Abstract
The current epidemic of antibiotic resistance has been facilitated by the wide and rapid horizontal dissemination of antibiotic resistance genes (ARGs) in microbial communities. Indeed, ARGs are often located on plasmids, which can efficiently shuttle genes across diverse taxa. While the existence conditions of plasmids have been extensively studied in a few model bacterial populations, their fate in complex bacterial communities is poorly understood. Here, we coupled plasmid transfer assays with serial growth experiments to investigate the persistence of the broad-host-range IncP-1 plasmid pKJK5 in microbial communities derived from a sewage treatment plant. The cultivation conditions combined different nutrient and oxygen levels, and were non-selective and non-conducive for liquid-phase conjugal transfer. Following initial transfer, the plasmid persisted in almost all conditions during a 10-day serial growth experiment (equivalent to 60 generations), with a transient transconjugant incidence up to 30%. By combining cell enumeration and sorting with amplicon sequencing, we mapped plasmid fitness effects across taxa of the microbial community. Unexpected plasmid fitness benefits were observed in multiple phylotypes of Aeromonas, Enterobacteriaceae, and Pseudomonas, which resulted in community-level plasmid persistence. We demonstrate, for the first time, that plasmid fitness effects across community members can be estimated in high-throughput without prior isolation. By gaining a fitness benefit when carrying plasmids, members within complex microbial communities might have a hitherto unrecognised potential to maintain plasmids for long-term community-wide access.
Subject terms: Microbial ecology, Microbial ecology, Environmental microbiology
Introduction
Plasmids—extra-chromosomal replicons—can support rapid bacterial adaptation by moving genes between phylogenetically diverse bacteria, a process known as horizontal gene transfer [1]. This is particularly important in the case of antibiotic resistance, where acquisition of plasmid-borne antibiotic resistance genes (ARGs) can instantly render a strain impervious to antibiotic treatment [2]. The current global antibiotic resistance crisis has been largely attributed to plasmid-mediated ARG dissemination [3].
Strategies to combat the antibiotic resistance epidemic, therefore, require an understanding of the mechanisms that underlie plasmid fate in microbial communities. Upon entering a microbial community, a plasmid will only persist if its original or secondary hosts (transconjugants) survive. If it persists, it will constitute a long-term reservoir of auxiliary genes for that community. In fact, persistence of IncP-1 plasmids and their derivatives has recently been observed in ex situ communities like anaerobic sludge microcosms [4] and an artificial multi-species system [5] over periods longer than 10 days, as well as in murine gut microbiota experiments [6]. However, by simply interpreting plasmid persistence as the frequency of plasmid occurrence, these observational studies did not explicitly examine the mechanisms that underlie plasmid persistence. Therefore, we are currently far from able to predict plasmid fate in a microbial community.
In contrast to our limited understanding of plasmid persistence at the community level, the fate of plasmids at the population level has received much more attention. There, theoretical and empirical studies have demonstrated that the conditions for plasmid persistence are governed by the interplay of segregational loss, conjugal transfer, and the effect of the plasmid on host fitness [7–9]. The rates of these processes depend on the plasmid, the host, and the environmental conditions, in a complex manner. First, although partitioning mechanisms exist to ensure ordered plasmid segregation prior to cell division [10], plasmid loss during vegetative growth can typically be detected [7]. Second, plasmid carriage often imposes a reduction in fitness to the host [11]. Therefore, in the absence of mechanisms countering segregational loss and fitness cost, plasmids would eventually be removed from bacterial populations. To persist, plasmids lacking stringent post-segregational killing systems thus depend on one or both of two mechanisms: fitness cost amelioration and horizontal transfer [9].
In the majority of long-term cultivation experiments, plasmid persistence is associated with a reduction in the burden of plasmid carriage through adaptation (mainly by compensatory mutations) [9]. This lessens selection against plasmid carriage, and therefore reduces the rate at which plasmids are removed from the population. For example, by experimentally evolving Pseudomonas fluorescens carrying a high-fitness-cost plasmid, Harrison et al. observed amelioration of the cost-of-carriage across a wide gradient of environmental conditions, even without positive selection for plasmid carriage [12]. In an evolutionary experiment conducted by Dionisio et al. [13], strong fitness-increase effects were noted of conjugative plasmid R1 both in the original host Escherichia coli and other bacterial lineages including Salmonella enterica. Recent studies on the genetic basis underlying such adaptations among different plasmid-host pairs suggest that poor plasmid persistence can be caused by a high cost involving helicase-plasmid interactions, which can be rapidly ameliorated [14]. Another explanation for plasmid persistence within a population is conjugal transfer. In contrast to conjugation-deficient pKJK5 derivatives, the ability of the original pKJK5 plasmid to conjugate played an essential role in its stable maintenance in populations of E. coli and Kluyvera sp [15, 16]. More recently, Lopatkin et al. [17] experimentally demonstrated that transfer rate of common conjugative plasmids was necessary and sufficient to maintain plasmids in an E. coli population.
Detailed investigations of plasmid fate have been limited to pure or defined cultures of isogenic or closely related strains of a few model species. Such research on species like E. coli and P. putida have confirmed the notion that plasmid persistence is strain dependent [18–20]. Strains of P. putida can differ largely in their ability to maintain the IncP-1 plasmid pB10 [21, 22]. The cause of this inter-strain variability is currently unknown and our ability to identify it is likely limited by the fact that testing single strains is labour intensive.
Since most bacteria exist within complex communities of hundreds to thousands of species, plasmid persistence is ideally examined within this context. However, tracking the fate of a plasmid and its hosts within a complex community poses a great challenge. Population-level experiments ignore the complex interactions within communities, which further prevents inferring plasmid fate in communities. For example, interspecies transfer enabled persistence of plasmid pQBR57, which would have been impossible within a single population [23]. Besides, multi-level ecological dynamics play a role in a community, since fitness differences do not only exist between plasmid-free and plasmid-bearing cells within populations of a singular strain, but also between strains. Therefore, a plasmid can be lost because of low intrinsic fitness of its hosts or even merely because of ecological drift.
Previous work on community permissiveness (i.e., the ability to take up a plasmid from an exogenous donor strain) has revealed that IncP-1 plasmids can readily be transferred to diverse members of complex environmental communities [24–26]. However, it is not clear to what extent the communities can maintain the plasmid and in which way the individual members can contribute to community-level plasmid persistence.
By coupling permissiveness assays using a gfp-tagged plasmid with serial growth experiments, we monitored the persistence of the IncP-1 plasmid pKJK5 not only at the community level but also at the level of individual phylotypes to identify the relative importance of the processes acting on plasmid persistence and how they vary across phylotypes. To reduce complexity, we chose conditions where conjugal transfer is limited, and thus, where segregational loss, plasmid fitness effects, and competition between phylotypes drive plasmid fate. We identified that plasmid fitness benefits in multiple phylotypes played an essential role in community-level plasmid persistence.
Materials and methods
Donor strain and recipient microbial community
E. coli MG1655 carrying pKJK5, was used as donor (further denoted E. coli (pKJK5)). pKJK5, originally recovered from the barley rhizosphere by exogenous isolation [27], is a 54 Kb low-copy-number conjugative plasmid. It encodes the maintenance/stability systems that are typically associated with IncP-1 plasmids (i.e., kle, klc, kor, and par). Despite an identified putative post-segregational killing system, we have frequently observed plasmid loss from transconjugant cells (data not included here), indicating its loose control on plasmid-free segregants. An accessory region carries genes encoding tetracycline and trimethoprim resistance, which is associated with transposons [28]. Neither these nor the other plasmid-encoded genes are known to provide a selective advantage to the host in the absence of antibiotics. pKJK5 transfer host range has been measured in different microbial communities; a broad transfer host range was observed in soil and wastewater communities [24–26]. In this experiment, pKJK5 is marked with gfpmut3b under the control of a LacIq repressible promoter PA1O4O3 [24]. The donor strain is chromosomally tagged with a constitutively expressed lacIq and mCherry using the mini-Tn7 tagging system [24]. Therefore, plasmid-encoded gfp expression is repressed in the donor strain, but de-repressed following successful transfer to a recipient strain. As a result, recipients that acquire the plasmid can be quantified and retrieved as green fluorescent cells by fluorescence-activated cell sorting (FACS). The donor strain was cultured overnight in LB medium supplemented with tetracycline, harvested by centrifugation and washed in 0.9% (w/v) NaCl solution. Cell density was adjusted to 3.0 × 107 cells per ml by direct measurement in a Thomas counting chamber, and used in filter mating assays as described previously [29]. Activated sludge (microbial biomass) grab samples were collected from a municipal wastewater treatment plant (WWTP) (Mølleåværket, Lyngby-Taarbæk, Denmark) in March 2018, then transported refrigerated to the laboratory and processed immediately. Bacteria were extracted from 50 ml activated sludge by washing (centrifuging at approx. 5000 × g for 10 min, carefully removing supernatant and resuspending the pellet in 0.9% NaCl) and sonication (500 J for 4 min with alternating pulses and pauses of equal duration every second, tip diameter 3.0 mm, Branson 250). After 10 min settling, 5 ml supernatant was used for counting and adjusting to 3.0 × 107 cells per ml before filter mating assays.
Filter mating assay and serial growth experiment
As the activated sludge microbial community is a mixture of enteric and non-enteric bacteria with distinct growth condition preferences, we used two media in both filter mating and serial growth experiments to selectively enrich the two community fractions (Fig. S1). MacConkey (MC) medium and synthetic wastewater (SW) medium [30] were used to favour enteric and non-enteric bacteria, respectively. In addition, we set low (100 RPM) and high (500 RPM) stirring rates to vary the degree of oxygen transfer during the serial growth experiment. The high stirring rate was sufficient to ensure fully oxic conditions while the low stirring rate would result in oxygen-limited to anoxic growth in the substrate rich medium (hereafter 500 and 100 RPM are referred to as oxic and anoxic conditions). Initial plasmid transfer in the activated sludge community was facilitated via filter mating assays. Donor and recipient suspensions of 3.0 × 107 cells per ml were mixed at 1:1 ratio (for matings on SW medium) or 1:5 ratio (for matings on MC medium, to compensate for the selectivity of MC media for enterobacteria), and immediately filtered onto a track-etched membrane (pore size: 0.2 µm, Whatman CycloporeTM, UK). Filters were placed on agar-solidified (15 g/l) SW or MC medium. After 48 h incubation at 25 °C, cells from triplicate filters were combined in 2 ml 0.9% NaCl solution and detached by vortexing at 3000 RPM for 3 min. Cell number was adjusted to 3.0 × 107 cells to initiate serial growth in 50 ml Erlenmeyer flasks containing 15 ml of appropriate media. The serial growth experiments were run in triplicate for 10 days under one out of four combinations of media type and stirring rate (i.e., SW100, SW500, MC100, and MC500), without antibiotic selection. Daily, 1% (v/v) of each culture was transferred to a new flask carrying fresh medium, ensuring average community-level growth of about six generations per day. Control experiments were run in parallel to assess the potential for plasmid transfer during the serial growth experiment. To this effect, donor and recipient cells were separately grown on filters (on either SW or MC medium) for 48 h at 25 °C. Then donor and recipient cells were mixed directly in flasks with liquid growth media; triplicate flasks were subjected to serial growth and treated exactly as the experimental groups under the four conditions.
Cell sorting and 16S rRNA gene amplicon sequencing
Throughout the serial growth experiment, the relative abundance of donor, recipient, and transconjugant cells were quantified daily by FACS (BD FACSAria III, USA) with the same gate settings as for sorting (see below). At selected time points during the serial growth experiment (day 2, 4, 6, 8, and 10), cell fractions were also subject to sorting and 16S rRNA gene amplicon sequencing. Transconjugant and recipient cells were sorted using FACS settings as described earlier [24]. In all sortings, we targeted a minimum of 10,000 cells. Samples with low transconjugant cell fractions (<0.1%) were not sorted, as those conditions would require excessive time to ensure purity. Sorted cells were concentrated by centrifugation, and subject to cell lysis and DNA extraction using GenePurgeDirectTM DNA releasing agent (NimaGen, NL). The hypervariable regions V3–V4 of the 16S rRNA gene were PCR amplified using primer set 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) originally published by Yu et al. [31] and modified as described in Sundberg et al. [32]. Tagging using loci-specific index primers for individual samples was done in a second PCR step. After purification using HighPrep PCR magnetic beads (MagBio, USA), sample-wise indexed PCR products were pooled in equimolar proportions and concentrated. Sequencing of pooled libraries was performed on a MiSeq platform using the v3 600 cycles sequencing kit in 300 bp paired-end mode (Illumina, USA). Based on index sequences, samples were directly demultiplexed by Casava 1.8 (Illumina, USA) on the MiSeq instrument to produce the paired-end FASTQ files for sequencing analysis.
Sequence data analysis
The DADA2 pipeline based on amplicon sequence variants (ASVs) was used for 16S rRNA gene amplicon sequence analysis [33]. Briefly, filtration and trimming (maxN = 0, maxEE = 2, truncQ = 2) were performed to clean raw sequences, which then went through error learning, de-replication, and merging. By discriminating biological sequences from sequencing errors, ASVs were then inferred. Taxonomy classification of chimera-free ASVs was done by searching against the SILVA non-redundant database (release 132) [34]. The generated sample-wise ASV abundance table was used for further analysis. Each ASV is hereafter referred to as a phylotype. Only ASVs with a relative abundance threshold above 0.1% were considered for downstream analysis to limit noise attributable to rare taxa. To better place the phylogeny of Aeromonas phylotypes, Aeromonas reference 16S rRNA gene sequences were retrieved from NCBI RefSeq database, and the V3–V4 region sequence was extracted using the same primer set (341F and 806R). The phangorn R-package [35] was used to construct a maximum-likelihood phylogenetic tree, using a neighbour-joining tree as initial tree. Results were visualised using the R-package ggplot2 [36]. All sequences obtained in this study were deposited in NCBI under accession number PRJNA515836.
Statistical analysis
Non-metric multidimensional scaling (NMDS) was carried out using Bray-Curtis distance matrices that were based on relative abundance of ASVs. Diversity indexes (Shannon, richness, and evenness) were calculated by the R-package Vegan [37]. Permutational multivariate analysis of variance (PERMANOVA) tests were conducted to evaluate the effect of the experimental treatments (e.g., oxygen level and medium type), since it is robust to deviation from parametric assumptions. Pearson correlation analysis was performed on individual phylotypes to investigate relationships between their absolute abundance and the total transconjugant density as estimated from FACS signals. Linear regression models were applied to estimate the temporal trends in the relative richness, relative fitness, and loss of unique phylotypes. Linear models were fitted because of the approximate linearity of most of data and because the limited number of data points made fitting more complex models irrelevant.
Identification of the fate of plasmid at the phylotype level
Flow cytometry allows one to quantify the density of three pools of community members and sort them: the transconjugants (cells with green fluorescence), the recipients (which we define here as all community members derived from the wastewater community, irrespective of whether or not they carry, or are permissive to, the pKJK5, i.e., cells with green fluorescence or without fluorescence), and the initial donors (cells with red fluorescence). The absolute abundance of each phylotype in the community was estimated by multiplying the cell densities of each pool by the relative abundance of the phylotype in the transconjugant or recipient pool based on the 16S rRNA gene amplicon sequencing. Phylotypes detected at least once in the transconjugant pool during any serial growth experiment were defined as permissive; phylotypes never detected in the transconjugant pools were defined as non-permissive.
The overall fitness of all permissive phylotypes relative to all recipient phylotypes was calculated by examining the temporal dynamics of the ratio of their absolute abundance (absolute abundance of all permissive phylotypes/absolute abundance of all recipient phylotypes). A positive or negative slope of this time-series ratio indicates an overall growth advantage or disadvantage of permissive phylotypes vs. recipient phylotypes during the experiment, respectively.
For each permissive phylotype, the temporal dynamics of the fraction of plasmid-bearing cells (transconjugants) is impacted by two processes (in the absence of its de novo formation by horizontal gene transfer): first, the positive or negative effect of the plasmid on the phylotype’s fitness relative to plasmid-free cells of the same phylotype; second, the segregational loss of the plasmid. These two processes cannot be distinguished in our experiments. To quantify the combination of these two processes, the temporal dynamics of the ratio of the abundance of each transconjugant phylotype relative to its the total abundance as measured in the recipient pool (absolute abundance of phylotype i in transconjugant pool/absolute abundance of phylotype i in recipient pool) was examined. The slope of this ratio as function of time (hereafter referred to as Sα+δ) is the combined result of plasmid segregational loss and plasmid fitness effects. In order to minimise noise and identify true positive/negative Sα+δ, we only consider slopes biologically significant when they exceed an arbitrary cutoff of 0.01 in absolute value.
Results
Plasmids persisted in microbial communities for more than 60 generations despite negligible horizontal transfer
We created a range of environmental conditions to study plasmid persistence in complex microbial communities. The composition of the recipient pools derived from activated sludge and propagated under different conditions became rapidly and significantly different (SW vs. MC media, oxic vs. anoxic) (PERMANOVA test on media and RPM effect, P value < 0.001) (Fig. S2). The effect of mixing on community composition was the strongest in the MC medium, probably due to the oxygen-sensitivity of enterobacteria, which was also evident in the enrichment of lactose fermenting Enterobacteriaceae under anoxic compared with oxic conditions.
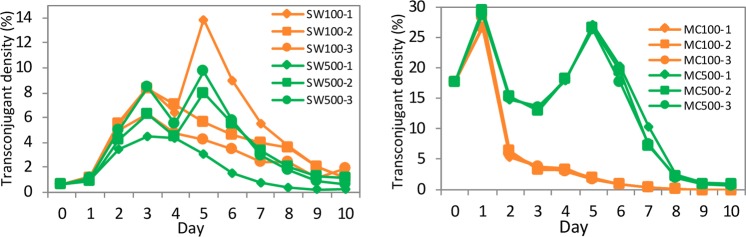
The plasmid persisted across all examined environmental conditions, except in anoxic conditions in MC media (Fig. 1). During the first 3 days in the SW medium, the transconjugant density increased from <1% in the initial filter mating to 4–8% on the 3rd cultivation day. Subsequently, transconjugant densities decreased except for treatments SW100-1, SW500-2, and SW500-3, where a transient increase was observed (8–14%) before densities gradually decreased to low but stable levels (<1%) on 10th day. In the MC medium, transconjugant densities increased from 18% of initial filter mating up to 30% on the 1st day. The degree of oxygen transfer had a significant impact on transconjugant density in the MC medium, but not in the SW medium. The transconjugant densities decreased monotonically to undetectable in the anoxic (MC100) condition, while they increased transiently to 25% under oxic conditions (MC500). Because of the significant variability among replicates, under all experimental conditions, data were presented for all three replicate groups rather than the means.
Fig. 1. Transconjugant density (%) detected by FACS in the initial filter mating samples (0 day) and in the daily samples through 10-day serial growth experiments (1–10 days).
Transconjugant density in serial growth in SW and MC media are shown on the left and right panels, respectively. Transconjugants are cells only with green fluorescence. Three replicates are shown for each experimental condition.
During the serial growth experiments, the observed transconjugant cells must derive either from new plasmid transfer events from donors to recipient cells or from persistent transconjugant cells that were formed during the initial filter mating. We included a control experiment to measure the extent of plasmid transfer during the serial growth experiment. Donor cells decreased rapidly in both control and experimental groups (e.g., from >10% on 1st day to <0.1% on 10th day) (Figs. S3 and S4). Transconjugant cells were absent in almost all control groups, except for 0.1% transconjugant density in MC media on the 1st day (Fig. S5). Hence, horizontal transfer of pKJK5 during the serial growth experiment was negligible.
Persistent transconjugants spanned multiple phylotypes
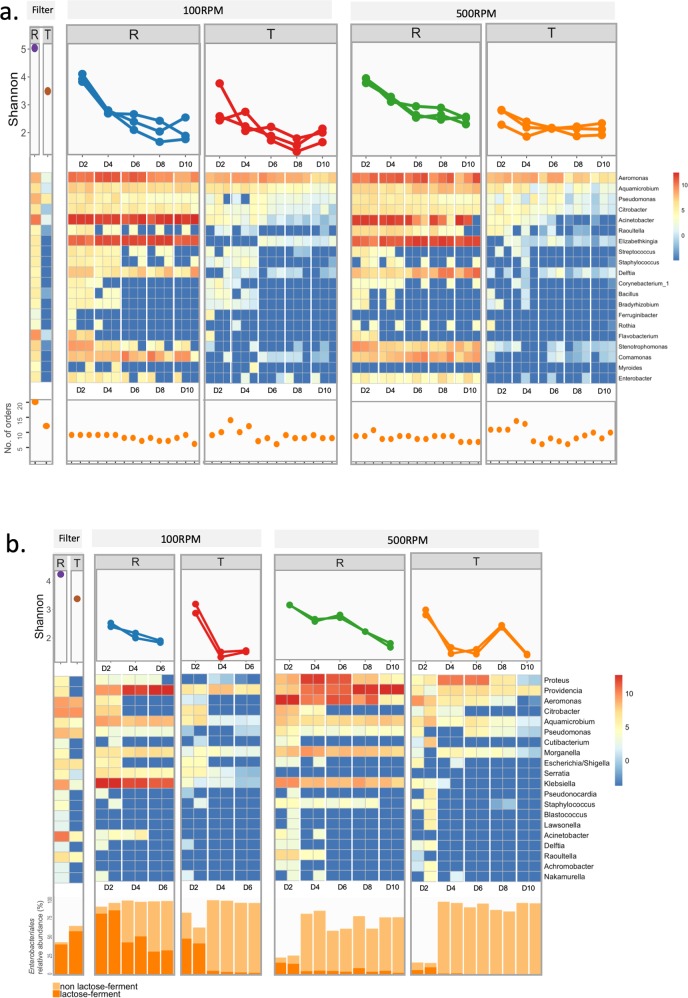
While the recipient community diversity decreased through the experiment (e.g., Shannon index decreased from 4.0 to 2.0 in SW; from 3.0 to 1.7 in MC), substantial diversity remained by the end of the experiment (12–16 genera of 8–10 orders in SW; 7–14 genera of 5–6 orders in MC (Fig. 2)). Although the loss of recipient diversity restricted potential plasmid persistence to the remaining phylotypes, we did not observe significant correlation between recipient and transconjugant community structures by Procrustes analysis of NMDS ordination plots (P value > 0.1).
Fig. 2. Diversity and absolute abundance of the dominant phylotypes in samples from initial filter mating and serial growth experiments.
The top and bottom panels present results from the serial growth experiments in SW (a) and MC (b) media, respectively. (R recipient pools; T transconjugant pools). Within each main panel, the top subpanel show the Shannon diversity index; the middle subpanel, the log2-transformed absolute abundance of the 20 most abundant genera; and the bottom subpanel, the number of unique orders in SW samples (a) and relative abundance of Enterobacteriales (with and without lactose fermenting ability) in MC samples (b). Sampling date is indicated as D2, D4, D6, D8, and D10 on X-axis. Shannon indexes of replicates were plotted along Y-axis and grouped by sampling date, otherwise each replicate data were plotted along X-axis and grouped by sampling date. Scale bar on the right is log2-transformed absolute abundance.
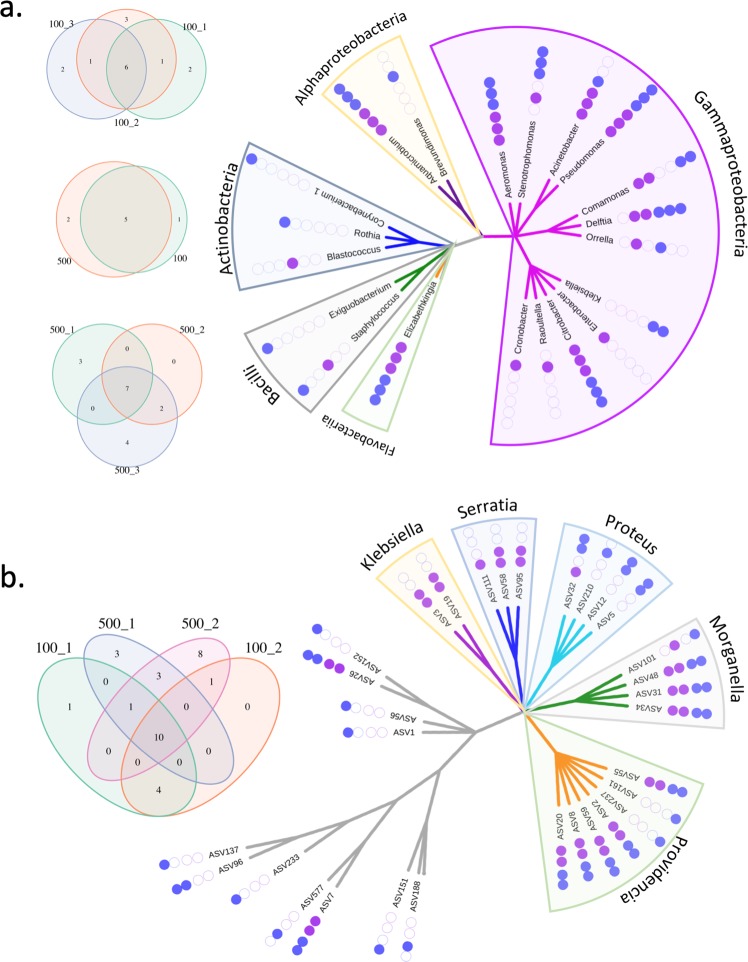
During the filter mating on SW medium, pKJK5 transferred to 29 genera (four phyla) (Fig. 2a). This diversity of transconjugants eroded gradually during serial growth in SW liquid medium (Shannon index decreased from 3.8 to 2.0). The overall composition of transconjugant pools under oxic and anoxic conditions became distinguishable (PERMANOVA, P value = 0.027). The dynamics of transconjugant density in the different serial growth experiments may mainly be driven by a few specific (abundant) phylotypes. We indeed identified multiple phylotypes with absolute abundance highly correlated with the transconjugant density profile during serial growth experiment (r > 0.9, Pearson correlation), e.g., phylotypes in Citrobacter and Enterobacter genera in SW100-1, and in Aeromonas and Stenotrophomonas in SW500-2 (Fig. S6). After 10 days, 62 phylotypes in 9–14 genera were detected across the transconjugant pools. A core of 12 persistent phylotypes in five genera (Aeromonas, Aquamicrobium, Citrobacter, Elizabethkingia, and Pseudomonas) were shared among all SW experiments (Fig. 3a). In each transconjugant pool, Aeromonas phylotypes became enriched (relative abundance > 50%) (Fig. S7). In total, 37 different Aeromonas phylotypes were detected in the recipient pools grown in SW, 12 of which persisted as transconjugants throughout at least one serial growth experiment (Fig. S8). These Aeromonas phylotypes spanned multiple species (Fig. S9), where the dominant ASV6 and ASV1 are closely related to the human opportunistic pathogens A. hydrophila and A. veronii [38], respectively.
Fig. 3. Persistent genera or phylotypes at the end of the serial growth experiment.
Given the higher diversity observed in SW compared with MC, genus level results are presented for SW (a) and phylotype-level results for MC (b). Venn diagrams show the shared and unique genera/phylotypes among groups. Phylogenetic trees show all persistent genera/phylotypes. Class names are used as label for SW, while genus names in Enterobacteriales order are used for MC.
In MC medium, both transconjugant and recipient pools differed significantly from those that developed in SW medium during the 10-day experiment (Figs. 2b and S2). On MC medium, pKJK5 was initially transferred to 18 genera (two phyla) in filter matings, mainly consisting of genera Citrobacter (48% relative abundance), Aeromonas (23%), and Pseudomonas (12%). In the serial growth experiment, transconjugant diversity continuously decreased, as indicated by Shannon index decreasing from 3.0 to 1.0. Oxygen conditions significantly affected the composition of transconjugant pools (PERMANOVA, P value < 0.001). Several phylotypes had abundances that were strongly correlated (r > 0.9, Pearson correlation) with the observed dynamic profile of transconjugant density, such as Proteus and Providencia under oxic condition (Fig. S6). Transconjugants of the Enterobacteriaceae family were more dominant under anoxic than oxic condition. At the end of the experiment, 31 transconjugant phylotypes in 13 genera were detected, in which 8 core phylotypes in Enterobacteriales were shared across all treatments, including 3 Morganella phylotypes and 5 Providencia phylotypes (Fig. 3b).
No general fitness deficit of permissive phylotypes relative to the rest of the community
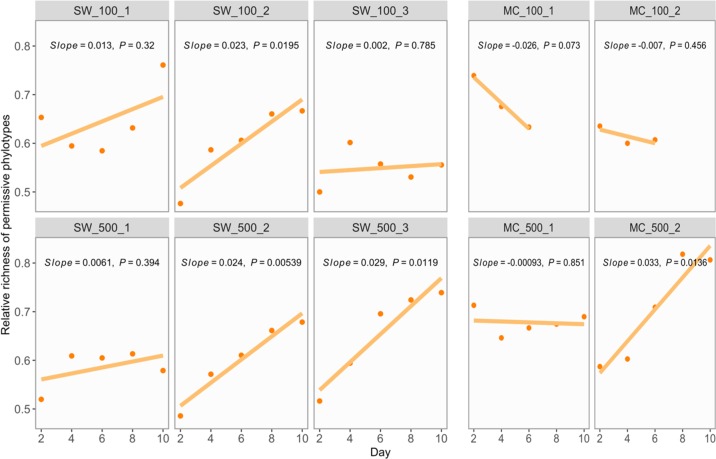
We noted an average 70% decrease in the richness of permissive phylotypes during all serial growth experiments (Fig. S10). In other words, (potential) plasmid hosts disappeared from the community because of insufficient fitness or ecological drift. For example, several Acinetobacter and Pseudomonas phylotypes, abundant at the start of the serial growth experiment in SW media, progressively and simultaneously became absent from both recipient and transconjugant pools. This process would have limited plasmid persistence if permissive phylotypes were lost faster than the rest of the community. The only condition where this might have been the case was MC100 (Fig. 4), although three data points are insufficient to test whether that decline is significant. MC100 was indeed the condition where the plasmid failed to persist. In this case, strong selection against some permissive phylotypes contributed to the disappearance of transconjugants, and thus, to plasmid loss. Elsewhere, the permissive phylotypes appeared to decline equally or less rapidly than the rest of the community, with a similar effect of richness loss on recipient and transconjugant pools within a community, and thus a neutral or positive effect on plasmid persistence.
Fig. 4. Richness of permissive phylotypes relative to all recipient phylotypes over time.
A linear regression model was applied to estimate the temporal trend of the relative richness. Linear model was fitted because of the approximate linearity of most of data and because the limited number of data points made fitting more complex models irrelevant. Solid lines represent regression lines labelled with slope and P value. Positive or negative slope indicated slower or faster richness loss trends in permissive phylotypes compared with recipient phylotypes.
As removal from the community is an extreme outcome of low fitness, we also compared average fitness of permissive and non-permissive phylotypes to look for an overall fitness differential. Across all experimental conditions, the average relative fitness of permissive phylotypes was similar or higher compared with the rest of the community (no significantly negative slopes in Fig. S11). Thus, we did not detect selection against the permissive fraction of the community.
Fitness benefit associated with plasmid carriage improved persistence in multiple phylotypes
The presence of the same phylotype in the plasmid-free and plasmid-carrying fractions of the community gave us the opportunity to assess their relative dynamics and explore the cumulated effect (denoted as Sα+δ) of plasmid segregational loss and plasmid-induced fitness cost/benefit at the phylotype level at high throughput (Fig. 5). A positive value of Sα+δ indicates a net fitness benefit of plasmid carriage since the contribution of plasmid segregation can only be neutral or negative. In contrast, a zero to negative value of Sα+δ makes it difficult to assess the strength and direction of the plasmid fitness effect because plasmid fitness cost/benefit and plasmid segregational loss are indistinguishable in this study. For example, Sα+δ = 0 can be caused by either (segregational loss (negative) + fitness benefit (positive)) or (segregational stability (neutral) + no fitness effect (neutral)).
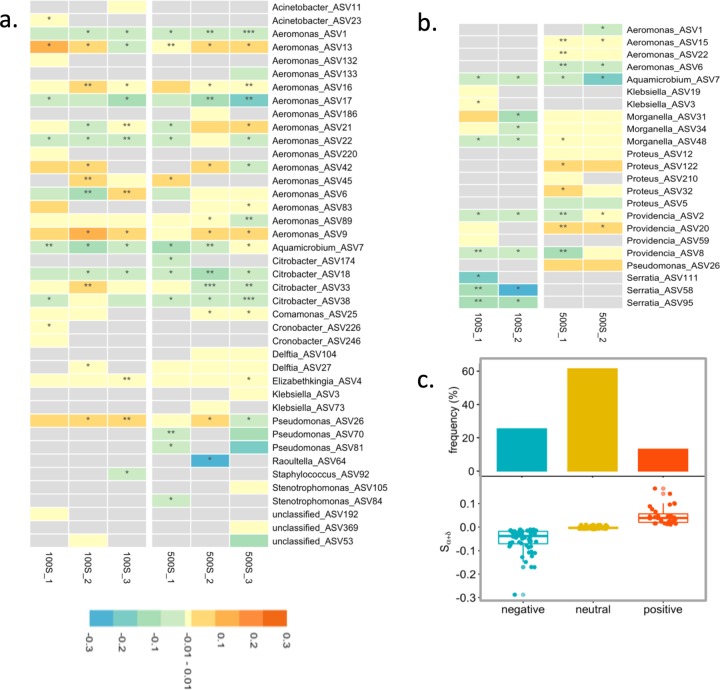
Fig. 5. Plasmid effect of permissive phylotypes during the serial growth experiments.
Sα+δ value profile in SW media (a), MC media (b), and all experimental groups (c). Sα+δ is an estimate of the combined plasmid fitness and segregation effects. −0.01 ≤ Sα+δ ≤ 0.01 or Sα+δ < −0.01: neutral or negative plasmid effect; Sα+δ > 0.01: positive plasmid effect. P values of slope: *<0.5; **<0.1; ***<0.01. Occurrence frequency of three Sα+δ groups with negative, neutral, and positive values in a and b are summarised in c.
Across different experimental conditions, we identified diverse Sα+δ profiles with both positive and negative values among transconjugant phylotypes (hereafter a phylotype under one specific experimental condition is indicated as ASVexpt, e.g., ASV1SW100-1) (Fig. 5a, b). In SW media, more than half of the ASVexpts were without any negative plasmid effect (79 out of 132 ASVexpts with Sα+δ ≥ 0, of which 25 with Sα+δ > 0). Especially, within the genus Aeromonas, there was no negative plasmid effect for 40 of 67 ASVexpts (Sα+δ ≥ 0), with obvious fitness benefit for 20 ASVexpts (Sα+δ > 0). In particular, three Aeromonas phylotypes (ASV9, ASV13, and ASV42) showed fitness benefits in the majority of the serial growth experiments, whereas some phylotypes of the same genus, such as ASV1 and ASV17, experienced plasmid fitness cost. Variability of plasmid effects within a genus was also observed in Pseudomonas, where the incidence of the plasmid in population ASV26 often decreased, in contrast to persistence in ASV70 and ASV81 populations. In other genera like Delftia and Elizabethkingia, there were a few phylotypes with neutral or even positive plasmid effects across experiments. In SW media, the plasmid had a similar effect on phylotypes irrespective of oxygen transfer conditions. However, in MC media, a neutral or positive plasmid effect was more often detected under oxic than anoxic conditions, e.g., 72% (22 out of 32) and 28% (6 out of 21) of ASVexpts with Sα+δ ≥ 0 under oxic and anoxic conditions, respectively. For example, the three phylotypes of Morganella were always associated with a positive plasmid effect under oxic condition but were more likely to suffer from fitness cost or plasmid loss under anoxic condition. Within the same genus, we observed phylotype-level variation of plasmid effects in Aeromonas (e.g., ASV15 vs ASV6) and Providencia (e.g., ASV2 vs ASV20). We identified 28 out of 53 ASVexpts with neutral (20 with Sα+δ = 0) or positive (8 with Sα+δ > 0) plasmid effects in MC media. Interestingly, Pseudomonas ASV26 and Aquamicrobium ASV7 presented a consistent response to plasmid carriage (respectively positive and negative) on the two growth media, suggesting limited impact of the experimental conditions on plasmid maintenance in the two phylotypes. Overall, across experimental conditions in both SW and MC media, more than half (107 out of 185) of the examined ASVexpts maintained the plasmid without any negative plasmid effect, and several (33 out of 185) experienced fitness benefits (Fig. 5c).
Discussion
We examined, for the first time, a complex environmental community to quantitatively monitor persistence of an IncP-1 plasmid pKJK5 among diverse taxa in serial growth experiments. Without any antibiotic pressure to select for transconjugants, we focused on the potential of the community to maintain the plasmid under different nutrient and oxygen conditions. We could discount the contribution of new transfer events to the plasmid fate, as supported by the negligible transfer of pKJK5 in the control experiments and consistent with the previously reported low transfer of pKJK5 in liquid conditions [16]. Indeed, plasmids (IncP/N/W) with genes encoding short and rigid pili only transfer efficiently on solid surfaces, unlike those with long and flexible pili (IncF/H/T/J), capable of transferring equally well in liquid and on solid surfaces [39, 40].
We experimentally demonstrate community-level plasmid persistence across different environmental conditions in the absence of intentional selection for plasmid carriage and in the absence of significant de novo transfer. In SW and MC media, time-series profiles of transconjugant density and composition became significantly different. The role of the environment in shaping transconjugant composition became obvious, especially under oxic and anoxic conditions in MC media. In fact, earlier observations have consistently identified the importance of the environment in determining plasmid fate, by either setting up microcosms under different oxygen conditions (e.g., aerobic and anaerobic sludge reactors) [4] or manipulating other environmental factors (e.g., spatial structure or presence/absence of antibiotics) [5].
By examining plasmid effects temporally across taxa in complex microbial communities, we dissected processes contributing to plasmid persistence within a community. We observed that a plasmid fitness benefit in multiple phylotypes contributed strongly to plasmid persistence at the community level. We expected plasmid pKJK5 to be a neutral or even detrimental extra-chromosomal element for the fitness of its host, as it does not encode apparent beneficial phenotypes under the applied conditions. It is possible that during the experimental period, co-adaptation between plasmid and hosts decreased their negative interactions, thus, improving plasmid maintenance in specific phylogenetic groups. Although such co-adaptation—mainly through plasmid fitness cost amelioration—is frequently observed in population-level studies over evolutionary time scale (>500 generations), it can also appear as early as within 50 generations, such as the rapidly improved stability of IncP-1 plasmid pBP136 in E. coli under non-selective conditions [41]. In experiments on plasmid-host compensatory coevolution, genomic changes frequently emerge within 100 generations [20, 42]. Therefore, early genomic adaptations might have occurred improving persistence within the time scale of the experiment. These adaptions might ameliorate fitness cost of plasmids, and even confer fitness advantages to the host [13]. Indeed, naturally occurring plasmids can confer a fitness advantage upon their hosts, even if the genetic basis of this fitness advantage is not clear [43]. It is also possible that the microbial community we used as potential recipients might already have been subjected to compensatory adaptation for hosting IncP-1 plasmids. Genomic and experimental studies have demonstrated a high prevalence of IncP-1 plasmids spanning all subgroups (IncP-1 α, β, γ, δ, and ε) in WWTP systems [44–46]. If this were the case, community members with prior experience of genetic interaction with IncP-1 plasmids might favour carrying cognate plasmids like pKJK5. While this could explain why pKJK5 might have caused minimal burden to some phylotypes, it does not explain why it would provide a fitness benefit. It is possible that experimental conditions might bias the observed pattern of plasmid effects. However, we observed frequent plasmid fitness benefits with two media and with two mixing conditions. Further experiments with specific knockout mutants of target plasmid genes will likely be necessary to provide a molecular explanation.
The wide range of Sα+δ values across phylotypes and conditions confirmed that the plasmid effect is a phylotype and context dependent feature. In fact, population-level studies have repeatedly revealed that plasmid effects are indeed determined by strain-specific factors [47, 48]. We identify here, for the first time, phylotype-level-specific plasmid effects within a complex microbial community. Although the underlying mechanisms are not yet clear, the heterogenous plasmid effect observed in this study might be the overall result of phylotype-level-specific compensatory adaptations, protective mechanisms, and/or regulatory networks. For example, as revealed in plasmid-host interactions in clinical isolates of E. coli and K. pneumoniae [20], differential adaptive evolutions among strains and species modify the plasmid effect on hosts in various ways.
Across the experimental conditions, plasmid persistence was especially observed in several phylotypes in the Aeromonas, Proteus, Providencia and Pseudomonas genera. In particular, several phylotypes of Aeromonas experienced fitness benefits from pKJK5 carriage under different experimental conditions. These genera have often been reported as carriers of antibiotic resistance plasmids in either environmental or clinical settings, such as plasmid-mediated carbapenem- and colistin-resistance in Proteus and Providencia isolates. Recent studies have demonstrated the high potential of Aeromonas for carrying multidrug-resistance plasmids [49–51], and transferring plasmids in various environmental communities [24–26]. Given the ubiquitous presence of Aeromonas in aquatic environments [52, 53], Aeromonas might play a significant role in facilitating transfer and maintenance of plasmid-mediated ARGs in environmental communities.
We demonstrated that complex microbial communities can maintain plasmids under non-selective conditions, and phylotype-level plasmid fitness benefits significantly facilitate community-level persistence. To further understand the role of the community in modulating plasmid behaviour, the distinct mechanisms of plasmid-host interactions in communities compared with populations remain to be elucidated.
Supplementary information
Acknowledgements
This work was supported by a Joint Programming Initiative-Antimicrobial Resistance grant (JPI-AMR; DARWIN project #7044-00004B) to BFS; and a fellowship under the H.C. Ørsted Postdoc programme co-funded by the Marie Skłodowska-Curie Actions awarded to LL.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Liguan Li, Arnaud Dechesne
Supplementary information
The online version of this article (10.1038/s41396-020-0596-4) contains supplementary material, which is available to authorized users.
References
- 1.Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet. 2015;16:472–82. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 2.Holmes AH, Moore LSP, Sundsfjord A, Steinbakk M, Regmi S, Karkey A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387:176–87. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 3.Crofts TS, Gasparrini AJ, Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol. 2017;15:422–34. doi: 10.1038/nrmicro.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merlin C, Bonot S, Courtois S, Block JC. Persistence and dissemination of the multiple-antibiotic-resistance plasmid pB10 in the microbial communities of wastewater sludge microcosms. Water Res. 2011;45:2897–905. doi: 10.1016/j.watres.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Cairns J, Ruokolainen L, Hultman J, Tamminen M, Virta M, Hiltunen T. Ecology determines how low antibiotic concentration impacts community composition and horizontal transfer of resistance genes. Commun Biol. 2018;1–8. 10.1038/s42003-018-0041-7. [DOI] [PMC free article] [PubMed]
- 6.Ronda C, Chen SP, Cabral V, Yaung SJ, Wang HH. Metagenomic engineering of the mammalian gut microbiome in situ. Nat Methods. 2019;16:167–70. doi: 10.1038/s41592-018-0301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergstrom CT, Lipsitch M, Levin BR. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics. 2000;155:1505–19. doi: 10.1093/genetics/155.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin BR, Stewart FM. The population biology of bacterial plasmids: a priori conditions for the existence of mobilizable nonconjugative factors. Genetics. 1980;94:425–43. doi: 10.1093/genetics/94.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison E, Brockhurst MA. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012;20:262–7. doi: 10.1016/j.tim.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Pinto UM, Pappas KM, Winans SC. The ABCs of plasmid replication and segregation. Nat Rev Microbiol. 2012;10:755–65. doi: 10.1038/nrmicro2882. [DOI] [PubMed] [Google Scholar]
- 11.San Millan A, MacLean RC. Fitness costs of plasmids: a limit to plasmid transmission. Microbiol Spectr. 2017;5:1–12. doi: 10.1128/microbiolspec.mtbp-0016-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison E, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr Biol. 2015;25:2034–9. doi: 10.1016/j.cub.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 13.Dionisio F, Conceicao IC, Marques AC, Fernandes L, Gordo I. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol Lett. 2005;1:250–2. doi: 10.1098/rsbl.2004.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loftie-Eaton W, Bashford K, Quinn H, Dong K, Millstein J, Hunter S, et al. Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat Ecol Evol. 2017;1:1354–63. doi: 10.1038/s41559-017-0243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundquist PD, Levin BR. Transitory derepression and the maintenance of conjugative plasmids. Genetics. 1986;113:483–97. doi: 10.1093/genetics/113.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bahl MI, Hansen LH, Sørensen SJ. Impact of conjugal transfer on the stability of IncP-1 plasmid pKJK5 in bacterial populations. FEMS Microbiol Lett. 2007;266:250–6. doi: 10.1111/j.1574-6968.2006.00536.x. [DOI] [PubMed] [Google Scholar]
- 17.Lopatkin AJ, Meredith HR, Srimani JK, Pfeiffer C, Durrett R, You L. Persistence and reversal of plasmid-mediated antibiotic resistance. Nat Commun. 2017;8:1689. doi: 10.1038/s41467-017-01532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall JPJ, Harrison E, Lilley AK, Paterson S, Spiers AJ, Brockhurst MA. Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects. Environ Microbiol. 2015;17:5008–22. doi: 10.1111/1462-2920.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heuer H, Ebers J, Weinert N, Smalla K. Variation in permissiveness for broad-host-range plasmids among genetically indistinguishable isolates of Dickeya sp. from a small field plot. FEMS Microbiol Ecol. 2010;73:190–6. doi: 10.1111/j.1574-6941.2010.00880.x. [DOI] [PubMed] [Google Scholar]
- 20.Porse A, Schønning K, Munck C, Sommer MOA. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol Biol Evol. 2016;33:2860–73. doi: 10.1093/molbev/msw163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heuer H, Fox RE, Top EM. Frequent conjugative transfer accelerates adaptation of a broad-host-range plasmid to an unfavorable Pseudomonas putida host. FEMS Microbiol Ecol. 2007;59:738–48. doi: 10.1111/j.1574-6941.2006.00223.x. [DOI] [PubMed] [Google Scholar]
- 22.De Gelder L, Ponciano JM, Joyce P, Top EM. Stability of a promiscuous plasmid in different hosts: No guarantee for a long-term relationship. Microbiology. 2007;153:452–63. doi: 10.1099/mic.0.2006/001784-0. [DOI] [PubMed] [Google Scholar]
- 23.Hall JPJ, Wood AJ, Harrison E, Brockhurst MA. Source–sink plasmid transfer dynamics maintain gene mobility in soil bacterial communities. Proc Natl Acad Sci. 2016;113:8260–5. doi: 10.1073/pnas.1600974113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klümper U, Riber L, Dechesne A, Sannazzarro A, Hansen LH, Sørensen SJ, et al. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2015;9:934–45. doi: 10.1038/ismej.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Dechesne A, He Z, Madsen JS, Nesme J, Sørensen SJ, et al. Estimating the transfer range of plasmids encoding antimicrobial resistance in a wastewater treatment plant microbial community. Environ Sci Technol Lett. 2018;5:260–5. [Google Scholar]
- 26.Jacquiod S, Brejnrod A, Morberg SM, Abu Al-Soud W, Sørensen SJ, Riber L. Deciphering conjugative plasmid permissiveness in wastewater microbiomes. Mol Ecol. 2017;26:3556–71. doi: 10.1111/mec.14138. [DOI] [PubMed] [Google Scholar]
- 27.Gstalder ME, Faelen M, Mine N, Top EM, Mergeay M, Couturier M. Replication functions of new broad host range plasmids isolated from polluted soils. Res Microbiol. 2003;154:499–509. doi: 10.1016/S0923-2508(03)00143-8. [DOI] [PubMed] [Google Scholar]
- 28.Bahl MI, Hansen LH, Goesmann A, Sørensen SJ. The multiple antibiotic resistance IncP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established α, β, and δ sub-groups. Plasmid. 2007;58:31–43. doi: 10.1016/j.plasmid.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Klümper U, Dechesne A, Smets BF. Protocol for evaluating the permissiveness of bacterial communities toward conjugal plasmids by quantification and isolation of transconjugants. In: McGenity TJ, Timmis KN, Balbina N, editors. Hydrocarbon and lipid microbiology protocols: genetic, genomic and system analyses of communities. Berlin Heidelberg: Springer; 2014. p. 275–88.
- 30.OECD. Test No. 303: simulation test – aerobic sewage treatment. In: OECD guidelines for the testing of chemicals, section 3. France, Paris: OECD Publishing; 2013. p. 50.
- 31.Yu Y, Lee C, Kim J, Hwang S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng. 2005;89:670–9. doi: 10.1002/bit.20347. [DOI] [PubMed] [Google Scholar]
- 32.Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, et al. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol. 2013;85:612–26. doi: 10.1111/1574-6941.12148. [DOI] [PubMed] [Google Scholar]
- 33.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schliep KP. Phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–3. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wickham H. ggplot2—elegant graphics for data analysis. 2nd ed. New York, NY: Springer; 2016. [Google Scholar]
- 37.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: community ecology package. R package version 2.5-4. 2019. https://CRANR-project.org/package=vegan.
- 38.Janda JM, Abbott SL. The genus aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev. 2010;23:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bradley DE. Determination of pili by conjugative bacterial drug resistance plasmids of incompatibility groups B, C, H, J, K, M, V, and X. J Bacteriol. 1980;141:828–37. doi: 10.1128/jb.141.2.828-837.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradley DE. Characteristics and function of thick and thin conjugative pili determined by transfer-derepressed plasmids of incompatibility groups I1, I2, I5, B, K and Z. Microbiology. 1984;130:1489–502. doi: 10.1099/00221287-130-6-1489. [DOI] [PubMed] [Google Scholar]
- 41.Sota M, Top EM. Host-specific factors determine the persistence of IncP-1 plasmids. World J Microbiol Biotechnol. 2008;24:1951–4. [Google Scholar]
- 42.Bottery MJ, Wood AJ, Brockhurst MA. Temporal dynamics of bacteria-plasmid coevolution under antibiotic selection. ISME J. 2018;13:559–62. doi: 10.1038/s41396-018-0276-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enne VI, Bennett PM, Livermore DM, Hall LMC. Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J Antimicrob Chemother. 2004;53:958–63. doi: 10.1093/jac/dkh217. [DOI] [PubMed] [Google Scholar]
- 44.Zhang T, Zhang X-X, Ye L. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS ONE. 2011;6:e26041. doi: 10.1371/journal.pone.0026041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlüter A, Szczepanowski R, Pühler A, Top EM. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol Rev. 2007;31:449–77. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- 46.Bahl MI, Burmølle M, Meisner A, Hansen LH, Sørensen SJ. All IncP-1 plasmid subgroups, including the novel ε subgroup, are prevalent in the influent of a Danish wastewater treatment plant. Plasmid. 2009;62:134–9. doi: 10.1016/j.plasmid.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 47.Vogwill T, Maclean RC. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl. 2015;8:284–95. doi: 10.1111/eva.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kottara A, Hall JPJ, Harrison E, Brockhurst MA. Variable plasmid fitness effects and mobile genetic element dynamics across Pseudomonas species. FEMS Microbiol Ecol. 2018;94:1–7. doi: 10.1093/femsec/fix172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olaniran AO, Nzimande SBT, Mkize NG. Antimicrobial resistance and virulence signatures of Listeria and Aeromonas species recovered from treated wastewater effluent and receiving surface water in Durban, South Africa. BMC Microbiol. 2015;15:234. doi: 10.1186/s12866-015-0570-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Popowska M. Occurrence and variety of β-lactamase genes among Aeromonas spp. isolated from urban wastewater treatment plant. Front Microbiol. 2017;8:1–12. doi: 10.3389/fmicb.2017.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hedges RW, Smith S, Brazil G. Resistance plasmids of aeromonads. J Gen Microbiol. 1985;131:2091–5.
- 52.Piotrowska M, Popowska M. The prevalence of antibiotic resistance genes among aeromonas species in aquatic environments. Ann Microbiol. 2014;921–34.
- 53.Ye L, Zhang T. Pathogenic bacteria in sewage treatment plants as revealed by 454 pyrosequencing. Environ Sci Technol. 2011;45:7173–9. doi: 10.1021/es201045e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.