Abstract
To determine antimicrobial resistance, 431 samples of retail foods purchased at different supermarkets in Northern Xinjiang were examined in this study. There were 112 Escherichia coli strains that were isolated, with approximately 26% of the samples contaminated by E. coli. The detection rate of E. coli isolated from pork was the highest (59.6%), followed by mutton (52.6%), retail fresh milk (52.4%), duck (36.4%), beef (35.3%), chicken (33.3%), and ready‐to‐eat food (12.9%); the E. coli detection rate for fish and vegetables was <11%. The result showed that the 112 isolates were mostly resistant to tetracycline (52%), followed by ampicillin (42%), compound trimethoprim/sulfamethoxazole (37%), amoxicillin (33%), and nalidixic acid (32%), imipenem resistance was not detected. One hundred isolates carried at least one antimicrobial resistance gene. The detection rate of resistance genes of our study was as follows: tetA (38%), tetB (27%), bla OXA (40%), bla TEM (20%), floR (20%), sul1 (16%), sul2 (27%), aad Ala (19%), aadB (11%), strA (28%), and strB (24%); tetC and bla PSE were not detected. Virulence genes fimC, agg, stx2, fimA, fyuA, papA, stx1, and eaeA were found in 52, 34, 21, 19, 6, 3, 2, and 2 isolates, respectively; papC was not detected. There was a statistically significant association between fimC and resistance to ciprofloxacin (p = .001), gentamicin (p = .001), amikacin (p = .001), levofloxacin (p = .001), and streptomycin (p = .001); between fimA and resistance to tetracycline (p = .001), ampicillin (p = .001), compound trimethoprim/sulfamethoxazole (p = .001), and amoxicillin (p = .003); between agg and resistance to gentamicin (p = .001), tetracycline (p = .001), ciprofloxacin (p = .017), and levofloxacin (p = .001); and between stx2 and resistance to ampicillin (p = .001), tetracycline (p = .001), compound trimethoprim/sulfamethoxazole (p = .002), and amoxicillin (p = .015).
Keywords: Escherichia coli, multidrug resistance, resistance gene, virulence gene
The drug resistance and virulence genes of Escherichia coli isolated from retail food in northern Xinjiang, China, were studied, and the relationship between them was analyzed to reveal the resistance of foodborne E. coli to veterinary and clinical commonly used drugs in the region. The paper provided reference materials to guide the rational use of antibiotics in clinical and animal feeding and control the spread of drug‐resistant strains in nature.
1. INTRODUCTION
It is well known that Escherichia coli mainly exists in the human and animal gastrointestinal tract. It also occurs in the natural environment, especially in soil, water, and plants (Katarzyna & Anna, 2016). Therefore, it is not surprising that some of the E. coli in the environment reinfects humans through vegetable‐ or animal‐derived foods.
Escherichia coli is a highly diverse virulent species that is widely distributed in open systems, is easy to spread in the environment, and can be harmful to human health (Tenaillon, Skurnik, Picard, & Denamur, 2010). Drug resistance genes carried by E. coli can be transferred to other pathogenic bacteria, and, due to the excessive use of antibiotics, selection pressure is very high, resulting in bacterial strains resistant to a variety of drugs. Multi‐drug‐resistant strains are characterized by the presence of multiple genes conferring drug resistance, which results in insensitivity to many different drug groups (Hu, Yang, & Li, 2016; Rasheed, Thajuddin, Ahamed, Teklemariam, & Jamil, 2014).
Genetic mutations or genetic acquisition of antibiotic resistance genes (ARG) through horizontal gene transfer might also result in the occurrence of antibiotic‐resistant bacteria (ARB) throughout the environment (Céline & David, 2015). This has resulted in the emergence of many different ARG, including the dfr and sul genes related to trimethoprim and sulfamethoxazole resistance, respectively (Chang, Lin, Chang, & Lu, 2007; Ho, Wang, Chow, & Que, 2009), and other genes, such as ampC, oxa2, and tetA.
The ever‐increasing threat of ARB may be associated with enhanced virulence (Guillard, Pons, Roux, Pier, & Skurnik, 2016; Roux et al., 2015), and with the increase in antibiotic resistance, an increase in virulence may naturally evolve. Therefore, when controlling the spread of antibiotic resistance, we must also control the spread of virulence (Meredith, Brooks, & Brooks, 2017). Although the profile of virulence and antimicrobial resistance genes of E. coli from foods has been reported (Luo, Ji, & Wang, 2016), the data elucidating the association between these two gene sets are lacking.
In Xinjiang, China, a previous study conducted antibiotic resistance research on foodborne E. coli based on samples from slaughterhouses, butcher shops, and farms (Xia, Xiang, & Guo, 2014; Yao, Long, Kuerbannaimu, Wang, & Xia, 2017). However, little is known about the resistance of those bacteria in retail foods.
There have been some reports describing the antimicrobial resistance and virulence of E. coli, such as Arisoy, Rad, Akin, and Akar (2008), who showed that the virulence genes afaI, pap, hly, aer, and sfa were increased in sensitive strains. However, detailed information on the relationship between antimicrobial resistance genes and virulence genes of E. coli isolated from retail foods in Xinjiang is scarce.
The purpose of this study was to evaluate the drug resistance of E. coli strains isolated from retail foods in northern Xinjiang, identify their virulence genes, and determine the possible relationship between the virulence genes and drug resistance.
2. MATERIALS AND METHODS
2.1. Sampling and E. coli isolation
A total of 431 food samples were purchased at supermarkets in Shihezi, Kuitun, and Urumqi, in northern Xinjiang, China, from 2014 to 2016, and each type of sample and its number are listed in Table 1. Each sample weighed 25 g and was placed in a sterile plastic bag containing 225 ml of sterilized sodium chloride solution (0.85%) and then homogenized for 90 s using a BagMixer 400 CC beating homogenizer. Lauryl Sulfate Tryptose (LST) broth was inoculated with 1 ml of homogenate and incubated for 48 hr at 37 ± 1°C. Gas‐positive tubes were inoculated into 100 ml of E. coli (EC) broth and incubated at 44 ± 0.5°C for 48 hr (Wang, Sun, & Ji, 2014). After that, one loopful from each gas‐positive tube was streaked onto eosin methylene blue agar. Presumptive E. coli colonies were streaked onto Luria–Bertani nutrient agar and incubated for 12–48 hr at 36 ± 1°C. Each culture was confirmed as E. coli through an IMViC test. E. coli ATCC 25922 was used as a positive control for polymerase chain reaction (PCR) of UidA. Template was prepared via the boiling method, for the amplification of selected UidA genes in E. coli using PCR (Heijnen & Medema, 2006). The oligonucleotide sequences used and the predicted sizes of PCR amplification products of genes are listed in Table 2.
Table 1.
The original number of samples
| Number | Sampling number | Origin | Number | Sampling number | Origin | Number | Sampling number | Origin |
|---|---|---|---|---|---|---|---|---|
| 1 | K1 | Pig heart | 145 | K3 | Celery | 289 | K15 | Duck |
| 2 | K2 | Pork | 146 | K5 | Broccoli | 290 | K16 | Duck |
| 3 | K4 | Pork liver | 147 | K7 | Lettuce | 291 | K17 | Duck leg |
| 4 | K6 | Pork | 148 | K11 | Tomato | 292 | K19 | Duck |
| 5 | K8 | Pork | 149 | K12 | Pepper | 293 | K20 | Duck |
| 6 | K9 | Pork | 150 | K14 | Cabbage | 294 | K24 | Duck |
| 7 | K10 | Pork stuffing | 151 | K21 | Ginger | 295 | K25 | Duck |
| 8 | K13 | Porcine blood | 152 | K22 | Celery | 296 | K27 | Duck |
| 9 | K18 | Pork | 153 | K23 | Pepper | 297 | K35 | Duck |
| 10 | K33 | Porcine blood | 154 | K26 | Cabbage | 298 | W7 | Duck |
| 11 | K34 | Pork | 155 | W1 | Broccoli | 299 | W12 | Duck |
| 12 | K40 | Pork liver | 156 | W4 | Lettuce | 300 | N4 | Fish |
| 13 | W2 | Pork intestine | 157 | W5 | Pepper | 301 | N5 | Fish |
| 14 | W3 | Pork liver | 158 | N1 | Ginger | 302 | N8 | Fish |
| 15 | W6 | Porcine blood | 159 | N2 | Broccoli | 303 | N14 | Fish |
| 16 | W8 | Pigtail | 160 | N3 | Eggplant | 304 | N15 | Fish |
| 17 | W9 | Pork | 161 | S18 | Spinach | 305 | N16 | Crustacean |
| 18 | W10 | Pork fillet | 162 | S19 | Celery | 306 | N17 | Fish |
| 19 | W11 | Pork liver | 163 | N6 | Shallot | 307 | W17 | Fish |
| 20 | W13 | Pork | 164 | N7 | Tomato | 308 | W18 | Fish |
| 21 | W14 | Pork | 165 | N9 | Lettuce | 309 | W61 | Fish |
| 22 | W15 | Pork | 166 | W21 | Tomato | 310 | W62 | Fish |
| 23 | W16 | Pork | 167 | H11 | Ginger | 311 | W63 | Fish |
| 24 | W19 | Pork | 168 | N52 | Cowpea | 312 | K36 | Fish |
| 25 | W20 | Pork | 169 | H14 | Spinach | 313 | K37 | Fish |
| 26 | W25 | Porcine blood | 170 | H15 | Broccoli | 314 | S1 | Fish |
| 27 | W26 | Porcine blood | 171 | H16 | Pepper | 315 | S2 | Fish |
| 28 | S5 | Pork | 172 | H17 | Shallot | 316 | S3 | Fish |
| 29 | S8 | Pig heart | 173 | Tomato | 317 | S4 | Fish | |
| 30 | S9 | Pork stuffing | 174 | W22 | Eggplant | 318 | W64 | Fish |
| 31 | S10 | Pork fillet | 175 | W23 | Spinach | 319 | W65 | Fish |
| 32 | S12 | Pork liver | 176 | W24 | Tomato | 320 | W66 | Fish |
| 33 | S14 | Pig hind leg | 177 | W67 | Celery | 321 | W69 | Fish |
| 34 | S15 | Pork | 178 | W68 | Ginger | 322 | W72 | Fish |
| 35 | S16 | Pork liver | 179 | W70 | Shallot | 323 | W73 | Fish |
| 36 | S17 | Pork | 180 | W71 | Cowpea | 324 | W75 | Fish |
| 37 | H2 | Pork intestine | 181 | W74 | Tomato | 325 | W54 | Fish |
| 38 | H4 | Pork | 182 | W76 | Pepper | 326 | W55 | Fish |
| 39 | H5 | Pork | 183 | K38 | Broccoli | 327 | W56 | Fish |
| 40 | H6 | Porcine blood | 184 | K39 | Ginger | 328 | S6 | Fish |
| 41 | H7 | Pig trotters | 185 | K41 | Shallot | 329 | S7 | Fish |
| 42 | H8 | Porcine blood | 186 | W77 | Lettuce | 330 | S11 | Brine shrimp |
| 43 | H9 | Pork | 187 | W78 | Cowpea | 331 | N10 | Bean curd skin |
| 44 | H12 | Porcine blood | 188 | W79 | Spinach | 332 | N11 | Marinated tofu |
| 45 | H13 | Pork | 189 | W80 | Eggplant | 333 | N12 | Stewed chicken leg |
| 46 | H23 | Porcine blood | 190 | S13 | Tomato | 334 | N13 | Stewed beef |
| 47 | H24 | Pork liver | 191 | H1 | Shallot | 335 | N51 | Red oil chicken gizzards |
| 48 | H27 | Pork | 192 | H3 | Celery | 336 | K42 | Hot and sour gluten |
| 49 | H28 | Pork | 193 | H10 | Ginger | 337 | K43 | Marinated chicken leg |
| 50 | H30 | Pork | 194 | W28 | Pepper | 338 | K45 | Cold bamboo shoots |
| 51 | H33 | Pork | 195 | W29 | Broccoli | 339 | K74 | Soy sauce pickles |
| 52 | H34 | Pork | 196 | W34 | Tomato | 340 | K75 | Spiced gizzard |
| 53 | K28 | Celery | 197 | H66 | Lettuce | 341 | K76 | Beef salad |
| 54 | K29 | Shallot | 198 | H67 | Shallot | 342 | K77 | Beef tendon in cold sauce |
| 55 | K30 | Spinach | 199 | H68 | Eggplant | 343 | K78 | Cold bamboo shoots |
| 56 | N46 | Potato | 200 | H69 | Ginger | 344 | K79 | Bean salad |
| 57 | N47 | Eggplant | 201 | H70 | Spinach | 345 | S22 | Fungus salad |
| 58 | N48 | Spinach | 202 | H71 | Cowpea | 346 | S23 | Kelp salad |
| 59 | N49 | Shallot | 203 | H72 | Tomato | 347 | K80 | Bean curd skin in cold sauce |
| 60 | W52 | Cowpea | 204 | H73 | Coriander | 348 | K81 | Kelp salad |
| 61 | W53 | Bitter gourd | 205 | H74 | Snow pea | 349 | W32 | Shredded lotus root slice |
| 62 | W57 | Eggplant | 206 | H75 | Lettuce | 350 | W33 | Spiced gizzard |
| 63 | S20 | Flammulina velutipes mushroom | 207 | N18 | Drumsticks | 351 | H18 | Pea noodles |
| 64 | S21 | Celery | 208 | N19 | Chicken wings | 352 | H19 | Dried bean curd |
| 65 | S24 | Zhaer root | 209 | N20 | Drumsticks | 353 | H20 | Bean curd |
| 66 | S25 | Lettuce | 210 | N21 | Chicken gizzard | 354 | H26 | Red ear silk |
| 67 | S26 | Chinese cabbage | 211 | N22 | Chicken | 355 | H29 | Chicken salad |
| 68 | S27 | Bok choy | 212 | H21 | Drumsticks | 356 | H30 | Sweet potato |
| 69 | S28 | Ginger | 213 | H22 | Chicken wings | 357 | S95 | Chinese wolfberries |
| 70 | S47 | Tomato | 214 | K44 | Chicken gizzard | 358 | S96 | Cold bean curd |
| 71 | S48 | Bitter gourd | 215 | K46 | Chicken | 359 | S97 | Bean curd skin |
| 72 | S49 | Black fungus | 216 | H23 | Chicken wing | 360 | S98 | Gluten |
| 73 | S50 | Garlic sprouts | 217 | S53 | Drumsticks | 361 | S99 | Cold pig ears |
| 74 | S51 | Chive | 218 | N53 | Chicken | 362 | S100 | Peanut salad |
| 75 | S52 | Coriander | 219 | N54 | Chicken wing | 363 | H76 | Cold bamboo shoots |
| 76 | N55 | Broccoli | 220 | S64 | Drumsticks | 364 | H77 | Marinated tofu |
| 77 | N56 | Celery | 221 | S65 | Chicken gizzard | 365 | H78 | Spicy dried tofu |
| 78 | S61 | Pepper | 222 | S66 | Chicken | 366 | K47 | Spicy dried tofu |
| 79 | S62 | Coriander | 223 | S67 | Drumsticks | 367 | K64 | Red oil ear silk |
| 80 | S63 | Green Chinese onion | 224 | S68 | Chicken wings | 368 | K65 | Cold bean curd stick |
| 81 | H24 | Bitter gourd | 225 | W35 | Drumsticks | 369 | K66 | Dried vegetables |
| 82 | H25 | Lentinus edodes mushroom | 226 | W38 | Chicken wings | 370 | K67 | Brine shrimp |
| 83 | H27 | Pepper | 227 | S69 | Drumsticks | 371 | K71 | Bean curd skin |
| 84 | H28 | Kelp | 228 | S70 | Chicken gizzard | 372 | K72 | Chicken skewer |
| 85 | H31 | Pepper | 229 | S71 | Chicken | 373 | K73 | Hot and sour gluten |
| 86 | S72 | Bean sprouts | 230 | S29 | Chicken wings | 374 | W36 | Marinated tofu |
| 87 | S73 | Coprinus comatus mushroom | 231 | S30 | Chicken | 375 | W37 | Stewed pork liver |
| 88 | S74 | Romaine lettuce | 232 | H41 | Chicken wings | 376 | S34 | Stewed beef |
| 89 | S75 | Coriander | 233 | H42 | Drumsticks | 377 | S35 | Stewed chicken leg |
| 90 | S76 | Tomatoes | 234 | H43 | Drumsticks | 378 | S36 | Marinated tofu |
| 91 | S77 | Pepper | 235 | H44 | Chicken wings | 379 | S54 | Brine shrimp |
| 92 | S78 | Celery | 236 | H60 | Chicken gizzard | 380 | S55 | Bean curd skin |
| 93 | S79 | Lotus root | 237 | S81 | Drumsticks | 381 | S56 | Chicken skewer |
| 94 | S80 | Cabbage | 238 | S82 | Chicken | 382 | S57 | Marinated chicken leg |
| 95 | S89 | Cucumber | 239 | S83 | Chicken gizzard | 383 | N34 | Marinated tofu |
| 96 | S90 | Celery | 240 | S84 | Chicken wings | 384 | N35 | Stewed beef |
| 97 | S91 | Garlic sprouts | 241 | S85 | Chicken gizzard | 385 | N36 | Stewed beef |
| 98 | S92 | Spinach | 242 | S86 | Drumsticks | 386 | N37 | Hot and sour gluten |
| 99 | S93 | Towel gourd | 243 | S87 | Drumsticks | 387 | N38 | Marinated chicken leg |
| 100 | S94 | Peas | 244 | S88 | Drumsticks | 388 | N45 | Stewed chicken leg |
| 101 | K48 | Chives | 245 | K32 | Chicken wings | 389 | N50 | Stewed pork liver |
| 102 | K49 | Garlic sprouts | 246 | W27 | Chicken | 390 | K61 | Marinated tofu |
| 103 | K52 | Lettuce | 247 | W30 | Drumsticks | 391 | K62 | Stewed pork liver |
| 104 | K68 | Pepper | 248 | W31 | Chicken wings | 392 | K63 | Lamb tripe |
| 105 | K69 | Cucumber | 249 | K53 | Chicken | 393 | K31 | Mutton |
| 106 | K70 | Lettuce | 250 | K54 | Chicken | 394 | W39 | Mutton |
| 107 | H40 | Cucumber | 251 | K59 | Drumsticks | 395 | W46 | Mutton |
| 108 | H45 | Pepper | 252 | K60 | Chicken gizzard | 396 | W51 | Sheep heart |
| 109 | H48 | Peas | 253 | W47 | Chicken gizzard | 397 | W63 | Mutton |
| 110 | H50 | Cucumber | 254 | W48 | Drumsticks | 398 | W64 | Mutton |
| 111 | H56 | Lettuce | 255 | K50 | Beef | 399 | W65 | Mutton |
| 112 | H57 | Towel gourd | 256 | K51 | Beef | 400 | W66 | Mutton |
| 113 | H58 | Pepper | 257 | W47 | Beef | 401 | S39 | Mutton |
| 114 | H59 | Peas | 258 | W48 | Beef stuffing | 402 | S40 | Mutton |
| 115 | W40 | Chives | 259 | N23 | Beef | 403 | S41 | Mutton |
| 116 | W43 | Spinach | 260 | N24 | Beef | 404 | S44 | Mutton |
| 117 | W45 | Pepper | 261 | N25 | Beef | 405 | S58 | Mutton |
| 118 | W60 | Towel gourd | 262 | N26 | Beef | 406 | S59 | Mutton |
| 119 | W61 | Spinach | 263 | N27 | Beef | 407 | S60 | Mutton |
| 120 | W62 | Cucumber | 264 | H32 | Beef | 408 | N31 | Mutton |
| 121 | S42 | Celery | 265 | H33 | Beef | 409 | N32 | Mutton |
| 122 | S43 | Chives | 266 | H34 | Beef | 410 | N33 | Mutton |
| 123 | N28 | Peas | 267 | H61 | Beef | 411 | R1 | Retail fresh milk |
| 124 | N29 | Lettuce | 268 | H62 | Beef | 412 | R2 | Retail fresh milk |
| 125 | N30 | Pepper | 269 | H63 | Beef | 413 | R3 | Retail fresh milk |
| 126 | S31 | Towel gourd | 270 | H64 | Beef | 414 | R4 | Retail fresh milk |
| 127 | S32 | Pepper | 271 | H65 | Beef | 415 | R5 | Retail fresh milk |
| 128 | S33 | Lettuce | 272 | W44 | Beef stuffing | 416 | R6 | Retail fresh milk |
| 129 | W41 | Cucumber | 273 | S37 | Beef stuffing | 417 | R7 | Retail fresh milk |
| 130 | W42 | Peas | 274 | S38 | Beef | 418 | R8 | Retail fresh milk |
| 131 | N39 | Lettuce | 275 | S45 | Beef | 419 | R9 | Retail fresh milk |
| 132 | N40 | Lettuce | 276 | S46 | Beef | 420 | R10 | Retail fresh milk |
| 133 | K55 | Pepper | 277 | S50 | Beef | 421 | R11 | Retail fresh milk |
| 134 | K57 | Chives | 278 | S51 | Beef | 422 | R12 | Retail fresh milk |
| 135 | S47 | Towel gourd | 279 | S53 | Beef | 423 | R13 | Retail fresh milk |
| 136 | S48 | Lettuce | 280 | K56 | Beef | 424 | R14 | Retail fresh milk |
| 137 | S52 | Cucumber | 281 | K58 | Beef | 425 | R15 | Retail fresh milk |
| 138 | N41 | Spinach | 282 | S49 | Beef | 426 | R19 | Retail fresh milk |
| 139 | N42 | Pepper | 283 | H36 | Beef | 427 | R20 | Retail fresh milk |
| 140 | N43 | Cucumber | 284 | H37 | Beef | 428 | R21 | Retail fresh milk |
| 141 | N44 | Cucumber | 285 | W59 | Beef | 429 | R23 | Retail fresh milk |
| 142 | W49 | Chives | 286 | W60 | Beef | 430 | R26 | Retail fresh milk |
| 143 | W50 | Spinach | 287 | H38 | Beef | 431 | R31 | Retail fresh milk |
| 144 | H35 | Towel gourd | 288 | H39 | Beef |
H, supermarket sampling in Shihezi; K, samples collected from Kuitun; N, sampling in cooperation with Inspection Institute; R, retail fresh milk collected from Shihezi; S, samples collected from Shihezi; W, samples collected from Urumqi.
Table 2.
Primers used for detection of genes encoding resistance to different antimicrobials
| Gene | Primer | DNA sequence (5′ → 3′) | Size (bp) | Thermocycling conditions | References |
|---|---|---|---|---|---|
| UidA | UidAF | 5′‐ATGGAATTTCGCCGATTTTGC‐3′ | 194 | 95°C for 5 min, 40 cycles of 95°C for 30 s, 60°C for 1 min, 72°C for 1 min, and final extension at 72°C for 7 min | Heijnen and Medema (2006) |
| UidAR | 5′‐ATTGTTTGCCTCCCTGCTGC‐3′ | ||||
| tetA | tetA‐F | 5′‐GCTACATCCTGCTTGCCTTC‐3′ | 210 | 95°C for 5 min, 30 cycles of 94°C for 30 s, 60°C for 1 min, 72°C for 1 min, and final extension at 72°C for 5 min | Ng, Martin, Alfo, and Mulvey (2001) |
| tetA‐R | 5′‐CATAGATCGCCGTGAAGAGG‐3′ | Ng et al. (2001) | |||
| tetB | tetB‐F | 5′‐TTGGTTAGGGGCAAGTTTTG‐3′ | 659 | Ng et al. (2001) | |
| tetB‐R | 5′‐GTAATGGGCCAATAACACCG‐3′ | Ng et al. (2001) | |||
| tetC | tetC‐F | 5′‐CTTGAGAGCCTTCAACCCAG‐3′ | 418 | Sáenz et al. (2004) | |
| tetC‐R | 5′‐ATGGTCGTCATCTACCTGCC‐3′ | Sáenz et al. (2004) | |||
| bla TEM | bla TEM‐F | 5′‐TTGGGTGCACGACTGGGT‐3′ | 503 | 95°C for 5 min, 30 cycles of 94°C for 1 min, 60°C for 1 min, 72°C for 1 min, and final extension at 72°C for 5 min | Knapp, Dolfing, Ehlert, and Graham (2010) |
| bla TEM‐R | 5′‐TAATTGTTGCCGGGAAGC‐3′ | Knapp et al. (2010) | |||
| bla PSE | bla PSE ‐F | 5′‐CGCTTCGGGTTAACAAGTAC‐3′ | 419 | Zhi, Xi, and Shen (2009) | |
| bla PSE ‐R | 5′‐CTGGTTCATTTCAGATAGCG‐3′ | Zhi et al. (2009) | |||
| bla OXA | bla OXA‐F | 5′‐AGCAGCGCCAGTGCATCA‐3′ | 708 | Guerra et al. (2003( | |
| bla OXA‐R | 5′‐ATTCGACCCCAAGTTTCC‐3′ | Guerra et al. (2003) | |||
| floR | floR‐F | 5′‐CACGTTGAGCCTCTATAT‐3′ | 868 | 95°C for 5 min, 30 cycles of 94°C for1 min, 52°C for 1 min, 72°C for 1 min, and final extension at 72°C for 10 min | Sáenz et al. (2004) |
| floR‐R | 5′‐ATGCAGAAGTAGAACGCG‐3′ | Sáenz et al. (2004) | |||
| sul1 | Sul1‐F | 5′‐CGGCGTGGGCTACCTGAACG‐3′ | 433 | 94°C for 5 min, 30 cycles of 94°C for 15 s, 69°C for 30 s, 72°C for 1 min, and final extension at 72°C for 7 min | Sáenz et al. (2004) |
| Sul1‐R | 5′‐GCCGATCGCGTGAAGTTCCG‐3′ | Sáenz et al. (2004) | |||
| sul2 | Sul2‐F | 5′‐GCGCTCAAGGCAGATGGCATT‐3′ | 285 | Sáenz et al. (2004) | |
| Sul2‐R | 5′‐GCGTTTGATACCGGCACCCGT‐3′ | Sáenz et al. (2004) | |||
| aad Ala | aad Ala ‐F | 5′‐AACGACCTTTTGGAAACTTCGG−3′ | 352 | 94°C for 10 min, 35 cycles of 94°C for 1 min, 60°C for 30 s, 72°C for 1 min, and final extension at 72°C for 10 min | Sáenz et al. (2004) |
| aad Ala ‐R | 5′‐TTCGCTCATCGCCAGCCCAG‐3′ | Sáenz et al. (2004) | |||
| aadB | AadB‐F | 5′‐GGGCGCGTCATGGAGGAGTT‐3′ | 329 | 94°C for 10 min, 35 cycles of 94°C for 1 min, 65°C for 30 s, 72°C for 1 min, and final extension at 72°C for 10 min | Rosengren, Waldner, and Reid‐Smith (2009) |
| aadB‐R | 5′‐TATCGCGACCTGAAAGCGGC‐3′ | Rosengren et al. (2009) | |||
| strA | StrA‐F | 5′‐CCTGGTGATAACGGCAATTC‐3′ | 546 | 95°C for 4 min, 35 cycles of 95°C for 1 min, 55°C for 1 min, 72°C for 1 min, and final extension at 72°C for 7 min | Rosengren et al. (2009) |
| StrA‐R | 5′‐CCAATCGCAGATAGAAGGC‐3′ | Rosengren et al. (2009) | |||
| strB | StrB‐F | 5′‐ATCGTCAAGGGATTGAAACC‐3′ | 509 | Rosengren et al. (2009) | |
| StrB‐R | 5′‐GGATCGTAGAACATATTGGC‐3′ | Rosengren et al. (2009) |
2.2. Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed utilizing the disk‐diffusion method as recommended by the Clinical and Laboratory Standards Institute (CLSI, 2015). The following antibiotics were used: ampicillin (AMP: 10 μg/p), cefotaxime (CTX: 30 μg/p), ceftazidime (CAZ: 30 μg/p), gentamicin (GEN: 10 μg/p), imipenem (IPM: 10 μg/p), ciprofloxacin (CIP: 5 μg/p), levofloxacin (LEV: 5 μg/p), tetracycline (TET: 30 μg/p), chloramphenicol (CHL: 30 μg/p), amikacin (AMK: 30 μg/p), piperacillin (PIP: 100 μg/p), compound trimethoprim/sulfamethoxazole (T/S: 23.75 μg/1.25 μg/p), erythromycin (ERY: 15 μg/p), amoxicillin (AMX: 10 μg/p), streptomycin (STR: 10 μg/p), nalidixic acid (NAL: 30 μg/p), and polymyxin B (PB: 300 μg/p). Standard strain E. coli ATCC 25922 was used as a quality control. Strains were classified as either susceptible, intermediate, or resistant strains (CLSI, 2015).
2.3. PCR amplification of antimicrobial resistance and virulence genes
Genomic DNA for PCR was extracted by the boiling method. Tables 2 and 3 list the oligonucleotide sequences of different antimicrobial genes and virulence genes in E. coli and the predicted sizes after PCR amplification.
Table 3.
Primers used for detection of genes encoding resistance to different virulence
| Gene | Primer | DNA sequence (5′ → 3′) | Size (bp) | Thermocycling conditions | References |
|---|---|---|---|---|---|
| stx1 | stx1‐F | 5′‐ACACTGGATGATCTCAGTGG‐3′ | 244 | 95°C for 5 min, 35 cycles of 94°C for 1 min, 60°C for 1 min, 72°C for 1 min, final extension at 72°C for 10 min | Moses, Garbati, and Egwu (2006) |
| stx1‐R | 5′‐CTGAATCCCCCTCCATTATG‐3′ | Moses et al. (2006) | |||
| stx2 | stx2‐F | 5′‐CCATGACAACGGACAGCAGTT‐3′ | 255 | Moses et al. (2006) | |
| stx2‐R | 5′‐CCTGTCAACTGAGCACTTTG‐3′ | Moses et al. (2006) | |||
| agg | agg‐F | 5′‐AAGAAAAAGAAGTAGACCAAC‐3′ | 400 | Pass, Odedra, and Batt (2000) | |
| agg‐R | 5′‐AAACGGCAAGACAAGTAAATA‐3′ | Pass et al. (2000) | |||
| eaeA | eae‐F | 5′‐AAGCGACTGAGGTCACT‐3′ | 384 | Lopez et al. (2003) | |
| eae‐R | 5′‐ACGCTGCTCACTAGATGT‐3′ | Lopez et al. (2003) | |||
| fyuA | fyu‐F | 5′‐ACACGGCTTTATCCTCTGGC‐3′ | 235 | 95°C for 5 min, 30 cycles of 94°C for 30 s, 52°C for 30 s, 72°C for 45 s, and final extension at 72°C for 10 min | Viktoria, Lionel, and Per (2008) |
| fyu‐R | 5′‐GGCATATTGACGATTAACGA‐3′ | Viktoria et al. (2008) | |||
| fimA | fimA‐F | 5′‐CTGTGAGTGGTCAGGCAAGCG‐3′ | 352 | Rawool et al. (2015) | |
| fimA‐R | 5′‐TAACCGTGTTGGCGTAAGAGC‐3′ | Rawool et al. (2015) | |||
| papC | papC‐F | 5′‐GACGGCTGTACTGCAGGGTCGGGCG‐3′ | 234 | 95°C for 5 min, 30 cycles of 94°C for 30 s, 47°C for 30 s, 72°C for 45 s, and final extension at 72°C for 10 min | Xia et al. (2011) |
| papC‐R | 5′‐ATATCCTTTCTGCAGGGATGCAATA‐3′ | Xia et al. (2011) | |||
| papA | papA‐F | 5′‐GGAACGAACGCAGAAACG‐3′ | 374 | 95°C for 5 min, 30 cycles of 94°C for 30 s, 52°C for 30 s, 72°C for 45 s, and final extension at 72°C for 10 min | Xia et al. (2011) |
| papA‐R | 5′‐CGCAATGGGCGAATACTT‐3′ | Xia et al. (2011) | |||
| fimC | fimC‐F | 5′TAAGGAAATCGCAGGAA‐3′ | 337 | 95°C for 5 min, 30 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 45 s, and final extension at 72°C for 10 min | Antonio et al. (2007) |
| fimC‐R | 5′‐GCTGTGGGATAATGGACT‐3′ | Antonio et al. (2007) |
The presence of genes associated with resistance to tetracycline (tetA, tetB, and tetC), β‐lactams (bla TEM, bla PSE, and bla OXA), aminoglycosides (aad A1a, aadB, strA, and strB), chloramphenicol (floR), and sulfonamide (Sul1 and Sul2), and virulence‐encoding genes were detected by PCR. The PCR products were electrophoresed for 40 min at 90 V in 1% agarose gel containing 0.5 µg/ml of ethidium bromide, and then, the gels were visualized on a Gel Doc 2000 transmittance apparatus (Kerrn, Klemmensen, Frimodt‐MØller, & Espersen, 2002). Target fluorescentbands were removed from the gel with a razor blade. The DNA fragments were purified with a MIDI gel purification kit and then sequenced. The DNA sequence data were compared with the data in the GenBank database.
2.4. Statistical analysis
SPSS v.17.0 software was used to analyze the data. Logistical regression analysis was used to analyze the correlation between variables. p < .05 was considered statistically significant.
3. RESULTS AND CONCLUSIONS
3.1. E. coli isolated from retail foods
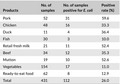
A total of 112 strains of E. coli were isolated from 431 random samples, with 26% of the samples testing positive for contamination. The overall incidence was higher than 14.7% reported elsewhere (Rasheed et al., 2014). As shown in Table 4, pork was most frequently contaminated with E. coli (59.6%). The detection rates of E. coli were 52.6%, 52.4%, 36.4%, 35.3%, and 33.3% in mutton, retail fresh milk, duck, beef, and chicken, respectively, followed by ready‐to‐eat food (12.9%), vegetables (11%), and fish (10%).
Table 4.
Samples and isolates from different food origins
| Products | No. of samples | No. of samples positive for E. coli | Positive rate (%) |
|---|---|---|---|
| Pork | 52 | 31 | 59.6 |
| Chicken | 48 | 16 | 33.3 |
| Duck | 11 | 4 | 36.4 |
| Fish | 30 | 3 | 10.0 |
| Retail fresh milk | 21 | 11 | 52.4 |
| Beef | 34 | 12 | 35.3 |
| Mutton | 19 | 10 | 52.6 |
| Vegetables | 154 | 17 | 11.0 |
| Ready‐to‐eat food | 62 | 8 | 12.9 |
| Total | 431 | 112 | 26.0 |
Several studies have documented antibiotic‐resistant E. coli and other coliforms in raw meat (Srinivasa, Gill, Ravi, & Sandeep, 2011), poultry (Nuno et al., 2016), eggs (Arathy, Vanpee, Belot, DeAllie, & Sharma, 2011), milk (Alharbi & Khaled, 2018), and vegetables (Rasheed et al., 2014). Whether there is a link between high contamination rates and high antibiotic resistance rates for E. coli in food remains to be determined.
In both developed and developing countries, antibiotic resistance has been recognized as a problem in the field of human and veterinary medicine (Bottacini et al., 2018; Zhang et al., 2017). There is ample evidence that the widespread use of antibiotics in agriculture and medicine is the main reason for the high resistance rate of Gram‐negative bacteria (Bothyna & Randa, 2018). Various food and environmental sources contain bacteria resistant to one or more antimicrobial agents used in human or veterinary medicine and animal food production (Hinthong, Pumipuntu, & Santajit, 2017).
3.2. Antimicrobial resistance profiles of E. coli isolates
Antibiotic resistance in E. coli is of particular concern because it is the most common Gram‐negative pathogen in humans, the most common cause of urinary tract infections, and a frequent cause of community and hospital‐acquired bacteremia (Bothyna & Randa, 2018) and diarrhea (Jessica, Lashaunda, & Levens, 2016).
Worldwide data have shown that resistance to traditional drugs is increasing, and resistance is also being encountered against newer and more effective antibiotics (Sara, Mohammad, & Sadegh, 2014). As in this study, the most frequent resistance was seen for third‐generation cephalosporin–ceftazidime (22%) and tetracyclines (52%; Table 5). A comparative study by Dominguez et al. (2018) showed that high resistance rates (76.5%–79.4%) were observed in oxyimino‐cephalosporins (cefotaxime, ceftriaxone, and ceftiofur) and cefepime (70.6%). This phenomenon requires additional study and sustained data support.
Table 5.
The reactions of E. coli to 17 antibacterial agents
| Antimicrobials | Resistant (n = 112) | Susceptible (n = 112, %) |
|---|---|---|
| AMP | 47 (42%) | 23 (20) |
| CTX | 12 (11%) | 34 (30) |
| CAZ | 25 (22%) | 38 (34) |
| IPM | 0 | 112 (100) |
| PIP | 31 (28%) | 40 (36) |
| AMX | 37 (33%) | 35 (31) |
| PB | 2 (2%) | 72 (64) |
| CIP | 18 (16%) | 48 (43) |
| LEV | 12 (11%) | 50 (45) |
| NAL | 36 (32%) | 34 (30) |
| GEN | 12 (11%) | 50 (45) |
| AMK | 10 (9%) | 55 (49) |
| STR | 24 (21%) | 44 (39) |
| TET | 58 (52%) | 22 (20) |
| CHL | 30 (27%) | 38 (34) |
| T/S | 41 (37%) | 32 (29) |
| ERY | 12 (11%) | 38 (34) |
n = 112: No. of samples positive for E. coli.
As shown in Table 5, our study revealed that 87 (77.7%) isolates (n = 112) were resistant to one or more antimicrobials, including tetracycline (52%), ampicillin (42%), compound trimethoprim/sulfamethoxazole (37%), amoxicillin (33%), and nalidixic acid (32%). No resistance to imipenem was observed. Among those isolates, two strains (E36, E37) isolated from chicken and one strain (E38) isolated from mutton were resistant to 13 antimicrobial agents. There were two strains (E24 and E53) isolated from chicken and one strain (E56) isolated from fish resistant to 11 antimicrobial agents. The specific multiple drug resistance rate is shown in Table 6, and the pattern of antibiotic resistance in those isolates is shown in Table 7.
Table 6.
Profile of multiple antibiotic‐resistant Escherichia coli isolates
| Resistance type | The number of multi‐drug‐resistant strain | The rate of multi‐drug‐resistant strains (%; n = 112) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CTX | GEN | CIP | LEV | TET | CHL | AMK | PIP | T/S | AMX | STR | NAL | E36 | 3 (2.7) |
| AMP | CTX | CAZ | GEN | CIP | LEV | TET | CHL | AMK | PIP | T/S | AMX | NAL | E37 | |
| AMP | CTX | CAZ | GEN | CIP | LEV | TET | CHL | AMK | PIP | T/S | AMX | NAL | E38 | |
| CAZ | CIP | LEV | TET | CHL | PIP | T/S | ERY | AMX | STR | NAL | E24 | 3 (2.7) | ||
| CTX | GEN | TET | CHL | AMK | PIP | T/S | ERY | AMX | STR | NAL | E53 | |||
| AMP | CTX | CAZ | CIP | LEV | TET | T/S | ERY | AMX | STR | NAL | E56 | |||
| AMP | CTX | GEN | CIP | TET | STR | AMK | PIP | AMX | T/S | F41 | 1 (0.9) | |||
| AMP | CTX | CAZ | CIP | TET | CHL | T/S | ERY | NAL | E48 | 1 (0.9) | ||||
| AMP | CAZ | TET | CHL | PIP | T/S | AMX | CIP | E28 | 5 (4.5) | |||||
| AMP | CAZ | TET | CHL | AMK | T/S | ERY | AMX | E31 | ||||||
| AMP | CAZ | TET | CHL | PIP | T/S | ERY | LEV | E42 | ||||||
| AMP | TET | T/S | CAZ | CHL | AMX | STR | NAL | E47 | ||||||
| AMP | CIP | LEV | TET | T/S | AMX | STR | NAL | F38 | ||||||
| AMP | TET | PIP | T/S | ERY | AMX | NAL | E9 | 6 (5.4) | ||||||
| AMP | CAZ | GEN | PIP | T/S | AMX | AMK | E23 | |||||||
| AMP | CAZ | TET | PIP | AMX | CIP | LEV | E41 | |||||||
| CAZ | TET | CHL | T/S | AMX | STR | NAL | E46 | |||||||
| CAZ | TET | PIP | T/S | AMX | STR | NAL | E49 | |||||||
| TET | NAL | T/S | AMP | PIP | AMX | CHL | F21 | |||||||
| AMP | CIP | TET | CHL | PIP | T/S | E2 | 12 (11) | |||||||
| AMP | TET | CHL | PIP | T/S | AMX | E6 | ||||||||
| AMP | CTX | CAZ | PIP | NAL | PB | E22 | ||||||||
| AMP | CTX | CAZ | TET | PIP | T/S | E32 | ||||||||
| AMP | CAZ | TET | PIP | NAL | CHL | E34 | ||||||||
| AMP | CAZ | TET | CHL | T/S | AMX | E44 | ||||||||
| AMP | TET | CHL | PIP | T/S | AMX | E52 | ||||||||
| AMP | CTX | CAZ | TET | T/S | NAL | E54 | ||||||||
| AMP | TET | CHL | AMK | T/S | NAL | E55 | ||||||||
| TET | NAL | T/S | AMP | PIP | AMX | F1 | ||||||||
| TET | NAL | T/S | AMP | PIP | AMX | F3 | ||||||||
| TET | NAL | T/S | AMP | PIP | AMX | F11 | ||||||||
| TET | CHL | T/S | NAL | CIP | E5 | 11 (10) | ||||||||
| AMP | TET | CHL | T/S | STR | E8 | |||||||||
| AMP | TET | PIP | AMX | NAL | E43 | |||||||||
| GEN | TET | CHL | T/S | AMX | E51 | |||||||||
| NAL | T/S | AMP | LEV | CHL | F10 | |||||||||
| TET | NAL | AMP | PIP | LEV | F18 | |||||||||
| TET | AMP | PIP | AMX | CHL | F19 | |||||||||
| TET | NAL | T/S | AMP | LEV | F24 | |||||||||
| AMP | PIP | AMX | CHL | STR | F30 | |||||||||
| TET | NAL | T/S | GEN | STR | F32 | |||||||||
| NAL | PIP | AMX | STR | ERY | F56 | |||||||||
| GEN | CIP | TET | AMX | E3 | 9 (8) | |||||||||
| AMP | TET | CHL | T/S | E12 | ||||||||||
| CAZ | TET | AMX | STR | E19 | ||||||||||
| CIP | ERY | AMX | NAL | E20 | ||||||||||
| TET | NAL | PIP | AMK | E26 | ||||||||||
| CAZ | TET | AMX | NAL | E27 | ||||||||||
| TET | T/S | CIP | AMK | E33 | ||||||||||
| TET | AMP | PIP | STR | F45 | ||||||||||
| TET | NAL | AMP | STR | F47 | ||||||||||
| CAZ | TET | CIP | E18 | 10 (9) | ||||||||||
| CTX | CAZ | CHL | E39 | |||||||||||
| TET | AMX | CHL | E40 | |||||||||||
| AMP | CTX | CAZ | E45 | |||||||||||
| TET | T/S | AMP | F9 | |||||||||||
| CHL | STR | ERY | F23 | |||||||||||
| TET | NAL | AMP | F35 | |||||||||||
| T/S | AMX | STR | F49 | |||||||||||
| CHL | ERY | STR | F53 | |||||||||||
| CHL | GEN | STR | F55 | |||||||||||
| TET | T/S | E1 | 16 (14) | |||||||||||
| AMP | CAZ | E15 | ||||||||||||
| AMP | CIP | E16 | ||||||||||||
| CAZ | NAL | E17 | ||||||||||||
| AMP | TET | E21 | ||||||||||||
| PB | CIP | E25 | ||||||||||||
| AMP | AMX | F4 | ||||||||||||
| AMP | PIP | F6 | ||||||||||||
| AMP | PIP | F15 | ||||||||||||
| AMP | STR | F17 | ||||||||||||
| TET | STR | F28 | ||||||||||||
| TET | NAL | F29 | ||||||||||||
| NAL | T/S | F31 | ||||||||||||
| AMP | GEN | F39 | ||||||||||||
| GEN | STR | F42 | ||||||||||||
| TET | STR | F44 | ||||||||||||
Table 7.
Phenotypic and genotypic resistance patterns of E. coli isolates
| Sampling number | Origin | Strain number | Resistance to antimicrobial agent | Resistance gene(s) |
|---|---|---|---|---|
| K2 | Pork | E1 | TET‐T/S | tetA, bla OXA, bla TEM |
| K13 | Pork tenderloin | E2 | AMP‐CIP‐TET‐CHL‐PIP‐T/S | tetA, floR |
| N19 | Chicken wings | E3 | GEN‐CIP‐TET‐AMX | tetA |
| K50 | Beef | E4 | — | bla OXA, floR |
| K34 | Pork | E5 | TET‐CHL‐T/S‐NAL‐CIP | tetA, bla OXA, floR, aad Ala, Sul1 |
| K46 | Chicken | E6 | AMP‐TET‐CHL‐PIP‐T/S‐AMX | bla OXA ,bla TEM ,, Sul1, sul2, strB |
| K51 | Beef | E7 | — | aadB |
| K17 | Duck leg | E8 | AMP‐TET‐CHL‐T/S‐STR | floR, Sul1, sul2, strA, strB |
| S24 | Zhaer root leaf vegetable | E9 | AMP‐TET‐PIP‐T/S‐ERY‐AMX‐NAL | tetA, floR, Sul1, strA |
| S99 | Cold pig ears | E10 | — | — |
| S100 | Peanut salad | E11 | — | — |
| H8 | Porcine blood | E12 | AMP‐TET‐CHL‐T/S | aadB, strA |
| H22 | Chicken wings | E13 | — | — |
| W41 | Mutton | E14 | — | strA |
| N23 | Beef | E15 | AMP‐CAZ | — |
| S25 | Lettuce | E16 | AMX‐CIP | strA |
| K14 | Chinese cabbage | E17 | CAZ‐NAL | tetA |
| H23 | Chicken wings | E18 | CAZ‐TET‐CIP | tetA |
| H76 | Cold bamboo shoots | E19 | CAZ‐TET‐AMX‐STR | tetB, Sul1, sul2, strA, strB |
| S65 | Chicken breast | E20 | CIP‐ERY‐AMX‐NAL | strA |
| S49 | Black fungus | E21 | AMP‐TET | tetA |
| H32 | Beef | E22 | AMP‐CTX‐CAZ‐PIP‐NAL‐PB | tetA, bla OXA, bla TEM, |
| W9 | Pork | E23 | AMP‐CAZ‐GEN‐PIP‐T/S‐AMX‐AMK | tetB, bla OXA, aad Ala |
| S55 | Chicken wings | E24 | CAZ‐CIP‐LEV‐TET‐CHL‐PIP‐T/S‐ERY‐AMX‐STR‐NAL | floR, Sul1, sul2, aad Ala, strA, strB |
| H33 | Beef | E25 | PB‐CIP | tetA, bla OXA, strA |
| W39 | Mutton | E26 | TET‐NAL‐PIP‐AMK | tetA, tetB, aadB |
| W46 | Mutton | E27 | CAZ‐TET‐AMX‐NAL | bla TEM, strA |
| K4 | Pork liver | E28 | AMP‐CAZ‐TET‐CHL‐PIP‐T/S‐AMX‐CIP | tetA, bla OXA, floR, sul2, aad Ala, strA,strB |
| H65 | Beef hind legs | E29 | AMX | — |
| H61 | Dried beef | E30 | — | bla TEM |
| H13 | Pork | E31 | AMP‐CAZ‐TET‐CHL‐AMK‐T/S‐ERY‐AMX | bla OXA, floR, aad Ala |
| N11 | Marinated tofu | E32 | AMP‐CTX‐CAZ‐TET‐PIP‐T/S | bla TEM |
| S66 | Chicken | E33 | TET‐T/S‐CIP‐AMK | tetA, aad Ala |
| H27 | Pork | E34 | AMP‐CAZ‐TET‐PIP‐NAL‐CHL | floR, bla OXA |
| K47 | Spicy dried tofu | E35 | TET | tetA, tetB |
| W38 | Chicken wings | E36 | AMP‐CTX‐GEN‐CIP‐LEV‐TET‐CHL‐AMK‐PIP‐T/S‐AMX‐STR‐NAL | bla TEM, bla OXA, floR, sul2, strA, strB, tetA |
| S70 | Chicken gizzard | E37 | AMP‐CTX‐CAZ‐GEN‐CIP‐LEV‐TET‐CHL‐AMK‐PIP‐T/S‐AMX‐NAL | tetA, tetB, floR, sul2, strA, strB |
| S39 | Mutton | E38 | AMP‐CTX‐CAZ‐GEN‐CIP‐LEV‐TET‐CHL‐AMK‐PIP‐T/S‐AMX‐NAL | aadB, tetA, tetB |
| K40 | Pork liver | E39 | CTX‐CAZ‐CHL | bla OXA |
| W2 | Pork | E40 | TET‐AMX‐CHL | tetA, bla TEM |
| S71 | Chicken | E41 | AMP‐CAZ‐TET‐PIP‐AMX‐CIP‐LEV | tetB, bla OXA, sul2, aadB, strA, strB |
| H24 | Pork liver | E42 | AMP‐CAZ‐TET‐CHL‐PIP‐T/S‐ERY‐LEV | tetA, tetB, bla OXA |
| H60 | Chicken gizzard | E43 | AMP‐TET‐PIP‐AMX‐NAL | tetA, tetB, bla TEM |
| K33 | Porcine blood | E44 | AMP‐CAZ‐TET‐CHL‐T/S‐AMX | tetA, bla TEM,floR |
| H78 | Spicy dried tofu | E45 | AMP‐CTX‐CAZ | tetA |
| H28 | Pork liver | E46 | CAZ‐TET‐CHL‐T/S‐AMX‐STR‐NAL | tetA, bla TEM, Sul1, sul2, aadB, strA, strB |
| H30 | Pork | E47 | AMP‐TET‐T/S‐CAZ‐CHL‐AMX‐STR‐NAL | tetA, tetB, Sul1, sul2, strB |
| H34 | Pork liver | E48 | AMP‐CTX‐CAZ‐CIP‐TET‐CHL‐T/S‐ERY‐NAL | tetA, tetB, Sul1, sul2, strA, strB |
| S10 | Pork fillet | E49 | CAZ‐TET‐PIP‐T/S‐AMX‐STR‐NAL | tetA, Sul1, sul2, strA, strB |
| N31 | Mutton | E50 | — | bla TEM |
| K10 | Pork stuffing | E51 | GEN‐TET‐CHL‐T/S‐AMX | tetA, bla TEM |
| W3 | Pork liver | E52 | AMP‐TET‐CHL‐PIP‐T/S‐AMX | tetA, tetB, bla TEM, aadAla |
| S30 | Chicken | E53 | CTX‐GEN‐TET‐CHL‐AMK‐PIP‐T/S‐ERY‐AMX‐STR‐NAL | tetA, tetB, bla TEM, Sul1, sul2, strA, strB |
| H64 | Beef hind legs | E54 | AMP‐CTX‐CAZ‐TET‐T/S‐NAL | tetA, tetB, strA, strB |
| K64 | Red oil ear silk | E55 | AMP‐TET‐CHL‐AMK‐T/S‐NAL | sul2 |
| N5 | Fish | E56 | AMP‐CTX‐CAZ‐CIP‐LEV‐TET‐T/S‐ERY‐AMX‐STR‐NAL | bla TEM, strA, strB, sul1, sul2, strB |
| N16 | Crustacean | F1 | TET‐NAL‐T/S‐AMP‐PIP‐AMX | strA, strB, bla OXA, tetA, floR, Sul1, sul2 |
| R1 | Retail fresh milk | F2 | — | tetB |
| S27 | Bok choy | F3 | TET‐NAL‐T/S‐AMP‐PIP‐AMX | strA, strB, sul2, bla OXA, tetA, bla TEM, aad Ala, floR |
| S56 | Broccoli | F4 | AMP‐AMX | tetB |
| S96 | Cold bean curd stick | F5 | — | — |
| W51 | Sheep heart | F6 | AMP‐PIP | strA, strB, bla TEM, aad Ala, floR, Sul1, sul2 |
| S72 | Bean sprouts | F7 | TET | bla OXA |
| H4 | Pork | F8 | TET | strA, strB, sul2, bla OXA,, tetA, bla TEM |
| H9 | Pork | F9 | TET‐T/S‐AMP | tetA |
| N22 | Chicken | F10 | NAL‐T/S‐AMP‐LEV‐CHL | strB, aadA1a, floR, Sul1, sul2 |
| R2 | Retail fresh milk | F11 | TET‐NAL‐T/S‐AMP‐PIP‐AMX | bla OXA |
| N30 | Pepper | F12 | — | — |
| W8 | Pig tail | F13 | T/S | bla OXA, tetB, aad Ala |
| R5 | Retail fresh milk | F14 | T/S | tetB |
| R7 | Retail fresh milk | F15 | AMP‐PIP | floR |
| R8 | Retail fresh milk | F16 | — | bla OXA, aadB |
| S38 | Beef | F17 | AMP‐STR | strB, sul2, bla OXA |
| K44 | Chicken gizzard | F18 | TET‐NAL‐AMP‐PIP‐LEV | bla OXA |
| W47 | Beef | F19 | TET‐AMP‐PIP‐AMX‐CHL | strA, strB, sul2, bla OXA, aad Ala |
| R8 | Retail fresh milk | F20 | — | bla OXA |
| H9 | Pork | F21 | TET‐NAL‐T/S‐AMP‐PIP‐AMX‐CHL | strA, strB, sul2, bla OXA, tetA, tetB, bla TEM, floR, aadB |
| K28 | Celery | F22 | — | bla OXA, |
| H33 | Pork | F23 | CHL‐STR‐ERY | strA, strB, bla OXA, aad Ala, Sul1, sul2, aadB |
| S68 | Chicken wings | F24 | TET‐NAL‐T/S‐AMP‐LEV | strA, strB, Sul1, sul2,, tetA, bla TEM, aad Ala |
| S79 | Lotus root | F25 | ERY | tetB |
| S80 | Cabbage | F26 | — | bla OXA |
| S89 | Cucumber | F27 | TET | bla OXA, tetA, tetB |
| S58 | Sheep fat | F28 | TET‐STR | bla OXA, tetB, aad Ala |
| K60 | Chicken gizzard | F29 | TET‐NAL | tetB |
| S8 | Pig heart | F30 | AMP‐PIP‐AMX‐CHL‐STR | strA, strB, bla OXA, tetA, bla TEM, aad Ala, Sul1 |
| W13 | Pork | F31 | NAL‐T/S | bla OXA |
| W14 | Pork | F32 | TET‐NAL‐T/S‐GEN‐STR | bla TEM, aad Ala, aadB |
| K26 | Carrot | F33 | — | — |
| R9 | Retail fresh milk | F34 | — | sul2, bla OXA |
| S60 | Mutton | F35 | TET‐NAL‐AMP | tetA, tetB, bla OXA |
| H34 | Beef | F36 | — | — |
| R3 | Retail fresh milk | F37 | — | bla OXA |
| S59 | Lamb tripe | F38 | AMP‐CIP‐LEV‐TET‐T/S‐AMX‐STR‐NAL | bla OXA, tetB, floR |
| R6 | Retail fresh milk | F39 | AMP‐GEN | bla OXA, tetA, aad Ala, floR |
| R7 | Retail fresh milk | F40 | — | — |
| S90 | Celery | F41 | AMP‐CTX‐GEN‐CIP‐TET‐STR‐AMK‐PIP‐T/S‐AMX | strA, strB, sul2, tetA, tetB, aad Ala, floR |
| R10 | Retail fresh milk | F42 | GEN‐STR | bla OXA, aadB |
| S45 | Beef | F43 | NAL | bla OXA |
| S12 | Pork liver | F44 | TET‐STR | bla OXA, tetA, tetB, aad Ala |
| S41 | Lamb tripe | F45 | TET‐AMP‐PIP‐STR | bla OXA, tetB |
| K66 | Dried vegetables | F46 | — | tetB |
| S91 | Garlic sprouts | F47 | TET‐NAL‐AMP‐STR | tetA, tetB, bla OXA |
| K32 | Chicken wings | F48 | — | bla OXA |
| W43 | Spinach | F49 | T/S‐AMX‐STR | sul2 |
| H12 | Porcine blood | F50 | — | — |
| N10 | Bean curd skin | F51 | — | bla OXA |
| S93 | Towel gourd | F52 | — | — |
| K19 | Duck | F53 | CHL‐ERY‐STR | floR,aadB |
| K25 | Duck | F54 | LEV | sul2 |
| W12 | Duck | F55 | CHL‐GEN‐‐STR | sul2, aad Ala |
| N4 | Fish | F56 | NAL‐PIP‐AMX‐STR‐ERY | strA |
—, not detected.
The incidence of multidrug resistance is a compelling issue, as there is a repository of antimicrobial resistance genes in the community, and drug resistance genes and plasmids can easily be transferred to other strains. The high resistance to tetracycline and ampicillin may be due to the easy availability and low cost of those medications. Although these antibiotics have been banned, the bans have not been effectively implemented by the relevant regulatory bodies. Another explanation for a strain's high resistance rate is its contact with environmental microorganisms that produce natural antibiotics, or with soil contaminated by wildlife feces carrying antibiotic‐resistant microorganisms.
3.3. Antimicrobial resistance genotypes of E. coli isolates
We detected 11 of the 13 resistance genes (tetA, tetB, bla tem, bla oxa, floR, aad Ala, aadB, sul1, sul2, strA, and strB), and one hundred isolates carried one or more antimicrobial genes. Resistance genes were not detected in twelve strains of E. coli. The resistance genotypes of E. coli isolates are shown in Table 7.
Among 58 tetracycline‐resistant E. coli isolates, tetA was found in 43 isolates and tetB in 30 isolates, although tetC was not detected in any. One of the beta‐lactam resistance genes, bla TEM, was detected in 23 E. coli isolates, bla OXA was detected in 45, and bla PSE was not detected. Other resistance genes such as floR, sul1, sul2, aad Ala, aadB, strA, and strB were detected in 22, 18, 30, 21, 12, 31, and 27 isolates, respectively. The detection rate of resistance genes of our study was as follows: tetA (38%, 43/112), tetB (27%, 30/112), bla OXA (40%, 45/112), bla TEM (20%, 23/112), floR (20%, 22/112), sul1 (16%, 18/112), sul2 (27%, 30/112), aad Ala (19%, 21/112), aadB (11%, 12/112), strA (28%, 31/112), and strB (24%, 27/112). These data suggest that retail foods may be a reservoir of multi‐drug‐resistant bacteria and contribute to the spread of drug‐resistant genes.
We found that the detection rate of pork was more than that of chicken, duck, and beef, but there are fewer resistance genes in pork as compared to chicken. Ayoyi, Bii, and Okemo (2008) showed that multidrug resistance is closely related to different farm management treatments, and statistical significance (p ≤ .001) was found between them.
Chickens are more likely to get sick than pigs, and in large‐scale chicken breeding operations, farmers will use a large number of antibiotic and antiviral drugs for the prevention and treatment of chicken diseases. The antibiotics used include enrofloxacin, amikacin, colistin, ciprofloxacin, azithromycin, doxycycline hydrochloride, levofloxacin, lincomycin, doxycycline, gentamicin, gentamicin, levofloxacin, neomycin sulfate, ceftriaxone sodium, cefotaxime sodium, penicillin, sulfachloropyridine, and sulfaquinoxaline sodium.
3.4. Virulence genes of E. coli isolates
Table 8 shows that among the nine tested virulence genes, fimC, agg, stx2, fimA, fyuA, papA, stx1, and eaeA were found in 52, 34, 21, 19, 6, 3, 2, and 2 isolates, respectively, papC was not detected. Two strains (F6, F52) carried five virulence genes, and six strains (F5, F11, F12, F14, F50, and F51) also carried four virulence genes. Detailed results are shown in Table 9.
Table 8.
The detection rate of strains and virulence genes
| Virulence genes | No. of positive strains | Number of positive strains | Positive rate (%; n = 112) |
|---|---|---|---|
| stx1 | F1, F11 | 2 | 1.8 |
| stx2 | F3, F4, F5, F6, F7, F11, F12, F14, F17, F18, F20, F29, F36, F39, F45, F47, F48, F49, F50, F51, F52 | 21 | 18.8 |
| eaeA | F6, F18 | 2 | 1.8 |
| agg | E2, E7, E13, E14, E24, E39, F1, F5, F6, F8, F10,F11,F12, F16, F17, F18, F19, F21, F22, F24, F27, F28, F29, F32, F33, F34, F37, F38, F43, F44, F49, F50, F51, F52 | 34 | 30.4 |
| fyuA | E6, E13, E53, F13, F14, F50 | 6 | 5.4 |
| papA | E24, F14, F52 | 3 | 2.7 |
| papC | — | 0 | 0 |
| fimA | E5、E23、E26、E29、E33、E50, F2, F3, F5, F6, F10, F11, F12, F24, F25, F28, F50, F51, F52 | 19 | 17.0 |
| fimC | E4, E5, E6, E7, E8, E12, E22, E24, E26, E28, E29, E30, E35, E38, E43, E45, E49, E52, E54, E56, F1, F2, F3, F4, F5, F6, F8, F12, F13, F14, F17, F19, F22, F23, F24, F25, F27, F28, F30, F31, F33, F34, F35, F36, F37, F38, F43, F45, F47, F49, F51F52 | 52 | 46.4 |
Table 9.
Profile of Escherichia coli isolates with multiple virulence genes
| Virulence genes | No. of strains with multiple virulence genes | The rate of strains with multiple virulence genes (%; N = 112) | ||||
|---|---|---|---|---|---|---|
| Stx2 | agg | papA | fimA | fimC | F52 | 2 (1.8) |
| Stx2 | agg | eaeA | fimA | fimC | F6 | |
| Stx1 | Stx2 | agg | fimA | F11 | 6 (5.4) | |
| Stx2 | fyuA | papA | fimC | F14 | ||
| Stx2 | agg | fimA | fimC | F51 | ||
| Stx2 | agg | fimA | fimC | F5 | ||
| Stx2 | agg | fimA | fimC | F12 | ||
| Stx2 | agg | fimA | fyuA | F50 | ||
| Stx1 | agg | fimC | F1 | 7 (6.3) | ||
| Stx2 | fimA | fimC | F5 | |||
| Stx2 | agg | fimC | F12 | |||
| agg | fimA | fimC | F24 | |||
| agg | fimA | fimC | F28 | |||
| Stx2 | agg | fimC | F49 | |||
| Stx2 | eaeA | agg | F18 | |||
| Stx2 | fimC | F4 | 23 (20.5) | |||
| Stx2 | agg | F18 | ||||
| Stx2 | fimC | F36 | ||||
| Stx2 | fimC | F45 | ||||
| Stx2 | fimC | F47 | ||||
| agg | fimC | E7 | ||||
| agg | fimC | E24 | ||||
| agg | fimC | F8 | ||||
| agg | fimC | F19 | ||||
| agg | fimC | F22 | ||||
| agg | fimC | F27 | ||||
| agg | fimC | F33 | ||||
| agg | fimC | F34 | ||||
| agg | fimC | F37 | ||||
| agg | fimC | F38 | ||||
| agg | fimC | F43 | ||||
| agg | fimA | E7 | ||||
| fyuA | fimC | E6 | ||||
| fyuA | fimC | F13 | ||||
| fimA | fimC | E5 | ||||
| fimA | fimC | E26 | ||||
| fimA | fimC | E29 | ||||
| fimA | fimC | F2 | ||||
The emergence of virulence is mainly due to the presence of multiple virulence genes in E. coli pathogenicity islands. fyuA is highly pathogenic and is often used as an indication of the presence or absence of high pathogenicity islands (HPI; Paniagua et al., 2017). We detected fyuA virulence genes in six isolates (5.4%), compared to 83.3% found by Laupland, Gregson, Church, Ross, and Pitout (2008).
Bacterial pili and fimbriae are important structures for bacterial pathogenicity, and it has been suggested that type I fimbriae function primarily in the initial pathogenic phase of avian pathogenic E. coli (APEC) infection. P‐type fimbriae are also thought to contribute to bacterial pathogenicity (Paniagua et al., 2017). The fimC virulence gene encodes a protein necessary for the biosynthesis of type I fimbriae. The papA virulence gene encodes the main protein component of P‐type fimbriae, and P‐type fimbriae are encoded by the nine‐gene pap operon, which includes papA, papB, papC, papD, papE, papF, papG, papH, and papI. Sequence analysis showed that there is sufficienthomology between P fimbriae in humans and chickens to indicate that they share some common antigen (Laupland, Kibsey, & Gregson, 2013). We detected the fimC gene in 46.4% of isolates, and the papA gene was detected in 2.7%; papC was not detected. This suggests that APEC in the Xinjiang region is mainly caused by a type I fimbriae.
3.5. The relationship between virulence genes and antibiotic resistance
Arisoy et al. (2008) showed that there was a correlation between antibiotic sensitivity and virulence factors (VFs) of E. coli isolates causing pyelonephritis. They reported an increased presence of virulence genes pap, sfa, afai, hly, and aer in sensitive strains. Horcajada et al. (2005) showed that a significant correlation was found between nalidixic acid resistance and the decreased prevalence of three VFs: sfa, hly, and cnf‐1.
In the current study, strong associations were found between the presence of fimC and resistance to ciprofloxacin, gentamicin, amikacin, levofloxacin, and streptomycin; between the presence of fimA and resistance to tetracycline, ampicillin, compound trimethoprim/sulfamethoxazole, and amoxicillin; between the presence of agg and resistance to gentamicin, tetracycline, ciprofloxacin, and levofloxacin; and between the presence of stx2 and resistance to ampicillin, tetracycline, compound trimethoprim/sulfamethoxazole, and amoxicillin.
Based on statistical analysis, the following correlations were identified: (a) expression of the fimC gene and resistance to ciprofloxacin (p = .001), gentamicin (p = .001), amikacin (p = .001), levofloxacin (p = .001), and streptomycin (p = .001); (b) expression of the fimA gene and resistance to tetracycline (p = .001), ampicillin (p = .001), compound trimethoprim/sulfamethoxazole (p = .001), and amoxicillin (p = .003); (c) expression of the agg gene and resistance to gentamicin (p = .001), tetracycline (p = .001), ciprofloxacin (p = .017), and levofloxacin (p = .001); and (d) expression of the stx2 gene and resistance to ampicillin (p = .001), tetracycline (p = .001), compound trimethoprim/sulfamethoxazole (p = .002), and amoxicillin (p = .015; Table 10).
Table 10.
Distribution of antimicrobial resistance among virulence factor a
| Antibiotic | AMP | TET | STR | GEN | CIP | LEV | AMK | T/S | AMX |
|---|---|---|---|---|---|---|---|---|---|
| fim C (n = 52) | |||||||||
| Positive, % | 23 (44.2) | 25 (48.1) | 12 (23.1) b | 1 (1.9) b | 6(11.5) b | 5 (9.6) b | 2 (3.8) b | 18(34.6) | 16 (30.8) |
| p Value | .592 | .352 | .001 | .001 | .001 | .001 | .001 | .224 | .056 |
| fim A (n = 19) | |||||||||
| Positive, % | 6 (31.6) b | 7 (36.8) b | 1 (5.2) | 1 (5.3) | 2 (10.5) | 2 (10.5) | 3(15.8) | 7 (36.8) b | 4 (21) b |
| p Value | .001 | .001 | .307 | .165 | 1.000 | .241 | .107 | .001 | .003 |
| agg (n = 34) | |||||||||
| Positive, % | 11 (32.4) | 15 (44.1) b | 7 (20.6) | 1 (2.9) b | 3(8.8) b | 5(14.7) b | 0 (0) | 10 (29.4) | 7 (20.6) |
| p Value | .051 | .001 | .169 | .001 | .017 | .001 | / | .204 | .566 |
| stx2 (n = 21) | |||||||||
| Positive, % | 8 (38.1) b | 7 (33.3) b | 4 (19) | 1 (4.8) | 0 (0) | 1(4.8) | 0 (0) | 4(19) b | 4 (19) b |
| p Value | .001 | .001 | .619 | .057 | / | .091 | / | .002 | .015 |
Abbreviations: AMK, amikacin; AMP, ampicillin; AMX, amoxicillin; CIP, ciprofloxacin; GEN, gentamicin; LEV, levofloxacin; STR, streptomycin; T/S, cotrimoxazole; TET, tetracycline.
Data are presented as No. (%).
Statistically significant.
4. CONCLUSIONS
Differences in the pathogenicity of E. coli and its susceptibility to antimicrobial agents were detected in different retail foods. This must be taken into account in developing guidelines for retail food management. Periodic review and formulation of antibiotic consumption policies are required to control the spread and acquisition of antibiotic resistance. Because most isolates express several types of VFs at the same time, it is necessary to further study the interaction between different VFs at the molecular level.
In conclusion, E. coli has become a potential source of foodborne illness due to the possibility of horizontal transfer of drug‐resistant genes, high drug resistance rate, and the correlation between the resistance to some antibiotics and several virulence factors. As those problems become more and more serious, we need to strengthen the supervision of veterinary drugs used in the raising of livestock. At the same time, the detection and monitoring of antimicrobial agents in animal foods can help to reveal the ongoing use of prohibited animal husbandry practices.
CONFLICT OF INTEREST
The authors declare that they do not have any conflicts of interest.
ETHICAL STATEMENTS
This study did not involve any human or animal testing.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China—Xinjiang Joint Fund Project (No. U1703119), National Natural Science Foundation of China (No. 31301469), Projects of Innovation and Development Pillar Program for Key Industries in Southern Xinjiang of Xinjiang Production and Construction Corps (No. 2018DB002), and Shihezi University Major Science and Technology Research Project (gxjs2015‐zdgg05).
Li Y, Zhang M, Luo J, et al. Antimicrobial resistance of Escherichia coli isolated from retail foods in northern Xinjiang, China. Food Sci Nutr. 2020;8:2035–2051. 10.1002/fsn3.1491
Funding information
This work was supported by grants from the National Natural Science Foundation of China—Xinjiang Joint Fund Project (No. U1703119), National Natural Science Foundation of China (No. 31301469), Projects of Innovation and Development Pillar Program for Key Industries in Southern Xinjiang of Xinjiang Production and Construction Corps (No. 2018DB002), and Shihezi University Major Science and Technology Research Project (gxjs2015‐zdgg05).
REFERENCES
- Alharbi, N. S. , & Khaled, J. M. (2018). Prevalence of Escherichia coli strains resistance to antibiotics in wound infections and raw milk. Saudi Journal of Biological Sciences, 26(7), 1557–1562. 10.1016/j.sjbs.2018.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio, C. , Elena, C. , Davide, G. , Donato, P. , Patrizia, B. , Evelyn, C. , … Silvia, T. (2007). Avian pathogenic Escherichia coli in Audouin gulls (Larus audouinii) could they affect the surviving of the bird colonies? Italian Journal of Animal Science, 6, 317–320. 10.4081/ijas.2007.317 [DOI] [Google Scholar]
- Arathy, D. S. , Vanpee, G. , Belot, G. , DeAllie, C. , & Sharma, R. (2011). Antimicrobial drug resistance in Escherichia coli isolated from commercial chicken eggs in Grenada, West Indies. West Indian Medical Journal, 60, 53–56. 10.2298/VSP1101081P [DOI] [PubMed] [Google Scholar]
- Arisoy, M. , Rad, A. Y. , Akin, A. , & Akar, N. (2008). Relationship between susceptibility to antimicrobials and virulence factors in paediatric Escherichia coli isolates. International Journal of Antimicrobial Agents, 31(S1), 4–8. 10.1016/j.ijantimicag.2007.07.030 [DOI] [PubMed] [Google Scholar]
- Ayoyi, A. O. , Bii, C. C. , & Okemo, P. (2008). Detection of antibiotic resistance and virulence related factors in Escherichia coli isolates from broiler chicken in Limuru, Kenya. International Journal of Infectious Diseases, 12(1), e109–e110. 10.1016/j.ijid.2008.05.274 [DOI] [Google Scholar]
- Bothyna, G. , & Randa, N. H. (2018). Multiple drug resistance and biocide resistance in Escherichia coli environmental isolates from hospital and household settings. Antimicrobial Resistance and Infection Control, 7, 47 10.1186/s13756-018-0339-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottacini, F. , Morrissey, R. , Roberts, R. J. , James, K. , van Breen, J. , Egan, M. , … van Sinderen, D. (2018). Comparative genome and methylome analysis reveals restriction/modification system diversity in the gut commensal Bifidobacterium breve . Nucleic Acids Research, 46(4), 1860–1877. 10.1093/nar/gkx1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Céline, B. A. , & David, M. (2015). Horizontal gene transfer in microbial ecosystems. Environmental Microbiology: Fundamentals and Applications, 48, 445–481. 10.1051/vetres:2001125 [DOI] [Google Scholar]
- Chang, L. L. , Lin, H. H. , Chang, C. Y. , & Lu, P. L. (2007). Increased incidence of class 1 integrons in trimethoprim/sulfamethoxazole‐resistant clinical isolates of Stenotrophomonas maltophilia . Journal of Antimicrobial Chemotherapy, 59, 1038–1039. 10.1093/jac/dkm034 [DOI] [PubMed] [Google Scholar]
- CL SI (2015). Performance Standards for Antimicrobial Susceptibility Testing (25th ed.). CLSI Supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA: Retrieved from https://clsi.org/standards/products/microbiology/documents/m100/ [Google Scholar]
- Dominguez, J. E. , Redondo, L. M. , Figueroa Espinosa, R. A. , Cejas, D. , Gutkind, G. O. , Chacana, P. A. , … Fernández Miyakawa, M. E. (2018). Simultaneous carriage of mcr‐1 and other antimicrobial resistance determinants in Escherichia coli from poultry. Frontiers in Microbiology, 9, 1–10. 10.3389/fmicb.2018.01679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra, B. , Junker, E. , Schroeter, A. , Malorny, B. , Lehmann, S. , & Helmuth, R. (2003). Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. International Journal of Infectious Diseases, 52(3), 489–492. 10.1093/jac/dkg362 [DOI] [PubMed] [Google Scholar]
- Guillard, T. , Pons, S. , Roux, D. , Pier, G. B. , & Skurnik, D. (2016). Antibiotic resistance and virulence: Understanding the link and its consequences for prophylaxis and therapy. BioEssays, 38, 682–693. 10.1002/bies.201500180 [DOI] [PubMed] [Google Scholar]
- Heijnen, L. , & Medema, G. (2006). Quantitative detection of E. coli, E. coli O157 and other shiga toxin producing E. coli in water samples using a culture method combined with real‐time PCR. Journal of Water and Health, 4(4), 487–498. 10.2166/wh.2006.0032 [DOI] [PubMed] [Google Scholar]
- Hinthong, W. , Pumipuntu, N. , & Santajit, S. (2017). Detection and drug resistance profile of Escherichia coli from subclinical mastitis cows and water supply in dairy farms in Saraburi Province, Thailand. Peer J, 5, e3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, P. L. , Wang, R. C. , Chow, K. H. , & Que, T. L. (2009). Distribution of integron‐associated trimethoprim‐sulfamethoxazole resistance determinants among Escherichia coli from humans and food‐producing animals. Letters in Applied Microbiology, 49(5), 627–634. 10.1111/j.1472-765X.2009.02717.x [DOI] [PubMed] [Google Scholar]
- Horcajada, J. P. , Soto, S. , Gajewski, A. , Smithson, A. , Jiménez de Anta, M. T. , Mensa, J. , … Johnson, J. R. (2005). Quinolone‐resistant uropathogenic Escherichia coli strains from phylogenetic group B2 have fewer virulence factors than their susceptible counterparts. Journal of Clinical Microbiology, 43(6), 2962–2964. 10.1128/JCM.43.6.2962-2964.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Yang, X. , & Li, J. (2016). The transfer network of bacterial mobile resistome connecting animal and human microbiome. Applied and Environmental Microbiology, 82, 6672–6681. 10.1128/AEM.01802-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessica, C. , Lashaunda, B. , & Levens, J. (2016). Longitudinal comparison of antibiotic resistance in diarrheagenic and non‐pathogenic Escherichia coli from young Tanzanian children. Frontiers in Microbiology, 7, 1420 10.3389/fmicb.2016.01420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katarzyna, W. K. , & Anna, L. B. (2016). Phenotypic and molecular assessment of drug resistance profile and genetic diversity of waterborne Escherichia coli . Water, Air, & Soil Pollution, 227, 146 10.1007/s11270-016-2833-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrn, M. B. , Klemmensen, T. , Frimodt‐MØller, N. , & Espersen, F. (2002). Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. Journal of Antimicrobial Chemotherapy, 50(4), 513–516. 10.1093/jac/dkf164 [DOI] [PubMed] [Google Scholar]
- Knapp, C. W. , Dolfing, J. , Ehlert, P. , & Graham, D. W. (2010). Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environmental Science & Technology, 44(2), 580–587. 10.1021/es901221x [DOI] [PubMed] [Google Scholar]
- Laupland, K. B. , Gregson, D. B. , Church, D. L. , Ross, T. , & Pitout, J. D. D. (2008). Incidence, risk factors and outcomes of Escherichia coli bloodstream infections in a large Canadian region. Clinical Microbiology & Infection, 14(11), 1041–1047. 10.1111/j.1469-0691.2008.02089.x [DOI] [PubMed] [Google Scholar]
- Laupland, K. B. , Kibsey, P. C. , & Gregson, D. B. (2013). Population‐based laboratory assessment of the burden of community‐onset bloodstream infection in Victoria. Epidemiology and Infection, 141(1), 174–180. 10.1017/S0950268812000428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, S. C. , Cerna, J. F. , Villegas, S. N. , Thompson, R. , Velazquez, F. R. , Torres, J. , … Estrada, G. T. (2003). Single multiplex polymerase chain reaction to detect diverse loci associated with diarrheagenic Escherichia coli . Emerging Infectious Diseases, 9, 127–131. 10.3201/eid0901.010507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J. , Ji, H. , & Wang, Q. L. (2016). Drug resistance of Escherichia coli isolated from different foods in Xinjiang. Modern Food Science and Technology, 32(8), 271–275. 10.13982/j.mfst.1673-9078.2016.8.041 [DOI] [Google Scholar]
- Meredith, S. , Brooks, B. D. , & Brooks, A. E. (2017). The complex relationship between virulence and antibiotic resistance. Genes, 8, 1–3. 10.3390/genes8010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses, A. E. , Garbati, M. A. , & Egwu, G. O. (2006). Detection of E. coli O157 and O26 serogroups in human immunodeficiency virus‐infected patients with clinical manifestation of diarrhea in Maiduguri, Nigeria. Journal of Research in Medical Science, 1(4), 140–145. [Google Scholar]
- Ng, L. K. , Martin, I. , Alfo, M. , & Mulvey, M. (2001). Multiplex PCR for the detection of tetracycline resistance genes. Molecular and Cellular Probes, 15, 209–215. 10.1006/mcpr.2001.0363 [DOI] [PubMed] [Google Scholar]
- Nuno, M. , Figueiredo, R. , Mendes, C. , Card, R. , Anjum, M. , & da Silva, G. (2016). Microarray evaluation of antimicrobial resistance and virulence of Escherichia coli isolates from Portuguese poultry. Antibiotics, 5(1), 4 10.3390/antibiotics5010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua, G. L. , Monroy‐Pérez, E. , Rodríguez‐Moctezuma, J. R. , Domínguez‐Trejo, P. , Vaca‐Paniagua, F. , & Vaca, S. (2017). Virulence factors, antibiotic resistance phenotypes and O‐serogroups of Escherichia coli strains isolated from community‐acquired urinary tract infection patients in Mexico. Journal of Microbiology, Immunology and Infection, 50(4), 478–485. 10.1016/j.jmii.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Pass, M. A. , Odedra, R. , & Batt, R. M. (2000). Multiplex PCRs for identification of Escherichia coli virulence genes. Journal of Clinical Microbiology, 38, 2001–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed, M. U. , Thajuddin, N. , Ahamed, P. , Teklemariam, Z. , & Jamil, K. (2014). Antimicrobial drug resistance in strains of Escherichia coli isolated from food sources. Revista do Instituto de Medicina Tropical de Sã, 56(4), 341–346. 10.1590/s0036-46652014000400012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawool, D. B. , Vergis, J. , Vijay, D. , Dhaka, P. , Negi, M. , Kumar, M. , … Barbuddhe, S. B. (2015). Evaluation of a PCR targeting fimbrial subunit gene (fimA) for rapid and reliable detection of Enteroaggregative Escherichia coli recovered from human and animal diarrhoeal cases. Journal of Microbiological Methods, 110, 45–48. 10.1016/j.mimet.2015.01.008 [DOI] [PubMed] [Google Scholar]
- Rosengren, L. B. , Waldner, C. L. , & Reid‐Smith, R. J. (2009). Associations between antimicrobial resistance phenotypes, antimicrobial resistance genes, and virulence genes of fecal Escherichia coli isolates from healthy grow‐finish pigs. Applied and Environmental Microbiology, 75(5), 1373–1380. 10.1128/AEM.01253-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, D. , Danilchanka, O. , Guillard, T. , Cattoir, V. , Aschard, H. , Fu, Y. , … Skurnik, D. (2015). Fitness cost of antibiotic susceptibility during bacterial infection. Science Translational Medicine, 7, 297 10.1126/scitranslmed.aab1621 [DOI] [PubMed] [Google Scholar]
- Sáenz, Y. , Briñas, L. , Domínguez, E. , Ruiz, J. , Zarazaga, M. , Vila, J. , & Torres, C. (2004). Mechanisms of resistance in multiple‐antibiotic‐resistant Escherichia coli strains of human, animal, and food origin. Antimicrobial Agents and Chemotherapy, 48(10), 3996–4001. 10.1128/AAC.48.10.3996-4001.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sara, A. , Mohammad, K. , & Sadegh, G. (2014). The association of virulence determinants of uropathogenic Escherichia coli with antibiotic resistance. Jundishapur Journal of Microbiology, 7(5), e9936 10.5812/jjm.9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasa, R. T. , Gill, J. P. S. , Ravi, K. , & Sandeep, G. (2011). Multi drug resistance patterns of Shiga toxin ‐ producing Escherichia coli (STEC) and non‐STEC isolates from meats, RTE meat foods, drinking water and human diarrhoeic samples of Punjab, India. Archives of Clinical Microbiology, 2, 1–12. [Google Scholar]
- Tenaillon, O. , Skurnik, D. , Picard, B. , & Denamur, E. (2010). The population genetics of commensal Escherichia coli . Nature Reviews Microbiology, 8(3), 207–217. 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- Viktoria, H. , Lionel, F. , & Per, K. (2008). The ferric yersiniabactin uptake receptor FyuA is required for efficient biofilm formation by urinary tract infectious Escherichia coli in human urine. Microbiology, 154, 167–175. 10.1099/mic.0.2007/011981-0 [DOI] [PubMed] [Google Scholar]
- Wang, J. W. , Sun, Y. , & Ji, X. (2014). Isolation, identification and drug resistance analysis of Escherichia coli isolated from pigs in Changchun, China. Journal of Veterinary Medicine, 48(11), 14–18. [Google Scholar]
- Xia, L. N. , Xiang, F. , & Guo, Q. Y. (2014). Drug resistance analysis of bovine Escherichia coli in different areas of Xinjiang. Chinese Animal Husbandry and Veterinary Medicine, 41(2), 203–207. [Google Scholar]
- Xia, X. , Meng, J. , Zhao, S. , Bodeis‐jones, S. , Gaines, S. A. , Ayers, S. L. , & McDermott, P. F. (2011). Identification and antimicrobial resistance of extraintestinal pathogenic Escherichia coli from retail meats. Journal of Food Protection, 74(1), 38–44. 10.4315/0362-028X.JFP-10-251 [DOI] [PubMed] [Google Scholar]
- Yao, X. H. , Long, M. J. , Kuerbannaimu, K. , Wang, K. , & Xia, L. N. (2017). Antibiotic resistance of Escherichia coli from different animal sources. Xinjiang Agricultural Science, 54(12), 2314–2319. 10.6048/j.issn.1001-4330.2017.12.019 [DOI] [Google Scholar]
- Zhang, P. , Shen, Z. , Zhang, C. , Song, L. I. , Wang, B. , Shang, J. , … Wu, C. (2017). Surveillance of antimicrobial resistance among Escherichia coli from chicken and swine, China, 2008–2015. Veterinary Microbiology, 203, 49–55. 10.1016/j.vetmic.2017.02.008 [DOI] [PubMed] [Google Scholar]
- Zhi, S. , Xi, M. L. , & Shen, J. L. (2009). The antibiotic resistance of food‐borne Escherichia coli . Acta Agriculturae Boreali‐occidentalis Sinica, 18(6), 377–381. [Google Scholar]


