Abstract
See Lemke (doi:10.1093/brain/awaa079) for a scientific commentary on this article.
Many rare and severe neurodevelopmental disorders are caused by de novo variants, but epidemiological estimates are available for very few of these disorders. Using a statistical approach based on genetic data, López-Rivera et al. provide more than 3000 incidence estimates for disorders caused by de novo variants.
Keywords: epilepsy, autism, neurodevelopmental disorder, incidence
Abstract
A large fraction of rare and severe neurodevelopmental disorders are caused by sporadic de novo variants. Epidemiological disease estimates are not available for the vast majority of these de novo monogenic neurodevelopmental disorders because of phenotypic heterogeneity and the absence of large-scale genomic screens. Yet, knowledge of disease incidence is important for clinicians and researchers to guide health policy planning. Here, we adjusted a statistical method based on genetic data to predict, for the first time, the incidences of 101 known de novo variant-associated neurodevelopmental disorders as well as 3106 putative monogenic disorders. Two corroboration analyses supported the validity of the calculated estimates. First, greater predicted gene-disorder incidences positively correlated with larger numbers of pathogenic variants collected from patient variant databases (Kendall’s τ = 0.093, P-value = 6.9 × 10−6). Second, for six of seven (86%) de novo variant associated monogenic disorders for which epidemiological estimates were available (SCN1A, SLC2A1, SALL1, TBX5, KCNQ2, and CDKL5), the predicted incidence estimates matched the reported estimates. We conclude that in the absence of epidemiological data, our catalogue of 3207 incidence estimates for disorders caused by de novo variants can guide patient advocacy groups, clinicians, researchers, and policymakers in strategic decision-making.
See Lemke (doi:10.1093/brain/awaa079) for a scientific commentary on this article.
Introduction
Neurodevelopmental disorders (NDDs) are a collection of severe neurological and neuropsychiatric conditions that manifest during childhood and persist throughout life. De novo mutations have been found to play a substantial causal role in the development of these disorders (de Ligt et al., 2013). Combined, NDDs are thought to affect ∼2–5% of children (Deciphering Developmental Disorders Study, 2017; Wilfert et al., 2017). However, the individual disease incidence of many rare NDDs remains unknown. Understanding the number of individuals affected by rare NDDs provides critical information for drug development and the planning of clinical trials (Auvin et al., 2018). Epidemiological information is also important for healthcare providers and researchers seeking to coordinate research studies, as well as patients and families organizing patient advocacy groups (Groft and Posada de la Paz, 2017). However, there are significant challenges in determining epidemiological estimates for rare disorders. These include low individual disease abundances, heterogeneous clinical presentations, and limitations in systems for reporting and tracking diagnoses. Therefore, epidemiological estimates for rare diseases—including the determination of incidence, the number of new cases in a given year—is oftentimes inexact or non-existent (Groft and Posada de la Paz, 2017).
The incidence of rare recessive Mendelian disorders can be estimated through the screening of healthy populations for carrier status (Schrodi et al., 2015; Fujikura, 2016). However, these methods are not applicable for epidemiological estimates for rare dominant monogenic disorders that can occur sporadically due to de novo variants (DNVs). This is particularly true for severe and early onset NDDs which can result in early mortality or reduced fecundity. In such disorders, the causal DNVs are under strong negative selection and will be rapidly eliminated from the population genetic pool (Eyre-Walker and Keightley, 2007).
DNVs are rare, occur sporadically, and are not always pathogenic (Shendure and Akey, 2015). However, the occurrence of DNVs in the same gene in several unrelated individuals with similar clinical phenotypes is considered strong support for the association of a gene with a disorder (Richards et al., 2015). A statistical model that calculates the expected number of DNVs for a given gene in parent-offspring trio cohorts was developed based on exome sequencing data (Samocha et al., 2014). In several research studies, significant deviations between the number of expected DNVs calculated for a gene and those observed in patients was used to identify over 100 new candidate genes for sporadic genetic conditions including NDDs, epilepsy, congenital hydrocephalus, and congenital heart disease (Homsy et al., 2015; Deciphering Developmental Disorders Study, 2017; Jin et al., 2017; Furey et al., 2018; Heyne et al., 2018). Notably, in trio cohorts with unaffected offspring, the number of expected de novo variants matched the number detected. Thus, the mutational model is well calibrated (Heyne et al., 2018).
Here, we adjust this mutational model to predict incidence estimates for 101 established DNV-associated neurological disorder genes with reported exome-wide significant DNV enrichment from four recent studies (Homsy et al., 2015; Deciphering Developmental Disorders Study, 2017; Furey et al., 2018; Heyne et al., 2018). Incidence estimates have never been reported for the vast majority of these individually rare monogenic NDDs, despite well-established associations with disease. Additionally, we predict incidence estimates for 3106 genes intolerant to variation with putative association to single gene disorders. We use variant reporting frequency data from public databases, diagnostic outcomes from gene panel testing, and epidemiological incidence estimates from the literature to evaluate and verify the predicted incidence estimates.
Materials and methods
Incidence estimation
Gene-specific, estimated incidences of missense and loss-of-function de novo variants in 100 000 births were calculated using expectation scores from the mutational model developed by Samocha et al. (2014) (Supplementary Table 1). For all human genes, benign population variants have been identified (http://gnomad.broadinstitute.org/; Lek et al., 2016). As not all DNVs are pathogenic, we adjusted our incidence estimates using the observed to expected ratios (o/e) for both missense and loss-of-function variants from the genome aggregation database (gnomAD) (Supplementary Table 1). A 90% confidence interval (CI) was considered for all estimates, based on the 90% CIs of the o/e scores provided by gnomAD (Karczewski et al., 2019). For genes that did not show intolerance to variants in the general population, incidence estimates were not calculated.
Retrieval of variant intolerant genes
To retrieve genes with putative association to monogenic disorders, we used two variant-intolerance gene metrics from gnomAD that quantify variant depletion in the general population, the missense Z-score and loss-of-function o/e metric. We acquired missense Z-score and loss-of-function o/e constraint metrics from the genome aggregation database for 19 705 canonical transcripts (Karczewski et al., 2019). A higher missense Z-score is indicative that a gene is more intolerant to missense variants whereas a lower loss-of-function o/e score indicates a gene more intolerant to protein truncating variants (PTVs). Established cut-offs for variant intolerance are a missense Z-score ≥ 3 standard deviations (Z = 3.09) for missense variant intolerance (Samocha et al., 2014) and an upper bound of the loss-of-function o/e score 90% CI ≤ 0.35 for PTV intolerance (Karczewski et al., 2019). Genes that met both cut-offs were considered intolerant to both missense variants and PTVs.
Assessment of gene essentiality
We acquired data regarding human gene essentiality from the Online GEne Essentiality (OGEE) database on 4 October 2018 (Chen et al., 2017). Data from OGEE were matched with corresponding data from gnomAD based on matching ensembl canonical transcript ID and HUGO Gene Nomenclature Committee (HGNC, https://www.genenames.org/) symbol.
Disease association and correlation analysis
Patient variants annotated as ‘pathogenic’ or ‘likely pathogenic’ were retrieved from ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accessed 01/2019) (Landrum et al., 2018). Similarly, patient variants annotated as ‘disease mutation’ were obtained from The Human Gene Mutation Database (HGMD, http://www.hgmd.cf.ac.uk/ac/index.php, 2018 release) (Stenson et al., 2017). Variants present in both databases were considered only once. Correlation analysis was performed in R using the Kendall ranked correlation test.
Literature review
We used the R package ‘RISmed’ to perform a series of searches on the PubMed database on 1 November 2018, using the search string ‘Incidence OR Prevalence’ and the HGNC symbols of the 3207 genes of interest. We only included articles with the terms ‘1 in’, ‘1:’, and ‘1:’ in their titles or abstracts (e.g. ‘1 in 50 000’, ‘1:50 000’, ‘1:50 000’). The identified articles were then manually evaluated at the title and abstract levels. Only incidence estimates acquired from epidemiological or large patient cohort studies for monogenic disorders that are dominantly inherited, early onset, and predominantly sporadic were retained in the analysis (see Supplementary Fig. 1 for details).
Statistical comparison of predicted incidence estimates
For each study identified, we calculated the expected number of de novo variants for the reported cohort size based on our predicted incidence rates. The expected number of de novo variants were rounded to the nearest integer and then compared to the number of observed cases using Fisher’s exact test. All statistical analyses were performed using R.
Data availability
All datasets and R scripts used to carry out this analysis are available at our GitHub repository (https://github.com/dlal-group/incidence_catalog).
Results
Estimation of de novo disorder incidence for 101 known neurological disorder genes
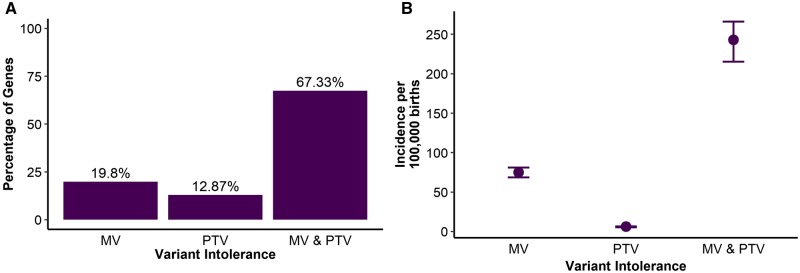
We selected 101 genes associated with NDDs, epilepsy, or congenital hydrocephalus with reported exome-wide significant, DNV enrichment from four recent studies and classified these as established DNV-associated monogenic disorder genes (Homsy et al., 2015; Deciphering Developmental Disorders Study, 2017; Furey et al., 2018; Heyne et al., 2018). For 20 of these 101 neurological disorder genes, only de novo missense variants were reported in studies that showed exome-wide significant DNV gene burden. We classified these genes as intolerant to missense variation. For 13 genes, only PTVs were reported and we classified these genes as intolerant to PTVs. For 68 genes, both types of DNVs were reported and we classified these as intolerant to both classes of variants (Fig. 1A and Supplementary material).
Figure 1 Gene-variant intolerance.
(A) Distribution of intolerance to protein-coding variation of established de novo NDD genes by intolerance type. MV = missense variant intolerant; MV & PTV = intolerant to both missense and truncating variants. (B) Total predicted sporadic disease incidence due to established de novo variant-associated disease genes. Error bars represent 90% CIs.
To predict disease incidence at birth for all DNV-associated monogenic disorders, we adjusted the mutational framework developed by Samocha et al. (2014) (see ‘Materials and methods’ section). The complete list of estimates and 90% CIs is provided in the Supplementary material. From these estimates, we predicted a global incidence of de novo brain disorders of 329:100 000 (90% CI: 291–360) due to established DNV-associated neurological disorder genes. Among the 101 genes, the greatest contribution to the predicted incidence originated from genes intolerant to both PTVs and missense variants (243:100 000; 90% CI: 215.2–266), followed by genes intolerant to missense variants (75:100 000; 90% CI: 68.6–80.8) and genes intolerant to PTVs (6.1:100 000; 90% CI: 5.3–6.4) (Fig. 1B). The two established monogenic disorders with the highest predicted DNV incidence estimates were DYNC1H1 (Charcot-Marie-Tooth disease, OMIM: 614228; 19.2:100 000 births; 90% CI: 18.6–19.8) and KMT2A (Weidemann-Steiner syndrome, OMIM: 605130; 11.5:100 000 births; 90% CI: 10.8–12.2).
Retrieval of potential de novo disorder-associated genes
We used metrics for intolerance to genetic variation to retrieve putative DNV-associated genes from gnomAD (http://gnomad.broadinstitute.org/; see ‘Materials and methods’ section for details). Dominant DNV disorder-associated genes are expected to be significantly depleted for variants in the general population. Excluding the aforementioned 101 established DNV-associated neurological disorder genes, we retrieved 3106 additional variant intolerant genes that met one or both of the established cut-offs (Supplementary Fig. 1).
To support a putative role in rare DNV-associated disorders for the retrieved variant intolerant genes, we evaluated their essentiality and annotated disease associations in comparison to genes with no significant DNV burden or variant intolerance (n = 16 496). We found that variant intolerant genes were significantly enriched for essential genes [odds ratio (OR): 2.52, 95% CI: 1.79–3.49; P-value = 1.8 × 10−7] and conditionally essential genes (OR: 2.76, 95% CI: 2.55–2.98; P-value = 1.03 × 10−144) as well as depleted for non-essential genes (OR: 0.34, 95% CI: 0.32–0.37; P-value = 8.49 × 10−162). Variant intolerant genes as a group were also significantly enriched for pathogenic patient variants annotated in HGMD and ClinVar (n = 1014/3106 variant intolerant genes; OR: 1.96; 95% CI: 1.80–2.13; P-value = 2.2 × 10−16). Together, these findings further support the pathogenic potential of variant intolerant genes. Thus, we predicted disorder incidences for all 3106 variant intolerant genes (Supplementary material). These predictions include incidence estimates for many well-established brain disorder genes for which exome-wide significant DNV burden has yet to be established, such as SLC2A1 (Glut1-deficiency syndrome, OMIM: 606777), GRIN2D (early infantile epileptic encephalopathy, OMIM: 617162), CACNA1C (Timothy syndrome, OMIM: 601005), ZEB2 (Mowat-Wilson syndrome, OMIM: 235730), and GABRA1 (early infantile epileptic encephalopathy, OMIM: 615744), among others.
The predicted incidence estimates can be validated
Next, we evaluated whether the 3207 predicted DNV-disorder incidences were supported by observational data. First, we explored the correlation between the predicted de novo incidence of a gene and the number of pathogenic variants annotated for that gene in ClinVar and HGMD patient variant databases. We expected that genes with a greater predicted incidence would have been associated with diseases earlier than those with lower predicted incidence because of larger patient populations. Accordingly, these genes should have been clinically screened for a longer period of time. Therefore, genes with higher predicted incidences would be expected to have a greater number of pathogenic variants annotated in the public patient databases ClinVar and HGMD compared to genes with lower predicted incidence. Considering 94 established DNV-associated genes and 1014 variant intolerant genes with annotated pathogenic variants in ClinVar and HGMD, we observed a positive correlation between the predicted incidence estimate and the total number of reported pathogenic variants within these 1108 genes (Kendall’s τ = 0.093, P-value = 6.9 × 10−06). When considered separately, a significant positive correlation was still observed (established DNV-associated genes only: Kendall’s τ = 0.271, P-value = 1.16 × 10−4; variant intolerant genes only: Kendall’s τ = 0.053, P-value = 0.014).
Second, for a more direct comparison between our predicted incidence rates and epidemiologically derived incidence estimates for monogenic disorders, we performed an extensive systematic literature review (see the ‘Materials and methods’ section and Supplementary Fig. 1 for details). Epidemiological estimates for de novo monogenic disorders are sparse in the current literature. We found epidemiological estimates for seven monogenic disorders that were predominantly sporadic and de novo dominant diseases (SCN1A, SLC2A1, SALL1, STXBP1, TBX5, KCNQ2, and CDKL5) (Martinez-Frias et al., 1999; Barisic et al., 2014; Bayat et al., 2015; Larsen et al., 2015; Wu et al., 2015; Stamberger et al., 2016; Symonds et al., 2019). We also provide additional unpublished incidence estimates for five monogenic disorders with previously published estimates (SCN1A, SLC2A1, STXBP1, KCNQ2, and CDKL5).
We compared the reported incidence estimates with our predicted incidences for those monogenic disorders (Table 1). Of these seven reported epidemiological incidence estimates, only the reported incidence estimates for STXBP1 were significantly different from our predicted incidence (P-value = 0.003 and 0.0005, Supplementary Table 3). The best prediction was observed for the case of SCN1A (Dravet syndrome) with an expected incidence of 6.69–7.62:100 000 and an observed incidence of 5.90:100 000. This was followed by SALL1 (Townes-Brockes syndrome) with an expected incidence of 0.30–0.36:100 000 and an observed incidence of 0.42:100 000.
Table 1.
Comparison of calculated estimates to reported estimates
| Gene | Disease | Incidence per 100 000 births | Significant difference a | Source PMID | |
|---|---|---|---|---|---|
| Predicted | Reported | ||||
| SCN1A | Dravet syndrome (OMIM: 607208) | 6.69–7.62 | 4.78 | No | 26438699 |
| 4.54 | No | 25778844 | |||
| 5.90 | No | 31302675 | |||
| 4.10 | No | R.S. Møllerb | |||
| SLC2A1 | GLUT1 deficiency syndrome (OMIM: 606777) | 1.65–2.22 | 1.20 | No | 26537434 |
| 2.95 | No | 31302675 | |||
| TBX5 | Holt-Oram syndrome (OMIM: 142900) | 0.39–0.45 | 0.56 | No | 25344219 |
| STXBP1 | STXBP1 encephalopathy (OMIM: 612164) | 3.30–3.81 | 1.09 | Yes | 26865513 |
| 0.82 | Yes | R.S. Møllerb | |||
| SALL1 | Townes-Brocks syndrome (OMIM: 107480) | 0.30–0.36 | 0.42 | No | 10083645 |
| KCNQ2 | KCNQ2 encephalopathy (OMIM: 613720) | 2.93–3.59 | 1.18 | No | 31302675 |
| 1.23 | No | R.S. Møllerb | |||
| CDKL5 | CDKL5 deficiency disorder (OMIM: 300672) | 1.81–2.49 | 1.77 | No | 31302675 |
| 0.96 | No | R.S. Møllerb | |||
Significant difference is based on Fisher’s exact test with a Bonferroni corrected cut-off of P ≤ 0.004. See Supplementary Table 2 for specific P-values.
Personal communication.
Structural variants play a minor role for most de novo variant-associated disorder genes
The mutational framework we used and adjusted to calculate incidence estimates is based on single nucleotide mutations and thus, does not account for the contribution of de novo structural variants. To understand if our gene-level predicted incidence estimates were deflated, we evaluated the relative frequency of pathogenic single-nucleotide variants (SNVs) compared to copy number variants (CNVs) in clinically tested NDD-associated genes. The dataset included targeted testing data (gene panels) from 18 334 epilepsy and NDD patients published by two large commercial diagnostic testing companies (Lindy et al., 2018; Truty et al., 2019). Thirteen established DNV-associated neurological disorder genes and 15 variant intolerant genes (Supplementary Table 2) were tested in all 18 334 patients. CNVs played a minor role compared to SNVs in these clinically tested DNV-disorder associated genes. SNVs made up a median 91.5% (interquartile range: 86.25–100) of pathogenic variants in these 28 genes. Additionally, 28.6% of these genes (8/28) did not have any pathogenic CNVs.
Discussion
Epidemiological estimates are needed to optimize planning of healthcare services such as training of specialists, hospital and support services provided, and implementation of public health programmes. Establishing the true incidence of many rare NDDs, which include a large number of de novo severe monogenic disorders, is challenging. The vast majority of such disorders have been linked to a causal gene only within the past decade because of the evolution of sequencing technology and clinical genetic testing. Thus, our understanding of the pathophysiology and natural history for most rare disorders is still in its infancy. In this study, we present monogenic disorder incidence estimates for 101 established DNV-associated NDD genes as well as 3106 putative DNV-associated genes based on a well-established mutational model (Samocha et al., 2014).
Supporting the predicted incidence rates, we observed a significant correlation between the number of reported pathogenic patient variants in publicly available databases and the calculated disease estimates. This observation was encouraging and unexpected, given the large variety of possible data entry confounders in patient variant databases (Richards et al., 2015; Thorogood et al., 2017), e.g. variability in the frequency that individual genes are screened for. Nonetheless, the significant correlation is supportive that the predicted disease incidences are associated with true disease incidences.
To evaluate our estimates further, we performed a gene-level comparison with literature-derived estimates. We observed a significant difference between epidemiologically-derived and predicted incidence in only one of seven comparisons. Our predicted incidence for STXBP1 disorders was the only significantly different estimate, 3.43-fold and 4.33-fold higher than the reported incidences. However, the epidemiologically-derived incidence estimates for STXBP1 encephalopathy were not prospective, population-based estimates (Stamberger et al., 2016). Rather, they are based on the number of patients with a positive diagnosis of epileptic encephalopathy referred to one lead national epilepsy centre in Denmark over a period of time. Furthermore, it has been reported that STXBP1’s phenotypic spectrum includes patients with non-epileptic syndromes that may not have been ascertained in the Danish incidence estimation (Hamdan et al., 2009, 2011). Potentially, patients with milder clinical representations of the disease may have remained undiagnosed and without genetic testing, leading to an underestimation of the total disease incidence. Conversely, our gene-specific incidence estimation for STXBP1 would include the entire phenotypic spectrum associated with mutations in a gene.
Still, we cannot rule out that our predicted incidence estimates might be overestimates due to accounting for a proportion of variants that may be incompatible with human life (Li et al., 2017). However, the majority of the 101 NDDs for which we predicted incidence do not have heterozygous embryonically lethal orthologues in mice or are essential for human cell viability as characterized by CRISPR screens (Supplementary material). In the specific case of STXBP1, its orthologue is not heterozygous embryonically lethal in mice and it is reported to be non-essential for human cell viability in CRISPR screens. Therefore, embryonic lethal mutations are unlikely to be the cause for the discrepancy between our predicted incidences and reported incidence estimates for STXBP1.
Nonetheless, our systematic literature review revealed a lack of epidemiological estimates for the vast majority of rare sporadic dominant monogenic disorders. These estimates are particularly important for NDDs, where de novo variants can account for 13–60% of diagnostic yield from clinical genetic testing, depending on the disease or diagnostic criteria (Wilfert et al., 2017). Rare and severe developmental and epileptic encephalopathies in particular demonstrate an enrichment of causal de novo point mutations (Hamdan et al., 2017).
Our method is based on mutation rate and can only account for de novo SNVs. Therefore, we assessed the contribution of structural variants and found that the median contribution of such variants to clinically tested pathogenic variants was only ∼10%, in agreement with prior reported findings (Truty et al., 2019). Recent studies have also associated other types of variants with NDDs. For example, repeat expansion disorders have recently been associated with new epilepsy genes (Ishiura et al., 2018; Florian et al., 2019; Lei et al., 2019; Yeetong et al., 2019) and so-called ‘poison’ exons have been identified in epilepsy genes such as SCN1A (Carvill et al., 2018). Currently, these types of variants are extremely rare and not well studied because of limitations of targeted and short read sequencing. In the future, as the field advances and knowledge of these variants increases, we will incorporate these types of variants in our incidence model. However, for most disorders, our calculated estimated incidences represent a valid approximation of the true incidence. Thus, our catalogue of predicted incidence estimates can inform strategic decision-making, particularly in cases of predominantly sporadic disorders where epidemiological data are not available because of low disease abundance.
Funding
This work was supported by the Molecular Medicine Ph.D. Program of Cleveland Clinic and Case Western Reserve University and the National Institute of General Medical Sciences (Award Number: T32GM088088). E.P-P. was supported by a Dravet Syndrome Foundation research grant awarded to D.L.
Competing interests
D.A.M. and A.S.L. are employees of GeneDx, a subsidiary of OPKO Health. The remaining authors report no competing interests.
Supplementary Material
Glossary
- DNV =
de novo variant
- NDD =
neurodevelopmental disorder
- PTV =
protein truncating variant
References
- Auvin S, Irwin J, Abi-Aad P, Battersby A.. The problem of rarity: estimation of prevalence in rare disease. Value Health 2018; 21: 501–7. [DOI] [PubMed] [Google Scholar]
- Barisic I, Boban L, Greenlees R, Garne E, Wellesley D, Calzolari E, et al. Holt Oram syndrome: a registry-based study in Europe. Orphanet J Rare Dis 2014; 9: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayat A, Hjalgrim H, Moller RS.. The incidence of SCN1A-related Dravet syndrome in Denmark is 1:22,000: a population-based study from 2004 to 2009. Epilepsia 2015; 56: e36–9. [DOI] [PubMed] [Google Scholar]
- Carvill GL, Engel KL, Ramamurthy A, Cochran JN, Roovers J, Stamberger H, et al. Aberrant inclusion of a poison exon causes Dravet syndrome and related SCN1A-associated genetic epilepsies. Am J Hum Genet 2018; 103: 1022–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Lu G, Chen X, Zhao XM, Bork P.. OGEE v2: an update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res 2017; 45: D940–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ligt J, Veltman JA, Vissers LE.. Point mutations as a source of de novo genetic disease. Curr Opin Genet Dev 2013; 23: 257–63. [DOI] [PubMed] [Google Scholar]
- Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017; 542: 433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD.. The distribution of fitness effects of new mutations. Nat Rev Genet 2007; 8: 610–8. [DOI] [PubMed] [Google Scholar]
- Florian RT, Kraft F, Leitão E, Kaya S, Klebe S, Magnin E, et al. Unstable TTTTA/TTTCA expansions in MARCH6 are associated with familial adult myoclonic epilepsy type 3. Nat Commun 2019; 10: 4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura K. Global carrier rates of rare inherited disorders using population exome sequences. PloS One 2016; 11: e0155552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey CG, Choi J, Jin SC, Zeng X, Timberlake AT, Nelson-Williams C, et al. De novo mutation in genes regulating neural stem cell fate in human congenital hydrocephalus. Neuron 2018; 99: 302–14.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groft SC, Posada de la Paz M.. Rare diseases: joining mainstream research and treatment based on reliable epidemiological data. Adv Exp Med Biol 2017; 1031: 3–21. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Dobrzeniecka S, Lortie A, Mottron L, Vanasse M, et al. Intellectual disability without epilepsy associated with STXBP1 disruption. Eur J Hum Genet 2011; 19: 607–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Myers CT, Cossette P, Lemay P, Spiegelman D, Laporte AD, et al. High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am J Hum Genet 2017; 101: 664–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Piton A, Gauthier J, Lortie A, Dubeau F, Dobrzeniecka S, et al. De novo STXBP1 mutations in mental retardation and nonsyndromic epilepsy. Ann Neurol 2009; 65: 748–53. [DOI] [PubMed] [Google Scholar]
- Heyne HO, Singh T, Stamberger H, Abou Jamra R, Caglayan H, Craiu D, et al. De novo variants in neurodevelopmental disorders with epilepsy. Nat Genet 2018; 50: 1048–53. [DOI] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science (New York, NY) 2015; 350: 1262–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiura H, Doi K, Mitsui J, Yoshimura J, Matsukawa MK, Fujiyama A, et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat Genet 2018; 50: 581–90. [DOI] [PubMed] [Google Scholar]
- Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet 2017; 49: 1593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 2019; 531210: [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 2018; 46: D1062–d1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J, Johannesen KM, Ek J, Tang S, Marini C, Blichfeldt S, et al. The role of SLC2A1 mutations in myoclonic astatic epilepsy and absence epilepsy, and the estimated frequency of GLUT1 deficiency syndrome. Epilepsia 2015; 56: e203–8. [DOI] [PubMed] [Google Scholar]
- Lei XX, Liu Q, Lu Q, Huang Y, Zhou XQ, Sun HY, et al. TTTCA repeat expansion causes familial cortical myoclonic tremor with epilepsy. Eur J Neurol 2019; 26: 513–8. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Oehlert J, Snyder M, Stevenson DK, Shaw GM.. Fetal de novo mutations and preterm birth. PLoS Genet 2017; 13: e1006689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindy AS, Stosser MB, Butler E, Downtain‐Pickersgill C, Shanmugham A, Retterer K, et al. Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia 2018; 59: 1062–71. [DOI] [PubMed] [Google Scholar]
- Martinez-Frias ML, Bermejo Sanchez E, Arroyo Carrera I, Perez Fernandez JL, Pardo Romero M, Buron Martinez E, et al. The Townes-Brocks syndrome in Spain: the epidemiological aspects in a consecutive series of cases. An Esp Pediatr 1999; 50: 57–60. [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet 2014; 46: 944–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodi SJ, DeBarber A, He M, Ye Z, Peissig P, Van Wormer JJ, et al. Prevalence estimation for monogenic autosomal recessive diseases using population-based genetic data. Hum Genet 2015; 134: 659–69. [DOI] [PubMed] [Google Scholar]
- Shendure J, Akey JM.. The origins, determinants, and consequences of human mutations. Science (New York, NY) 2015; 349: 1478–83. [DOI] [PubMed] [Google Scholar]
- Stamberger H, Nikanorova M, Willemsen MH, Accorsi P, Angriman M, Baier H, et al. STXBP1 encephalopathy: a neurodevelopmental disorder including epilepsy. Neurology 2016; 86: 954–62. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet 2017; 136: 665–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds JD, Zuberi SM, Stewart K, McLellan A, O’Regan M, MacLeod S, et al. Incidence and phenotypes of childhood-onset genetic epilepsies: a prospective population-based national cohort. Brain 2019; 142: 2303–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorogood A, Cook-Deegan R, Knoppers BM.. Public variant databases: liability? Genet Med 2017; 19: 838–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truty R, Patil N, Sankar R, Sullivan J, Millichap J, Carvill G, et al. Possible precision medicine implications from genetic testing using combined detection of sequence and intragenic copy number variants in a large cohort with childhood epilepsy. Epilepsia Open 2019; 4: 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truty R, Paul J, Kennemer M, Lincoln SE, Olivares E, Nussbaum RL, et al. Prevalence and properties of intragenic copy-number variation in Mendelian disease genes. Genet Med 2019; 21: 114–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfert AB, Sulovari A, Turner TN, Coe BP, Eichler EE.. Recurrent de novo mutations in neurodevelopmental disorders: properties and clinical implications. Genome Med 2017; 9: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YW, Sullivan J, McDaniel SS, Meisler MH, Walsh EM, Li SX, et al. Incidence of Dravet syndrome in a US population. Pediatrics 2015; 136: e1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeetong P, Pongpanich M, Srichomthong C, Assawapitaksakul A, Shotelersuk V, Tantirukdham N, et al. TTTCA repeat insertions in an intron of YEATS2 in benign adult familial myoclonic epilepsy type 4. Brain 2019; 142: 3360–6. [DOI] [PubMed] [Google Scholar]
- Prevalence and architecture of de novo mutations in developmental disorders. Nature 2017; 542: 433–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets and R scripts used to carry out this analysis are available at our GitHub repository (https://github.com/dlal-group/incidence_catalog).