Abstract
Objective: The current study aimed to investigate the functional roles and clinical significance of microRNA-148a (miR-148a) in the progression of oral squamous cell carcinoma (OSCC).
Methods: Relative expression of miR-148a in OSCC cells and tissues were detected using quantitative real-time polymerase chain reaction (qRT-PCR). Chi-square test was performed to estimate the relationship between miR-148a expression and clinical characteristics of OSCC patients. Cell transfection was carried out using Lipofectamine® 2000. Biological behaviors of tumor cells were detected using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and transwell assays. Bioinformatics analysis and luciferase reporter assay were used to identify the target genes of miR-148a. Protein expression was detected through Western blot analysis.
Results: MiR-148a expression was obviously decreased in OSCC tissues and cells, and such down-regulation was closely correlated with lymph node metastasis (P=0.027) and tumor node metastasis (TNM) stage (P=0.001) of OSCC patients. miR-148a overexpression could significantly impair OSCC cell proliferation, migration and invasion in vitro (P<0.05 for all). Insulin-like growth factor-I receptor (IGF-IR) was a potential target of miR-148a. MiR-148a could inhibit ERK/MAPK signaling pathway through targeting IGF-IR.
Conclusion: MiR-148a plays an anti-tumor role in OSCC and inhibits OSCC progression through suppressing ERK/MAPK pathway via targeting IGF-IR.
Keywords: ERK/MAPK pathway, IGF-IR, MicroRNA, MiR-148a, Oral squamous cell carcinoma
Introduction
Oral squamous cell carcinoma (OSCC) is a frequently diagnosed cancer in oral cavity around the world, especially in men [1]. OSCC represents a leading cause of cancer-related deaths among the head and neck cancers [2]. Tobacco smoking, alcohol abuse and human papilloma virus (HPV) infection are conformed as major risk factors for the occurrence of OSCC [3,4]. With the cessation of tobacco smoking, the morbidity of OSCC exhibits a decreased trend [2]. However, OSCC still presents a great challenge for clinicians, due to its poor prognosis caused by drug resistance, metastasis and recurrence [5]. Therefore, comprehending molecular mechanisms underlying the etiology of OSCC could provide potential novel ways to inhibit metastasis and recurrence, thus improving survival rate.
The progression of OSCC is a complex process caused by the interactions of genetic and epigenetic alterations, and environmental factors [6]. Alteration in microRNAs (miRNAs) is a common epigenetic even in cancer development and progression [7]. MiRNAs are a group of endogenous single-stranded RNAs without protein-coding ability [8]. MiRNAs could bind to the 3′ untranslated regions (UTRs) of their target mRNAs through complementary interaction, thus playing regulatory roles in gene expression at post-transcriptional level [9]. It has been reported that miRNAs can regulate approximately 60% of human genes [10]. The expression profiles of miRNAs show close association with several biological processes, such as cell cycle, development, differentiation, metastasis, metabolism etc [11]. The dysregulation of miRNAs is frequently observed in human diseases, certainly including cancers [12]. The expression patterns of miRNAs are significantly associated with cancer development and progression, so they are considered as promising predictive biomarkers in diagnosing cancers and as therapeutic targets [13].
MiRNA-148a (MiR-148a) belongs to miR-148/miR-152 family which is characterized by a stem-loop structure [14]. The abnormal expression of miR-148a has been observed in several human cancers, including gastric cancer [15], esophageal squamous cell carcinoma [16], non-small cell lung cancer [17] etc. The function of miR-148a in OSCC was also reported in published articles. The study carried out by Min et al. [18] reported that the overexpression of miR-148a could obviously inhibit the migration and invasion of oral carcinoma cells in vitro. However, the molecular mechanisms of miR-148a functioning in the progression of OSCC remain unclear.
In the present study, we aimed to investigate the clinical significance and function of miR-148a in the progression of OSCC. Additionally, cell experiments were designed to explore the underlying molecular mechanisms of miR-148a functioning in OSCC.
Materials and methods
Patients and tissue collection
OSCC tissues and adjacent normal tissues were collected from 110 patients who were pathologically diagnosed with OSCC in The Chinese PLA General Hospital. None of the patients had received any treatments, such as surgery, radiotherapy, chemotherapy etc. Tissue specimens were immediately put in liquid nitrogen and then stored at −80°C. The clinical information of the patients was collected from their medical records. The present study was approved by the Ethic Committee of the hospital. All patients signed the written informed consents in advance.
Cell line and cell culture
OSCC cell line SCC-15 (ATCC® CRL-1623™) and human immortalized oral mucosa epithelial cell HOK (human oral keratinocytes, HOKs) (ATCC® PCS-200-014™) were obtained from American Type Culture Collection (ATCC). The cells were cultured in RPMI-1640 medium with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientifc, Inc., Waltham, MA, U.S.A.). The cells were incubated in a humid chamber at 37°C with 5% CO2.
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was extracted from prepared tissues and cells using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc.) following the instructions of the manufacturer. Then total RNA samples were used for cDNA synthesis which was performed using PrimerScript RT reagent kit (Takara, Dalian, China). Quantitative analysis for genes or mRNAs was carried out using quantitative real-time polymerase chain reaction (qRT-PCR), and reaction was constructed using SYBR Green PCR master mix (Applied Biosystems, U.S.A.) in 7300 Real-Time PCR System (Applied Biosystems, U.S.A.). Specific primer sequences were as follows: U6 forward: 5′-CTCGCTTCGGCAGCACA-3′; reverse: 5′-AACGCTTCACGAATTTGCGT-3′, miR-148a forward: 5′-GGCAGTCTCAGTGCACTACAG-3′; reverse: 5′-GTGCAGGGTCCGAGGT-3′; GAPDH forward: 5′-AAGGCTGGGGCTCATTTGCAGG-3′; reverse: 5′-AGTTGGTGGTGCAGGAGGCA-3′, insulin-like growth factor-I receptor (IGF-IR) forward: 5′-AGCCCCCATCTACCAACAAG-3′; reverse: 5′-GGTGGCATGTCACTCTTCACT-3′. U6 served as an internal reference in detecting miRNAs, while GAPDH was employed as an internal control for mRNA. Results were analyzed employing the method of 2−ΔΔCt. Each test was performed in triplicate.
Cell transfection
To investigate the functional roles of miR-148a in the progression of OSCC, miR-148a mimic and mimic NC were designed and synthesized in HANBIO Company (Shanghai, China). Cells were harvested at logarithmic phase and digested using 0.25% typsin. Then, the cells were seeded into a six-well plate at a density of 1 × 105 cells/ml. Subsequently, cell transfection was performed with Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and corresponding procedures were carried out according to the manufacturer’s instructions. Cell medium was maintained in a humid chamber at 37°C with 5% CO2 for 48 h. Then, the cells were harvested and qRT-PCR method was used to detect the expression of miR-148a in cells to estimate transfection efficacy.
Cell proliferation
Cell proliferation ability was estimated using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cells were adjusted to a density of 1 × 104 cells/ml. Then 200 μl medium was added into 96-well plate and incubated in a humid chamber at 37°C with 5% CO2. And 20 μl of MTT (Sigma) was added into cell medium every 24 h (0 24, 48 and 72 h), and incubated for additional 4 h. Later, 150 μl DMSO was added and incubated at dark to stop reaction. Then absorbance at 490 nm was detected using a Microplate Reader (TECAN, Salzburg, Austria) to estimate cell proliferation ability. Each test was repeated three times.
Cell migration and invasion
In our study, we investigated cell migration and invasion abilities using Transwell assays (8.0 µm pore size, Costar, Shanghai, China). The upper chamber was coated with 200 μl RPMI-1640 medium and 500 μl RPMI-1640 medium with 10% FBS was added to the lower chamber. In addition, Matrigel (Corning Glass Works, Corning, N.Y., U.S.A.) was added to the upper chamber for invasion analysis. A total of 200 μl cell suspension solution with a density of 5 × 104 cells/ml was seeded to the upper chamber, and the cells were incubated in a humid chamber at 37°C with 5% CO2 for 48 h. Then the cells in the lower chamber were stained by Crystal Violet and counted under an inverted microscope (IX31; Olympus Corporation, Tokyo, Japan). Five random files were selected for each sample.
Luciferase reporter assay
Bioinformatics analysis demonstrated that miR-148a might bind to IGF-IR gene in OSCC. Thus, luciferase reporter assay was used to confirm targeting relationship between miR-148a and IGF-IR. The fragment of IGF-IR gene containing the complementary sequence of miR-148a (IGF-IR wt), and the fragment with mutated binding site (IGF-IR mt) were amplified adopting PCR method, and linked to luciferase reporter vector pGL3 (Promega, U.S.A.) according to the instructions of the manufacturer. Then, pGL3-IGF-IR-wt or pGL3-IGF-IR-mt and miR-148a mimic or mimic NC were co-transfected into OSCC cells using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were cultured in a humid chamber at 37°C with 5% CO2 for 48 h. Dual-Luciferase Reporter Assay System (Promega Corporation) was used to detect the luciferase activity of the transfected cells.
Western blot analysis
Protein analysis was completed using Western blot analysis. Protein samples were extracted from cells using RIPA Lysis and Extraction Buffer (Thermo Scientific, Waltham, MA, U.S.A.), and then BCA Protein Assay Kit (Thermo Scientific, Waltham, MA, U.S.A.) was used for the quantitative analysis of protein. Twenty micrograms of protein lysate samples were separated adopting 10% SDS/PAGE. Then the protein samples were transferred on to polyvinylidene fluoride membrane (PVDF) (0.45 µm pore size; EMD Millipore, Billerica, MA, U.S.A.), and then blocked by 5% skim milk powder at room temperature for 2 h. Next, the membrane was incubated with specific primary antibodies at 4°C overnight. Adopted primary antibodies were from Abcam: anti-p-MEK1 antibody (1:5000, rabbit monoclonal antibody, ab96379), anti-p-ERK antibody (1:1000, rabbit polyclonal antibody, ab74032), anti-p38 MAPK antibody (1:10000, rabbit polyclonal antibody, ab197348), anti-p-JNK antibody (1:1000, rabbit polyclonal antibody, ab4821), anti-MEK1 antibody (1:5000, rabbit monoclonal antibody, ab32091), anti-ERK antibody (1:100, rabbit polyclonal antibody, ab137619), anti-MAPK antibody (1:1000, mouse monoclonal antibody, ab185145), anti-JNK antibody (1:1000, rabbit monoclonal antibody, ab179461) and anti-GAPDH antibody (1:10000, mouse monoclonal antibody, ab8245). GAPDH was employed as an internal control. Later, the membranes were incubated with secondary anti-rabbit IgG antibody (1:2000, Abcam, ab190492) at room temperature for 2 h. Band gray was analyzed using Chemi Genius gel imaging system.
Statistical analysis
Continuous variables were expressed as mean ± standard deviation (SD), and their comparison between two groups was performed via Student’s t test. Classification variables were recorded as case number and percentage, and compared between groups using chi-square test. All data analyses were performed using SPSS 18.0 software (SPSS, Inc., Chicago, IL, U.S.A.), and figures were plotted in GraphPad Prism version 5.0 (GraphPad, San Diego, CA, U.S.A.). P-values less than 0.05 meant that results were statistically significant.
Results
Baseline characteristics of the study subjects
A total of 110 OSCC patients including 64 males and 46 females were selected in our study, and their mean age was 61.25 ± 10.15 years. Fifty-eight patients had smoking history, while 56 cases had a history of drinking. Forty-five cases exhibited low differentiation, and lymph node metastasis was observed in 40 patients. In addition, according to tumor node metastasis (TNM) staging, 66 patients were classified into stages I–II and 44 cases into stages III–IV. Detailed characteristics of the included patients are summarized in Table 1.
Table 1. The association of miR-148a expression with clinical characteristics of OSCC patients.
| Characteristics | N (n=110) | miR-148a low expression (n=48) | miR-148a high expression (n=62) | P |
|---|---|---|---|---|
| Age (years) | 0.400 | |||
| ≥60 | 60 | 24 | 36 | |
| <60 | 50 | 24 | 26 | |
| Gender | 0.696 | |||
| Male | 64 | 32 | 32 | |
| Female | 46 | 26 | 30 | |
| Smoking | 0.300 | |||
| Yes | 58 | 28 | 30 | |
| No | 52 | 20 | 32 | |
| Drinking | 0.349 | |||
| Yes | 56 | 22 | 34 | |
| No | 54 | 26 | 28 | |
| Differentiation | 0.522 | |||
| High-moderate | 65 | 30 | 35 | |
| Low | 45 | 18 | 27 | |
| Lymph node metastasis | 0.027 | |||
| Yes | 40 | 23 | 17 | |
| No | 70 | 25 | 45 | |
| TNM stage | 0.001 | |||
| I–II | 66 | 20 | 46 | |
| III–IV | 44 | 28 | 16 |
Expression patterns of miR-148a in OSCC
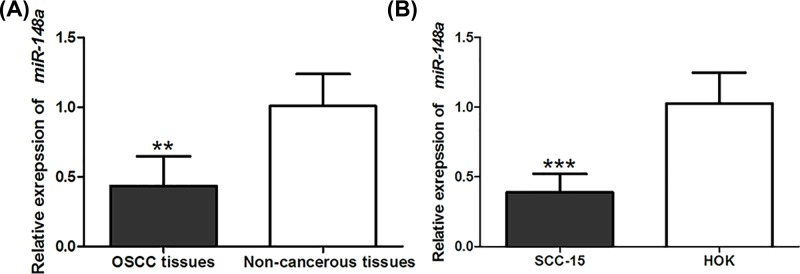
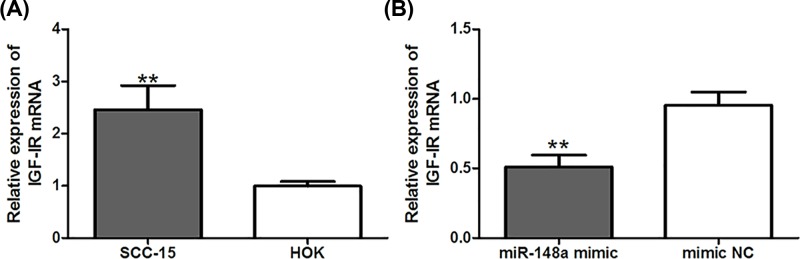
qRT-PCR was performed to investigate the expression profile of miR-148a in OSCC tissues and cells. The results shown in Figure 1 demonstrated that the expression of miR-148a was significantly down-regulated in OSCC tissues and cells, compared with non-cancerous specimens and HOK cell line (P<0.01 for both).
Figure 1. miR-148 expression level in OSCC tissues and cells.
The expression of miR-148a was down-regulated in OSCC tissues (A) and cells (B), compared with the non-cancerous specimens. ***P<0.001, **P<0.01.
Relationship between miR-148a and clinical characteristics of OSCC patients
The included patients were divided into high (n=62) and low (n=48) expression groups based on their mean miR-148a expression in OSCC tissues. Chi-square test was used to estimate the association of miR-148a with clinical characteristics of OSCC patients. The results suggested that the expression of miR-148a showed negative association with lymph node metastasis (P=0.027) and TNM stage (P=0.001). But the expression of miR-148a had no close association with patients’ age, gender, smoking habit, drinking history or differentiation (P>0.05 for all) (Table 1).
Enforced expression of miR-148a could suppress OSCC cell proliferation, migration and invasion
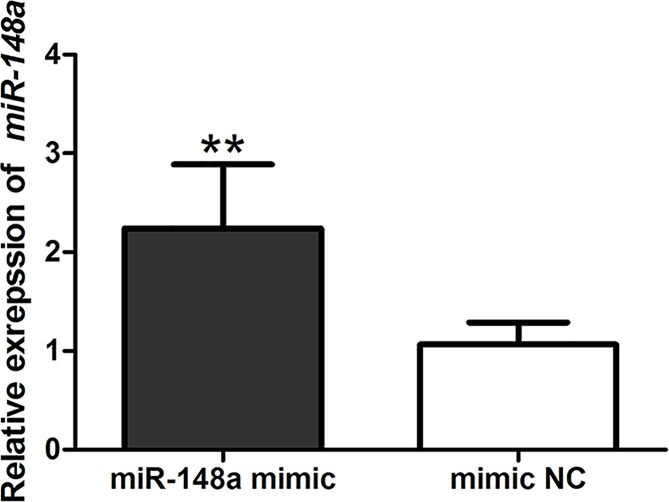
Cell experiments were designed to investigate the functional roles of miR-148a in OSCC progression. SCC-15 cells were transfected by miR-148a mimic and mimic NC. qRT-PCR method was performed to estimate the expression of miR-148a in transfected cells. The expression of miR-148a was obviously increased in miR-148a mimic transfection cells, compared with mimic NC group (P<0.01) (Figure 2).
Figure 2. Detection of transfection efficiency.
The expression of miR-148a was significantly enhanced after the transfection of miR-148a mimic. **: P<0.01.
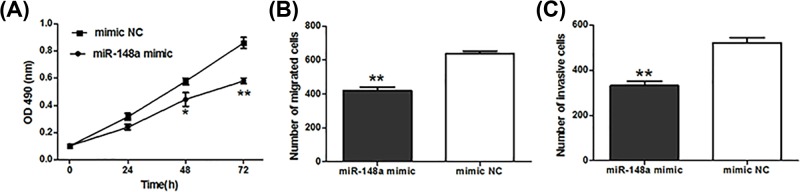
MTT assay suggested that the enforced expression of miR-148a obviously suppressed cell proliferation (P<0.05, Figure 3A). Moreover, cell migration and invasion abilities were also significantly inhibited in OSCC cells with enforced miR-148a expression (P<0.01, Figure 3B,C).
Figure 3. Effect of abnormal miR-148a expression on the behaviors of OSCC cells.
Enforced expression of miR-148a could suppress cell proliferation (A), migration (B) and invasion (C). *: P<0.05, **: P<0.01.
IGF-IR was a potential target gene of miR-148a
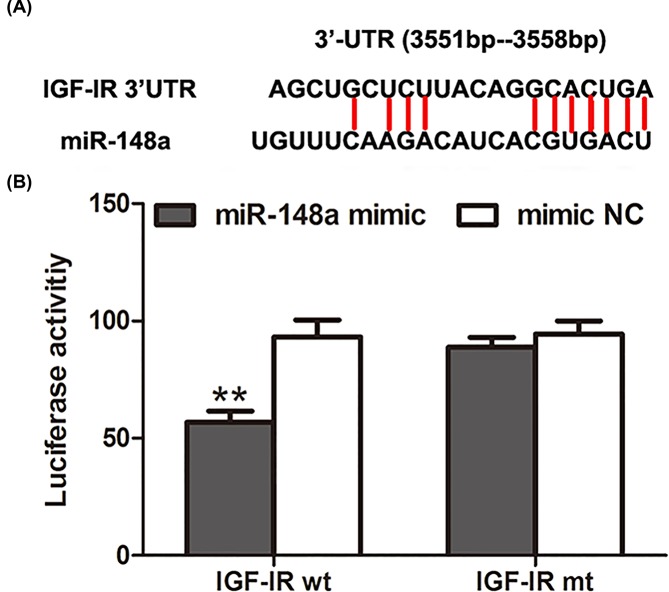
It is known to all that miR-148a has no protein-coding ability, and takes part in biological processes through its target genes. Biological analysis suggested that the 3′UTR of IGF-IR gene had the complementary sequences of miR-148a (Figure 4A). Then, luciferase reporter assay was used to investigate targeting relationship between miR-148a and IGF-IR. Corresponding results showed that the co-transfection by miR-148a mimic and IGF-IR wt could significantly reduce luciferase activity, while the co-transfection by miR-148a mimic and IGF-IR mt had no obvious influenceon luciferase activity, compared with the controls (Figure 4B). These results suggested that miR-148a could bind to the 3′UTR of IGF-IR.
Figure 4. Targeted relationship of miR-148a with IGF-IR.
Biological analysis suggested that the 3′UTR of IGF-IR gene had the complementary sequences of miR-148a (A). Luciferase reporter assay demonstrated that miR-148a could bind to the 3′UTR of IGF-IR (B). **: P<0.01.
qRT-PCR method was used to investigate the expression of IGF-IR in OSCC cells. The results demonstrated that the expression level of IGF-IR was significantly increased in OSCC cells, compared with normal cells (P<0.01, Figure 5A). Moreover, the enforced expression of miR-148a could obviously inhibit the expression of IGF-IR in OSCC cells (P<0.01, Figure 5B). The expression of IGF-IR was negatively correlated with the levels of miR-148a. All data revealed that IGF-IR was a target gene of miR-148a in OSCC.
Figure 5. miR-148a expression is negatively associated with IGF-IR.
Expression of IGF-IR was significantly increased in OSCC cells (A), but the enforced expression of miR-148a could obviously inhibit the expression of IGF-IR (B). **: P<0.01.
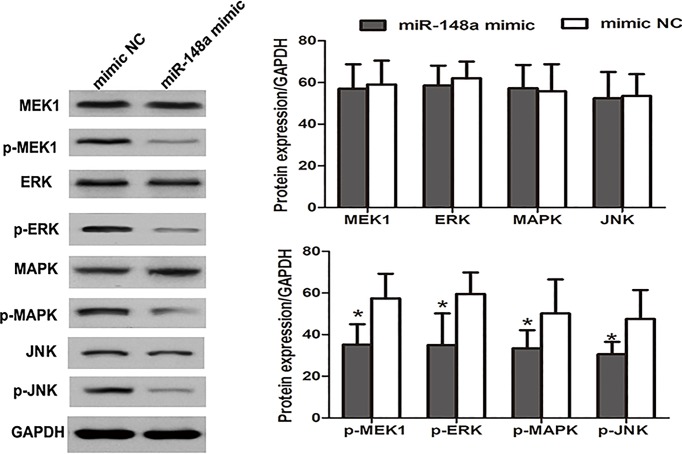
MiR-148a could suppress ERK/MAPK signaling pathway
Several published articles have suggested that IGF-IR could regulate the activity of ERK/MAPK signaling pathway [19,20]. Given the targeting relationship between miR-148a and IGF-IR, we hypothesized that miR-148 might influence ERK/MAPK pathway. Western blot analysis was performed to investigate the expression of proteins in ERK/MAPK pathway. As displayed in Figure 6, the levels of MEK1, ERK, MAPK and JNK did not show significant changes after enhancing miR-148a expression, while miR-148a mimic transfection could significantly reduce their phosphorylation levels, and the levels of p-MEK1, p-ERK, p-MAPK and p-JNK were significantly decreased (P<0.05 for all). The data revealed that the overexpression of miR-148a could inhibit the activity of ERK/MAPK pathway.
Figure 6. Influence of miR-148a expression on the expression of ERK/MAPK signaling pathway relative proteins.
Western blot analysis was performed for protein analysis in ERK/MAPK signaling pathway. The enforced expression of miR-148a could obviously suppress the expression of p-MEK1, p-ERK, p-MAPK and p-JNKL, revealing the decreased activity of ERK/MAPK pathway. *: P<0.05.
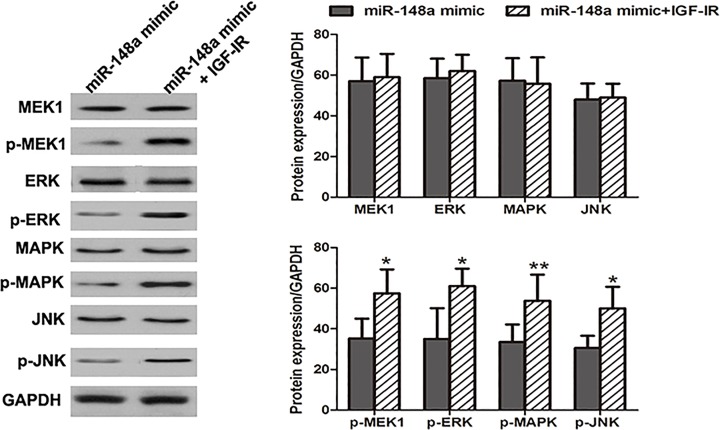
IGF-IR could reverse the anti-tumor action of miR-148a in OSCC
To further explore the molecular mechanisms of miR-148a functioning in OSCC progression, SCC-15 cells were co-transfected by miR-148a mimic and IGF-IR overexpression vector. OSCC cells transfected by miR-148a mimic were employed as the controls. Western blot analysis demonstrated that compared with the controls, co-transfection by miR-148a mimic and IGF-IR overexpression could significantly enhance the expressions of p-MEK1, p-ERK, p-MAPK and p-JNK, suggesting the activation of ERK/MAPK pathway (Figure 7).
Figure 7. miR-148a regulated ERK/MAPK signaling pathway through targeting IGF-IR.
Compared with miR-148a transfection, the co-transfection of miR-148a mimic and IGF-IR overexpression could significantly enhance the expression of p-MEK1, p-ERK, p-MAPK and p-JNKL, suggesting the activation of ERK/MAPK pathway. *: P<0.05, **: P<0.01.
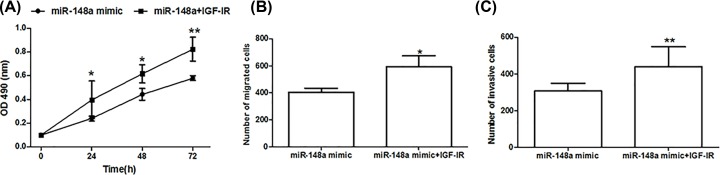
The biological behaviors of OSCC cells after the co-transfection by miR-148a mimic and IGF-IR overexpression vector were also detected. Relevant results demonstrated that enhanced IGF-IR expression could obviously enhance cell proliferation (Figure 8A), migration (Figure 8B) and invasion (Figure 8C) (P<0.05 for all) of OSCC cells with miR-148a overexpression. IGF-IR could reverse anti-tumor action induced by miR-148a overexpression in OSCC (Supplementary Figure S1).
Figure 8. miR-148a impacted the progression of OSCC cells though regulating IGF-IR expression.
Co-transfection of miR-148a mimic and IGF-IR overexpression could obviously enhance cell proliferation (A), migration (B) and invasion (C). *: P<0.05, **: P<0.01.
Discussion
MiRNAs are a group of non-coding RNAs and regulate gene expression through binding to the 3′UTR of their target genes. The dysregulation of miRNAs may lead to alterations in genes which are important in cellular signaling pathways, thus leading to pathological conditions, especially cancers [21]. In cancer development, miRNAs can serve as oncogenes or tumor suppressors. The expression patterns of miRNAs can be employed as potential biomarkers for early detection, screening, monitoring and target treatment in different cancers [22]. Therefore, to explore the function of miRNAs in cancer may improve cancer management. For OSCC, a variety of miRNAs have been focused on. For example, miR-182-5p might promote the growth of OSCC through suppressing the expression of CAMK2N1 [23]. MiR-211 could activate EGFR/MAPK signaling pathway through targeting BIN1 expression, thus contributing to malignant progression of OSCC [24]. In the current study, we investigated the function and clinical significance of miR-148a in the progression of OSCC. We found that miR-148a played an anti-tumor role against OSCC progression through IGF-IR/ERK/MAPK axis.
MiR-148a is a member of miR-148/152 family and its dysregulatiuon has been observed in several human cancers [15–17]. In this study, we found that the expression of miR-148a was significantly down-regulated in OSCC tissues and cells. Furthermore, its down-regulation predicted positive lymph node metastasis and advanced TNM stages among OSCC patients. Cell experiments demonstrated that the enforced expression of miR-148a could obviously suppress the proliferation, migration and invasion of OSCC cells. All data revealed that miR-148a might act as a tumor suppressor against OSCC. The conclusion was consistent with that from previously published article. Min et al. [18] reported that the down-regulation of miR-148a expression might increase the migration and invasion of OSCC cells in vitro. MiR-148a might be a target in treating OSCC.
MiRNAs are considered to take part in biological processes through binding to target genes. In our study, biological analysis and luciferase reporter assay confirmed that IGF-IR might be a target of miR-148a in OSCC. IGF-IR is a transmembrane tyrosine kinase receptor and imposes an important influence on the growth, development and metabolism of cells through binding to IGF-I ligands [25,26]. Growing evidences have demonstrated that the overexpression of IGF-IR may lead to uncontrolled cell proliferation and metastasis, and to drug resistance [27]. The inhibition of IGF-IR may lead to massive apoptosis of tumor cells, realizing anti-tumor action. IGF-IR is confirmed to be a therapeutic target in a variety of cancers [28,29]. In our study, we found that the expression of IGF-IR was obviously increased in OSCC cells, and that its expression was negatively regulated by miR-148a. MiR-148a wielded an anti-tumor action through targeting IGF-IR in OSCC.
ERK/MAPK signaling pathway is an important signaling pathway in mediating cell proliferation, migration and invasion. The activation of ERK/MAPK is frequently observed in human cancers, such as breast cancer [30], gallbladder cancer [31] and OSCC [32]. ERK/MAPK signaling pathway could be activated by RNA-binding proteins and miRNAs at post-transcriptional level [33]. Moreover, the regulatory relationship between miRNAs and ERK/MAPK pathway has been confirmed in several cancers. For example, Li et al. [34] reported that miR-130b promoted glioma progression through activating ERK/MAPK pathway. Wang et al. [35] indicated that miR-16 acted as a tumor suppressor against pituitary adenoma via suppressing ERK/MAPK signaling pathway. In our study, we found that miR-148a could regulate the activity of ERK/MAPK signaling pathway through IGF-IR in OSCC. IGF-IR could reverse the anti-tumor action of miR-148a in OSCC. Despite those encouraging results, several limitations in the current study should be addressed. First, the sample size was relatively small that might influence the statistical power of our results. Second, only one OSCC cell line was adopted in our study, and the expression pattern and function of miR-148a in other OSCC cell lines remained unclear. Third, miR-148a might be involved in tumorigenesis through multiple target genes, but only IGF-IR was confirmed in our study. Further investigations are required to verify and improve our findings.
In conclusion, miR-148a expression is down-regulated in OSCC and it may inhibit the progression of OSCC through inactivating ERK/MAPK signaling pathway via targeting IGF-IR.
Supplementary Material
Abbreviations
- ATCC
American Type Culture Collection
- BIN1
Bridging integrator 1 protein
- CAMK2N1
Calcium/calmodulin-dependent protein kinase II inhibitor 1
- EGFR
Epidermal growth factor receptor
- ERK
extracellular regulated protein kinases
- HOK
human oral keratinocyte
- HPV
human papilloma virus
- IGF-IR
insulin-like growth factor-I receptor
- MAPK
mitogen-activated protein kinase
- MEK1
Mitogen-activated
- MiR-148a
microRNA-148a
- miRNA
microRNA
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NC
Negative control
- OSCC
oral squamous cell carcinoma
- qRT-PCR
quantitative real-time polymerase chain reaction
- RIPA
Radio Immunoprecipitation Assay
- TNM
tumor node metastasis
- UTR
untranslated region
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Innovation Foundation for Junior Researchers of the Chinese PLA General Hospital [grant number 17KMM09]; the Clinical Investigation Foundation of the Chinese PLA General Hospital [grant number 2017FC-TSYS-2013]; and the Special Scientific Research Projects in Public Welfare Industry [grant number 201502018].
Author Contribution
T.J., Y.R., F.W. and R.Z. conceived and designed the experiments, analyzed the data and wrote the article. B.Q., L.X., B.G. and L.O. performed the experiments. All authors read and approved the final manuscript.
Ethics Approval
The present study was supported by the Ethics Committee of The Chinese PLA General Hospital and the experimental procedures were carried out in accordance with the World Medical Association Declaration of Helsinki. All study subjects signed written informed consents.
References
- 1.Siegel R.L., Miller K.D. and Jemal A. (2017) Cancer Statistics, 2017. CA Cancer J. Clin. 67, 7–30 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J. and Jemal A. (2015) Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87–108 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 3.Sathish N., Wang X. and Yuan Y. (2014) Human Papillomavirus (HPV)-associated oral cancers and treatment strategies. J. Dent. Res. 93, 29S–36S 10.1177/0022034514527969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russo D., Merolla F., Varricchio S., Salzano G., Zarrilli G., Mascolo M. et al. (2018) Epigenetics of oral and oropharyngeal cancers. Biomed. Rep. 9, 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gharat S.A., Momin M. and Bhavsar C. (2016) Oral squamous cell carcinoma: current treatment strategies and nanotechnology-based approaches for prevention and therapy. Crit. Rev. Ther. Drug Carrier Syst. 33, 363–400 10.1615/CritRevTherDrugCarrierSyst.2016016272 [DOI] [PubMed] [Google Scholar]
- 6.Boscolo-Rizzo P., Furlan C., Lupato V., Polesel J. and Fratta E. (2017) Novel insights into epigenetic drivers of oropharyngeal squamous cell carcinoma: role of HPV and lifestyle factors. Clin. Epigenetics 9, 124 10.1186/s13148-017-0424-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hema K.N., Smitha T., Sheethal H.S. and Mirnalini S.A. (2017) Epigenetics in oral squamous cell carcinoma. J. Oral Maxillofac. Pathol. 21, 252–259 10.4103/jomfp.JOMFP_150_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z., Xu R. and Li N. (2018) MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr. Metab. (Lond) 15, 68 10.1186/s12986-018-0305-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michlewski G. and Caceres J.F. (2019) Post-transcriptional control of miRNA biogenesis. RNA 25, 1–16 10.1261/rna.068692.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Acunzo M., Romano G., Wernicke D. and Croce C.M. (2015) MicroRNA and cancer–a brief overview. Adv. Biol. Regul. 57, 1–9 10.1016/j.jbior.2014.09.013 [DOI] [PubMed] [Google Scholar]
- 11.Treiber T., Treiber N. and Meister G. (2019) Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 20, 5–20 10.1038/s41580-018-0059-1 [DOI] [PubMed] [Google Scholar]
- 12.Hata A. and Lieberman J. (2015) Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci. Signal. 8, re3 10.1126/scisignal.2005825 [DOI] [PubMed] [Google Scholar]
- 13.Lan H., Lu H., Wang X. and Jin H. (2015) MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed. Res. Int. 2015, 125094 10.1155/2015/125094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Song Y.X. and Wang Z.N. (2013) The microRNA-148/152 family: multi-faceted players. Mol. Cancer 12, 43 10.1186/1476-4598-12-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H., Chen X., Jiang H., Wang X., Yu H., Sun P. et al. (2018) miR-148a suppresses cell invasion and migration in gastric cancer by targeting DNA methyltransferase 1. Oncol. Lett. 15, 4944–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Q., Luo G. and Zhang X. (2017) MiR-148a modulates HLA-G expression and influences tumor apoptosis in esophageal squamous cell carcinoma. Exp. Ther. Med. 14, 4448–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu D., Li J., Lin H., Zhang X., Pan H., Liu L. et al. (2018) Quantitative proteomic analysis of the miR-148a-associated mechanisms of metastasis in non-small cell lung cancer. Oncol. Lett. 15, 9941–9952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min A., Zhu C., Peng S., Shuai C., Sun L., Han Y. et al. (2016) Downregulation of microrna-148a in cancer-associated fibroblasts from oral cancer promotes cancer cell migration and invasion by targeting Wnt10b. J. Biochem. Mol. Toxicol. 30, 186–191 10.1002/jbt.21777 [DOI] [PubMed] [Google Scholar]
- 19.Hollier B.G., Kricker J.A., Van Lonkhuyzen D.R., Leavesley D.I. and Upton Z. (2008) Substrate-bound insulin-like growth factor (IGF)-I-IGF binding protein-vitronectin-stimulated breast cell migration is enhanced by coactivation of the phosphatidylinositide 3-Kinase/AKT pathway by alphav-integrins and the IGF-I receptor. Endocrinology 149, 1075–1090 10.1210/en.2007-0740 [DOI] [PubMed] [Google Scholar]
- 20.Di X., Yu L., Moore A.B., Castro L., Zheng X., Hermon T. et al. (2008) A low concentration of genistein induces estrogen receptor-alpha and insulin-like growth factor-I receptor interactions and proliferation in uterine leiomyoma cells. Hum. Reprod. 23, 1873–1883 10.1093/humrep/den087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupaimoole R. and Slack F.J. (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 22.Tutar L., Tutar E., Ozgur A. and Tutar Y. (2015) Therapeutic targeting of microRNAs in cancer: future perspectives. Drug Dev. Res. 76, 382–388 10.1002/ddr.21273 [DOI] [PubMed] [Google Scholar]
- 23.Li N., Nan C.C., Zhong X.Y., Weng J.Q., Fan H.D., Sun H.P. et al. (2018) miR-182-5p promotes growth in oral squamous cell carcinoma by inhibiting CAMK2N1. Cell. Physiol. Biochem. 49, 1329–1341 10.1159/000493411 [DOI] [PubMed] [Google Scholar]
- 24.Zheng J., Wang J., Jia Y., Liu T., Duan Y., Liang X. et al. (2019) microRNA-211 promotes proliferation, migration, and invasion ability of oral squamous cell carcinoma cells via targeting the bridging integrator 1 protein. J. Cell. Biochem. 120, 4644–4653 10.1002/jcb.27753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qu X., Wu Z., Dong W., Zhang T., Wang L., Pang Z. et al. (2017) Update of IGF-1 receptor inhibitor (ganitumab, dalotuzumab, cixutumumab, teprotumumab and figitumumab) effects on cancer therapy. Oncotarget 8, 29501–29518 10.18632/oncotarget.15704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bieghs L., Johnsen H.E., Maes K., Menu E., Van Valckenborgh E., Overgaard M.T. et al. (2016) The insulin-like growth factor system in multiple myeloma: diagnostic and therapeutic potential. Oncotarget 7, 48732–48752 10.18632/oncotarget.8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Batth I.S., Qu X., Xu L., Song N., Wang R. et al. (2017) IGF-IR signaling in epithelial to mesenchymal transition and targeting IGF-IR therapy: overview and new insights. Mol. Cancer 16, 6 10.1186/s12943-016-0576-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motallebnezhad M., Aghebati-Maleki L., Jadidi-Niaragh F., Nickho H., Samadi-Kafil H., Shamsasenjan K. et al. (2016) The insulin-like growth factor-I receptor (IGF-IR) in breast cancer: biology and treatment strategies. Tumour Biol. 37, 11711–11721 10.1007/s13277-016-5176-x [DOI] [PubMed] [Google Scholar]
- 29.Janssen J.A. and Varewijck A.J. (2014) IGF-IR targeted therapy: past, present and future. Front. Endocrinol. (Lausanne) 5, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park S., Jung H.H., Park Y.H., Ahn J.S. and Im Y.H. (2011) ERK/MAPK pathways play critical roles in EGFR ligands-induced MMP1 expression. Biochem. Biophys. Res. Commun. 407, 680–686 10.1016/j.bbrc.2011.03.075 [DOI] [PubMed] [Google Scholar]
- 31.Buchegger K., Silva R., Lopez J., Ili C., Araya J.C., Leal P. et al. (2017) The ERK/MAPK pathway is overexpressed and activated in gallbladder cancer. Pathol. Res. Pract. 213, 476–482 10.1016/j.prp.2017.01.025 [DOI] [PubMed] [Google Scholar]
- 32.Tsui I.F., Poh C.F., Garnis C., Rosin M.P., Zhang L. and Lam W.L. (2009) Multiple pathways in the FGF signaling network are frequently deregulated by gene amplification in oral dysplasias. Int. J. Cancer 125, 2219–2228 10.1002/ijc.24611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whelan J.T., Hollis S.E., Cha D.S., Asch A.S. and Lee M.H. (2012) Post-transcriptional regulation of the Ras-ERK/MAPK signaling pathway. J. Cell. Physiol. 227, 1235–1241 10.1002/jcp.22899 [DOI] [PubMed] [Google Scholar]
- 34.Li B., Liu Y.H., Sun A.G., Huan L.C., Li H.D. and Liu D.M. (2017) MiR-130b functions as a tumor promoter in glioma via regulation of ERK/MAPK pathway. Eur. Rev. Med. Pharmacol. Sci. 21, 2840–2846 [PubMed] [Google Scholar]
- 35.Wang D.W., Wang Y.Q. and Shu H.S. (2018) MiR-16 inhibits pituitary adenoma cell proliferation via the suppression of ERK/MAPK signal pathway. Eur. Rev. Med. Pharmacol. Sci. 22, 1241–1248 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.