. 7). Surprisingly, the mycelial radial growth of Blastobotrys sp. (RFE 192A) was significantly higher at 36 than at 23°C, whereas C. globosum (RFE 174) grew equally well at both temperatures. The growth of the remaining nine isolates at 36°C was significantly less than at 23°C. Interestingly, the two isolates of Trichoderma asperellum and Trichoderma harzianum respectively grew at 36°C whereas no growth was observed for the two T. viride isolates (Supplementary Fig. 7).
DISCUSSION
Host genotype influences the abundance of dominant species in the mycobiome
Plant genotype plays a major role in shaping the fungal endophytic communities of tomato roots, since significant differences were found in both alpha and beta diversity analyses by comparing the cultivars Castlemart and UC82B. A significant role of the host genotype in the structure of the fungal endophytic communities was previously reported using an amplicon sequencing approach only in A. thaliana (Urbina et al. 2018). It is, therefore, still unclear which factors drive the community differences between cultivars, but it has been shown that plant loci involved in the defence response affected the communities of leaf-associated bacteria and fungi (Horton et al. 2014). Phytohormones are known to be involved in the signalling of plant defence responses (Pieterse et al. 2012), therefore the clear difference in the phytohormone profiles between Castlemart and UC82B may be linked to the differences in the endophytic communities of these two cultivars. In our study, the genera whose relative abundances were significantly different between Castlemart and UC82B are mostly comprised of species previously described as plant pathogens (Thielaviopsis, Pyrenochaeta, Fusarium and Plectosphaerella), whose pathogenic lifestyle was also confirmed by our in planta assay. This suggests that changes in the phytohormone balance may affect the plant immune response and therefore influence the abundance of pathogenic taxa in the host plant. Vice versa, the changes in the abundance of these pathogenic genera may also be responsible for the differences in the hormone profiles between the cultivars. This highlights the need for more experiments in order to clarify whether differences in the hormone profiles drove the mycobiome structure or they were a consequence of the mycobiome composition in the two cultivars. Nonetheless, other factors may have affected the structure of the mycobiome in the two tomato cultivars. For example, the composition of the root exudates, known to affect the structure of the root microbiome (Sasse, Martinoia and Northen 2018), may differ between the cultivars, as previously shown for different A. thaliana accessions (Mönchgesang et al. 2016).
Lower levels of JA increase species richness of fungal endophytes in tomato roots
The JA-mediated changes in the diversity of the endophytic fungal community found in this study have not previously been reported for any plant species, to the best of our knowledge. However, studies on bacterial root endophytes showed no effect of JA in A. thaliana (Kniskern, Traw and Bergelson 2007), while Santhanam et al. (2014) found a minor influence of JA on structuring bacterial root communities in N. attenuata and MeJA treatment was shown to decrease the diversity of the bacterial endophytic communities in wheat (Liu et al. 2017). By quantification of phytohormones in root and leaf subsamples from plants harvested for mycobiome analysis, we could link hormone profiles and endophytic fungal community structure to further explore the role of phytohormones. The separation of phytohormone profiles of Castlemart and def1 in both leaves and roots suggests that changes in the JA levels affect the whole phytohormone balance. In this context, the enrichment in rare species of def1 may be the result of both the decreased JA levels and the change in the general hormone balance of the host. The only statistically significant difference between Castlemart and def1 was found in the level of ABA in the leaves. However the increase of SA in both leaves and roots of def1 as well as the changes in IAld and tZR in roots could contribute to the differences found in the alpha diversity between these two genotypes, despite the fact that these changes were not significant. JA, SA and ABA are known to be involved in plant resistance against several pathogens, as well as auxins and cytokinins (Jameson 2000; Kazan and Manners 2009; De Vleesschauwer, Xu and Höfte 2014; Spence and Bais 2015). For this reason, reduced levels of JA together with the minor changes in the level of the hormones mentioned above could increase the species richness in the roots due to changes/imbalances in the defence mechanisms of the host. Moreover, previous studies in Arabidopsis showed that the level of JA in the host plant can influence the composition of the root exudates and thereby the composition of the rhizosphere microbiome (Carvalhais et al. 2015; Carvalhais, Schenk and Dennis 2017). It is likely that the effect of JA on the fungal communities of tomato roots may also be linked to changes in the composition of the tomato root exudates.
Furthermore, the phytohormone profiling was performed on samples harvested together with the samples analysed for amplicon sequencing and endophyte isolations, therefore it represents a snapshot of the hormonal status at that specific growth stage. However, the effect of the hormones on endophytic fungi can differ at different stages of the interactions, as demonstrated for JA (Jacobs et al. 2011) and ET (Khatabi et al. 2012), both involved in colonization of Arabidopsis roots by S. indica. Therefore, time course experiments combining microbiome analysis and phytohormone profiling are needed to elucidate further the role of phytohormones in sculpting the endophytic communities of tomato roots during the different developmental stages of the host.
Tomato roots harbour tomato pathogens, non-host pathogens, beneficial and commensal taxa
The amplicon sequencing analysis combined with the isolation of fungal endophytes revealed the presence of species previously described as tomato pathogens, non-pathogens (pathogens in other host species) as well as endophytes known as beneficial or commensal. However, even more intriguing, we also found taxa not previously reported as beneficial to plant performance and/or to have an endophytic lifestyle. For example, the genus Pseudeurotium was one of the most abundant and P. bakeri was the most abundant species found in this genus. This species was previously found associated with the rhizosphere of different plants, such as potato (Hannula, de Boer and van Veen 2010) and soybean (Liu et al. 2011), but never as an endophyte. Furthermore, we demonstrated a putative plant growth promoting effect of a Pseudeurotium sp. isolate, making this species an interesting candidate for further in planta experiments. Likewise, Pseudogymnoascus verrucosus, the most abundant species found in the genus Pseudogymnoascus, has not previously been described to colonize plant tissues, although other species belonging to this genus are reported as root endophytes in various plant species (Sharples et al. 2000; Huang et al. 2015; Granzow et al. 2017). In addition, the yeast genera Solicoccozyma, Candida, Apiotrichum and Saitozyma were highly abundant in the tomato root endosphere. Interestingly, Saitozyma podzolica (formerly Cryptococcus podzolicus) was previously found to be part of the root endosphere of different plant species (Sánchez Márquez et al. 2010; Urbina et al. 2018), whereas Apiotrichum xylopini (formerly Trichosporon xylopini) and Solicoccozyma terricola (formerly Cryptococcus terricola) were never reported as root endophytes. Candida subhashii was the most abundant species detected in its genus and its presence was previously reported in the root endosphere of faba bean and wheat (Granzow et al. 2017).
Among the endophytic species known to have a beneficial effect on the host plant we found several OTUs assigned to different species belonging to the genera Trichoderma and Penicillium. Trichoderma species are frequently reported as endophytic fungi capable of colonizing a big range of host plants, improving plant growth or acting as biocontrol agents against plant pathogens (Harman et al. 2004; López-Bucio, Pelagio-Flores and Herrera-Estrella 2015; Card et al. 2016). Also the genus Penicillium comprehends several species capable of giving beneficial effects to the host plant, such as growth promotion and biocontrol of plant pathogens (Larena et al. 2003; Khan et al. 2011; Elsharkawy et al. 2012; Ahmad et al. 2014). The beneficial effect of these two genera was also shown in this study by the significant increase in the fresh weight of plants inoculated with Penicillium sp. (RFE 84) and Trichoderma viride (RFE 177) compared to the non-inoculated plants.
In addition, species belonging to the genus Mortierella were also abundant in tomato roots. A few studies reported Mortierella species as root endophytes in different plant species (Sánchez Márquez et al. 2010; Urbina et al. 2018). Recently, it was demonstrated that M. hyalina is able to colonize the roots of A. thaliana, promoting plant growth and conferring tolerance to Alternaria brassicae (Johnson et al. 2019).
As previously mentioned, several species previously described as tomato pathogens were among the most abundant species detected in the mycobiome of tomato roots, such as Plectosphaerella cucumerina (Raimondo and Carlucci 2018), Fusarium oxysporum (Takken and Rep 2010; McGovern 2015), Colletotrichum coccodes (Garibaldi et al. 2008; Gilardi et al. 2014), Thielaviopsis basicola (Blancard 2012) and Pyrenochaeta lycopersici (Giotis et al. 2009; Blancard 2012). In fact, the pathogenic lifestyle of the isolates belonging to these species was confirmed by the in planta assay in this study. Finally, the species Ilyonectria mors-panacis, Olpidium brassicae and Nectria ramulariae (also known as Neonectria ramulariae) were the dominant species found in the respective genera and they were previously reported as plant pathogens in different plant species (Hirooka et al. 2012; Farh et al. 2017; Lay, Hamel and St-Arnaud 2018), but never in tomato.
Interestingly, despite tomato being a mycorrhizal plant species, the phylum Glomeromycota was not detected in this root mycobiome study. The absence of this phylum in our dataset could be due to several factors, such as the primer pair and/or marker gene choice, which may have impaired the detection of this phylum. In fact, it was recently shown that the phylum Glomeromycota was in extremely low abundance in the root endosphere of tomato plants grown in a greenhouse when the ITS2 region was used as a molecular marker (Lee et al. 2019). On the other hand, the arbuscular-mycorrhizal communities have been successfully analysed, both in tomato plants (Higo et al. 2020) and other species (Hassan et al. 2014), by next generation sequencing approaches targeting the 18S region using arbuscular mycorrhiza-specific primers. These studies together confirm that the choice of both target region and primer pair influences significantly the detection of Glomeromycota species in the complex endophytic mycobiome community.
The equilibrium between beneficial and pathogenic fungi may maintain plant health
The combined approach of isolations and amplicon sequencing enabled us to get a deeper understanding of the lifestyle/function of specific taxa in the root mycobiome, especially since there were noticeable similarities in the results of the two methods. Firstly, all the 15 genera isolated were detected by amplicon sequencing. Moreover, the high frequency of isolates belonging to Plectosphaerella sp., Fusarium sp. and Pyrenochaeta lycopersici in UC82B compared to Castlemart and vice versa for T. basicola, confirmed the statistically significant differences found by amplicon sequencing for these species. The functional characterization of these isolates showed that they most likely were present as latent pathogens in the soil-grown plants.
Additionally, we also obtained isolates of A. infectoria, Blastobotrys sp., C. globosum, C. sphaerospermum, Mucor sp., P. atrogriseum, P. inflatum and Umbelopsis isabellina that belong to genera detected in low abundance by the amplicon sequencing analysis (rare fraction of the mycobiome). These genera were previously reported as endophytes in roots of several plant species (Hoff et al. 2004; Naik, Shashikala and Krishnamurthy 2009; Sánchez Márquez et al. 2010; Huang et al. 2015; Granzow et al. 2017) and, in the in planta assays, we confirmed that these isolates actually can colonize tomato roots endophytically.
Taken together, our findings suggest that the equilibrium between pathogenic and endophytic or beneficial fungi in the root mycobiome contributes to keeping the host plant healthy. In line with this, recent work on the root microbiome of A. thaliana showed that the presence of fungal pathogens in healthy plants was balanced by other members of the microbiome, such as beneficial and commensal bacterial taxa (Durán et al. 2018). Therefore, it remains to further elucidate how these inter-kingdom interactions sustain the equilibrium in the root endosphere. Moreover, our complementary in vitro characterization of the isolates showed that differences between the fungal species can also be found in their ability to cope with osmotic and temperature stress, further highlighting the complexity of the fine balance between microorganisms in healthy tomato plants.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Associate Professor Frank L. W. Takken from Faculty of Science, Swammerdam Institute for Life Sciences, University of Amsterdam (The Netherlands) for providing the seeds of the tomato genotypes used in this study. We are grateful to Dr. Verena Jeschke from Department of Plant and Environmental Sciences, University of Copenhagen (Denmark) and Professor Helle Sørensen from Department of Mathematical Sciences, University of Copenhagen (Denmark) for the important advices on data analysis and statistics. The stereomicroscope images were collected with the Leica Stereo-Fluorescence system MZ FLII at the Center for Advanced Bioimaging (CAB) Denmark, University of Copenhagen (Denmark).
FUNDING
This work was supported by the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 676480 and by the Danish National Research Foundation DNRF grant 99.
AUTHOR CONTRIBUTIONS
A.M. designed the experiments with the advices from all the authors. M.B. analysed the phytohormone extracts by UHPLC/TQ-MS and A.M. performed the rest of the experiments presented in the paper. A.M. performed the analysis and visualization of the data with the support of A.B., M.B., H.J.L.J. and B.J. A.B. produced the OTU table from the amplicon sequencing raw data and T.C. provided experimental advice while preparing the amplicon libraries. A.M. wrote the manuscript and all the authors provided valuable input that improved it.
Conflicts of interest None declared.
REFERENCES
- Ahmad A, Shafique S, Shafique S et al.. Penicillium oxalicum directed systemic resistance in tomato against Alternariaalternata. Acta Physiol Plant. 2014;36:1231–40. [Google Scholar]
- Altschul SF, Gish W, Miller W et al.. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–10. [DOI] [PubMed] [Google Scholar]
- Blancard D. Tomato Diseases. 2nd Editio London: CRC Press, 2012. [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ et al.. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcard D, Gillet F, Legendre P. Numerical Ecology with R. New York, NY: Springer New York, 2011. [Google Scholar]
- Brader G, Compant S, Vescio K et al.. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu Rev Phytopathol. 2017;55:61–83. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K et al.. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91–5. [DOI] [PubMed] [Google Scholar]
- Busby PE, Peay KG, Newcombe G. Common foliar fungi of Populustrichocarpa modify Melampsora rust. New Phytol. 2016;209:1681–92. [DOI] [PubMed] [Google Scholar]
- Busby PE, Ridout M, Newcombe G. Fungal endophytes: modifiers of plant disease. Plant Mol Biol. 2016;90:645–55. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al.. correspondence QIIME allows analysis of high- throughput community sequencing data Intensity normalization improves color calling in SOLiD sequencing. Nature. 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Card S, Johnson L, Teasdale S et al.. Deciphering endophyte behaviour: the link between endophyte biology and efficacious biological control agents. FEMS Microbiol Ecol. 2016;92:1–19. [DOI] [PubMed] [Google Scholar]
- Carvalhais LC, Dennis PG, Badri D V et al.. Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol Plant Microbe Interact. 2015;28:1049–58. [DOI] [PubMed] [Google Scholar]
- Carvalhais LC, Schenk PM, Dennis PG. Jasmonic acid signalling and the plant holobiont. Curr Opin Microbiol. 2017;37:42–7. [DOI] [PubMed] [Google Scholar]
- Collinge DB, Jørgensen HJL, Latz MAC et al.. Searching for novel fungal biological control agents for plant disease control among endophytes. In: Hodkinson T, Doohan F, Saunders M et al. (eds.). Endophytes for a Growing World. Cambridge University Press, 2019, 25–51. [Google Scholar]
- Cubeta MA, Echandi E, Abernethy T et al.. Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology. 1991;81:1395–400. [Google Scholar]
- De Vleesschauwer D, Xu J, Höfte M. Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Front Plant Sci. 2014;5:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–30. [Google Scholar]
- du Jardin P. Plant biostimulants: Definition, concept, main categories and regulation. Sci Hortic (Amsterdam). 2015;196:3–14. [Google Scholar]
- Durán P, Thiergart T, Garrido-Oter R et al.. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell. 2018;175:973–83..e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsharkawy MM, Shimizu M, Takahashi H et al.. Induction of systemic resistance against Cucumber mosaic virus by Penicilliumsimplicissimum GP17-2 in Arabidopsis and tobacco. Plant Pathol. 2012;61:964–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh ME-A, Kim Y-J, Singh P et al.. Cross interaction between Ilyonectriamors-panacis isolates infecting Korean ginseng and ginseng saponins in correlation with their pathogenicity. Phytopathology. 2017;107:561–9. [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–8. [DOI] [PubMed] [Google Scholar]
- Garibaldi A, Baudino M, Minuto A et al.. Effectiveness of fumigants and grafting against tomato brown root rot caused by Colletotrichumcoccodes. Phytoparasitica. 2008;36:483–8. [Google Scholar]
- Gilardi G, Colla P, Pugliese M et al.. Control of Colletotrichumcoccodes on tomato by grafting and soil amendments. J Phytopathol. 2014;162:116–23. [Google Scholar]
- Giotis C, Markelou E, Theodoropoulou A et al.. Effect of soil amendments and biological control agents (BCAs) on soil-borne root diseases caused by Pyrenochaetalycopersici and Verticilliumalbo-atrum in organic greenhouse tomato production systems. Eur J Plant Pathol. 2009;123:387–400. [Google Scholar]
- Granzow S, Kaiser K, Wemheuer B et al.. The effects of cropping regimes on fungal and bacterial communities of wheat and faba bean in a greenhouse pot experiment differ between plant species and compartment. Front Microbiol. 2017;8:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannula SE, de Boer W, van Veen JA. In situ dynamics of soil fungal communities under different genotypes of potato, including a genetically modified cultivar. Soil Biol Biochem. 2010;42:2211–23. [Google Scholar]
- Hardoim PR, van Overbeek LS, Berg G et al.. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79:293–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman GE, Howell CR, Viterbo A et al.. Trichoderma species - Opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2:43–56. [DOI] [PubMed] [Google Scholar]
- Hassan SE-D, Bell TH, Stefani FOP et al.. Contrasting the community structure of arbuscular mycorrhizal fungi from hydrocarbon-contaminated and uncontaminated soils following willow (Salix spp. L.) planting. PLoS One. 2014;9:e102838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo M, Azuma M, Kamiyoshihara Y et al.. Impact of phosphorus fertilization on tomato growth and arbuscular mycorrhizal fungal communities. Microorganisms. 2020;8:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirooka Y, Ichihara Y, Masuya H et al.. Seed rot, a new disease of beech tree caused by Neonectriaramulariae (anamorph: Cylindrocarponobtusiusculum). J Phytopathol. 2012;160:504–6. [Google Scholar]
- Hoff JA, Klopfenstein NB, McDonald GI et al.. Fungal endophytes in woody roots of Douglas-fir (Pseudotsugamenziesii) and ponderosa pine (Pinusponderosa). For Pathol. 2004;34:255–71. [Google Scholar]
- Horton MW, Bodenhausen N, Beilsmith K et al.. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat Commun. 2014;5:5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Lightner J, Browse J et al.. An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell. 1996;8:2067–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, An H, Song H et al.. Diversity and biotransformative potential of endophytic fungi associated with the medicinal plant Kadsuraangustifolia. Res Microbiol. 2015;166:45–55. [DOI] [PubMed] [Google Scholar]
- Jacobs S, Zechmann B, Molitor A et al.. Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformosporaindica. Plant Physiol. 2011;156:726–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson PE. Cytokinins and auxins in plant-pathogen interactions - An overview. Plant Growth Regul. 2000;32:369–80. [Google Scholar]
- Johnson JM, Ludwig A, Furch ACU et al.. The beneficial root-colonizing fungus Mortierella hyalina promotes the aerial growth of Arabidopsis and activates calcium-dependent responses which restrict Alternaria brassicae-induced disease development in roots. Mol Plant-Microbe Interact. 2019;32:1–13. [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. Linking development to defense: auxin in plant–pathogen interactions. Trends Plant Sci. 2009;14:373–82. [DOI] [PubMed] [Google Scholar]
- Khan AL, Hamayun M, Kim YH et al.. Ameliorative symbiosis of endophyte (Penicilliumfuniculosum LHL06) under salt stress elevated plant growth of Glycine max L. Plant Physiol Biochem. 2011;49:852–61. [DOI] [PubMed] [Google Scholar]
- Khatabi B, Molitor A, Lindermayr C et al.. Ethylene supports colonization of plant roots by the mutualistic fungus Piriformosporaindica. PLoS One. 2012;7:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ, Hayford MB, Kretzmer KA et al.. Control of ethylene synthesis by expression of a bacterial enzyme in transgenic tomato plants. Plant Cell. 1991;3:1187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniskern JM, Traw MB, Bergelson J. Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana. Mol Plant-Microbe Interact. 2007;20:1512–22. [DOI] [PubMed] [Google Scholar]
- Kogel KH, Franken P, Hückelhoven R. Endophyte or parasite - what decides? Curr Opin Plant Biol. 2006;9:358–63. [DOI] [PubMed] [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K et al.. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–7. [DOI] [PubMed] [Google Scholar]
- Larena I, Sabuquillo P, Melgarejo P et al.. Biocontrol of fusarium and verticillium wilt of tomato by Penicilliumoxalicum under greenhouse and field conditions. J Phytopathol. 2003;151:507–12. [Google Scholar]
- Lay CY, Hamel C, St-Arnaud M. Taxonomy and pathogenicity of Olpidiumbrassicae and its allied species. Fungal Biol. 2018;122:837–46. [DOI] [PubMed] [Google Scholar]
- Lebeis SL, Paredes SH, Lundberg DS et al.. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science (80-). 2015;349:860–4. [DOI] [PubMed] [Google Scholar]
- Lee SA, Kim Y, Kim JM et al.. A preliminary examination of bacterial, archaeal, and fungal communities inhabiting different rhizocompartments of tomato plants under real-world environments. Sci Rep. 2019;9:9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Carvalhais LC, Schenk PM et al.. Effects of jasmonic acid signalling on the wheat microbiome differ between body sites. Sci Rep. 2017;7:41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang G, Jin J et al.. Effects of different concentrations of phosphorus on microbial communities in soybean rhizosphere grown in two types of soils. Ann Microbiol. 2011;61:525–34. [Google Scholar]
- Long HH, Sonntag DG, Schmidt DD et al.. The structure of the culturable root bacterial endophyte community of Nicotianaattenuata is organized by soil composition and host plant ethylene production and perception. New Phytol. 2010;185:554–67. [DOI] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH et al.. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J, Pelagio-Flores R, Herrera-Estrella A. Trichoderma as biostimulant: exploiting the multilevel properties of a plant beneficial fungus. Sci Hortic (Amsterdam). 2015;196:109–23. [Google Scholar]
- McGovern RJ. Management of tomato diseases caused by Fusariumoxysporum. Crop Prot. 2015;73:78–92. [Google Scholar]
- McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mönchgesang S, Strehmel N, Schmidt S et al.. Natural variation of root exudates in Arabidopsis thaliana-linking metabolomic and genomic data. Sci Rep. 2016;6:29033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik BS, Shashikala J, Krishnamurthy YL. Study on the diversity of endophytic communities from rice (Oryzasativa L.) and their antagonistic activities in vitro. Microbiol Res. 2009;164:290–6. [DOI] [PubMed] [Google Scholar]
- Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Schwartz R (ed.). Bioinformatics. 2019;35:526–8. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C et al.. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012;28:489–521. [DOI] [PubMed] [Google Scholar]
- Porras-Alfaro A, Bayman P. Hidden fungi, emergent properties: endophytes and microbiomes. Annu Rev Phytopathol. 2011;49:291–315. [DOI] [PubMed] [Google Scholar]
- Raimondo ML, Carlucci A. Characterization and pathogenicity assessment of Plectosphaerella species associated with stunting disease on tomato and pepper crops in Italy. Plant Pathol. 2018;67:626–41. [Google Scholar]
- Ritpitakphong U, Falquet L, Vimoltust A et al.. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016;210:1033–43. [DOI] [PubMed] [Google Scholar]
- Rognes T, Flouri T, Nichols B et al.. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam R, Groten K, Meldau DG et al.. Analysis of plant-bacteria interactions in their native habitat: Bacterial communities associated with wild tobacco are independent of endogenous jasmonic acid levels and developmental stages. PLoS One. 2014;9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse J, Martinoia E, Northen T. Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 2018;23:25–41. [DOI] [PubMed] [Google Scholar]
- Sharples JM, Chambers SM, Meharg AA et al.. Genetic diversity of root-associated fungal endophytes from Callunavulgaris at contrasting field sites. New Phytol. 2000;148:153–62. [DOI] [PubMed] [Google Scholar]
- Spence C, Bais H. Role of plant growth regulators as chemical signals in plant-microbe interactions: a double edged sword. Curr Opin Plant Biol. 2015;27:52–8. [DOI] [PubMed] [Google Scholar]
- Stingl N, Krischke M, Fekete A et al.. Analysis of defense signals in Arabidopsis thaliana leaves by ultra-performance liquid chromatography/tandem mass spectrometry: jasmonates, salicylic acid, abscisic acid. In: Munnik T, Heilmann I (eds.). Plant Lipid Signaling Protocols. Vol 1009. Totowa, NJ: Humana Press, 2013, 103–13. [DOI] [PubMed] [Google Scholar]
- Sánchez Márquez S, Bills GF, Domínguez Acuña L et al.. Endophytic mycobiota of leaves and roots of the grass Holcuslanatus. Fungal Divers. 2010;41:115–23. [Google Scholar]
- Takken F, Rep M. The arms race between tomato and Fusariumoxysporum. Mol Plant Pathol. 2010;11:309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toju H, Tanabe AS, Yamamoto S et al.. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One. 2012;7:e40863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina H, Breed MF, Zhao W et al.. Specificity in Arabidopsis thaliana recruitment of root fungal communities from soil and rhizosphere. Fungal Biol. 2018;122:231–40. [DOI] [PubMed] [Google Scholar]
- van den Berg RA, Hoefsloot HC, Westerhuis JA et al.. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S et al.. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. San Diego, CA: Academic Press, 1990, 315–22. [Google Scholar]
- Xu L, Wu C, Oelmüller R et al.. Role of phytohormones in Piriformosporaindica-induced growth promotion and stress tolerance in plants: more questions than answers. Front Microbiol. 2018;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Hochholdinger F. The role of host genetic signatures on root–microbe interactions in the rhizosphere and endosphere. Front Plant Sci. 2018;9:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.