Abstract
See Veldsman (doi:10.1093/brain/awaa075) for a scientific commentary on this article.
There is both clinical and neuroanatomical variability at the single-subject level in Alzheimer’s disease. Using single-subject atrophy maps and the human connectome, Tetreault et al. show that clinical, cognitive, and neuropsychiatric symptoms localize to specific brain networks.
Keywords: Alzheimer’s disease, delusion, memory, functional connectivity, brain atrophy
Abstract
There is both clinical and neuroanatomical variability at the single-subject level in Alzheimer’s disease, complicating our understanding of brain-behaviour relationships and making it challenging to develop neuroimaging biomarkers to track disease severity, progression, and response to treatment. Prior work has shown that both group-level atrophy in clinical dementia syndromes and complex neurological symptoms in patients with focal brain lesions localize to brain networks. Here, we use a new technique termed ‘atrophy network mapping’ to test the hypothesis that single-subject atrophy maps in patients with a clinical diagnosis of Alzheimer’s disease will also localize to syndrome-specific and symptom-specific brain networks. First, we defined single-subject atrophy maps by comparing cortical thickness in each Alzheimer’s disease patient versus a group of age-matched, cognitively normal subjects across two independent datasets (total Alzheimer’s disease patients = 330). No more than 42% of Alzheimer’s disease patients had atrophy at any given location across these datasets. Next, we determined the network of brain regions functionally connected to each Alzheimer’s disease patient’s location of atrophy using seed-based functional connectivity in a large (n = 1000) normative connectome. Despite the heterogeneity of atrophied regions at the single-subject level, we found that 100% of patients with a clinical diagnosis of Alzheimer’s disease had atrophy functionally connected to the same brain regions in the mesial temporal lobe, precuneus cortex, and angular gyrus. Results were specific versus control subjects and replicated across two independent datasets. Finally, we used atrophy network mapping to define symptom-specific networks for impaired memory and delusions, finding that our results matched symptom networks derived from patients with focal brain lesions. Our study supports atrophy network mapping as a method to localize clinical, cognitive, and neuropsychiatric symptoms to brain networks, providing insight into brain-behaviour relationships in patients with dementia.
See Veldsman (doi:10.1093/brain/awaa075) for a scientific commentary on this article.
Introduction
Neuroimaging measures of cortical thickness can be used as in vivo markers of neurodegeneration in patients with Alzheimer’s disease (Jack et al., 2010). At the group level, atrophy in specific neuroanatomical locations is associated with the clinical diagnosis (Du et al., 2007; Dickerson et al., 2009), progression (Dickerson and Wolk, 2013; Leung et al., 2013), and cognitive and neurobehavioural symptoms of Alzheimer’s disease (Dickerson et al., 2004; Darby et al., 2017a, 2019). Newly developed quantitative methods using structural MRI can measure brain atrophy at the single-subject level (Jack et al., 1997; La Joie et al., 2012; Ossenkoppele et al., 2015a, b; Perry et al., 2017), providing a potential bridge between these group-level neuroimaging findings and clinical care of individual dementia patients. However, a major challenge is that there is significant heterogeneity at the single-subject level, leading to different locations of atrophy (Lehmann et al., 2013; Noh et al., 2014; Byun et al., 2015; Dong et al., 2017; Dickerson et al., 2017; Poulakis et al., 2018; Torok et al., 2018) and different symptoms (Lehmann et al., 2013; Byun et al., 2015; Dickerson et al., 2017; Darby et al., 2019) in different patients. This clinical and neuroanatomical heterogeneity makes it difficult to localize clinical dementia syndromes and specific cognitive and neuropsychiatric symptoms, complicating our fundamental understanding of brain-behaviour relationships (Darby and Fox, 2019; Darby et al., 2019).
A promising solution to this problem is to map both clinical dementia syndromes and specific neurological symptoms to brain networks (Seeley et al., 2009; Fox, 2018). Prior studies have found that group-level atrophy in patients with neurodegenerative disorders occurs within syndrome-specific, intrinsically connected brain networks (Seeley et al., 2009; Greicius and Kimmel, 2012; Zhou et al., 2012; Seeley, 2017; Darby et al., 2019). Similarly, a recent technique called lesion network mapping has identified symptom-specific brain networks for a range of complex lesion-induced symptoms, including hallucinations (Boes et al., 2015), delusions (Darby and Fox, 2017; Darby et al., 2017b, 2019), criminal behaviour (Darby et al., 2018a), disordered free-will perception (Darby et al., 2018b), and memory (Ferguson et al., 2019). Collectively, this prior work has localized clinical, cognitive, and neuropsychiatric symptoms to specific brain networks. However, it remains unknown whether these same symptom-specific brain networks can account for the observed clinical and neuroanatomical heterogeneity in Alzheimer’s disease patients at the single-subject level.
Here, we use a new technique, atrophy network mapping, which uses the human connectome to test whether clinical, cognitive, and neuropsychiatric symptoms in patients with Alzheimer’s disease map onto specific brain networks. First, we define single-subject atrophy maps by comparing cortical thickness in each Alzheimer’s disease subject against estimates from a normative model of cortical thickness in cognitively normal subjects. Next, we use each patient’s single-subject atrophy map as a seed location in functional connectivity analysis with a large normative connectome (Yeo et al., 2011; Holmes et al., 2015) to determine the network of brain regions functionally connected to each single-subject atrophy map. This approach is similar to lesion network mapping (Boes et al., 2015; Darby and Fox, 2017; Darby et al., 2017b, 2018a, b; Fox, 2018) with the exception that single-subject atrophy maps are used as seeds instead of brain lesions. We then use atrophy network mapping to define symptom-specific brain networks for Alzheimer’s disease, impaired memory, and delusions and compare our results against symptom networks derived from patients with focal brain lesions.
Materials and methods
Subjects
Data used in this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of the ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease. Subjects in the study include cognitively normal subjects and those with a clinical diagnosis of MCI or Alzheimer’s disease between the ages of 55–90. For up-to-date information regarding these specific protocols, please see www.adni-info.org.
The present study included 184 subjects with a diagnosis of Alzheimer’s disease and 227 cognitively normal subjects from the first part of the ADNI study (ADNI-1), and 146 subjects with a diagnosis of Alzheimer’s disease and 201 cognitively normal subjects from the second part of the ADNI study (ADNI-2) (Table 1). Per the ADNI protocol, a diagnosis of Alzheimer’s disease required a subjective memory complaint, clinical dementia rating score of 0.5–1, impaired performance on the logical memory task part II delayed recall below the education-adjusted cut-off, Mini-Mental State Examination (MMSE) score between 20–26 (inclusive), and clinician’s diagnosis of probable Alzheimer’s disease using the NINCDS/ADRDA criteria. Memory impairment was defined using delayed recall and recognition scores on the auditory verbal learning task (AVLT). The presence or absence of delusions was obtained from the neuropsychiatric inventory (NPI) assessment. Patients reporting delusions within 6 months of baseline MRI scans were included in the delusions group (n = 39), while our non-delusions group was defined as patients who never reported delusions at any time point during the ADNI study (n = 121).
Table 1.
Demographics
| ADNI-1 | ADNI-2 | |||
|---|---|---|---|---|
| Alzheimer’s disease | Cognitively normal | Alzheimer’s disease | Cognitively normal | |
| n | 184 | 227 | 146 | 201 |
| Age | 75.3 ± 7.6 | 76.0 ± 5.0 | 74.6 ± 8.2 | 73.3 ± 6.4 |
| Gender, male | 96 (52.2%) | 118 (52.0%) | 81 (54.4%) | 95 (47.0%) |
| Delusions | 23 (12.5%) | 16 (11.0%) | ||
| AVLT-recognition | 7.11 ± 3.96 | 6.62 ± 3.85 | ||
| AVLT- recall | 0.70 ±1.60 | 0.62 ± 1.16 | ||
| MMSE | 23.2 ± 2.0 | 29.1 ± 1.0 | 23.1 ± 2.1 | 29.1 ± 1.3 |
| CDR-global | ||||
| 0 | 0 | 227 (100%) | 0 | 199 (100%) |
| 0.5 | 101 (54.9%) | 0 | 64 (43.8%) | 0 |
| 1 | 82 (44.6%) | 0 | 81 (55.5%) | 0 |
| 2 | 0 | 0 | 1 (0.7%) | 0 |
| CDR-SOB | 4.3 ± 1.6 | 0.0 ± 0.1 | 4.5 ± 1.7 | 0.0 ± 0.1 |
| Education | 14.6 ± 3.2 | 16.0 ± 2.9 | 15.7 ± 2.8 | 16.6 ± 2.6 |
Results are presented as mean ± SD (%). CDR = Clinical Dementia Rating score; MMSE = Mini-Mental State Examination score; n = number of subjects; SOB = sum of boxes.
MRI and analysis
MRI scans for ADNI-1 were collected on a 1.5 T scanner using a standardized MPRAGE protocol: sagittal plane, repetition time/echo time/inversion time, 2400/3/1000 ms, flip angle 8°, 24 cm field of view, 192 × 192 in-plane matrix, 1.2 mm slice thickness (Jack et al., 2008). MRI scans for ADNI-2 were collected on a 3 T scanner using a standardized MPRAGE protocol: sagittal plane, repetition time/echo time/inversion time 2300/2.95/900 ms, flip angle 9°, 26 cm field of view, 256 × 256 in-plane matrix, 1.2 mm slice thickness.
Quantitative morphometric analysis was performed using FreeSurfer version 6.0 (Dale et al., 1999). After spatial and intensity normalization and skull stripping, the resulting volume was segmented into grey matter, white matter, and CSF, and a deformable surface algorithm was used to identify the pial surface. Cortical thickness was determined by measuring the distance between the white matter and pial surfaces at ∼160 000 points (vertices). Each subject’s reconstructed brain was then morphed and registered to an average spherical space, enabling the accurate matching of cortical locations among individuals across the entire cortical surface.
Quality control analysis of Freesurfer reconstruction included automatic detection of recon-all processing errors and visual inspection for segmentation, intensity normalization, and skull stripping errors. One Alzheimer’s disease subject and one control subject from ADNI-1 were excluded for recon-all errors, and three Alzheimer’s disease subjects were excluded from ADNI-2 for segmentation errors noted with visual inspection.
Single-subject atrophy maps
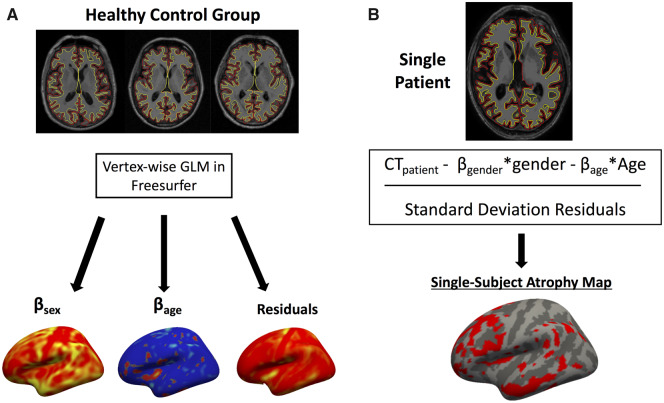
We performed a vertex-wise general linear model (GLM) for cortical thickness for the cognitively normal subjects from each dataset using age and gender as covariates (Fig. 1A). Normative models were computed separately for ADNI-1 and ADNI-2 given different scanner acquisition protocols between the two datasets. Next, we used the beta-term maps for age and gender, as well as the maps of the residuals from these normative models, to calculate a vertex-wise w-score for cortical thickness in each patient (a w-score is a z-score adjusted for covariates such as age and sex; Fig. 1B). To calculate the w-scores we used the formula: w-score = (actual − expected)/RSD, where actual is the patient’s observed cortical thickness, expected is the predicted cortical thickness based on the control GLM, and RSD is the residual standard deviation from the control GLM, similar to prior methods (La Joie et al., 2012; Ossenkoppele et al., 2015a, b; Perry et al., 2017). Atrophy w-maps were binarized at a w-score < −2, corresponding to cortical thickness 2 standard deviations (SD) below the mean of the population of healthy control subjects, controlling for age and gender. We repeated all analyses with more stringent (w < −2.5) and less stringent (w < −1.5) atrophy thresholds in control analyses to ensure that results did not change according to the selected atrophy threshold (Supplementary Fig. 1). We also performed two additional control analyses. First, we thresholded w-maps at the patient’s highest 5% of atrophied voxels in order to control for single-subject atrophy size across patients. Second, we created an 8-mm radius spherical atrophy seed at each subject’s peak location of atrophy in order to determine whether an atrophy seed constrained to a single location resulted in more or less robust results compared with a distributed atrophy seed. We overlaid binarized atrophy w-maps from all patients to identify regions consistently showing atrophy in the greatest number of patients.
Figure 1.
Individualized atrophy mapping method. (A) GLM using cortical thickness from controls is used to generate a normative model for cortical thickness based on a patient’s age and gender. (B) Cortical thickness from each individual patient is compared against model estimates to generate a vertex-wise w-map (z-map controlling for age and gender) for cortical atrophy.
Atrophy network mapping
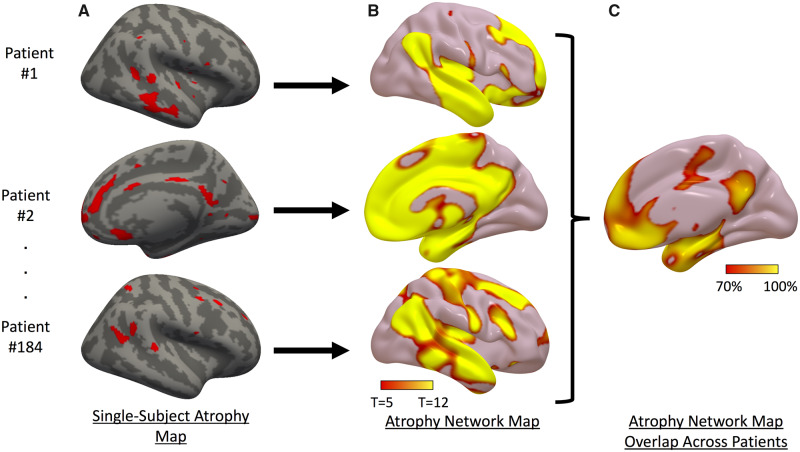
Next, we derived an ‘atrophy network map’ for each patient, defined as the brain regions functionally connected to each patient’s binarized single-subject atrophy map. First, single-subject atrophy maps in surface space from each hemisphere were combined and converted to MNI volume space (Fig. 2A). Using a publicly available normative functional connectivity dataset of 1000 healthy subjects from the Genome Superstruct Project (GSP) (Yeo et al., 2011; Holmes et al., 2015), we computed the average blood oxygen level-dependent (BOLD) time course for all voxels within each patient’s distributed single-subject atrophy map. Next, we correlated this mean time course within each single-subject atrophy map with the BOLD time course at every other brain voxel. Resulting r-values were converted to a normal distribution using Fisher’s r-to-z transform and were used to compute a single-group, voxel-wise t-test across the 1000 subjects in the normative connectome dataset to generate the unthresholded atrophy network t-maps (Fig. 2B) (Darby et al., 2018a). For visualization, we thresholded and binarized each patient’s atrophy network map at a voxel-wise family-wise error (FWE) corrected P < 0.05 (uncorrected P < 10−6). We overlaid the thresholded atrophy network maps from all patients to identify regions connected to locations of atrophy in most patients (Fig. 2C).
Figure 2.
Atrophy network mapping method. (A) Single-subject atrophy maps in three Alzheimer’s disease subjects. (B) Regions functionally connected to each patient’s atrophy map. (C) Overlap showing percentage of patients with atrophy functionally connected to the same regions.
Comparing atrophy network maps in Alzheimer’s disease versus control subjects
Unthresholded atrophy network maps from Alzheimer’s disease patients were compared to atrophy network maps derived from age-matched, cognitively normal subjects. We defined atrophy in these control subjects using the same approach as above, comparing every subject against the normative model and binarizing the resulting w-maps at w < −2. We expected these locations in control subjects to be random. We compared atrophy network maps between groups (Alzheimer’s disease versus control) at every voxel using a two-group t-test, correcting for multiple comparisons using permutation testing implemented by Statistical nonParametric Mapping (SnPM13, http://warwick.ac.uk/snpm, 10 000 simulations, voxel-wise FWE corrected P < 0.05).
Replication of atrophy network mapping results across two independent datasets
We performed all initial analyses using the ADNI-1 cohort as a discovery dataset. We then repeated all analyses in the ADNI-2 cohort as a replication dataset. All analyses were performed using identical methods to those in the original test dataset. To compare results quantitatively across the two datasets, we measured the voxel-wise spatial correlation between the atrophy network overlap maps and the group level t-test maps from ADNI-1 and ADNI-2.
Atrophy network mapping of cognitive and neuropsychiatric symptoms
We performed voxel-wise analyses comparing atrophy network maps to memory scores or the presence or absence of delusions. We chose these symptoms because we had strong a priori hypotheses for symptom-specific brain networks derived from patients with focal brain lesions. Because of the small number of subjects with delusions in each dataset, we combined ADNI-1 and ADNI-2 for these analyses. Memory recall was defined using delayed recall scores on the AVLT. Memory recognition was defined using delayed recognition scores from the AVLT. The presence or absence of delusions was obtained from the NPI assessment. Patients reporting delusions within 6 months of baseline MRI scans were included in the delusions group (n = 39), while our non-delusions group was defined as patients who never reported delusions at any time point during the ADNI study (n = 121). For delusions, we performed a voxel-wise two-group t-test of atrophy network mapping results comparing patients with versus without delusions. For memory, we performed a voxel-wise regression of memory scores against atrophy network mapping results. In each analysis, we corrected for multiple comparisons using permutation testing implemented by SnPM13 (http://warwick.ac.uk/snpm, 10 000 simulations, voxel-wise FWE corrected P < 0.05).
To quantify the effect sizes of our atrophy network mapping results and allow comparison with the lesion network mapping results for delusions and memory (below), we generated spherical seeds at peak locations corresponding with the t-test for delusions (MNI coordinates 12, 20, 68) and regressions for AVLT delayed recognition (MNI coordinates −18, −34, −2) and AVLT delayed recall (MNI coordinates 28, −40, 6). T-maps for each analysis were masked to voxels with at least 25% a priori probability of grey matter (= 63.75 intensity) according to FSL tissue priors. The peak voxel (as measured by T-value) falling within any of these grey matter clusters was selected. Thereafter, each peak was used to generate spherical seeds of radius 8 mm; each resulting spherical seed was then masked to the brain.
We then measured the functional connectivity strength between each patient’s single-subject atrophy map and this peak location seed using the normative connectome. We computed the average BOLD time course for all voxels within each single-subject atrophy map and for all voxels within each 8-mm radius region of interest. We then computed the time course correlations between the mean signal from each single-subject atrophy map and each region of interest to determine functional connectivity strength. We performed a linear regression analysis to determine the association between AVLT delayed recall scores and functional connectivity strength to the memory seed. We limited our analysis to patients with AVLT scores >0 to avoid floor effects (n = 93). We also repeated this analysis controlling for hippocampal volume. We performed a two-tailed t-test comparing functional connectivity strength to our delusions peak in Alzheimer’s disease patients with versus without delusions.
Comparing cognitive and neuropsychiatric symptom networks
We directly compared our atrophy network mapping results for memory and delusions with networks for these symptoms derived from patients with focal brain lesions using lesion network mapping. We defined lesion network maps by generating an 8 mm spherical seed at the peak lesion network mapping overlap location for memory (MNI coordinates 8, −39, 3) and delusions (MNI coordinates 48, 32, −4) from prior studies (Darby and Fox, 2017; Darby et al., 2017b; Ferguson et al., 2019). We defined atrophy network maps by generating an 8-mm spherical seed at the peak grey matter coordinate for each analysis as above. We then performed seed-based functional connectivity to determine the BOLD functional MRI time course correlations between each seed location and all other brain voxels. Finally, we measured the spatial correlation (excluding voxels outside of the MNI brain mask) between the symptom-specific brain networks generated using seeds from lesion network mapping and atrophy network mapping as a measure of similarity.
Data availability
Data used in this study are available from the ADNI website (www.adni-info.org).
Results
Single-subject atrophy maps in Alzheimer’s disease subjects are heterogeneous
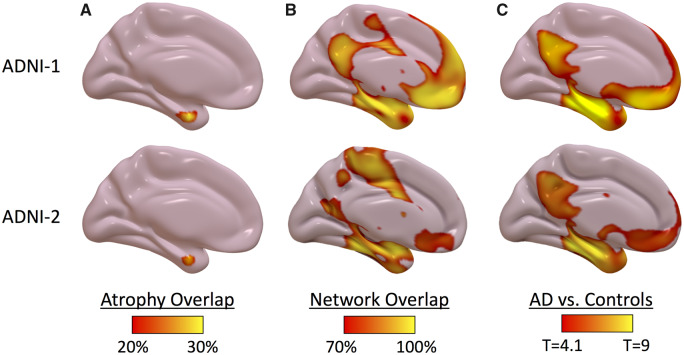
To derive single-subject atrophy maps, we compared each patient’s cortical thickness to cognitively normal subjects in the ADNI-1 dataset, defining atrophy as cortical thickness 2 SD below expected after controlling for age and gender (Fig. 1). There was significant heterogeneity in the locations of atrophy across individual patients, with only 42% of Alzheimer’s disease patients in ADNI-1 and 36% of Alzheimer’s disease patients in ADNI-2 having atrophy in the same location (Fig. 3A).
Figure 3.
Atrophy network mapping results in Alzheimer’s disease replicate across independent datasets. (A) Percentage of single-subject atrophy maps (w score < −2) overlapping in the same location. (B) Percentage of atrophy network maps (thresholded to an FWE corrected P < 0.05) overlapping in the same location. (C) Voxel-wise t-test comparing atrophy network maps in Alzheimer’s disease versus control subjects (voxel-wise FWE corrected P < 0.05). Results in discovery dataset (ADNI-1, top) are replicated in replication dataset (ADNI-2, bottom).
Atrophy network mapping identifies a common Alzheimer’s disease atrophy network
We hypothesized that cortical atrophy in Alzheimer’s disease would localize to a common brain network. To test this hypothesis using single-subject atrophy maps, we utilized a new technique called atrophy network mapping to determine the brain regions functionally connected to each patient’s location of atrophy (Fig. 2). While the locations of atrophy were heterogeneous, 100% of Alzheimer’s disease patients in ADNI-1 and 100% of Alzheimer’s disease patients in ADNI-2 had atrophy functionally connected to common brain regions in the mesial temporal cortex, precuneus/posterior cingulate cortex (PCC), and angular gyrus, a finding that was replicated across both datasets (Fig. 3B). While there were some differences noted in the atrophy network mapping results between ADNI-1 and ADNI-2, with a higher percentage of patients showing atrophy connected to the medial frontal cortex in ADNI-1, the results were nevertheless highly similar (spatial correlation r = 0.9). Group level t-tests comparing atrophy network maps in Alzheimer’s disease patients versus control subjects showed an even stronger replication of results across the two datasets (spatial correlation r = 0.97) (Fig. 3C and Supplementary Table 1). Results did not change significantly using different thresholds to define atrophy or controlling for the spatial extent of atrophy across subjects (Supplementary Fig. 1). Distributed atrophy seeds resulted in more reproducible localization of atrophy network maps across Alzheimer’s disease subjects and stronger group level effects when compared with using only the peak atrophied location as a seed (Supplementary Fig. 1).
Symptom-specific networks for memory impairment and delusions in Alzheimer’s disease
We next used atrophy network mapping to define symptom-specific networks for impaired memory and delusions. To define our impaired memory network, we regressed atrophy network mapping results with delayed recall scores on the AVLT. To define our delusions network, we compared atrophy network mapping in patients with versus without delusions as measured by the NPI assessment. For each comparison, we performed a voxel-wise analysis of atrophy network mapping connectivity strength, using permutation correction for multiple comparisons to an FWE-corrected P < 0.05.
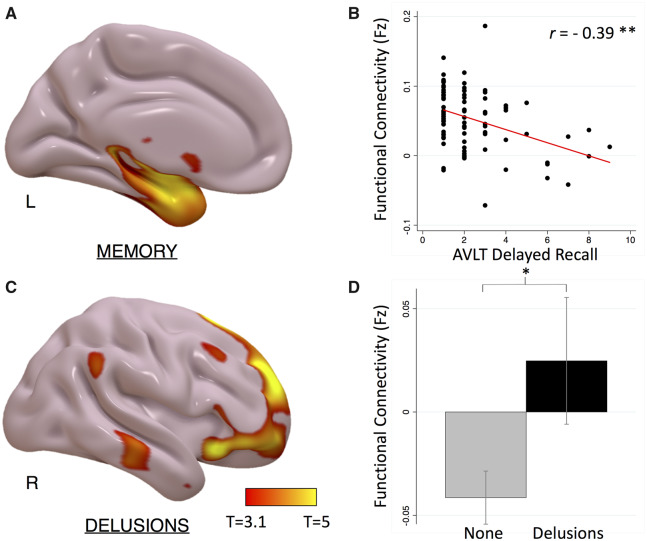
Atrophy network mapping identified a memory network for delayed recall that included regions in the mesial temporal lobes (Fig. 4A and Supplementary Table 2). Functional connectivity to the peak location identified in this analysis was strongly correlated with memory delayed recall scores (Pearson’s correlation r = −0.39, P < 0.0001, Fig. 4B), an effect that remained significant after controlling for hippocampal volume (partial correlation r = −0.37, P < 0.0001). The memory network for delayed recognition localized more posteriorly in the mesial temporal lobe (Supplementary Fig. 2).
Figure 4.
Atrophy network mapping of cognitive and neuropsychiatric symptoms in Alzheimer’s disease. (A) Voxel-wise GLM regression of memory recall scores with atrophy network mapping (cluster-wise FWE-corrected P < 0.05). (B) Scatter plot showing relationship between functional connectivity strength to the peak atrophy network mapping result and delayed recall scores. (C) Voxel-wise t-test of atrophy network mapping in Alzheimer’s disease patients with versus without delusions (FWE-corrected P < 0.05). (D) Bar graph showing functional connectivity strength to peak atrophy network mapping result in Alzheimer’s disease patients with versus without delusions. *P < 0.001; **P < 0.0001.
Atrophy network mapping identified a delusions network that included regions in the bilateral ventrolateral frontal, orbitofrontal frontal, and superior frontal cortices (Fig. 4C and Supplementary Table 2). Functional connectivity to the peak location identified in this analysis was significantly higher in Alzheimer’s disease patients with versus without delusions (P < 0.001, Fig. 4D).
Symptom networks identified using atrophy network mapping and lesion network mapping are similar
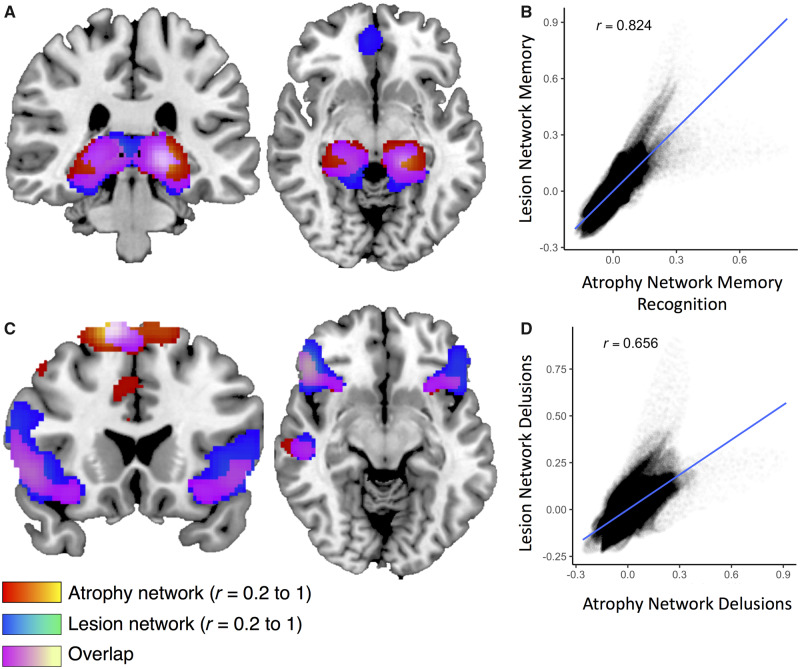
We compared symptom networks for memory and delusions derived using single-subject atrophy maps in Alzheimer’s disease patients with symptom networks derived in patients with focal brain lesions. Lesion network maps were derived using seeds generated at the peak locations of overlap from prior lesion network mapping studies in memory (Ferguson et al., 2019) and delusions (Darby and Fox, 2017; Darby et al., 2017b). The lesion network map for memory was highly similar to the atrophy network map for memory recognition (spatial correlation r = 0.82) (Fig. 5A and B). The lesion network map for memory was also similar to the atrophy network map for memory recall (spatial correlation r = 0.46), but there were also differences, particularly more anterior versus posterior localization within the mesial temporal lobe when comparing atrophy network mapping versus lesion network mapping results (Supplementary Fig. 3). Lesion network mapping of delusions was highly similar to atrophy network mapping for delusions (spatial correlation r = 0.66), although there was greater extension into the orbitofrontal lobes when comparing atrophy network mapping versus lesion network mapping (Fig. 5C and D).
Figure 5.
Comparing atrophy network mapping and lesion network mapping of memory and delusions. The atrophy network map for memory recognition is similar to the lesion network map for memory (A), with a spatial correlation of r = 0.82 (B). Similarly, the atrophy network map for delusions is similar to the lesion network map for delusions (C), with a spatial correlation of r = 0.66 (D).
Discussion
Here, we used atrophy network mapping to localize single-subject atrophy maps to functionally connected brain networks in Alzheimer’s disease patients. First, we used atrophy network mapping to localize single-subject atrophy maps to an Alzheimer’s disease atrophy network across two independent datasets, matching extensive prior literature showing that atrophy in clinical dementia syndromes occurs within syndrome-specific functionally connected brain networks (Seeley et al., 2009; Zhou et al., 2012; Darby et al., 2019). Second, we used atrophy network mapping to localize cognitive and neuropsychiatric symptoms in Alzheimer’s disease to symptom-specific brain networks, matching symptom-specific brain networks derived from patients with focal brain lesions (Darby and Fox, 2017; Darby et al., 2017b; Ferguson et al., 2019). Taken together, our study validates a new method called atrophy network mapping to localize single-subject atrophy maps to common brain networks associated with a specific syndrome, symptom, or behaviour in neurodegenerative disorders.
Single-subject atrophy maps in Alzheimer’s disease patients are neuroanatomically heterogeneous
Individualized biomarkers of neurodegeneration are critical to translating research findings at the group level to impact the clinical care of individual patients and may serve as important secondary end points for clinical trials aimed at slowing the progression of neurodegeneration in Alzheimer’s disease and related dementias (Darby et al., 2019; Staffaroni et al., 2019). Quantified methods to assess structural brain MRIs have led to the technical development of such biomarkers (Jack et al., 1997; La Joie et al., 2012; Ossenkoppele et al., 2015a, b; Perry et al., 2017). However, a major challenge in the usefulness of these measures is the spatial heterogeneity of atrophied locations at the individual subject level (Lehmann et al., 2013; Noh et al., 2014; Byun et al., 2015; Dong et al., 2017; Dickerson et al., 2017; Poulakis et al., 2018; Torok et al., 2018). Our results show that this heterogeneity can be accounted for by defining structural neuroimaging biomarkers at the network level.
An alternative approach is to use the average cortical thickness across a predefined, spatially distributed a priori brain network. This method also allows for comparison of atrophy across patients despite heterogeneity and is associated with diagnosis (Dickerson et al., 2009; Ossenkoppele et al., 2018), progression (Bakkour et al., 2009; Dickerson et al., 2011; Dickerson and Wolk, 2012, 2013), and specific symptoms (Bickart et al., 2014) in neurodegenerative disorders. However, atrophy network mapping offers several advantages over average cortical thickness within a distributed region of interest. First, atrophy network mapping does not require an a priori region of interest, as the results themselves can identify brain regions related to a specific clinical syndrome or cognitive/neuropsychiatric symptom. This may be particularly useful for neuropsychiatric or behavioural symptoms with poorly understood neuroanatomical correlates. Second, atrophy averaged across a distributed region of interest will be biased against atrophy in locations that are small with respect to the size of the region of interest but are nevertheless clinically meaningful due to that location’s pattern of connectivity. Our approach provides a balance between what is unique to each individual patient (that patient’s specific locations of atrophy), but also what is common to all patients with a similar symptom (where atrophy is functionally connected to). As such, atrophy network mapping can be seen as a complementary and potentially synergistic approach to average atrophy across distributed regions of interest.
Localizing clinical dementia syndromes to brain networks
Prior studies at the group level have supported the hypothesis that atrophy in neurodegenerative disorders occurs in disease-specific functionally connected brain networks (Seeley et al., 2009; Raj et al., 2012; Zhou et al., 2012). For instance, if one takes the peak location of atrophy at the group level in Alzheimer’s disease and other dementias, patients will also tend to have atrophy in brain regions functionally connected to this peak location (Seeley et al., 2009). A neuroimaging meta-analysis approach called coordinate-based network mapping also found syndrome-specific atrophy networks for different clinical dementia syndromes (Darby et al., 2019). Further work found that the seed regions most strongly connected to the distributed atrophy maps for Alzheimer’s disease and other dementias occurred at the locations of maximal atrophy (Zhou et al., 2012). This led to the hypothesis that Alzheimer’s disease and other dementias begin with atrophy in these disease ‘epicentres’, with spreading to functionally connected locations in other regions of each disease-specific network over time (Zhou et al., 2012).
Later work found that such epicentres derived from individual Alzheimer’s disease patients show significant variability, and that individual, rather than group-level, epicentre seeds best predicted atrophy patterns (Torok et al., 2018). Here, we similarly found that single-subject atrophy maps show variability in Alzheimer’s disease. However, we further show that these variable regions of atrophy occur within the same Alzheimer’s disease-specific brain network. Our method illustrates a mechanism by which regional variability in atrophy at the single-subject level can nevertheless lead to the consistent network neurodegeneration findings at the group level. Methods such as atrophy network mapping may therefore be useful in determining different spatial patterns of expected progression based on individual variability in the locations of atrophy at baseline.
It is important to note that we analysed patients with a clinical diagnosis of Alzheimer’s syndrome but not a pathological diagnosis of Alzheimer’s disease. It is possible (even likely) that some of the patients included with a clinical diagnosis of Alzheimer’s syndrome have a different pathological diagnosis, such as limbic-predominant age-related TDP-43 encephalopathy (LATE) (Nelson et al., 2019). As such, our current results are best interpreted as localizing the clinical symptoms of Alzheimer’s syndrome, rather than localizing Alzheimer’s disease pathology specifically.
Localizing cognitive and neuropsychiatric symptoms to brain networks
The observed heterogeneity between locations of atrophy and symptoms makes it challenging to establish brain-behaviour relationships. Methods that localize symptoms to brain networks may provide a potential solution to this problem. In prior work, lesion network mapping has been used to localize abnormal motor symptoms (Laganiere et al., 2016; Fasano et al., 2017; Joutsa et al., 2018; Corp et al., 2019), cognitive symptoms (Boes et al., 2015; Sutterer et al., 2016; Ferguson et al., 2019), and neurobehavioural symptoms (Boes et al., 2015; Darby and Fox, 2017; Darby et al., 2017b, 2018a, b) to symptom-specific brain networks. Symptoms-specific brain networks identified using these methods have also been found to be dysfunctional in psychiatric patients with similar symptoms (Darby et al., 2018b). Symptom networks for memory and delusions identified in the current study using atrophy network mapping matched symptom networks from these prior lesion studies. Taken together, our results support a trans-diagnostic localization of complex cognitive and neuropsychiatric symptoms to similar symptom-specific brain networks.
While there were strong similarities between lesion network mapping and atrophy network mapping results, there were also some notable differences. We found that atrophy network maps for memory recognition, localizing in the posterior mesial temporal lobe structures, matched lesion network maps for memory more closely than atrophy network maps for memory recall, which localized to more anterior mesial temporal lobe structures and regions beyond the mesial temporal lobes. This may reflect a reporting bias in lesion cases because the loss of memory recognition presents as a more severe form of memory impairment that is more likely to be published in case reports and case series. We also found slight differences in our delusion networks, with atrophy network mapping identifying a network that was more symmetric (versus right-hemisphere predominant) and with greater extension into the orbitofrontal lobes versus lesion network mapping. This may be due to seed differences between typically symmetric atrophy patterns in Alzheimer’s disease patients versus typically unilateral focal brain lesions.
The mechanistic interpretation of network localization remains unclear. One possibility is that clinical symptoms in neurological diseases can result from dysfunction in a connected but undamaged brain region through a mechanism termed diaschisis (Carrera and Tononi, 2014). Alternatively, network localization may suggest that a complex symptom localizes to the interactive functioning of the entire network, rather than any specific location or node within that network. Future studies investigating the functional effects of atrophy on connected regions may help to differentiate between these and other potential mechanisms.
Limitations
There are several important limitations to the current study. First, our study was limited to one neurodegenerative disorder, Alzheimer’s disease. Confirmation of our method in other neurodegenerative disorders will be important. Further, while we replicated our results in two independent datasets of Alzheimer’s disease subjects, cohorts had similar recruitment strategies, methods, and inclusion criteria. Because there is likely to be even more variability between different study populations of Alzheimer’s disease patients, our method of network localization to account for inter-subject heterogeneity might be even more useful in this context.
Second, our study uses a normative connectome to determine regions functionally connected to each patient’s location of atrophy. The use of a normative connectome, rather than the patient’s functional connectivity, is advantageous for our method because local dysfunction in atrophied brain locations could reduce or alter functional connectivity strength to other brain regions. While the large size of this normative connectome (n = 1000) increases the confidence of our connectivity estimates, a normative connectome is unable to account for important individual subject differences in functional connectivity. Functional connectivity from individual patients is likely to be important in other contexts, such as modulating the relationship between structural brain atrophy and clinical symptoms. As such, methods using normative and patient-specific connectivity are both likely to be useful in understanding neural mechanisms leading to clinical symptoms in neurodegenerative disorders.
Third, there are differences between atrophy network mapping and lesion network mapping. Unlike lesions, which are by definition binarized, determining the threshold to define atrophy in single subjects is admittedly challenging. While our results did not change significantly using different thresholds to define atrophy, it is possible that more subtle differences based on atrophy threshold could emerge. It would therefore be recommended to repeat analyses at different atrophy thresholds in future analyses. Atrophy is also slower in onset, more diffuse, and more specific to cortical regions than focal brain lesions. These differences could lead to different effects of disease pathology on connected brain regions that may not be accounted for using similar network mapping approaches.
Fourth, we used the average time course for all voxels within each single-subject atrophy map to derive our atrophy network map. We found that using distributed atrophy maps as seeds, rather than peak locations of atrophy, resulted in more consistent results. Because patients can have atrophy within regions typically associated with different intrinsic resting state connectivity networks, atrophy network maps for a given individual may also identify connected regions from different networks. This same issue can occur in lesion network mapping when lesions extend across spatially adjacent but functionally distinct brain regions. In both cases, it is advantageous to account for all possible connected regions because connectivity to different networks may account for different symptoms. For example, we identified atrophy connected to the ventral frontal cortices in Alzheimer’s disease patients with delusions, a region outside of the network identified with the Alzheimer’s clinical syndrome. However, an overly extensive atrophy map (or lesion) runs the risk of producing a ‘watered-down’ mean signal that may be uninformative. Further methodological advances could incorporate more refined approaches to atrophy network mapping, such as weighted single-subject atrophy seeds or modelling the effects of atrophy on remote regions using networks derived using graph theory approaches.
Finally, the network neurodegeneration hypothesis has often been used as a model to explain the spread of neurodegeneration over time in Alzheimer’s disease (Seeley et al., 2009; Raj et al., 2012; Zhou et al., 2012; Torok et al., 2018). Our current study was cross-sectional and could not assess progressive atrophy over time. However, our method of defining individual atrophy maps and deriving atrophy network maps would allow one to test the network neurodegeneration hypothesis for atrophy over time while accounting for individual variability in baseline atrophy between patients.
Conclusions
There is both clinical and neuroanatomical heterogeneity in Alzheimer’s disease at the single-subject level. Atrophy network mapping is a new method that uses the human connectome to localize single subject atrophy maps to syndrome- and symptom-specific brain networks, providing insights into brain-behaviour relationships in dementia patients.
Supplementary Material
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for NeuroImaging at the University of Southern California.
Funding
This work was funded by grants from the Alzheimer’s Association (R.D.), BrightFocus Foundation (R.D.), and the Vanderbilt Institute for Clinical and Translational Research (R.D.).
Competing interests
The authors report no competing interests.
Glossary
- AVLT =
auditory verbal learning task
References
- Bakkour A, Morris JC, Dickerson BC.. The cortical signature of prodromal AD: regional thinning predicts mild AD dementia. Neurology 2009; 72: 1048–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickart KC, Brickhouse M, Negreira A, Sapolsky D, Barrett LF, Dickerson BC.. Atrophy in distinct corticolimbic networks in frontotemporal dementia relates to social impairments measured using the Social Impairment Rating Scale. J Neurol Neurosurg Psychiatry 2014; 85: 438–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boes AD, Prasad S, Liu H, Liu Q, Pascual-Leone A, Caviness VS, et al. Network localization of neurological symptoms from focal brain lesions. Brain 2015; 138: 3061–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun MS, Kim SE, Park J, Yi D, Choe YM, Sohn BK, et al. Heterogeneity of regional brain atrophy patterns associated with distinct progression rates in Alzheimer’s disease. PLoS One 2015; 10: e0142756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Tononi G.. Diaschisis: past, present, future. Brain 2014; 137: 2408–22. [DOI] [PubMed] [Google Scholar]
- Corp DT, Joutsa J, Darby RR, Delnooz CCS, van de Warrenburg BPC, Cooke D, et al. Network localization of cervical dystonia based on causal brain lesions. Brain 2019; 1660–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI.. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 1999; 9: 179–94. [DOI] [PubMed] [Google Scholar]
- Darby RR, Brickhouse M, Wolk DA, Dickerson BC; Alzheimer’s Disease Neuroimaging Initiative. Effects of cognitive reserve depend on executive and semantic demands of the task. J Neurol Neurosurg Psychiatry 2017a; 88: 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby RR, Fox MD.. Reply: Capgras syndrome: neuroanatomical assessment of brain MRI findings in an adolescent patient. Brain 2017; 140: e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby RR, Fox MD.. Reply: Heterogeneous neuroimaging findings, damage propagation and connectivity: an integrative view. Brain 2019; 142: e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby RR, Horn A, Cushman F, Fox MD.. Lesion network localization of criminal behavior. Proc Natl Acad Sci USA 2018a; 115: 601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby RR, Joutsa J, Fox MD.. Lesion network localization of free will. Proc Natl Acad Sci USA 2018b; 115: 10792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby RR, Joutsa J, Fox MD.. Network localization of heterogeneous neuroimaging findings. Brain 2019; 142: 70–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby RR, Laganiere S, Pascual-Leone A, Prasad S, Fox MD.. Finding the imposter: brain connectivity of lesions causing delusional misidentifications. Brain 2017b; 140: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Bakkour A, Salat DH, Feczko E, Pacheco J, Greve DN, et al. The cortical signature of Alzheimer’s disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb Cortex 2009; 19: 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Brickhouse M, McGinnis S, Wolk DA.. Alzheimer’s disease: the influence of age on clinical heterogeneity through the human brain connectome. Alzheimer’s Dement 2017; 6: 122–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Salat DH, Bates JF, Atiya M, Killiany RJ, Greve DN, et al. Medial temporal lobe function and structure in mild cognitive impairment. Ann Neurol 2004; 56: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Stoub TR, Shah RC, Sperling RA, Killiany RJ, Albert MS, et al. Alzheimer-signature MRI biomarker predicts AD dementia in cognitively normal adults. Neurology 2011; 76: 1395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Wolk DA.. MRI cortical thickness biomarker predicts AD-like CSF and cognitive decline in normal adults. Neurology 2012; 78: 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Wolk DA.. Biomarker-based prediction of progression in MCI: comparison of AD signature and hippocampal volume with spinal fluid amyloid-β and tau. Front Aging Neurosci 2013; 5: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A, Toledo JB, Honnorat N, Doshi J, Varol E, Sotiras A, et al. Heterogeneity of neuroanatomical patterns in prodromal Alzheimer’s disease: links to cognition, progression and biomarkers. Brain 2017; 140: 735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du A-T, Schuff N, Kramer JH, Rosen HJ, Gorno-Tempini ML, Rankin K, et al. Different regional patterns of cortical thinning in Alzheimer’s disease and frontotemporal dementia. Brain 2007; 130: 1159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano A, Laganiere SE, Lam S, Fox MD.. Lesions causing freezing of gait localize to a cerebellar functional network. Ann Neurol 2017; 81: 129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson MA, Lim C, Cooke D, Darby RR, Wu O, Rost NS, et al. A human memory circuit derived from brain lesions causing amnesia. Nat Commun 2019; 10: 3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD. Localizing symptoms to brain networks using the human connectome. N Engl J Med 2018; 2237–45. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Kimmel DL.. Neuroimaging insights into network-based neurodegeneration. Curr Opin Neurol 2012; 25: 727–34. [DOI] [PubMed] [Google Scholar]
- Holmes AJ, Hollinshead MO, O’Keefe TM, Petrov VI, Fariello GR, Wald LL, et al. Brain Genomics Superstruct Project initial data release with structural, functional, and behavioral measures. Sci Data 2015; 2: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 2008; 27: 685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol 2010; 9: 119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Xu YC, Waring SC, O’Brien PC, Tangalos EG, et al. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer’s disease. Neurology 1997; 49: 786–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutsa J, Horn A, Hsu J, Fox MD.. Localizing parkinsonism based on focal brain lesions. Brain 2018; 141: 2445–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Joie R, Perrotin A, Barre L, Hommet C, Mezenge F, Ibazizene M, et al. Region-specific hierarchy between atrophy, hypometabolism, and -amyloid (A) load in Alzheimer’s disease dementia. J Neurosci 2012; 32: 16265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganiere S, Boes AD, Fox MD.. Network localization of hemichorea-hemiballismus. Neurology 2016; 86: 2187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Madison CM, Ghosh PM, Seeley WW, Mormino E, Greicius MD, et al. Intrinsic connectivity networks in healthy subjects explain clinical variability in Alzheimer’s disease. Proc Natl Acad USA 2013; 110: 11606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KK, Bartlett JW, Barnes J, Manning EN, Ourselin S, Fox NC, et al. Cerebral atrophy in mild cognitive impairment and Alzheimer disease: rates and acceleration. Neurology 2013; 80: 648–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Dickson DW, Trojanowski JQ, Jack CR, Boyle PA, Arfanakis K, et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain 2019; 142: 1503–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh Y, Jeon S, Lee JM, Seo SW, Kim GH, Cho H, et al. Anatomical heterogeneity of Alzheimer disease: based on cortical thickness on MRIs. Neurology 2014; 83: 1936–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenkoppele R, Cohn-Sheehy BI, La Joie R, Vogel JW, Möller C, Lehmann M, et al. Atrophy patterns in early clinical stages across distinct phenotypes of Alzheimer’s disease. Hum Brain Mapp 2015a; 36: 4421–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenkoppele R, Pijnenburg YAL, Perry DC, Cohn-Sheehy BI, Scheltens NME, Vogel JW, et al. The behavioural/dysexecutive variant of Alzheimer’s disease: clinical, neuroimaging and pathological features. Brain 2015b; 138: 2732–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenkoppele R, Rabinovici GD, Smith R, Cho H, Scholl M, Strandberg O, et al. Discriminative accuracy of [18F]flortaucipir positron emission tomography for Alzheimer disease vs other neurodegenerative disorders. JAMA 2018; 320: 1151–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry DC, Brown JA, Possin KL, Datta S, Trujillo A, Radke A, et al. Clinicopathological correlations in behavioural variant frontotemporal dementia. Brain 2017; 140: 3329–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulakis K, Pereira JB, Mecocci P, Vellas B, Tsolaki M, Kłoszewska I, et al. Heterogeneous patterns of brain atrophy in Alzheimer’s disease. Neurobiol Aging 2018; 65: 98–108. [DOI] [PubMed] [Google Scholar]
- Raj A, Kuceyeski A, Weiner M.. A network diffusion model of disease progression in dementia. Neuron 2012; 73: 1204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW. Mapping neurodegenerative disease onset and progression. Cold Spring Harb Perspect Biol 2017; 9: a023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD.. Neurodegenerative diseases target large-scale human brain networks. Neuron 2009; 62: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staffaroni AM, Ljubenkov PA, Kornak J, Cobigo Y, Datta S, Marx G, et al. Longitudinal multimodal imaging and clinical endpoints for frontotemporal dementia clinical trials. Brain 2019; 142: 443–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterer MJ, Bruss J, Boes AD, Voss MW, Bechara A, Tranel D.. Canceled connections: lesion-derived network mapping helps explain differences in performance on a complex decision-making task. Cortex 2016; 78: 31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok J, Maia PD, Powell F, Pandya S, Raj A; Alzheimer’s Disease Neuroimaging Initiative. A method for inferring regional origins of neurodegeneration. Brain 2018; 45: 846–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 2011; 106: 1125–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW.. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron 2012; 73: 1216–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this study are available from the ADNI website (www.adni-info.org).