Abstract
Neural development requires the orchestration of dynamic changes in gene expression to regulate cell fate decisions. This regulation is heavily influenced by epigenetics, heritable changes in gene expression not directly explained by genomic information alone. An understanding of the complexity of epigenetic regulation is rapidly emerging through the development of novel technologies that can assay various features of epigenetics and gene regulation. Here, we provide a broad overview of several commonly investigated modes of epigenetic regulation, including DNA methylation, histone modifications, non-coding RNAs, as well as epitranscriptomics that describe modifications of RNA, in neurodevelopment and diseases. Rather than functioning in isolation, it is being increasingly appreciated that these various modes of gene regulation are dynamically interactive and coordinate the complex nature of neurodevelopment along multiple axes. Future work investigating these interactions will likely utilize “multi-omic” strategies that assay cell fate dynamics in a high-dimensional and high-throughput fashion. Novel human neurodevelopmental models including iPSC and cerebral organoid systems may provide further insight into human-specific features of neurodevelopment and diseases.
Keywords: Neurogenesis, Developmental Disorder, DNA methylation, Chromatin Remodelling, Epitranscriptomics
1. Introduction
1.1. Epigenetics of neurodevelopment: regulation of cellular identity
The development of the mammalian brain requires a complex orchestration of dynamic gene expression patterns in order to generate the diversity of cell types necessary for neural function (1). Despite all cells in the brain containing the same genetic material in the form of DNA, how cells specialize into different cell types and maintain distinct functions relies upon a complex regulation of cellular transitions from a more immature to differentiated state. During these transitions, asymmetric divisions may be necessary to maintain a more “stem-like” progenitor population while expanding more differentiated cells (2).
Cortical neurogenesis occurs through the action of radial glia, specialized cells that are able to generate cortical neurons as well as supporting glial cells. Unlike rodent cortex, the embryonic human cortex harbours a large number of outer radial glia cells, which lie outside of the ventricular zone during development and are thought to enable the increased size and complexity of the human brain (3). The adult mammalian brain contains continuous neurogenic niches in the subventricular zone, which generate new-born neurons to populate the olfactory bulb (4), and the subgranular zone of the dentate gyrus, which generate new-born granule neurons in the hippocampus (5) (Figure 1A). The degree to which continuous neurogenesis occurs in the adult human brain remains an area of active investigation and debate (6).
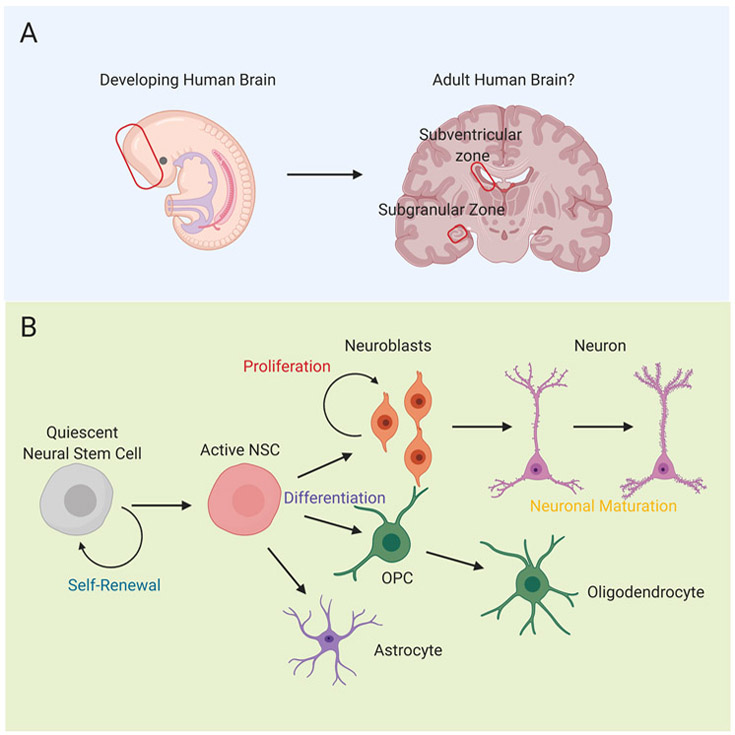
Figure 1. Regions of Mammalian Neurodevelopment.
(A) Embryonic neurodevelopment requires a complex orchestration of various cell types to generate the mammalian brain (red rectangle). As much of the brain becomes fully mature, enduring regions of adult neurogenesis have been characterized in the subventricular zone (red rectangle: top) and subgranular zone of the hippocampus (red rectangle: bottom). The degree to which this occurs in adult humans is an area of active investigation and debate. (B) Neural stem cells have several features including the process of self-renewal from stem cells that can become quiescent. Active neural stem cells can differentiate into multiple progeny including cells of neuronal, oligodendrocyte, and astrocyte lineages. Neuronal lineage cells (Neural progenitor cells) often undergo cellular expansion prior to neuron differentiation and subsequent maturation.
The spatiotemporal patterning of the mammalian brain undergoes several stages of development from embryogenesis to lifelong neurogenesis, with evidence of a common embryonic origin for these stem cells (7,8). Multipotent neural stem cells generate the diversity of cell types in the brain, including neurons, astrocytes and oligodendrocytes (9-11). While the nomenclature may vary depending on the particular neurogenic niche, neurodevelopment undergoes a common pattern to generate the cellular diversity of the mammalian brain. Specialized regions of the brain contain a population of self-renewing stem cells, which can divide to repopulate this niche (Figure 1B). These cells subsequently also undergo asymmetric division into neural progenitor cells of either neuronal or glial fate, which can further expand via cell division. These cells then migrate to their final destination and eventually mature and functionally integrate into the brain (12-14).
The mechanisms by which one cell either transitions to a new cellular identity (differentiation) or retains its original identity (self-renewal and/or proliferation) are epigenetic by nature. The central dogma of molecular biology refers to the process by which the general flow of sequence information starts as DNA, becomes RNA, and subsequently becomes protein (15). Historically, proteins are regarded as the primary functional molecules that carry out the biological processes necessary for life. However, it is now widely acknowledged that multiple levels of regulation as well as inherently functional properties also occur at the DNA and RNA levels. Epigenetics typically refers to the heritable changes in gene expression not directly explained by the DNA code (16). While originally studied in the context of heritable DNA methylation patterns, the term epigenetics now encompasses a vast network of different levels of gene regulation that spans the DNA-RNA-protein axis. Here, we provide an overview of the role of epigenetics by way of the hierarchical structure of organization and regulation. In particular, we review the role of DNA methylation, chromatin regulation, non-coding functional RNAs, and epitranscriptomics in neural development and their potential roles in brain diseases (Figure 2).
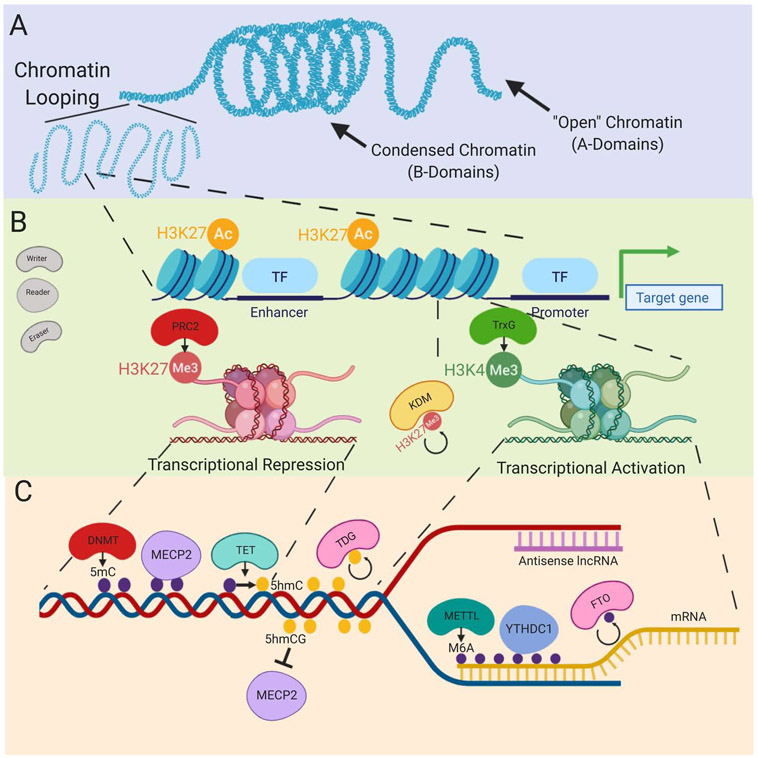
Figure 2. Epigenetic Mechanisms of Gene Regulation.
(A) Epigenetic regulation of higher ordered structures begins on the level of accessibility of chromatin. A-type chromatin (euchromatin) refers to open chromatin that is accessible for transcription, while B-type (heterochromatin) chromatin refers to closed chromatin that is typically association with gene inactivity. These areas of open chromatin can form chromatin loops that generate 3-dimensional structures that allow for long-range interactions that are not directly observable from the 2-dimensional sequence. (B) Long range interactions generated by 3-dimensional chromatin loops can form enhancer complexes, wherein transcription factors can bind to enhancer regions that are distal to the promoter region in 2-dimensions to influence gene expression. These enhancer regions are associated with histone modifications including H3K27ac (pictured), H3K4me1, as well as the enhancer-associated p300 protein. Promoter regions can be either inactive, which is associated with the Polycomb Repressive Complex-group catalysed H3K27me3 (pictured) or active, which is associated with the Trithorax group catalysed H3K4me3 (pictured). Regions with both H3K27me3 and H3K4me3 are considered inactive but poised for transcription. Removal of H3K27me3 by histone demethylases (KDM family proteins) can lead to transcriptional activation. (C) DNA methylation is written by DNA methyltransferases (DNMTs) and is traditionally associated with gene silencing due to interactions with MECP2. 5mC DNA methylation is removed through oxidation to 5hmC by TET family proteins and subsequent TDG-mediated excision, leading to base excision repair. MECP2 does not bind to 5hmCG, which is associated with regions of transcriptional activation. Transcription can be regulated by m6A modification on the RNA strand. This modification is written by METTL family proteins, read by various readers including YTHDC1, and enzymatically removed by various erases including FTO. m6A modification has diverse roles including regulating mRNA metabolism and translation and mediating RNA nuclear transport. Non-coding RNAs, including long-noncoding RNAs (lncRNAs), frequently occur on the anti-sense strand of mRNAs. These lncRNAs can operate locally in cis or distal to the area of transcription in trans in order to regulate gene expression.
2. Epigenetic modifications
2.1. DNA methylation
2.1.1. DNA methylation and gene regulation through writers and readers
Historically, DNA methylation was one of the first appreciated regulators of gene transcription (17-19), including its role as a key feature in X-chromosome inactivation and genomic imprinting (20). Methylation patterns are also key features of cellular identity; single-cell methylomes have been used to identify neuronal subtypes in the mammalian cortex (21). The most robustly studied type of DNA methylation comes in the form of methylation of the fifth position of cytosine (5mC), which has profound effects on gene expression (22). This modification is classically associated with transcriptional repression through the placement of 5mC along CpG dinucleotides at promoter regions. However, genome-wide analysis of promoter occupancy with transcription factors has shown differential binding of key promoters based upon variable methylation patterns (23), with evidence of enhanced promoter binding in the presence of DNA methylation at distinct promoter sequences (24). Additionally, in contrast to the repressive nature of methylation of promoter regions, DNA methylation is also found within gene bodies of actively transcribed genes (25). Intriguingly, non-promoter regions of DNA methylation have been associated with neurogenesis, highlighting the complexity of this epigenetic regulation (26). Therefore, rather than simply a mark of gene silencing, DNA methylation varies based on context with multiple functional roles deriving from this epigenetic mark (27).
DNA methylation has traditionally been detected via three types of methods: methylation-sensitive restriction enzyme-based DNA digestion, bisulfite sequencing, and affinity purification-based sequencing techniques (28). Currently, bisulfite sequencing is the most widely used of these methods (27). Utilizing this technique, bisulfite treatment deaminates cytosine into uracil, which would subsequently be read as thymine upon sequencing. 5mC samples are resistant to deamination and therefore do not change their base composition. This is then compared to untreated samples for determination of methylated cytosine (29). Affinity purification techniques employing antibodies against 5mC (30) followed by high throughput sequencing of enriched DNA (MeDIP-seq) are also utilized (30,31). Additionally, methods have been developed to profile 5-hydroxymethylcytosine (5hmC), an alternative form of DNA methylation, including Tet-assisted bisulfite sequencing (TAB-seq) (32) and oxidative bisulfite sequencing (oxBS-seq) (33).
DNA methylation is a dynamically-regulated process with diverse functions throughout development (34). In order for this regulation to occur, there are various writers, readers, and erasers of DNA methylation. DNA Methyltransferases (DNMTs) are key “writers” of this process, with different DNMTs holding various roles. For instance, DNMT1 is a maintenance methyltransferase which is believed to act during cell division in order to maintain methylation patterns onto daughter cells (35). Alternate methyltransferases have been found to establish novel patterns of methylation during development: deletion of DNMT3A and DNMT3B leads to deficits in de novo methylation in embryonic stem cells (ESCs) without any deficits in the maintenance of imprinted methylation patterns, indicating that these methyltransferases establish new patterns of methylation upon cell division (36).
DNA methylation regulates gene expression in part through the “readers” of DNA methylation, including methyl-CpG binding protein 2 (MeCP2) and other methyl-CpG binding domain family members, such as MBD1, MBD2, and MBD4 (37). However, recent evidence suggests that these proteins may not simply be mediators of transcriptional repression. For example, MeCP2 was originally described as a transcriptional repressor due to its ability to bind 5mC and repress transcription of methylated promoters in vitro (38), but in vivo loss of MeCP2 has been shown to cause both up-regulation and down-regulation of many genes, a finding inconsistent with a typical transcriptional repressor (39). Although there is strong in vitro evidence suggesting MeCP2 indeed functions to repress transcription via recruitment of the NCoR/SMRT co-repressor complex (40), the molecular function of MeCP2 in vivo remains elusive.
In addition to CpG methylation, non-CpG methylation is a brain-enriched DNA modification that plays critical roles in neuronal development and maturation. Profiling DNA methylation in the brain at the single-nucleotide resolution has revealed that non-CpG methylation (CpA, CpT, CpC) accumulates during postnatal brain development, reaching high levels during adulthood (1). Emphasizing the functional relevance of this modification, multiple studies have shown that MeCP2 binds non-CpG methylated DNA in addition to 5mCG (41-44). Non-CpG methylation in the brain has been shown to be written by DNMT3A during postnatal development (41,42). One study has linked the functions of DNMT3A and MeCP2 within the developing postnatal brain, demonstrating that DNMT3A binds within gene bodies of low-expression genes to establish non-CpG methylation marks that are later bound by MeCP2, influencing the expression of these genes (45).
Mutations to both the writers of DNA methylation (DNA methyltransferases) as well as the protein binders of DNA methylation (such as MeCP2) lead to developmental disorders. Gain-of-function mutations in DNMT3A have been associated with microcephalic dwarfism, leading to hypermethylation in regions normally regulated by Polycomb-mediated histone modification (46). Loss-of-function mutations in DNMT3A can also cause an overgrowth syndrome with intellectual disability (47). Additionally, mutations in DNMT3B have been associated with immunodeficiency, centromere instability, facial anomalies (ICF) syndrome, also leading to global histone modification changes (48). Furthermore, single nucleotide polymorphisms (SNPs) in DNMTs have been associated with autism spectrum disorder (ASD) (49). Mutations to the “readers” of DNA methylation also have neurodevelopmental consequences, particularly in the case of MeCP2: MECP2 loss-of-function mutations have been found to cause Rett syndrome (50), a progressive childhood neurological disorder characterized by developmental stagnation and subsequent regression, in which previously-learned motor, communicative, and social skills are lost (51).
2.1.2. DNA demethylation and 5-hydroxymethylcytosine: a brain-enriched base pair modification
“Erasing” of DNA methylation is thought to occur through both active and passive means. This process is most notable during early embryogenesis, when global demethylation occurs following the formation of the zygote and allows for the subsequent development of the vast array of cellular subtypes from a common cell of origin (52). Passive DNA demethylation refers to the dilution of 5mC during DNA replication without maintenance by DNA methyltransferases, while active DNA demethylation refers to enzymatic alterations of 5mC paired with base excision repair, wherein methylated cytosines are actively removed and replaced by their non-methylated counterparts. The process of active DNA demethylation has been demonstrated to occur through iterative oxidation of 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) via the action of TET family proteins (53-55). The active DNA demethylation process is completed either by dilution of these oxidized forms of 5mC by DNA replication (56), or by thymine DNA glycosylase-mediated excision of these modified bases, which is then coupled with base excision repair, wherein methylated cytosines are actively removed and replaced by their non-methylated counterparts (57,58).
5hmC is an alternative base pair modification that is enriched in the brain compared to other mammalian tissues (59-61). While initially found to be enriched in Purkinje cells (61), it was subsequently revealed to also be enriched in the hippocampus and cortex and increases in abundance with age (60). 5hmC is generated via conversion from 5mC through the TET family of enzymes In neurodevelopment, TET1 has been identified as a key gene in the process of DNA demethylation (62). Deficiency in TET1 leads to impaired hippocampal neurogenesis as well as hypermethylation and downregulation of genes involved in neural progenitor proliferation (63).
The mechanism by which 5hmC leads to these phenotypic changes is thought to be regulated in part by impairing the “readers” of 5mC, such as MeCP2, and therefore allowing for gene activation. 5hmC has been found to accumulate in neurons at key genetic loci during development and to be inversely associated with MeCP2 dosage (59). MeCP2 has been subsequently found to bind to 5hmCA but not 5hmCG; therefore, the presence of 5hmCG (the predominant form of 5hmC in the brain) can result in “functional demethylation” with subsequent increases in transcription (64).
2.1.3. Genomic imprinting: DNA methylation and allele-specific inheritance in neurodevelopmental diseases
In humans, approximately three billion base pairs of DNA are organized into 23 paired chromosomes, including the X and Y sex chromosomes. One copy of each chromosome is inherited from the mother and the father, leading to allelic variability. It is increasingly recognized that preferential expression of either a paternal or maternal allele can be observed and is epigenetically regulated (65). Genomic imprinting refers to the processes by which paternal or maternal genes are preferentially expressed in an allele-specific manner. Several imprinted genes have been found to have key roles in neurodevelopment.
This allelic specificity also has elements of stochasticity and region-specific silencing. A mouse model containing a tdTomato/GFP reporter of either the maternally- or paternally-expressed Dlk-Dio3 intergenic differentially DNA-methylated region (IG-DMR) revealed that, while many cells faithfully maintained allelic expression, changes in methylation patterns were present in a tissue-specific and cell type-specific manner (66). Intriguingly, neurogenic regions, such as the subventricular zone and subgranular zone of the dentate gyrus, have biallelic hypermethylation of the IG-DMR region, suggesting tissue-type specificity and allowing for neuronal heterogeneity (66). DNA methylation was first thought to drive this preferential expression of genomic imprinting (67). However, it is increasingly appreciated that other mechanisms, such as transcription factors (68) and histone modifications (69), also regulate this process.
Perhaps the most apparent consequence of genomic imprinting is through its manifestation and distinct inheritance pattern (70) in human diseases (71). Two of the most commonly described diseases of genomic imprinting include Prader-Willi Syndrome (PWS) and Angelman Syndrome. In the case of PWS, which is most often due to mutations in chromosome 15, the paternal copy is preferentially expressed. Therefore, mutations to the paternal gene or the aberrant inheritance of two maternal copies lead to disease states, causing a characteristic syndrome including facial anomalies, hyperphagia and intellectual disability (72). Mechanistically, small nucleolar-long non-coding RNAs (sno-lncRNAs), a class of nuclear-enriched, intron-derived long non-coding RNAs that are processed on both ends by the small nucleolar RNA (snoRNA) machinery, have been found to be deleted in PWS (73). These sno-lncRNAs normally interact with the Fox family of splicing regulators, wherein deletion (as in PWS) leads to aberrant splicing activity and subsequent neurodevelopmental phenotypes (73). In contrast, Angelman syndrome primarily involves UBE3A, an E3 ubiquitin ligase which is also located in chromosome 15 but in a region of the chromosome where the paternal gene is typically silenced. Disease-causing states occur due to mutations on the maternal chromosome or if two paternal copies are inherited. Angelman syndrome typically manifests as cognitive disability, seizures, microcephaly, and speech impairment (74).
2.1.4. Environmental alterations of DNA methylation in neurogenesis
Environmental factors are also known to alter epigenetic states to impact development and disease pathogenesis (75). Environmental factors can alter DNA methylation signatures and contribute significantly to the regulation of neurogenesis. Gadd45b was found to be upregulated following electroconvulsive therapy (ECT), with a role in promoting neural progenitor proliferation and dendritic growth of new-born neurons in the adult hippocampus. This study further showed that Gadd45b was required for DNA demethylation at key neurogenic promoters, indicating an epigenetic role in neurogenesis (76). Conflicting results from studies of Gadd45b knockdown in mice demonstrated a selective deficit in fear conditioning (77) versus enhanced memory in motor performance, aversive conditioning, and spatial navigation (78). Neuronal activity has also been linked to changes in DNA methylation (79). Intriguingly, differential DNA methylation patterns in Alzheimer’s disease patients compared to controls were preferentially associated with genes involved in neurodevelopment and neurogenesis (80).
Similarly, glucocorticoids have a known effect on DNA demethylation (81). With regards to neurodevelopment, glucocorticoids have been found to alter DNA methylation in the hippocampus and prime future stress responses (82). More recently, a novel modification, N(6)-methyladenine (6mA) has been identified as another form of DNA methylation in mammalian cells (83). While its role continues to be elucidated, one study has found that 6mA methylation is elevated in the brain in a chronic stress mouse model. These methylation changes overlap with genes involved in depression, schizophrenia, and ASD (84).
2.2. Histone modifications
2.2.1. Chromatin-based regulation
2.2.1.1. Chromatin structure
Chromatin refers to the higher-ordered structure through which DNA is organized to form chromosomes. Eukaryotic DNA wraps around histone proteins to form nucleosomes, the fundamental repeating unit of chromatin (85). Nucleosomes are comprised of approximately 147 base pairs DNA wrapped around an octamer of histone proteins, with two copies of the core histones H2A, H2B, H3, and H4 (86,87). A linker sequence of DNA of approximately 53 base pairs binds to the H1 histone and further contributes to chromatin compaction (88,89). These histone proteins hold not only a structural importance, but also a functional role as accessibility of DNA for RNA transcription can be regulated by the histone state, with binding affinities to histone proteins being altered by covalent, but reversible modifications to the histone proteins. Euchromatin refers to DNA that is not tightly bound to histone proteins and is believed to be accessible for transcription, meanwhile heterochromatin refers to DNA that is compacted and is consequently transcriptionally inactive (Figure 2A). During neurodevelopment, dynamic changes are necessary in order to transition from a more stem cell-like state to more differentiated progeny. Chromatin dynamics has been found to be among the key regulators for cell-state transition.
2.2.1.2. Chromatin accessibility
A common method to analyse the general epigenetic state is through an assessment of chromatin accessibility. Fundamental to this assessment is the notion that various molecules are able to interact with open chromatin (euchromatin) over compacted chromatin (heterochromatin) and that identifying regions of open chromatin can help identify active or readily activatable genes. Multiple methods have leveraged the differential accessibility of euchromatin compared to heterochromatin in order to assess the general chromatin landscape. Formaldehyde-assisted isolation of Regulatory Elements (Faire-Seq) utilizes differential rates of crosslinking (90), while DNAse-seq relies upon differential sensitivity to DNAse I. Assay for transposase-accessible chromatin using sequencing (ATAC-seq) is an increasingly utilized method to probe DNA accessibility. Rather than relying upon differential fixation, this method utilizes a hyperactive Tn5 transposase, which inserts sequencing primers to accessible DNA (91). These segments are then amplified for high throughput sequencing and are able to identify segments of open chromatin. Such methods have led to a better understanding of the dynamic changes underlying neurodevelopment.
Utilizing ATAC-seq, neuronal activity was found to result in genome-wide changes in chromatin accessibility one hour after activation and was mediated in large part at cFos binding sites (92). Unlike CpG methylation, in which 31% of activity related changes at 4 hours were maintained at 24 hours (79), the changes seen by ATAC-seq were rather transient, such that only 5.1% of gained-open regions detected at 1 hour were seen at 24 hours (92). Using ATAC-seq, the neuronal-associated transcription factor FoxP2 was found to alter the chromatin accessibility during neuronal differentiation, leading to repression of non-neuronal gene regions and activation of neuronal maturation genes (93). This further supports the notion that gene regulation by transcription factors involves changes in chromatin accessibility.
2.2.1.3. Histone modifications
Among the more studied modes of epigenetic regulation are histone modifications. Modifications to the H3 histone, including methylation and acetylation of lysine residues along the histone protein, have been found to be associated with transcriptional regulation (94). Histone modifications as well as direct interactions with histone modifiers are typically assayed through chromatin immunoprecipitation (ChIP) (95,96). In this method, cells are typically fixed using formaldehyde in order to maintain DNA-protein interactions prior to shearing of chromatin into approximately 300 base pair fragments. Chromatin of interest is enriched using antibodies against histone modifications, histone modifiers, or transcription factors, and then subsequently analysed using qPCR (97) or high-throughput sequencing based methods (98). This allows for profiling of the chromatin state at key genetic loci that may regulate gene expression.
One of the more prevalent models of chromatin regulation, particularly during neurodevelopment (99-101), is the notion of histone modification “bivalency” (102). H3K4me3 is a chromatin regulation mark commonly associated with gene activation (103,104). Acetylation of H3K9 (H3K9ac) is similarly associated with gene activation, and has been found to mediate the switch from H3K4me3-mediated transcriptional initiation to elongation (105). Meanwhile, H3K27me3 is a chromatin mark associated with gene silencing. Promoter regions that contain both H3K4me3 and H3K27me3 are thought to be transcriptionally inactive, but “poised” for activation. Dynamic changes in the histone state, particularly at promoter regions, are thought to be fundamental to transcriptomic dynamics during development. Chromatin profiling of neural progenitor cells reinforces this notion that resolution of histone marks influences cellular identity (99).
2.2.1.4. Histone modifications in neurodevelopment
The regulation of these histone modifications, including H3K4me3 and H3K27me3, relies upon enzymatic genes that either place (histone methyltransferases) or remove (histone demethylases) histone marks. Trithorax group proteins represent a class of proteins that regulate histone methylation, including H3K4me3, a marker of gene activation. Mll1-1 is a Trithorax group H3K4me3 methyltransferase that was originally found to be necessary for postnatal SVZ neurogenesis, wherein a conditional knockout of Mll-1 leads to failure to properly activate neurogenic transcription factors including Dlx2 (106) and Brn4 (107). Conditional deletion of a common H3K4 methyltransferase subunit, Dpy30, leads to both neurogenic and gliogenic defects (108). Other histone H3K4 methyltransferases include the SET family of proteins, such as SET1A, SET1B and MLL2-4 (109).
Among the most commonly appreciated mechanisms of gene regulation is through the Polycomb Repressive Complex (PRC), including both PRC1 and PRC2. The core PRC2 complex is composed of EZH1/2, EED164, SUZ12, and RBBP4/7, and this can be in complex with various PRC2 accessory proteins including PCL1-PCL3, JARID2, AEBP2, EPOP, and LCOR. PRC1 contains both a canonical and non-canonical complex. The core PRC1 is comprised of RING1A/B and PCGF-PCGF6. This core PRC1 forms a complex in the canonical PRC1 with CBX-family proteins, PHC1-PHC3, and SCMH1/2. Meanwhile, the non-canonical PRC1 includes RYBP/YAF2, KDM2B, DCAF7, and WDR5. These two complexes are thought to serve as an H3K27me3 methyltransferase and mono-ubiquitinate H2AK119, respectively (110,111). Knockout of polycomb components Ring1b (PRC1), Ezh2, or Eed (PRC2) lead to defects in neurogenic to astrocytic transitions during cortical development (112). Mechanistically, Ring1b was found to regulate timed termination of subcerebral projection neurons (SCPNs) via binding and subsequently decreasing expression of Fezf2, a fate determinant of SCPNs (113). The ubiquitin-ligase activity of Ring1b acts primarily in early-stage repression of neurogenic genes in neural stem cells and NPCs but does not act to repress neuronal genes in late-stage astrogliogenic cells. Meanwhile, ubiquitin-independent PRC1-mediated repression relies on histone deacetylation and Phc2-mediated clustering (114). Ezh2 was found to be an essential regulator of developmental cortical neurogenesis (115) as well as postnatal neurogenesis in the adult SVZ (116) and hippocampus (117). One key role of Ezh2 in the developing neocortex is the prevention of early gliogenesis through suppression of GFAP expression via interaction with chromodomain helicase DNA-binding protein 4 (Chd4) (118). In the postnatal SVZ, Ezh2 promotes neurogenesis in part through suppression of the Ink4a/Arf locus to promote cellular proliferation, temporal-specific repression of Olig2 during neuronal differentiation, and also through persistent suppression of non-SVZ neuronal subtypes to confer SVZ lineage specificity (116).
Removers of these histone marks, or histone demethylases, have also been shown to be essential for neurogenesis. Jmjd3 (also known as KDM6B) is an H3K27me3 demethylase that opposes the action of the PRC2 complex by removing the H3K27me3 mark (119). It has been found to be important on a genetic level to remove repressive marks at key neurogenic loci during neuronal differentiation. Conditional deletion of Jmjd3 results in a neurogenic defect in the subventricular zone of postnatal mice, and JMJD3 acts at both promoter and enhancer regions of Dlx2 to promote neurogenesis (120). UTX (also known as KDM6a) is another H3K27me3 demethylase that has been associated with neurodevelopment. Deletion of Utx impairs hippocampal function through reductions in long-term potentiation and amplitude of miniature excitatory postsynaptic currents, aberrant dendrite development and synapse formation. This phenotype is mediated in part through reduced expression of 5-hydroxytryptamine receptor 5b (Htr5b) (121). In another example, PHF2, a histone demethylase of H3K9me3, regulates the cell cycle of neural progenitors during DNA damage and genome instability (122).
Histone acetylation is typically known as a marker of active gene transcription (123). Liver-driven alcohol metabolism has been associated with elevated levels of acetate that acts on brain histone acetylation (124). However, much of what is understood of histone acetylation is via the role of histone deacetylases (HDACs), which remove histone acetylation. HDAC1 and HDAC3 are key drivers of embryonic neurogenesis, mediating H3K9 acetylation (125). HDAC inhibitors, such as valproic acid, are commonly used to treat neurologic and psychiatric diseases, such as epilepsy, bipolar disorder, and depression (126).
2.2.1.5. Histone modification-based dysregulation in diseases
Mutations in histone modifiers have been identified to cause various developmental syndromes (127). Mutations in MLL complex genes are associated with Wiedemann-Steiner syndrome, which is characterized by hypertrichosis cubiti, short stature, intellectual disability, and a distinct facial appearance (128). Mutations and deletions of UTX are associated with Kabuki syndrome, a rare genetic disease that causes developmental delay, craniofacial, and limb anomalies (129,130). Mutations to the histone acetyltransferase KAT6A have been identified as putative pathogenic variants to a rare neurodevelopmental disorder that includes global developmental delay, impaired speech development, and facial dysmorphism (131).
Genetic variants in genes involved in chromatin remodelling have also been associated with neurological disorders. For example, large exome sequencing studies have found that a number of ASD candidate genes encode chromatin remodellers, with genetic variants in CHD8 being particularly common (132,133). CHD8 normally interacts with the transcription factor REST, with haploinsufficiency causing autism-like phenotypes (134) and altered brain development in mice (135). AUTS2, encoded by another ASD susceptibility gene, interacts with the PRC1 repressive complex to paradoxically activate gene expression (136). Interestingly, chromatin-remodelling factors were also identified as a significant set of mutated genes when analysing the somatic landscape of glioblastoma (137). In one study, 135 of 291 (46%) of patients were found to have mutations in genes associated with chromatin remodelling, suggesting a significant epigenetic contribution to glioblastoma. The PRC2 complex is also notably found to be a key component of the tumorigenic phenotype of the H3K27M mutation (138,139), which is found in diffuse intrinsic pontine gliomas (DIPGs) and other midline gliomas. Inhibition of EZH2 in a mouse model of H3K27M-associated DIPG abolished cell growth through induction of the tumor suppressor p16INK4A, implicating PRC2 as a possible therapeutic target in H3K27M-associated DIPG (140).
2.3. Higher-order chromatin structure
While promoter elements are a key part of genetic regulation, it is now increasingly appreciated that enhancer elements are also a contributing mechanism of chromatin context-specific gene regulation (141). These enhancer elements act in the context of three-dimensional chromatin architecture, operating in cis to increase gene expression by binding specific transcription factors and stabilizing transcription factor-promoter interactions (142). Though typically distal to the gene promoter in a two-dimensional map, enhancers interact directly with gene promoters through chromosomal looping in a three-dimensional fashion (143). However, more recent studies of neural differentiation have suggested that the Sonic Hedgehog (Shh) gene can be activated through increased physical separation, suggesting that looping may not be the only mechanism involved in the function of enhancer elements (144).
Enhancers have been thought to mediate cell type-specific gene expression programs with temporal specificity by recruiting specific transcription factors and chromatin remodelling complexes (145,146). Since enhancers carry important functions, a significant amount of effort has gone into identifying genome-wide enhancer-promoter interactions with a variety of different techniques. Enhancers have traditionally been identified with reporter assays, but the advent of next-generation sequencing technologies has allowed for genome-wide predictions of putative enhancers. Some of these techniques involve profiling epigenetic marks associated with enhancers by leveraging the association of enhancers with particular chromatin states. In contrast to promoter regions, enhancers are commonly identified by association with H3K4 monomethylation (H3K4me1), H3K27 acetylation (H3K27ac), and p300 binding at non-promoter gene regions (147-150). Intriguingly, these enhancer regions respond to neural activity to regulate transcription (149).
Some of the most powerful techniques to understand enhancer-promoter dynamics are those that allow for the study of the three-dimensional architecture of the genome. These methods typically rely upon fixation of the three-dimensional state and the capture of this interaction through DNA ligation reactions. The fundamental concept of these chromosome conformation capture (3C) assays is to capture long-range DNA interactions through fixation and subsequent DNA enrichment (151). 3C technology has been combined with next-generation DNA sequencing in a technique called Hi-C to capture genome-wide long-range DNA interactions (152). These methods have not only allowed for identification of enhancers and their targets, but also provide fundamental information about higher-order chromatin architecture and how the genome is organized in three-dimensions. It has become clear that the genome is hierarchically organized into different features at different resolutions, with chromosomal compartments (the spaces occupied by each individual chromosome in the nucleus) as the largest organizational feature.
Hi-C has revealed that, on the subchromosomal scale, the genome exists as two compartments, denoted A and B compartments (152). The A compartment is enriched for actively transcribed genes and is thought to represent open chromatin, while the B compartment is broadly inactive and is thought to represent closed chromatin (Figure 2A) (153). On a finer scale, Hi-C maps have revealed that a large proportion of the genome is organized into topologically- associated domains (TADs), which are regions of DNA that associate preferentially with other sequences in the TAD and have boundaries demarcated by CTCF binding sites (154). Sub-TADs have also been identified, representing an even finer level of genome-wide organization (155). Chromatin loops have been identified as the organizational feature at the finest level of detail and may mediate enhancer-promoter interactions (156). These high-dimensional features have been studied extensively in mouse neural development, which demonstrated dynamic changes of TADs during neuronal differentiation, including disruption of a polycomb network and the appearance of neural transcription factor interactions (157).
As many of the disease-associated SNPs in neurodevelopmental disorders are found in non-coding regions of the genome, it is thought that one mechanism of functional consequence is the aberrant regulation of enhancer elements (158). As a neural stem cell marker, Sox2, has been recently found to be a key transcriptional factor that is bound to both promoter and enhancer elements in order to regulate gene transcription. Deletion of Sox2 leads to disruption of the long-range interactions between enhancer and promoter regions with a subsequent decrease in gene expression. Exogenous rescue of Socs3, a gene affected by Sox2 disruption, restored the self-renewal defect of Sox2-deleted NSCs (159). Meanwhile, enhancer-gene interactions in neural development unique to humans are enriched in outer-radial glia, while GWAS analysis found genetic variants for educational attainment, neuropsychiatric disease, and brain volume to be enriched within regulatory elements involved in cortical neurogenesis (160). This work further elucidates the interaction between transcription factors and enhancer elements in the maintenance of cellular function and identity.
2.4. Non-coding RNA-based regulation
2.4.1. Non-coding RNA
In addition to chromatin-based regulation, it is being increasingly appreciated that RNA-based mechanisms also influence cellular identity. Non-coding RNAs have functional roles in regulating gene transcription. Anti-sense RNAs have been found throughout the genome and are associated with regulating nearby gene transcription (161). Long non-coding RNA (lncRNAs) are polyadenylated RNAs that have been found to operative in trans and interact closely with epigenetic regulators, such as chromatin modifying factors. LncRNAs carry out a variety of different functions, and have been identified as key regulators of pluripotency and differentiation, with knockdown of lncRNAs having similar consequences on transcription as known ESC regulators (162).
2.4.2. Non-coding RNA in neurodevelopment
As the first functional non-coding RNA identified in neurodevelopment, Evf2 was initially identified as a lncRNA that regulates the transcription of Dlx5 and Dlx6 in the developing mouse forebrain, the knockout of which leads to reduced early production of GABAergic interneurons and reduced synaptic inhibition despite a normalization of neuronal levels in the adult hippocampus (163). Evf2 was noted to regulate transcription in trans through inhibition of CpG methylation of Dlx5 and Dlx6 enhancer regions (164). A direct interaction of chromatin remodelling inhibition was demonstrated with Evf2 co-localizing with SWI/SNF-related chromatin remodellers Brahma-related gene 1 (Brg1) and Brahama-associated factor (Baf170) in the developing mouse forebrain (165). These findings implicated lncRNAs as a key functional component of various mechanisms of epigenetic regulation.
With the advent of next generation sequencing, a catalogue of lncRNA expression was generated for adult SVZ neural stem cells with some evidence of a functional role for lineage specification (166). Pnky was identified to have a critical role in neurogenesis wherein knockdown of Pnky led to an increase in neuronal progenitors primarily through direct action on the splicing regulator PTBP1 (167). Conditional deletion of Pnky from the developing cortex altered cortical lamination and a BAC transgene rescued this defect, indicating that Pnky regulates development in trans (168). Transcriptomic characterization of lncRNAs during embryonic development similarly demonstrated strong correlation of expression of lncRNAs with neurogeneic genes. Miat is a lncRNA determined to affect brain development and regulate the splicing of Wnt7b (169).
Other lncRNAs have been found to regulate cortical development in cis and interact with histone modifying enzymes. LncKdm2b acts in cis to activate Kdm2b, a H36me2 and H3K4me3 demethylase, allowing for proper differentiation and migration of cortical projection neurons (170). Single-cell RNA sequencing analysis of lncRNAs in the developing human neocortex indicated that despite being detected at low levels in bulk sequencing, lncRNAs can be abundantly expressed in single cells suggesting cell-type specificity (171). Utilizing both primate and human cerebral organoid models and performing RNA sequencing over time, lncRNA conservation was observed across species with transient expression in a cell-type specific manner (172). While the diversity of expression has been established, the functional roles of many lncRNAs are still unclear. Traditional analysis for lncRNAs rely upon either RNA knockdown (167,173) or genetic deletion and in vivo characterization (163,168). However, with the advent of CRISPR-CAS9 (174) high-throughput methods have also been developed for assessing lncRNA function (175). Future work may allow for screening on specific cell types during neurodevelopment.
2.4.3. lncRNAs in diseases
LncRNAs have been implicated in the pathogenesis of various diseases normally associated with other modes of epigenetic regulation. LncRNA FMR4, which is located at the Fragile X mental retardation 1 (FMR1) gene, is a chromatin-associated transcript that is enriched in genes related to neural development and cellular proliferation (176). FMR4 has also been shown to regulate methyl-CpG-binding domain protein 4 (MBD4) (177). LncRNAs have also been associated with ASD. SHANK2-AS is upregulated in ASD, and exogenous overexpression leads to inhibition of neuronal proliferation and promotion of apoptosis (178). lnc-NR2F1 was recently identified as a candidate gene in a cohort of children with ASD/intellectual disability. lnc-NR2F1 enhances neuronal cell maturation and regulates transcription of neuronal genes including ASD-associated genes (179).
2.5. Epitranscriptomics
In addition to functional roles of non-coding RNAs, modifications of RNA transcripts through various nucleotide modifications have also been found to be a key component of gene regulation. Epitranscriptomics refers to the various posttranscriptional changes in mRNA and lncRNA in order to regulate gene expression (180). Over 100 chemical modifications have been identified in RNA. These reversible modifications have multiple roles, but act in part through altering mRNA metabolism and translation. Perhaps the most well studied form of epitranscriptomic modification is the methylation of adenosine at the sixth nitrogen position (m6A), which has a myriad of roles at various levels of gene regulation (181).
2.5.1. N6-methyladenosine
m6A modification to RNA has been found to be a key feature of post-transcriptional regulation of the dynamic changes necessary for neurodevelopment. Similar to histone marks, this modification has known machinery including the “writers” (METTL3, METTL14, and WTAP), “erasers” (FTO (182) and ALKBH5 (183)), and “readers”, such as FMRP, YTHDF1, and YTHDF2. m6A modification in particular is known to have multiple roles, including the promotion of mRNA decay, translation, regulation of epigenetic histone modification, and promoting nuclear RNA export (184).
Detection of m6A can be performed by utilizing antibody based methods against modified RNA such as MeRIP-seq (185) and m6A-seq (186). More recent work has determined that m6A can be detected through an analysis of increased systematic errors and decreased base-calling qualities. This method can detect m6A with approximately 90 percent accuracy in synthetic sequences and with 87 percent accuracy in vivo (187).
2.5.2. Multiple roles of m6A in neurodevelopment
Analysis of m6A signalling on cortical neurogenesis using an embryonic METTL14 knockout model demonstrated a prolonged cell cycle of radial glia and extension of cortical neurogenesis into the postnatal stages. Furthermore, m6A was found to be associated with mRNA turnover, with m6A tagging promoting mRNA decay (188). Loss of Ythdf2, a reader of m6A, has been associated with delayed degradation of neuron differentiation-related genes, with a failure to produce normally functioning neurites (189). m6A has been found to act locally upon axons, with inhibition of m6A demethylase FTO leading to increased m6A levels and consequent decreased translation of GAP-43 mRNA (190). Similarly, the m6A reader YTHDF1 binds to mRNA of Robo 3.1, an axon guidance regulator, in order to promote protein translation. Deletion of Ythdf1 results in axon guidance defects (191).
m6A was also determined to be a key feature of mRNA transport. Utilizing a Fmr1 knockout mouse, Fragile X mental retardation protein (FMRP) was determined to bind m6A and promotes nuclear transport of methylated mRNA targets during neural differentiation. Mettl14 conditional knockout mice are unable to obtain m6A modifications and have a nuclear export deficit for some mRNAs (192).
m6A modification has also been shown to be involved in the regulation of histone modifying enzymes. Knockdown of Mettl3 led to a decrease in both Ezh2 expression and H3K27me3 levels, leading to a neurogenic defect that could be rescued by Ezh2 overexpression (193).
In the adult brain, m6A has been shown to regulate phenotypic changes in the hippocampus. Deletion of Ythdf1 results in learning and memory deficits as well impaired synaptic transmission and long-term potentiation. Transcriptome-wide analysis of m6A RNA interaction with its reader YTHDF1 showed enrichment for key neuronal plasticity genes (194).
2.5.3. Epitranscriptomics in diseases
While the role of m6A is still being elucidated, mutations in genes associated with epitranscriptomic functions have been identified in diseases. Although FTO is named for its association with obesity (195), genetic associations have also noted that mutations in FTO are related to risk for bipolar disorder. In silico analysis indicated that these mutations disturb binding sites of SP1 and SP2, which are distinct from the obesity-associating binding site perturbation of FOXP1 (196). Direct mutation of the enzymatic dioxygenese-encoding portion FTO was identified in a single family, leading to an autosomal recessive lethal syndrome associated with postnatal growth retardation, microcephaly, severe psychomotor delay, functional brain deficits, and characteristic facial dysmorphism (197). A murine model of focal ischaemia found that ischaemic pathology leads to a decrease in FTO and associated increase in m6A abundance. Differentially expressed m6A methylated genes were related to inflammation, apoptosis, and transcriptional regulation (198). These varied phenotypes related to FTO support the notion of the vast diversity of function in epitranscriptomics.
3. Epigenetic crosstalk: systems in combination rather than isolation
As the various components of epigenetic regulation become elucidated, it becomes increasingly obvious that these various mechanisms have significant interactions. As one example, Fragile X Syndrome is classically described as an expansion of CGG nucleotide, which leads to hypermethylation of the FMR1 promoter (199). However, recent analyses have also shown that patients with Fragile X, as well as other repeat expansion disorders, have alterations in higher-ordered chromatin structure in the form of boundaries between chromatin domains. The extent of disruption of these chromatin domains correlates with FMR1 silencing (156). FMR5 and FMR6 are two lncRNAs that have been identified to be expressed within the FMR1 locus, with FMR6 being silenced in patients with both full mutations and pre-mutations, indicating its potential as a predictive biomarker (200). FMR1 protein interacts directly with TUG1, a lncRNA distal to the FMR1 gene, to decrease its stability. Knockdown of TUG1, promotes axon development in cultured hippocampal neurons, with overexpression leading to axonal growth defects during embryonic cortical development. Knockdown of TUG1 rescues axonal developmental defects in FMRP-deficient neurons, further emphasizing this dynamic interplay of epigenetic regulation (201). FMRP also regulates m6A-marked mRNA targets. Loss of FMRP leads to widespread changes in the m6A landscape, and FMRP was found to normally maintain stability of m6A mRNAs through interaction with the m6A reader, YTHDF2 (202). Therefore, developmental diseases can have various modes of epigenetic dysfunction as both cause and consequence that encompasses DNA methylation, chromatin structure, lncRNAs, and epitranscriptomics.
4. Challenges and future directions in epigenetics in neurodevelopment
4.1. Multi-omic analysis: combining novel technologies
While many of the components of epigenetics have traditionally been investigated in isolation, emerging technologies allow for the study of multiple aspects of epigenetic regulation in a “multi-omic” fashion and on a single-cell level (203). Methods have been developed to simultaneously sequence genomes, DNA methylation and transcription (204,205), chromatin state/nucleosome positioning, DNA methylation, copy number variation and ploidy (206), and chromatin accessibility, DNA methylation, and transcription (207). Developing a single-cell understanding of the coordination of these multiple processes may shed light on the multifaceted nature of neurodevelopment.
One of the fundamental challenges of the recently appreciated role of non-coding RNAs is the need to better identify the functional roles of this newly identified class of molecules. An emerging method to identify non-coding RNA function is through the use of CRISPR/CAS9 high-throughput screening efforts (175). More recently, CRISPR/CAS9 screening has been combined with single-cell epigenomic profiling in order to understand the epigenetic effects of CRISPR perturbations (208). As lncRNAs have been associated with many elements of epigenetic regulation, combining lncRNA-targeting CRISPR screens with single-cell epigenomic profiling could provide key insights into the degree to which lncRNAs may contribute to the epigenetic landscape.
4.2. Neurodevelopmental models
4.2.1. Mouse models of neurodevelopment
Mus musculus models have been a predominant means to study neurodevelopment. These animal models provide an avenue for genetic manipulation, which can facilitate the understanding of neurodevelopment in the context of knock-in/knock-out phenotypes of known disease-associated genes (209). These phenotypes can then be studied across many levels including molecular, developmental, structural, and behavioural changes upon genetic alteration (209). However, one potential limitation is the applicability of such findings in humans. For instance, murine and human m6A profiling have distinct features such that the species is a higher determinant of methylome similarity than the tissue type (210). Therefore, alternative approaches have been developed to study the mechanisms of neurodevelopment in human cells.
4.2.2. Induced pluripotent stem cells (iPSCs)
iPSCs are multipotent stem cells that are generated from the “de-differentiation” of fully differentiated cells, commonly fibroblasts (211). iPS cells allow for the study of human cells rather than other animal systems, which may not have equivalent regulatory systems. This can be particularly important in situations wherein human-specific features might need to be studied. For instance, an emerging line of work has been the use of neurons differentiated from iPS cells derived from families with known neurodevelopmental diseases to study the effects of inherited mutations in control versus affected individuals in the same family. This has been utilized in families with Rett Syndrome demonstrating impaired neuronal maturation and function (212), and in mental disorders associated with DISC1 (disrupted-in-schizophrenia-one) wherein mutations in DISC1 lead to impaired interaction with Activating Transcription Factor 4 (ATF4) resulting in transcriptional and synaptic dysregulation (213). However, the iPSC system lacks the capacity to model in vivo phenotypes. Furthermore, the three-dimensional cellular architecture that occurs in animal models is difficult to recapitulate in these cell-based monolayer culture systems.
4.2.3. Cerebral organoids: A three-dimensional culture model
Unlike traditional iPSC-derived monolayer cultures, cerebral organoids have been recently developed that grow in three-dimensional space and maintain a three-dimensional cellular architecture that in many aspects mimics human development, including cells with markers of outer radial glia (214-217). This has been leveraged to study neurodevelopmental structural disorders of the developing human brains such as lissencephaly (218) and the developmental consequences of known mutations associated with psychiatric disorders such as schizophrenia (219). One prominent example has been the use of cerebral organoids to understand the neurodevelopmental effects of Zika virus, which is associated with microcephaly following fetal exposure. Zika virus was found to preferentially affect neural progenitor cells and consequently alter cellular architecture and neuron generation (217). Future work can leverage both multi-omic strategies and these three-dimensional cerebral organoid models to elucidate the dynamic epigenetic changes occurring in the developing human brain.
5. Conclusion
Neurodevelopment relies upon multiple layers of temporal and spatial gene regulation in order to generate the diversity of cellular subtypes appreciated in the adult mammalian brain. Epigenetic mechanisms guide this diversity of gene expression through a variety of modifications to DNA, histone proteins, and subsequent RNA. While the central dogma of biology has largely held true for many biological processes, there is a growing appreciation of the complexity of the transition from DNA to RNA to protein. Regulatory roles of non-coding DNA, such as enhancer elements and functional non-coding RNA, such as lncRNAs, also further complicate this perhaps overly simplistic model. DNA methylation, histone modifications, non-coding RNA, and RNA modifications are among the various features of gene regulation that help coordinate proper cellular identity and function. These features do not act in isolation, but rather interact together in order to generate defined cellular identities during neural differentiation and subsequent maturation. Future work may utilize novel neurodevelopmental models and assay these various levels of gene regulation simultaneously and on the single cell level, in order to gain a more precise understanding of the dynamic interaction between these various players in neurodevelopment.
Acknowledgement:
We thank Kimberly Christian for comments. The work in the authors’ laboratory was supported by grants from National Institutes of Health (R37NS047344, R01AG057497, and P01NS097206 to H.S.) and Simons Foundation (to H.S.).
Footnotes
Conflicts of Interest: R.D.S., D.R.C., and H.J. declare that they have no conflicts of interest that might be relevant to the content of this manuscript.
References
- 1.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013. August 9;341(6146):1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LaMonica BE, Lui JH, Wang X, Kriegstein AR. OSVZ progenitors in the human cortex: an updated perspective on neurodevelopmental disease. Current Opinion in Neurobiology. 2012. October 1;22(5):747–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen DV, Lui JH, Parker PRL, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010. March;464(7288):554–61. [DOI] [PubMed] [Google Scholar]
- 4.Lim DA, Alvarez-Buylla A. The Adult Ventricular-Subventricular Zone (V-SVZ) and Olfactory Bulb (OB) Neurogenesis. Cold Spring Harb Perspect Biol. 2016. May 2;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kempermann G, Song H, Gage FH. Neurogenesis in the Adult Hippocampus. Cold Spring Harb Perspect Biol. 2015. September 1;7(9):a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kempermann G, Gage FH, Aigner L, Song H, Curtis MA, Thuret S, et al. Human Adult Neurogenesis: Evidence and Remaining Questions. Cell Stem Cell. 2018. July 5;23(1):25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg DA, Su Y, Jimenez-Cyrus D, Patel A, Huang N, Morizet D, et al. A Common Embryonic Origin of Stem Cells Drives Developmental and Adult Neurogenesis. Cell. 2019. April 18;177(3):654–668.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delgado RN, Lim DA. Embryonic Nkx2.1-expressing neural precursor cells contribute to the regional heterogeneity of adult V-SVZ neural stem cells. Dev Biol. 2015. November 15;407(2):265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonaguidi MA, Song J, Ming G, Song H. A unifying hypothesis on mammalian neural stem cell properties in the adult hippocampus. Current Opinion in Neurobiology. 2012. October 1;22(5):754–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A. Subventricular Zone Astrocytes Are Neural Stem Cells in the Adult Mammalian Brain. Cell. 1999. June 11;97(6):703–16. [DOI] [PubMed] [Google Scholar]
- 11.Gage FH. Mammalian Neural Stem Cells. Science. 2000. February 25;287(5457):1433–8. [DOI] [PubMed] [Google Scholar]
- 12.Gage FH. Neurogenesis in the Adult Brain. J Neurosci. 2002. February 1;22(3):612–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kintner C. Neurogenesis in Embryos and in Adult Neural Stem Cells. J Neurosci. 2002. February 1;22(3):639–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvarez-Buylla A, García-Verdugo JM. Neurogenesis in Adult Subventricular Zone. J Neurosci. 2002. February 1;22(3):629–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crick F. Central Dogma of Molecular Biology. Nature. 1970. August;227(5258):561–3. [DOI] [PubMed] [Google Scholar]
- 16.Deans C, Maggert KA. What Do You Mean, “Epigenetic”? Genetics. 2015. April;199(4):887–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985. January;40(1):91–9. [DOI] [PubMed] [Google Scholar]
- 18.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980. May;20(1):85–93. [DOI] [PubMed] [Google Scholar]
- 19.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975. January 24;187(4173):226–32. [PubMed] [Google Scholar]
- 20.Monk M, Holliday R, Monk M, Pugh JE. Changes in DNA methylation during mouse embryonic development in relation to X-chromosome activity and imprinting. Philosophical Transactions of the Royal Society of London B, Biological Sciences. 1990. January 30;326(1235):299–312. [DOI] [PubMed] [Google Scholar]
- 21.Luo C, Keown CL, Kurihara L, Zhou J, He Y, Li J, et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017. 11;357(6351):600–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010. March;11(3):204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017. 05;356(6337). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen HN, et al. DNA methylation presents distinct binding sites for human transcription factors. Elife. 2013. September 3;2:e00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009. November 19;462(7271):315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010. July 23;329(5990):444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012. May 29;13(7):484–92. [DOI] [PubMed] [Google Scholar]
- 28.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010. March;11(3):191–203. [DOI] [PubMed] [Google Scholar]
- 29.Kurdyukov S, Bullock M. DNA Methylation Analysis: Choosing the Right Method. Biology (Basel). 2016. January 6;5(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005. August;37(8):853–62. [DOI] [PubMed] [Google Scholar]
- 31.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010. July 8;466(7303):253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu M, Hon GC, Szulwach KE, Song C-X, Zhang L, Kim A, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012. June 8;149(6):1368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012. May 18;336(6083):934–7. [DOI] [PubMed] [Google Scholar]
- 34.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002. January 1;16(1):6–21. [DOI] [PubMed] [Google Scholar]
- 35.Leonhardt H, Page AW, Weier H-U, Bestor TH. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992. November 27;71(5):865–73. [DOI] [PubMed] [Google Scholar]
- 36.Okano M, Bell DW, Haber DA, Li E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell. 1999. October 29;99(3):247–57. [DOI] [PubMed] [Google Scholar]
- 37.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998. November;18(11):6538–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nan X, Campoy FJ, Bird A. MeCP2 Is a Transcriptional Repressor with Abundant Binding Sites in Genomic Chromatin. Cell. 1997. February 21;88(4):471–81. [DOI] [PubMed] [Google Scholar]
- 39.Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, et al. MeCP2, a Key Contributor to Neurological Disease, Activates and Represses Transcription. Science. 2008. May 30;320(5880):1224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lyst MJ, Ekiert R, Ebert DH, Merusi C, Nowak J, Selfridge J, et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci. 2013. July;16(7):898–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014. February;17(2):215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabel HW, Kinde B, Stroud H, Gilbert CS, Harmin DA, Kastan NR, et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015. June 4;522(7554):89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen L, Chen K, Lavery LA, Baker SA, Shaw CA, Li W, et al. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc Natl Acad Sci USA. 2015. April 28;112(17):5509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lagger S, Connelly JC, Schweikert G, Webb S, Selfridge J, Ramsahoye BH, et al. MeCP2 recognizes cytosine methylated tri-nucleotide and di-nucleotide sequences to tune transcription in the mammalian brain. PLoS Genet. 2017. May;13(5):e1006793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stroud H, Su SC, Hrvatin S, Greben AW, Renthal W, Boxer LD, et al. Early-Life Gene Expression in Neurons Modulates Lasting Epigenetic States. Cell. 2017. November 16;171(5):1151–1164.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heyn P, Logan CV, Fluteau A, Challis RC, Auchynnikava T, Martin C-A, et al. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nature Genetics. 2019. January;51(1):96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tatton-Brown K, Seal S, Ruark E, Harmer J, Ramsay E, Del Vecchio Duarte S, et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014. April;46(4):385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin B, Tao Q, Peng J, Soo HM, Wu W, Ying J, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet. 2008. March 1;17(5):690–709. [DOI] [PubMed] [Google Scholar]
- 49.Alex AM, Saradalekshmi KR, Shilen N, Suresh PA, Banerjee M. Genetic association of DNMT variants can play a critical role in defining the methylation patterns in autism. IUBMB Life. 2019;71(7):901–7. [DOI] [PubMed] [Google Scholar]
- 50.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2 , encoding methyl-CpG-binding protein 2. Nature Genetics. 1999. October;23(2):185–8. [DOI] [PubMed] [Google Scholar]
- 51.Neul JL, Kaufmann WE, Glaze DG, Christodoulou J, Clarke AJ, Bahi‐Buisson N, et al. Rett syndrome: Revised diagnostic criteria and nomenclature. Annals of Neurology. 2010;68(6):944–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santos F, Hendrich B, Reik W, Dean W. Dynamic Reprogramming of DNA Methylation in the Early Mouse Embryo. Developmental Biology. 2002. January 1;241(1):172–82. [DOI] [PubMed] [Google Scholar]
- 53.Guo JU, Su Y, Zhong C, Ming G, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011. April 29;145(3):423–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009. May 15;324(5929):930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011. September 2;333(6047):1300–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013. October 24;502(7472):472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weber AR, Krawczyk C, Robertson AB, Kuśnierczyk A, Vågbø CB, Schuermann D, et al. Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism. Nat Commun. 2016. March 2;7:10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He Y-F, Li B-Z, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011. September 2;333(6047):1303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szulwach KE, Li X, Li Y, Song C-X, Wu H, Dai Q, et al. 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011. December;14(12):1607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Münzel M, Globisch D, Brückl T, Wagner M, Welzmiller V, Michalakis S, et al. Quantification of the Sixth DNA Base Hydroxymethylcytosine in the Brain. Angewandte Chemie International Edition. 2010;49(31):5375–7. [DOI] [PubMed] [Google Scholar]
- 61.Kriaucionis S, Heintz N. The Nuclear DNA Base 5-Hydroxymethylcytosine Is Present in Purkinje Neurons and the Brain. Science. 2009. May 15;324(5929):929–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming G, et al. TET1 Controls CNS 5-Methylcytosine Hydroxylation, Active DNA Demethylation, Gene Transcription, and Memory Formation. Neuron. 2013. September 18;79(6):1086–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang R-R, Cui Q-Y, Murai K, Lim YC, Smith ZD, Jin S, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013. August 1;13(2):237–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mellén M, Ayata P, Heintz N. 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc Natl Acad Sci USA. 2017. 12;114(37):E7812–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011. 18;12(8):565–75. [DOI] [PubMed] [Google Scholar]
- 66.Stelzer Y, Wu H, Song Y, Shivalila CS, Markoulaki S, Jaenisch R. Parent-of-Origin DNA Methylation Dynamics during Mouse Development. Cell Rep. 2016. 20;16(12):3167–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993. November 25;366(6453):362–5. [DOI] [PubMed] [Google Scholar]
- 68.Takahashi N, Coluccio A, Thorball CW, Planet E, Shi H, Offner S, et al. ZNF445 is a primary regulator of genomic imprinting. Genes Dev. 2019. 01;33(1–2):49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu Q, Xiang Y, Wang Q, Wang L, Brind’Amour J, Bogutz AB, et al. SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat Genet. 2019;51(5):844–56. [DOI] [PubMed] [Google Scholar]
- 70.Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC, Erice Imprinting Group. Genomic Imprinting and Physiological Processes in Mammals. Cell. 2019. 21;176(5):952–65. [DOI] [PubMed] [Google Scholar]
- 71.Monk D, Mackay DJG, Eggermann T, Maher ER, Riccio A. Genomic imprinting disorders: lessons on how genome, epigenome and environment interact. Nat Rev Genet. 2019;20(4):235–48. [DOI] [PubMed] [Google Scholar]
- 72.Holm VA, Cassidy SB, Butler MG, Hanchett JM, Greenswag LR, Whitman BY, et al. Prader-Willi Syndrome: Consensus Diagnostic Criteria. Pediatrics. 1993. February;91(2):398–402. [PMC free article] [PubMed] [Google Scholar]
- 73.Yin Q-F, Yang L, Zhang Y, Xiang J-F, Wu Y-W, Carmichael GG, et al. Long Noncoding RNAs with snoRNA Ends. Molecular Cell. 2012. October 26;48(2):219–30. [DOI] [PubMed] [Google Scholar]
- 74.Margolis SS, Sell GL, Zbinden MA, Bird LM. Angelman Syndrome. Neurotherapeutics. 2015. July;12(3):641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu L, Li Y, Tollefsbol TO. Gene-Environment Interactions and Epigenetic Basis of Human Diseases. Curr Issues Mol Biol. 2008;10(1–2):25–36. [PMC free article] [PubMed] [Google Scholar]
- 76.Ma DK, Jang M-H, Guo JU, Kitabatake Y, Chang M, Pow-anpongkul N, et al. Neuronal Activity–Induced Gadd45b Promotes Epigenetic DNA Demethylation and Adult Neurogenesis. Science. 2009. February 20;323(5917):1074–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leach PT, Poplawski SG, Kenney JW, Hoffman B, Liebermann DA, Abel T, et al. Gadd45b knockout mice exhibit selective deficits in hippocampus-dependent long-term memory. Learn Mem. 2012. August 1;19(8):319–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic Deletion of gadd45b, a Regulator of Active DNA Demethylation, Enhances Long-Term Memory and Synaptic Plasticity. J Neurosci. 2012. November 28;32(48):17059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo JU, Ma DK, Mo H, Ball MP, Jang M-H, Bonaguidi MA, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011. August 28;14(10):1345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Altuna M, Urdánoz-Casado A, Sánchez-Ruiz de Gordoa J, Zelaya MV, Labarga A, Lepesant JMJ, et al. DNA methylation signature of human hippocampus in Alzheimer’s disease is linked to neurogenesis. Clin Epigenetics. 2019. June 19;11(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thomassin H, Flavin M, Espinás M-L, Grange T. Glucocorticoid-induced DNA demethylation and gene memory during development. The EMBO Journal. 2001. April 17;20(8):1974–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Provençal N, Arloth J, Cattaneo A, Anacker C, Cattane N, Wiechmann T, et al. Glucocorticoid exposure during hippocampal neurogenesis primes future stress response by inducing changes in DNA methylation. Proc Natl Acad Sci USA. 2019. August 9; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016. April 21;532(7599):329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yao B, Cheng Y, Wang Z, Li Y, Chen L, Huang L, et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat Commun. 2017. 24;8(1):1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974. May 24;184(4139):868–71. [DOI] [PubMed] [Google Scholar]
- 86.Arents G, Moudrianakis EN. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci USA. 1993. November 15;90(22):10489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997. September 18;389(6648):251–60. [DOI] [PubMed] [Google Scholar]
- 88.Bednar J, Garcia-Saez I, Boopathi R, Cutter AR, Papai G, Reymer A, et al. Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1. Mol Cell. 2017. May 4;66(3):384–397.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Syed SH, Goutte-Gattat D, Becker N, Meyer S, Shukla MS, Hayes JJ, et al. Single-base resolution mapping of H1-nucleosome interactions and 3D organization of the nucleosome. Proc Natl Acad Sci USA. 2010. May 25;107(21):9620–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007. June;17(6):877–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015. January 5;109:21.29.1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Su Y, Shin J, Zhong C, Wang S, Roychowdhury P, Lim J, et al. Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat Neurosci. 2017. March;20(3):476–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hickey SL, Berto S, Konopka G. Chromatin Decondensation by FOXP2 Promotes Human Neuron Maturation and Expression of Neurodevelopmental Disease Genes. Cell Reports. 2019. May 7;27(6):1699–1711.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Current Opinion in Cell Biology. 2001. June 1;13(3):263–73. [DOI] [PubMed] [Google Scholar]
- 95.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1(1):179–85. [DOI] [PubMed] [Google Scholar]
- 96.Orlando V Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends in Biochemical Sciences. 2000. March 1;25(3):99–104. [DOI] [PubMed] [Google Scholar]
- 97.Kim TH, Dekker J. ChIP-Quantitative Polymerase Chain Reaction (ChIP-qPCR). Cold Spring Harb Protoc. 2018. 01;2018(5). [DOI] [PubMed] [Google Scholar]
- 98.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007. August;4(8):651–7. [DOI] [PubMed] [Google Scholar]
- 99.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007. August;448(7153):553–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burney MJ, Johnston C, Wong K-Y, Teng S-W, Beglopoulos V, Stanton LW, et al. An epigenetic signature of developmental potential in neural stem cells and early neurons. Stem Cells. 2013. September;31(9):1868–80. [DOI] [PubMed] [Google Scholar]
- 101.Albert M, Kalebic N, Florio M, Lakshmanaperumal N, Haffner C, Brandl H, et al. Epigenome profiling and editing of neocortical progenitor cells during development. EMBO J. 2017. 01;36(17):2642–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006. April 21;125(2):315–26. [DOI] [PubMed] [Google Scholar]
- 103.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002. September 26;419(6905):407–11. [DOI] [PubMed] [Google Scholar]
- 104.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA. 2002. June 25;99(13):8695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gates LA, Shi J, Rohira AD, Feng Q, Zhu B, Bedford MT, et al. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem. 2017. July 17;jbc.M117.802074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lim DA, Huang Y-C, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009. March 26;458(7237):529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Potts MB, Siu JJ, Price JD, Salinas RD, Cho MJ, Ramos AD, et al. Analysis of Mll1 deficiency identifies neurogenic transcriptional modules and Brn4 as a factor for direct astrocyte-to-neuron reprogramming. Neurosurgery. 2014. October;75(4):472–82; discussion 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shah K, King GD, Jiang H. A chromatin modulator sustains self-renewal and enables differentiation of postnatal neural stem and progenitor cells. J Mol Cell Biol. 2019. May 8; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shilatifard A The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016. June 3;352(6290):aad9780. [DOI] [PubMed] [Google Scholar]
- 111.Schuettengruber B, Bourbon H-M, Di Croce L, Cavalli G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell. 2017. September 21;171(1):34–57. [DOI] [PubMed] [Google Scholar]
- 112.Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009. September 10;63(5):600–13. [DOI] [PubMed] [Google Scholar]
- 113.Morimoto-Suzki N, Hirabayashi Y, Tyssowski K, Shinga J, Vidal M, Koseki H, et al. The polycomb component Ring1B regulates the timed termination of subcerebral projection neuron production during mouse neocortical development. Development. 2014. November;141(22):4343–53. [DOI] [PubMed] [Google Scholar]
- 114.Tsuboi M, Kishi Y, Yokozeki W, Koseki H, Hirabayashi Y, Gotoh Y. Ubiquitination-Independent Repression of PRC1 Targets during Neuronal Fate Restriction in the Developing Mouse Neocortex. Dev Cell. 2018. 17;47(6):758–772.e5. [DOI] [PubMed] [Google Scholar]
- 115.Pereira JD, Sansom SN, Smith J, Dobenecker M-W, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci USA. 2010. September 7;107(36):15957–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hwang WW, Salinas RD, Siu JJ, Kelley KW, Delgado RN, Paredes MF, et al. Distinct and separable roles for EZH2 in neurogenic astroglia. Elife. 2014. May 27;3:e02439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang J, Ji F, Liu Y, Lei X, Li H, Ji G, et al. Ezh2 regulates adult hippocampal neurogenesis and memory. J Neurosci. 2014. April 9;34(15):5184–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sparmann A, Xie Y, Verhoeven E, Vermeulen M, Lancini C, Gargiulo G, et al. The chromodomain helicase Chd4 is required for Polycomb-mediated inhibition of astroglial differentiation. EMBO J. 2013. May 29;32(11):1598–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007. October;17(10):850–7. [DOI] [PubMed] [Google Scholar]
- 120.Park DH, Hong SJ, Salinas RD, Liu SJ, Sun SW, Sgualdino J, et al. Activation of neuronal gene expression by the JMJD3 demethylase is required for postnatal and adult brain neurogenesis. Cell Rep. 2014. September 11;8(5):1290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tang G-B, Zeng Y-Q, Liu P-P, Mi T-W, Zhang S-F, Dai S-K, et al. The Histone H3K27 Demethylase UTX Regulates Synaptic Plasticity and Cognitive Behaviors in Mice. Front Mol Neurosci. 2017;10:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pappa S, Padilla N, Iacobucci S, Vicioso M, Álvarez de la Campa E, Navarro C, et al. PHF2 histone demethylase prevents DNA damage and genome instability by controlling cell cycle progression of neural progenitors. Proc Natl Acad Sci USA. 2019. September 24;116(39):19464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, et al. Tetrahymena Histone Acetyltransferase A: A Homolog to Yeast Gcn5p Linking Histone Acetylation to Gene Activation. Cell. 1996. March 22;84(6):843–51. [DOI] [PubMed] [Google Scholar]
- 124.Mews P, Egervari G, Nativio R, Sidoli S, Donahue G, Lombroso SI, et al. Alcohol metabolism contributes to brain histone acetylation. Nature. 2019. October 23;1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Večeřa J, Bártová E, Krejčí J, Legartová S, Komůrková D, Rudá‐Kučerová J, et al. HDAC1 and HDAC3 underlie dynamic H3K9 acetylation during embryonic neurogenesis and in schizophrenia-like animals. Journal of Cellular Physiology. 2018;233(1):530–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008. February;8(1):57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013. May;14(5):347–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jones WD, Dafou D, McEntagart M, Woollard WJ, Elmslie FV, Holder-Espinasse M, et al. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am J Hum Genet. 2012. August 10;91(2):358–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lederer D, Grisart B, Digilio MC, Benoit V, Crespin M, Ghariani SC, et al. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am J Hum Genet. 2012. January 13;90(1):119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Miyake N, Mizuno S, Okamoto N, Ohashi H, Shiina M, Ogata K, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat. 2013. January;34(1):108–10. [DOI] [PubMed] [Google Scholar]
- 131.Millan F, Cho MT, Retterer K, Monaghan KG, Bai R, Vitazka P, et al. Whole exome sequencing reveals de novo pathogenic variants in KAT6A as a cause of a neurodevelopmental disorder. American Journal of Medical Genetics Part A. 2016;170(7):1791–8. [DOI] [PubMed] [Google Scholar]
- 132.Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014. November 13;515(7526):216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014. November 13;515(7526):209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Katayama Y, Nishiyama M, Shoji H, Ohkawa Y, Kawamura A, Sato T, et al. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016. 29;537(7622):675–9. [DOI] [PubMed] [Google Scholar]
- 135.Gompers AL, Su-Feher L, Ellegood J, Copping NA, Riyadh MA, Stradleigh TW, et al. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat Neurosci. 2017. August;20(8):1062–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gao Z, Lee P, Stafford JM, von Schimmelmann M, Schaefer A, Reinberg D. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature. 2014. December 18;516(7531):349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013. October 10;155(2):462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DTW, Kool M, et al. Reduced H3K27me3 and DNA Hypomethylation Are Major Drivers of Gene Expression in K27M Mutant Pediatric High-Grade Gliomas. Cancer Cell. 2013. November 11;24(5):660–72. [DOI] [PubMed] [Google Scholar]
- 139.Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013. May 17;340(6134):857–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mohammad F, Weissmann S, Leblanc B, Pandey DP, Højfeldt JW, Comet I, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med. 2017. April;23(4):483–92. [DOI] [PubMed] [Google Scholar]
- 141.Carullo NVN, Day JJ. Genomic Enhancers in Brain Health and Disease. Genes (Basel). 2019. 14;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Khoury G, Gruss P. Enhancer elements. Cell. 1983. June;33(2):313–4. [DOI] [PubMed] [Google Scholar]
- 143.Dekker J Gene Regulation in the Third Dimension. Science. 2008. March 28;319(5871):1793–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, et al. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Molecular Cell. 2019. November 7;76(3):473–484.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kim T-K, Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell. 2015. August 27;162(5):948–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015. March;16(3):144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gray JM, Kim T-K, West AE, Nord AS, Markenscoff-Papadimitriou E, Lomvardas S. Genomic Views of Transcriptional Enhancers: Essential Determinants of Cellular Identity and Activity-Dependent Responses in the CNS. J Neurosci. 2015. October 14;35(41):13819–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wu H, Nord AS, Akiyama JA, Shoukry M, Afzal V, Rubin EM, et al. Tissue-Specific RNA Expression Marks Distant-Acting Developmental Enhancers. PLOS Genetics. 2014. September 4;10(9):e1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010. May;465(7295):182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.De Gobbi M, Anguita E, Hughes J, Sloane-Stanley JA, Sharpe JA, Koch CM, et al. Tissue-specific histone modification and transcription factor binding in alpha globin gene expression. Blood. 2007. December 15;110(13):4503–10. [DOI] [PubMed] [Google Scholar]
- 151.Sati S, Cavalli G. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma. 2017. February 1;126(1):33–44. [DOI] [PubMed] [Google Scholar]
- 152.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009. October 9;326(5950):289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rowley MJ, Corces VG. Organizational principles of 3D genome architecture. Nat Rev Genet. 2018;19(12):789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012. April 11;485(7398):376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013. June 6;153(6):1281–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sun JH, Zhou L, Emerson DJ, Phyo SA, Titus KR, Gong W, et al. Disease-Associated Short Tandem Repeats Co-localize with Chromatin Domain Boundaries. Cell. 2018. 20;175(1):224–238.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017. October 19;171(3):557–572.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Short PJ, McRae JF, Gallone G, Sifrim A, Won H, Geschwind DH, et al. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature. 2018. March;555(7698):611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bertolini JA, Favaro R, Zhu Y, Pagin M, Ngan CY, Wong CH, et al. Mapping the Global Chromatin Connectivity Network for Sox2 Function in Neural Stem Cell Maintenance. Cell Stem Cell. 2019. March 7;24(3):462–476.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.de la Torre-Ubieta L, Stein JL, Won H, Opland CK, Liang D, Lu D, et al. The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell. 2018. 11;172(1–2):289–304.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense Transcription in the Mammalian Transcriptome. Science. 2005. September 2;309(5740):1564–6. [DOI] [PubMed] [Google Scholar]
- 162.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011. August 28;477(7364):295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bond AM, Vangompel MJW, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009. August;12(8):1020–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Berghoff EG, Clark MF, Chen S, Cajigas I, Leib DE, Kohtz JD. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development. 2013. November 1;140(21):4407–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cajigas I, Leib DE, Cochrane J, Luo H, Swyter KR, Chen S, et al. Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent inhibition of chromatin remodeling. Development. 2015. August 1;142(15):2641–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ramos AD, Diaz A, Nellore A, Delgado RN, Park K-Y, Gonzales-Roybal G, et al. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell. 2013. May 2;12(5):616–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz C, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015. April 2;16(4):439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Andersen RE, Hong SJ, Lim JJ, Cui M, Harpur BA, Hwang E, et al. The Long Noncoding RNA Pnky Is a Trans-acting Regulator of Cortical Development In Vivo. Dev Cell. 2019. May 20;49(4):632–642.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Aprea J, Prenninger S, Dori M, Ghosh T, Monasor LS, Wessendorf E, et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013. December 11;32(24):3145–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li W, Shen W, Zhang B, Tian K, Li Y, Mu L, et al. Long non-coding RNA LncKdm2b regulates cortical neuronal differentiation by cis-activating Kdm2b. Protein Cell. 2019. July 17; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016. April 14;17:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Field AR, Jacobs FMJ, Fiddes IT, Phillips APR, Reyes-Ortiz AM, LaMontagne E, et al. Structurally Conserved Primate LncRNAs Are Transiently Expressed during Human Cortical Differentiation and Influence Cell-Type-Specific Genes. Stem Cell Reports. 2019. 12;12(2):245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ng S-Y, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013. August 8;51(3):349–59. [DOI] [PubMed] [Google Scholar]
- 174.Jiang F, Doudna JA. CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys. 2017. 22;46:505–29. [DOI] [PubMed] [Google Scholar]
- 175.Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017. 06;355(6320). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Peschansky VJ, Pastori C, Zeier Z, Wentzel K, Velmeshev D, Magistri M, et al. The long non-coding RNA FMR4 promotes proliferation of human neural precursor cells and epigenetic regulation of gene expression in trans. Mol Cell Neurosci. 2016;74:49–57. [DOI] [PubMed] [Google Scholar]
- 177.Peschansky VJ, Pastori C, Zeier Z, Motti D, Wentzel K, Velmeshev D, et al. Changes in expression of the long non-coding RNA FMR4 associate with altered gene expression during differentiation of human neural precursor cells. Front Genet. 2015;6:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Luo T, Liu P, Wang X-Y, Li L-Z, Zhao L-P, Huang J, et al. Effect of the autism-associated lncRNA Shank2-AS on architecture and growth of neurons. J Cell Biochem. 2018. August 30; [DOI] [PubMed] [Google Scholar]
- 179.Ang CE, Ma Q, Wapinski OL, Fan S, Flynn RA, Lee QY, et al. The novel lncRNA lnc-NR2F1 is pro-neurogenic and mutated in human neurodevelopmental disorders. Elife. 2019. 10;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Peer E, Rechavi G, Dominissini D. Epitranscriptomics: regulation of mRNA metabolism through modifications. Current Opinion in Chemical Biology. 2017. December 1;41:93–8. [DOI] [PubMed] [Google Scholar]
- 181.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011. October 16;7(12):885–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013. January 10;49(1):18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013. February;29(2):108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012. June 22;149(7):1635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012. April 29;485(7397):201–6. [DOI] [PubMed] [Google Scholar]
- 187.Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, et al. Accurate detection of m 6 A RNA modifications in native RNA sequences. Nat Commun. 2019. September 9;10(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yoon K-J, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, et al. Temporal Control of Mammalian Cortical Neurogenesis by m6A Methylation. Cell. 2017. November 2;171(4):877–889.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, et al. Ythdf2-mediated m6A mRNA clearance modulates neural development in mice. Genome Biol. 2018. 31;19(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yu J, Chen M, Huang H, Zhu J, Song H, Zhu J, et al. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2018. 16;46(3):1412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhuang M, Li X, Zhu J, Zhang J, Niu F, Liang F, et al. The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 2019. May 21;47(9):4765–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H, et al. FMRP Modulates Neural Differentiation through m6A-Dependent mRNA Nuclear Export. Cell Reports. 2019. July 23;28(4):845–854.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chen J, Zhang Y-C, Huang C, Shen H, Sun B, Cheng X, et al. m6A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2. Genomics Proteomics Bioinformatics. 2019. April;17(2):154–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shi H, Zhang X, Weng Y-L, Lu Z, Liu Y, Lu Z, et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563(7730):249–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007. May 11;316(5826):889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Saucedo-Uribe E, Genis-Mendoza AD, Díaz-Anzaldúa A, Martínez-Magaña JJ, Tovilla-Zarate CA, Juárez-Rojop I, et al. Differential effects on neurodevelopment of FTO variants in obesity and bipolar disorder suggested by in silico prediction of functional impact: An analysis in Mexican population. Brain Behav. 2019;9(6):e01249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GSH, et al. Loss-of-Function Mutation in the Dioxygenase-Encoding FTO Gene Causes Severe Growth Retardation and Multiple Malformations. The American Journal of Human Genetics. 2009. July 10;85(1):106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chokkalla Anil K., Mehta Suresh L., Kim TaeHee, Chelluboina Bharath, Kim Jooyong, Vemuganti Raghu. Transient Focal Ischemia Significantly Alters the m6A Epitranscriptomic Tagging of RNAs in the Brain. Stroke. 2019. October 1;50(10):2912–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, et al. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992. September;1(6):397–400. [DOI] [PubMed] [Google Scholar]
- 200.Pastori C, Peschansky VJ, Barbouth D, Mehta A, Silva JP, Wahlestedt C. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum Genet. 2014. January;133(1):59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Guo Y, Chen X, Xing R, Wang M, Zhu X, Guo W. Interplay between FMRP and lncRNA TUG1 regulates axonal development through mediating SnoN-Ccd1 pathway. Hum Mol Genet. 2018. 01;27(3):475–85. [DOI] [PubMed] [Google Scholar]
- 202.Zhang F, Kang Y, Wang M, Li Y, Xu T, Yang W, et al. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum Mol Genet. 2018. 15;27(22):3936–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Macaulay IC, Ponting CP, Voet T. Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends in Genetics. 2017. February 1;33(2):155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nature Methods. 2016. March;13(3):229–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hu Y, Huang K, An Q, Du G, Hu G, Xue J, et al. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 2016. May 5;17:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Guo F, Li L, Li J, Wu X, Hu B, Zhu P, et al. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Research. 2017. August;27(8):967–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Clark SJ, Argelaguet R, Kapourani C-A, Stubbs TM, Lee HJ, Alda-Catalinas C, et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nature Communications. 2018. February 22;9(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Rubin AJ, Parker KR, Satpathy AT, Qi Y, Wu B, Ong AJ, et al. Coupled Single-Cell CRISPR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks. Cell. 2019. January 10;176(1):361–376.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Crawley JN. Translational animal models of autism and neurodevelopmental disorders. Dialogues Clin Neurosci. 2012. September;14(3):293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Liu J, Li K, Cai J, Zhang M, Zhang X, Xiong X, et al. Landscape and Regulation of m6A and m6Am Methylome across Human and Mouse Tissues. Mol Cell. 2019. October 18; [DOI] [PubMed] [Google Scholar]
- 211.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006. August 25;126(4):663–76. [DOI] [PubMed] [Google Scholar]
- 212.Marchetto MCN, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A Model for Neural Development and Treatment of Rett Syndrome Using Human Induced Pluripotent Stem Cells. Cell. 2010. November 12;143(4):527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wang X, Ye F, Wen Z, Guo Z, Yu C, Huang W-K, et al. Structural interaction between DISC1 and ATF4 underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders. Mol Psychiatry. 2019. August 23; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013. September;501(7467):373–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell–derived neocortex. PNAS. 2013. December 10;110(50):20284–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015. July;12(7):671–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016. May 19;165(5):1238–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bershteyn M, Nowakowski TJ, Pollen AA, Lullo ED, Nene A, Wynshaw-Boris A, et al. Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell. 2017. April 6;20(4):435–449.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ye F, Kang E, Yu C, Qian X, Jacob F, Yu C, et al. DISC1 Regulates Neurogenesis via Modulating Kinetochore Attachment of Ndel1/Nde1 during Mitosis. Neuron. 2017. December 6;96(5):1041–1054.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]