Abstract
Many commensal oral streptococci generate H2O2 via pyruvate oxidase (SpxB) to inhibit the growth of competing bacteria like Streptococcus mutans, a major cariogenic species. In Streptococcus sanguinis SK36 (SK36) and Streptococcus gordonii DL1 (DL1), spxB expression and H2O2 release are subject to carbon catabolite repression by the catabolite control protein A (CcpA). Surprisingly, ccpA deletion mutants of SK36 and DL1 fail to inhibit S. mutans despite their production of otherwise inhibitory levels of H2O2. Using H2O2-deficient spxB deletion mutants of SK36 and DL1, it was subsequently discovered that both strains confer protection in trans to other bacteria when H2O2 is added exogenously. This protective effect depends on the direct detoxification of H2O2 by the release of pyruvate. The pyruvate dependent protective effect is also present in other spxB-encoding streptococci, such as the pneumococcus, but is missing from spxB-negative species like S. mutans. Targeted and transposon-based mutagenesis revealed Nox (putative H2O-forming NADH dehydrogenase) as an essential component required for pyruvate release and oxidative protection, while other genes such as sodA and dps play minor roles. Furthermore, pyruvate secretion is only detectable in aerobic growth conditions at biofilm-like cell densities and is responsive to CcpA-dependent catabolite control. This ability of spxB-encoding streptococci reveals a new facet of the competitive interactions between oral commensals and pathobionts and provides a mechanistic basis for the variable levels of inhibitory potential observed among H2O2-producing strains of commensal oral streptococci.
Subject terms: Bacterial genetics, Bacterial genetics, Microbial ecology
Introduction
The oral cavity is a habitat for more than 700 different microbial taxa [1]. Oral microbes are exposed to saliva flow and nutrition intake causing mechanical removal, in addition to changing conditions in temperature, oxygen tension, and pH levels. To cope with the permanent environmental stresses oral bacteria are mostly organized in matrix enclosed communities called biofilms, which provide protection against all these factors [2]. Moreover, biofilm-dwelling oral microbes are in symbiosis with the host and provide protection against invading pathogens [3, 4]. However, in case of an increasing biofilm volume and/or an alteration of the species composition to a dysbiotic state, these mixed species biofilms cause oral diseases like caries, chronic gingivitis, periodontitis, and others [5–7].
Streptococci are among the first colonizers of the oral hard tissue; they provide specific lectin-like attachment sides for later colonizers and are essential for initial biofilm formation [8]. In their role as initial colonizers, oral streptococci compete with other microorganisms by releasing significant amounts of hydrogen peroxide (H2O2), which is generated by the pyruvate oxidase SpxB in an oxygen dependent manner [9–13]. The ability to release H2O2 as well as to deal with its toxicity influences the composition of the mature biofilm and is therefore of high relevance for the ecology of the mixed-species oral biofilm. Interestingly, despite many investigations into streptococcal reactive oxygen species stress mechanisms [14–17], it is still not fully understood how oral streptococci manage H2O2 stress, since they do not encode enzymes for direct detoxification of H2O2 (i.e., catalase) [17–19].
Streptococcus sanguinis SK36 (SK36) and S. gordonii DL1 (DL1) are abundant and well characterized oral commensals [8]. Their H2O2 dependent antagonism against caries associated S. mutans has been demonstrated [12, 13, 20–23]. In both species, the expression of spxB is under carbon catabolite control mediated by catabolite control protein A (CcpA) repression [24, 25]. As a result, H2O2 release and consequently its inhibitory capacity is influenced by different carbohydrates [24]. Interestingly, the CcpA dependent repression of spxB expression works differently in SK36 and DL1: while an increased glucose concentration causes a reduced release of H2O2 in DL1, there is almost no glucose effect on H2O2 release in SK36. Even under low-glucose concentrations, spxB is under strong CcpA dependent catabolite control. Under these conditions, H2O2 release is significantly increased in SK36 ΔccpA but mostly unaffected in DL1 ΔccpA compared with its parental wild-type strain [24, 25].
Accordingly, glucose concentration affects the inhibitory capacity of DL1, but exerts a minimal impact upon SK36 [11]. Recently, we made the surprising observation that ccpA mutants exhibit a dramatic reduction in their inhibitory capacities, despite producing inhibitory levels of H2O2. The present study aimed to elucidate the underlying mechanism of this intriguing phenotype. Here, we provide the first evidence demonstrating prokaryotic pyruvate secretion as a potent antioxidant for H2O2 production, which has potentially far reaching consequences for our understanding of the competitive interactions between members of the oral microbiome.
Materials and methods
Bacterial strains and media
All bacterial strains (Table S1) used in this study were grown in liquid or on agar solidified Bacto Brain Heart Infusion (BHI, Becton Dickinson & Co., Sparks, MD, USA) at 37 °C under aerobic (5% CO2) or anaerobic (90% N2, 5% CO2, 5% H2) conditions. Glucose was added to the media from a filter sterilized 20% (w/v) stock solution where indicated. The following antibiotics were supplemented for selection: kanamycin (300 µg ml−1), erythromycin (10 µg ml−1), and spectinomycin (200 µg ml−1).
Planktonic and high-cell-density growth conditions
The release of the protective component was tested under planktonic and high-cell-density growth conditions. Twenty milliliters of BHI was inoculated with S. sanguinis ΔccpA/ΔspxB cells from a fresh overnight culture (PBS washed) to a final OD600 = 0.05 and incubated aerobically (37 °C, 5% CO2). After 2 h intervals, 200 μl were applied on sterile membrane filters (47 mm, 0.45-μm pore size, Millipore) being placed on BHI plates and allowed to dry for 30 min. Subsequently, filters were removed and the plates were stored at 4 °C. For high-cell-density growth conditions (on agar solidified medium [26]), 200 μl of S. sanguinis ΔccpA/ΔspxB cells from a fresh overnight culture (PBS washed, OD600 = 0.5) was applied to sterile membrane filters, placed on BHI plates, dried for 30 min, and incubated aerobically (37 °C, 5% CO2). After 2 h intervals, filters were removed and the plates were stored at 4 °C. The next day, H2O2 protection assays were performed as described below.
DNA manipulations
Standard nucleic acid recombinant protocols were use [27]. Gene deletions as well as complementation of the respective genes were performed via allelic replacement with previously described transformation protocols [28–31]. For a detailed description please refer to the Supplementary material section. Primers are listed in Table S2.
Accession number(s)
GenBank accession numbers for sequences determined by comC locus sequencing (Supplementary material) are as follows: MN257610 (S. oralis subsp. tigurinus J22, comC), MN257611 (S. infantis LZ2, comC), and MN257612 (S. mitis 12261, comC).
Measurement of relative H2O2 production
To detect and quantify H2O2, BHI/Prussian-blue indicator plates were prepared as previously described [32]. Cultivation and determination of relative H2O2 release was performed as described earlier [29, 33]. Briefly, overnight-cultures of the investigated strains were washed with PBS and adjusted to an OD600 of 0.5. Twelve microliters was applied to indicator plates and incubated overnight at 37 °C under oxic conditions (5% CO2). H2O2 release was determined by measuring the distance between the edge of the colony and the end of the Prussian blue precipitation zone. For each colony, arithmetic means were calculated from four measurements at different positions. At least two colonies were measured per experiment with the average serving as a single data-point. Each experiment was repeated four times.
Growth inhibition assay
Deferred antagonism assays were performed following an earlier study [12]. Briefly, BHI plates were inoculated with 12 μl of washed cells (PBS, OD600 = 0.5) and incubated for 24 h at 37 °C (5% CO2). Subsequently, 12 μl of a similarly prepared S. mutans culture was spotted in close proximity followed by 24 h of incubation at 37 °C. Inhibition zones were measured at three different positions and averaged for each colony (indicated in Fig. 1a). Arithmetic means of at least two colonies served as a single data-point. Each experiment was repeated at least three times.
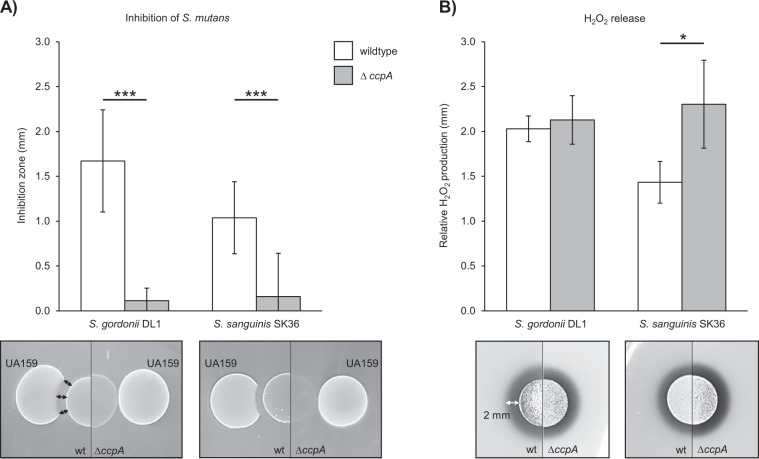
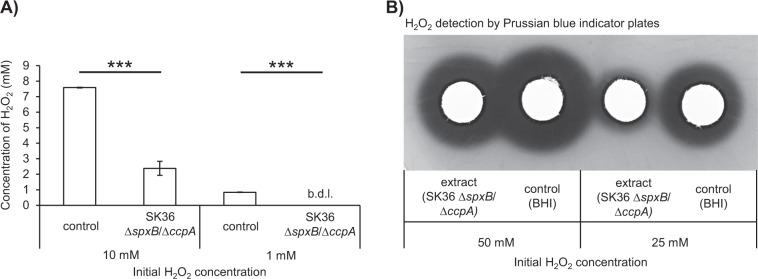
Fig. 1. H2O2 release and growth inhibition characteristics of S. gordonii DL1 and S. sanguinis SK36 wild-type vs. ccpA deletion mutants.
a Growth inhibition of S. mutans UA159 is displayed as the mean distance between the edge of the oral streptococci colony and the S. mutans colony. Below the graph, representative pictures of the inhibitory effect are shown. b H2O2 production is shown as relative H2O2 release of cells grown aerobically on Prussian blue H2O2 indicator plates, as previously described [33]. Below the graph, representative colonies are displayed forming a blue halo on Prussian blue indicator plates. Averages, standard deviations, and representative images are derived from at least three independent experiments. Significance levels: *p < 0.05; ***p < 0.001 (t-test, two-tailed, and unpaired).
H2O2 protection assay
The H2O2 oxidative protection assay was performed as follows: BHI plates were inoculated with 12 μl of washed cells from overnight-cultures (PBS, OD600 = 0.5) and incubated for 24 h at 37 °C (5% CO2). The next day, an overnight-culture of the indicator species (S. mutans/S. epidermidis/E. coli) was washed and adjusted to an OD600 of 0.5. Four milliliters of molten BHI-soft-agar (0.6% agar, 60 °C) was thoroughly mixed with 200 μl of the indicator species cell suspension and immediately poured onto the agar plate. Plates were kept at room temperature for 30 min to allow the soft agar layer to solidify. Subsequently, a filter disc (cotton fiber, 0.84 mm × 7.3 mm diameter, Thick Blot Filter Paper, Bio-Rad) containing H2O2 was added in close proximity to the colony of the test strain followed by 24 h of incubation at 37 °C. Inhibition zones were measured at three different positions and averaged to serve as a single data-point. Each experiment was repeated at least three times. A schematic description is given in the Supplementary material section (Fig. S1).
Transposon mutagenesis and screening
The MarC9/magelan6-based in vitro transposition system was utilized to generate random mutations in S. sanguinis ΔspxB as previously described [34]. All resulting transformants were screened by a modified H2O2 protection assay under the following assumptions: (i) the protective effect guards the H2O2 challenged indicator strain, leading to visible growth of indicator strain colonies; (ii) mutation of a gene related to the protective effect results in a reduced protective effect; and (iii) the H2O2 challenged indicator strain exhibits reduced or no growth. All candidates resulting in a reduced growth of the H2O2 challenged indicator strain were verified by an additional H2O2 protection assay. Affected genes were identified by a modified SiteFinding-PCR as described elsewhere [35]. For a detailed description of the screening process and SiteFinding-PCR protocol please refer to the respective Supplementary material section.
Extraction of the protective substance from solidified BHI
For the characterization and identification of the protective component, raw extracts were obtained as followed: a sterile filter membrane (47 mm, 0.45-μm pore size, Millipore) was placed on a BHI agar plate. A fresh overnight culture of each respective strain was washed (PBS) and adjusted to an OD600 = 0.5. One hundred and fifty microliters was applied to the membrane, allowed to dry, and incubated aerobically for 24 h at 37 °C (or otherwise stated). The membrane was then removed and the underlying agar was cut out, placed into a 10 ml syringe, and pressed through a sterile filter (0.2 μm cellulose acetate membrane, VWR). Liquid extracts were stored at 4 °C or tested immediately.
H2O2 depletion assay
Twenty five microliters of the liquid extracts described above were mixed 1:1 with the same volume of various concentrations of H2O2 and incubated for 30 min at room temperature. Forty microliters of the mixture was applied to a filter disc (cotton fiber, 0.84 mm × 7.3 mm diameter, Thick Blot Filter Paper, Biorad) and placed on a BHI/Prussian blue indicator plate for visualization of the remaining H2O2. Alternatively, H2O2 concentrations were determined with the Amplex Red Peroxide/Peroxidase Assay (Invitrogen) following manufacturer’s instructions.
Chemical characterization and identification of the protective compound
Extracts of SK36 ΔspxB/ΔccpA were collected from solidified BHI as described above and either incubated at 100 °C (30 min), treated with protease (alkaline protease, Promega), lipase (5%, MP Biomedicals), ultrafiltered with a weight cutoff of 1 kDa (Macrosep, Pall Corporation), or mixed with chloroform as described below. Subsequently, the H2O2 depleting activity was tested as described above (H2O2 depletion assay).
For chemical identification, 400 μl of extracts was mixed with chloroform by adding an equal volume followed by vigorous shaking (Bertin Precellys homogenizer, 5 min, 5000 rpm). Phases were separated by centrifugation (21,000 × g, 15 min, 4 °C). The aqueous phase, containing the active substance, was then analyzed by liquid chromatography–information-dependent acquisition–mass spectrometry (LC–IDA–MS; Oregon State University Mass Spectrometry, Corvallis, OR, USA) using the IROA library of metabolite standards (IROA Technologies, Bolton, MA, USA).
Enzymatic elimination of pyruvate and testing of H2O2 depletion activity
Pyruvate was depleted from extracts following a previously published procedure [36] with modifications by the addition of pyruvate oxidase (PoxB, MP Biomedicals). The reaction leads to the formation of acetate and H2O2, which was subsequently eliminated by the addition of catalase. Catalase was subsequently heat-inactivated.
The elimination of pyruvate resulted in a 1:5 dilution of the extracts. Thus, for H2O2 depletion activity testing, untreated extracts were diluted accordingly. Treated and untreated extract solutions were mixed with an equal amount of H2O2 (2 mM) and incubated at room temperature for 30 min. Subsequently, H2O2, pyruvate, and acetate concentrations were determined with the Amplex Red Peroxide/Peroxidase Assay (Invitrogen), EnzyChrom Pyruvate Assay (BioAssays Systems), and EnzyChrom Acetate Assay (BioAssays Systems) respectively, following the manufacturer’s instructions. For a detailed description of the procedure, please refer to the Supplementary material section.
Results
SK36 and DL1 excrete an antioxidant substance
While working with S. gordonii DL1 ΔccpA and S. sanguinis SK36 ΔccpA mutants, we were surprised to discover that these strains lose their antagonistic abilities against S. mutans (Fig. 1a), despite producing significant amounts of inhibitory H2O2 (Fig. 1b). We previously demonstrated that the deletion of spxB in S. sanguinis and S. gordonii as well as the addition of catalase to the wild-type strains abolished any inhibitory effect on S. mutans, which verified that no other inhibitory component is secreted under the tested conditions [20].
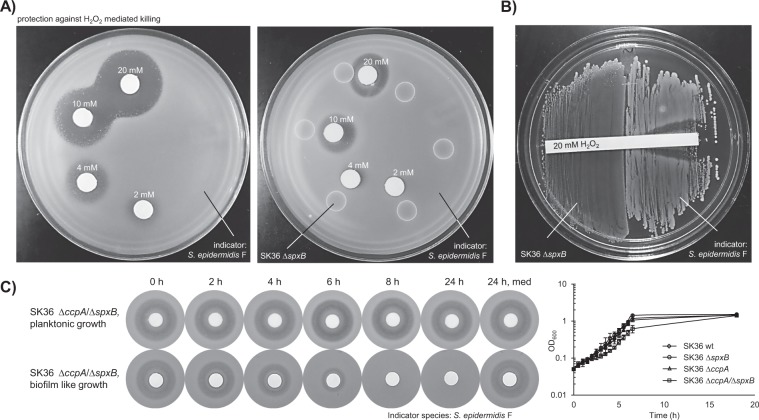
To test the hypothesis that the diminished inhibition of S. mutans by the ccpA mutants is due to the potential secretion of a protective compound, we developed a new antagonism assay (Fig. S1). Initially a set of clinical Staphylococcus isolates and S. mutans were evaluated for their suitability as indicator strains. S. epidermidis F was chosen for being the most H2O2 susceptible strain (Fig. S2). A dose-dependent inhibition was observed when filter disks infused with different H2O2 concentrations ranging from 2 to 20 mM were placed on a soft agar plate containing S. epidermidis F (Fig. 2a, left). However, when SK36 ΔspxB was inoculated on the plate first, the observed inhibition of S. epidermidis was reduced, especially near the border closest to SK36 ΔspxB (Fig. 2a, right). This observation could be repeated when both S. epidermidis F and SK36 ΔspxB were inoculated next to each other and a filter-strip with 200 μl H2O2 (20 mM) was placed over the strains. Inhibition of S. epidermidis by H2O2 could only be observed at the distal side, but not at the proximal side where both strains came into close contact. This observation points to the existence of a diffusible protective component, which prevents or reduces the effectiveness of H2O2 mediated killing (Fig. 2b). Exemplarily, wild-type strains and the isogenic ccpA mutants of S. sanguinis SK36 and S. gordonii DL1 are shown in Supplementary Fig. S3. Due to the autologous release of H2O2 in these strains, the protective effect is mostly masked. Notably, slight differences in the inhibition zone are visible comparing wild-type and ccpA mutants.
Fig. 2. S. sanguinis ΔspxB protects against H2O2 mediated killing.
a A representative H2O2 protection assay is shown. SK36 was grown on BHI agar plates, overlaid with soft agar containing S. epidermidis F (indicator strain), and challenged with different concentrations of H2O2 delivered by previously soaked filter discs. The left panel shows the respective control plate without S. sanguinis. b S. sanguinis ΔspxB was grown on BHI agar. Subsequently, the indicator strain was inoculated in close proximity to SK36 ΔspxB and challenged by an H2O2-soaked filter strip. The concentrations of H2O2 are indicated. c The appearance of the protective effect was tested with SK36 ΔspxB/ΔccpA under planktonic and high-cell-density conditions over a time course. Upper panel: 200 μl of supernatant derived from a liquid culture was sterile filtered at the indicated time points and added to BHI plates. Lower panel: bacteria were applied to sterile filters and then placed onto BHI plates. At the indicated time points, the filters containing the bacteria were removed from the plates. Afterward, the plates were overlaid with soft agar containing S. epidermidis F and then challenged with H2O2-soaked filter discs. Presented is also the planktonic growth monitored by optical density. SK36 wt, spxB, and ccpA single mutants serve as reference.
High-cell-density conditions promote the protective effect
The capacity of supernatants of planktonically grown SK36 ΔccpA/ΔspxB as well as cells grown under high-cell-density conditions on BHI plates were tested in H2O2 protection assays. Supernatants of planktonic cells demonstrated no protective capacity, while cells grown on BHI agar plates (representing high-density growth conditions similar to biofilms [26]) released significant amounts of the protective substance into the surrounding media within 4 h of incubation (Fig. 2c). Thus, the protective effect seems to be relevant under high-cell-density conditions of SK36.
Protection against H2O2 mediated killing is CcpA dependent
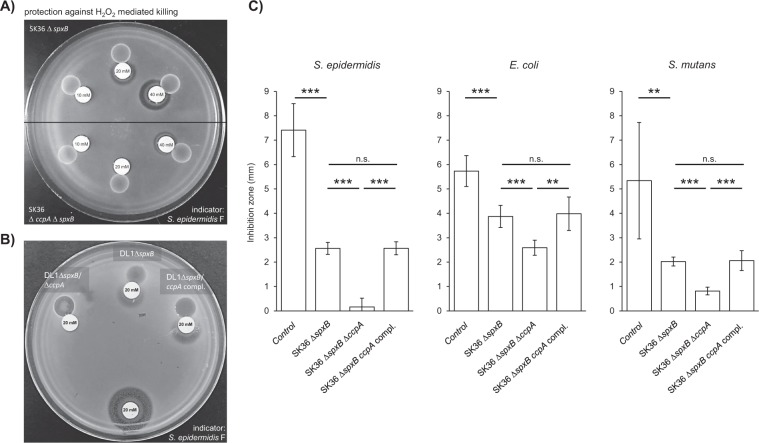
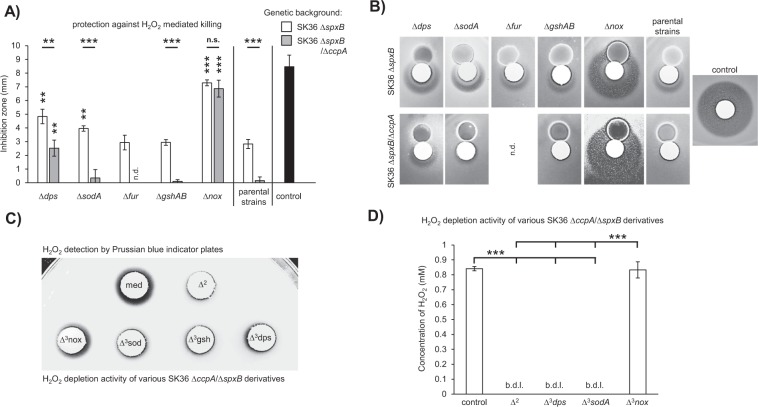
Considering the discrepancy between the relatively high amount of released H2O2 and the weak antagonistic ability of S. sanguinis SK36 ΔccpA and S. gordonii DL1 ΔccpA (Fig. 1), it was hypothesized that H2O2 protection might be CcpA regulated. H2O2 protection assays confirmed our hypothesis: inhibition zones around filter discs placed in close proximity to the spxB mutants were significantly wider (i.e., less protection) when compared with the spxB/ccpA double mutants (Fig. 3a, b). This effect was independent from the tested indicator species and was fully restored by complementation of ccpA in both SK36 and DL1 (Fig. 3b, c).
Fig. 3. CcpA dependency of the protective effect.
a A representative plate is shown to illustrate the differences in inhibition zones caused by various concentrations of H2O2 in close proximity to the SK36 spxB mutant (upper half of the plate) and to the spxB/ccpA double mutant (lower half of the plate). b A representative plate of DL1 is shown demonstrating the differences in the protective capacities of the spxB mutant, the spxB/ccpA double mutant, and the ccpA complemented strain, which exhibits the same protective capacity as the spxB single mutant. An additional H2O2 filter disc was placed on the opposite side of the plate to serve as a control. c SK36 wt, spxB mutant, ccpA/spxB double mutant, and the ccpA complemented strain were analyzed in a H2O2 protection assay. The indicator strains were challenged with different concentrations of H2O2 (S. epidermidis—20 mM; E. coli—100 mM; S. mutans—100 mM). The averages and standard deviations of at least five independent experiments are displayed. Significance levels: *p < 0.05; **p < 0.01; ***p < 0.001 (t-test, two-tailed, and unpaired).
Glucose inhibits the protective effect
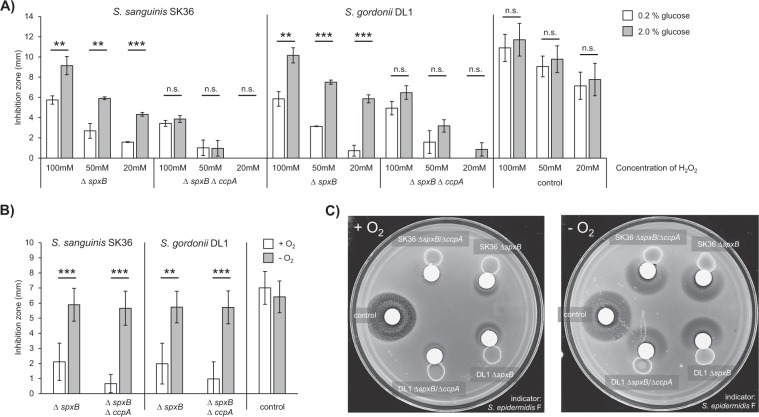
CcpA is a pleotropic regulator that controls gene expression in a carbohydrate dependent manner. Thus, to verify the observed CcpA dependency of the protective effect, H2O2 protection assays were performed with spxB single and spxB/ccpA double mutants of SK36 and DL1 grown in the presence of different glucose concentrations (2% vs. 0.2% glucose). Comparing both conditions, a significant increase in the inhibition zones could be observed in close proximity to the spxB single mutants of both species incubated with 2% glucose (p < 0.01). Thus, in CcpA+ strains, the protective effect is reduced due to the presence of higher glucose concentrations, consistent with CcpA-dependent catabolite repression. In the spxB/ccpA double mutants, glucose concentration had no impact on H2O2 protection (Fig. 4a).
Fig. 4. Glucose and oxygen dependency of the protective effect.
SK36 and DL1 wt, spxB mutants, and ccpA/spxB double mutants were analyzed in H2O2 protection assays. a The assay was performed on BHI agar with different glucose concentrations (2% and 0.2%). The indicator species was challenged with different concentrations of H2O2 (as displayed). An additional H2O2 filter disc was placed on the opposite side of the plate to serve as a control. b H2O2 protection assays were performed in the presence or absence of oxygen (20 mM H2O2). The averages and standard deviations of at least three independent experiments are shown. Significance levels: **p < 0.01; ***p < 0.001 (t-test, two-tailed, unpaired). c Representative plates are shown to illustrate the different sizes of the inhibition zones in close proximity to the indicated strains ± oxygen.
Oxygen is essential for H2O2 protection
In the wild-type background of SK36 and DL1, growth-inhibiting amounts of H2O2 are only produced (and thus released) in the presence of oxygen [13]. H2O2 protection assays revealed, that the protective effect similarly responds to oxygen availability. Comparing aerobic and anaerobic growth conditions, H2O2 inhibition zones were significantly increased on anaerobically incubated plates compared with their aerobic counterparts. Thus, oxygen is essential for the appearance of the protective effect. Furthermore, no differences could be detected in anoxic conditions between the spxB and spxB/ccpA mutants of SK36 and DL1 (Fig. 4b, c). Thus, the absence of oxygen renders SK36 and DL1 unable to produce the protective effect, irrespective of CcpA function.
SK36 eliminates H2O2 by releasing a protective component
Cell-free extracts from BHI agar inoculated with SK36 ccpA/spxB and from sterile BHI agar (serving as control) were co-incubated with different amounts of H2O2 in a H2O2 depletion assay. In the presence of the extract, final H2O2 concentrations decreased from 10 mM to 2.38 mM (±0.45 mM). With an initial concentration of 1 mM, H2O2 levels dropped below the detection limit of 26 nM (Fig. 5a). Comparable results were also observed on Prussian blue indicator plates (Fig. 5b). Blue halos, which indicate the presence of H2O2 in a concentration dependent manner, were considerably reduced in size in the presence of extract compared with the control. In both assays, the final H2O2 concentrations were significantly decreased when incubated with extracts, indicating the protective effect likely depends upon the direct elimination of H2O2, rather than through an oxidative stress response induced in the indicator organisms.
Fig. 5. Elimination of H2O2.
Conditioned medium extracts of the SK36 ccpA/spxB double mutant as well as a BHI growth medium control were mixed 1:1 with various H2O2 solutions and incubated at room temperature for 30 min. The estimated final concentrations of H2O2 are indicated. a H2O2 concentrations were determined with the Amplex Red Peroxide/Peroxidase Assay. The averages and standard deviations are shown from three independent experiments. b Filter discs were soaked with H2O2 and placed on Prussian blue indicator plates. The sizes of the resulting halos are proportional to the amounts of H2O2. Representative images are shown. b.d.l. below detection limit.
Identification of genes related to the protective phenotype
Initially, genes with a known connections to oxidative or H2O2 specific stress resistances were deleted, including gshAB (SSA_2186, glutathione biosynthesis bifunctional protein) [37–39], dps (SSA_0644, DNA-binding ferritin-like protein, oxidative damage protectant) [14, 17, 40], sodA (SSA_0721, Mn/Fe-dependent superoxide dismutase) [14, 17], and the peroxide stress response transcriptional regulator fur (SSA_0686, perR analog, Fe2+/Zn2+ uptake regulation protein) [14, 40]. All mutants were tested in the H2O2 protection assay described above. Of these, only the dps mutant exhibited a significant increase in the H2O2 inhibition zone (i.e., decreased protection) for both SK36 ΔspxB and SK36 ΔspxB/ΔccpA (Fig. 6a, b). The deletion of sodA modestly reduced the protective affect for SK36 ΔspxB, but did not affect the ΔspxB/ΔccpA mutant.
Fig. 6. Genes related to the protective effect.
H2O2 protection assays were performed with the indicated mutants and controls. a Mutations of dps, sodA, fur, gshAB, and nox were tested in the SK36 ΔspxB background, while dps, sodA, gshAB, and nox mutations were assayed in the SK36 ΔccpA/ΔspxB background. Paired t-tests were performed for each mutant relative to its parental stain. The respective significance levels are indicated by asterisks above each column. Significance levels between mutants of a respective gene in the SK36 ΔspxB and SK36 ΔccpA/ΔspxB backgrounds were tested with unpaired t-tests and are indicated above each pair of columns. b Representative examples of the inhibition zones for each tested mutant are presented. c, d Conditioned medium extracts of the SK36 ccpA/spxB double mutant, its derivatives (as indicated), and BHI (control) were tested in an H2O2 depletion assay. Initial H2O2 concentrations were 20 mM (c) and 1 mM (d). c Filter discs were soaked with the extract/H2O2 mixture and placed on Prussian-blue-indicator plates. The sizes of the resulting halos are proportional to the amount of H2O2 present. A representative image is shown. d H2O2 concentrations were determined with the Amplex Red Peroxide/Peroxidase Assay. Averages and standard deviations from at least three independent experiments are presented. Significance levels: **p < 0.01; ***p < 0.001; n.d. not determined, n.s. not significant, b.d.l. below detection limit, med medium control, Δ2—SK36 ΔccpA/ΔspxB, Δ3nox—SK36 ΔccpA/ΔspxB/Δnox, Δ3sod—SK36 ΔccpA/ΔspxB/ΔsodA, Δ3gsh—SK36 ΔccpA/ΔspxB/ΔgshAB, Δ3dps—SK36 ΔccpA/ΔspxB/Δdps.
Since the CcpA dependency of the protective phenotype could be demonstrated, numerous CcpA regulated genes were targeted. Forty genes from 19 operons were deleted due to their predicted regulation by CcpA in the RegPrecise database [41]. In addition, 24 neighboring genes were targeted, since they potentially share the respective palindromic CcpA recognition site (cre, catabolite responsive element) or are potentially co-transcribed with respective CcpA regulated genes. Among these mutants (given in Table S3; Supplementary material section), only the nox mutation (SSA_1127, putative H2O-forming NADH dehydrogenase) caused an obvious defect in H2O2 protection (Fig. 6a, b).
In addition, random transposon mutagenesis was performed to identify additional genes required for H2O2 protection. In total 6445 colonies were screened by a two-step H2O2 protection assay (see Supplementary materials). Eighteen mutants were identified showing a significantly increased inhibition zone compared with the parental strain. The genes SSA_1938 to SSA_1940 (fatty acid biosynthesis operon) were not considered further due to an obvious growth defect as previously described [42]. The most prominent H2O2 protection defect was caused by an insertion into nox (SSA_1127). Besides nox, dps was also identified by the transposon mutagenesis screen. However, the transposon insertion was located in the dps promoter, which might explain its relatively weak effect compared with the dps deletion mutant (Table S4, Fig. S4; Supplementary material section).
Confirmation of the impact of nox on the protective effect
BHI agar extracts derived from SK36 ΔspxB/ΔccpA, SK36 Δdps/ΔspxB/ΔccpA, SK36 ΔgshAB/ΔspxB/ΔccpA, SK36 ΔsodA/ΔspxB/ΔccpA, and SK36 Δnox/ΔspxB/ΔccpA as well as from plain BHI agar plates (control) were tested via the H2O2 depletion assay. A positive reaction on Prussian blue agar plates (i.e., H2O2) occurred only around the filter discs soaked with the H2O2/extract-mix derived from the control and SK36 Δnox/ΔspxB/ΔccpA (Fig. 6c). No Prussian blue reaction was detectable around the other discs, indicating that the other mutants were still capable of eliminating H2O2 from the mixture. As an independent verification, final concentrations of H2O2 (initial concentration: 1 mM) were enzymatically determined in the H2O2 depletion assay. Only the extract of SK36 Δnox/ΔspxB/ΔccpA and the control failed to eliminate H2O2 from the mixture. All other strains were able to deplete H2O2 with final concentrations falling below the 26 nM detection limit (Fig. 6d).
Identification and evaluation of pyruvate as the source of H2O2 protection
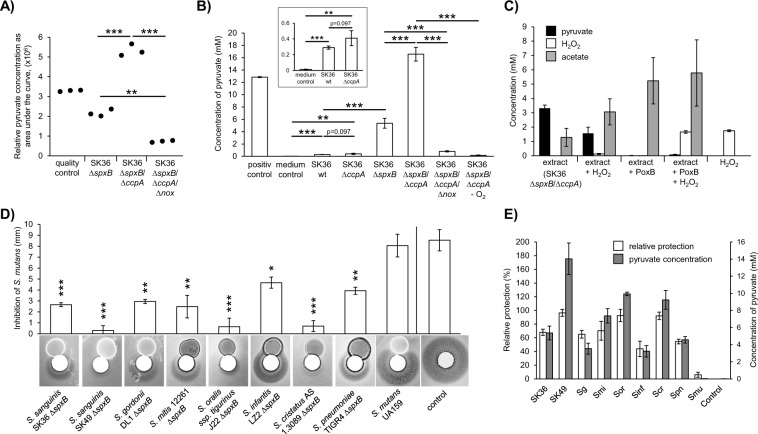
The extract of SK36 ΔspxB/ΔccpA is able to confer protection against H2O2 after lipase, protease, and heat treatment (data not shown). Furthermore, H2O2 protection activity was demonstrated for the aqueous phase after chloroform extraction and ultrafiltration (1 kDa cutoff), suggesting a water soluble small molecule. Therefore, mass spectrometry-based metabolite profiling was performed on extracts derived from the following mutants: SK36 ΔspxB (containing the protective component), SK36 ΔspxB/ΔccpA (increased production of the protective component), and SK36 ΔspxB/ΔccpA/Δnox (lacking the protective component). Of the metabolites detected, only pyruvate matched the expected abundances for each of the mutants (Fig. 7a). The concentrations of pyruvate were also determined enzymatically and mirrored the mass spectrometry results (Fig. 7b). In addition, almost no pyruvate was detected in the anaerobic control (Fig. 7b). This agrees with the oxygen dependency of the protective effect (Fig. 4b, c). As further evidence, pyruvate was depleted from SK36 ΔspxB/ΔccpA extracts using the PoxB enzyme, resulting in a total loss of H2O2 depleting activity (Fig. 7c). The concentrations of pyruvate and H2O2 also decreased in a stoichiometric manner. The initial pyruvate concentration in the extracts decreased from 3.31 mM (±0.22 mM) before H2O2 treatment to 1.56 mM (±0.43 mM) after treatment (i.e., 1.75 mM reduction). The decrease in pyruvate concentration corresponds precisely to the measured initial concentration of H2O2 (1.75 ± 0.07 mM). In addition we determined the concentration of acetate, the corresponding end product of the reaction of pyruvate and H2O2. Its concentration increased from 1.28 mM (±0.63 mM) before H2O2 treatment to 3.01 mM (±0.91 mM) after treatment. This increase also mimics the concentration of the initial H2O2 concentration. The concentration of acetate after pyruvate depletion corresponds approximately to the sum of initial acetate and pyruvate concentration. H2O2 concentrations were largely unaffected by treatment with pyruvate depleted extracts, which resulted in a final H2O2 concentration of 1.66 mM (±0.09 mM). From these data, we conclude that the observed H2O2 protective effect is specifically dependent upon the presence of extracellular pyruvate secreted by the streptococci.
Fig. 7. Pyruvate and H2O2 protection.
a Relative amounts of pyruvate in the extracts of SK36 ΔspxB, SK36 ΔccpA/ΔspxB, and SK36 ΔccpA/ΔspxB/Δnox. Metabolites were identified by liquid chromatography–information-dependent acquisition–mass spectrometry (LC–IDA–MS; Oregon State University Mass Spectrometry, Corvallis, OR, USA) using the IROA library of metabolite standards (IROA Technologies, Bolton, MA, USA). The integrated areas under the curve of extracted mass-to-charge-ratios (m/z) for pyruvate are shown. b Pyruvate concentrations were determined enzymatically in cell free extracts. Extracts were derived from aerobically grown SK36 wild-type strain, SK36 ΔccpA, SK36 ΔspxB, SK36 ΔccpA/ΔspxB, SK36 ΔccpA/ΔspxB/Δnox, and anaerobically grown SK36 ΔccpA/ΔspxB (−O2). Extract from a BHI agar plate served as a negative control (BHI medium). A 10 mM pyruvate solution served as positive control. A section of the graph is displayed with a different scale to highlight pyruvate concentrations of SK 36 wild-type and SK 36 ccpA mutant in comparison with the medium control. c Untreated and pyruvate depleted extracts of SK36 ΔccpA/ΔspxB were tested in an H2O2 depletion assay (2 mM H2O2). Concentrations of pyruvate, H2O2, and acetate were determined enzymatically before and after the assay. (“extract”—untreated extracts before H2O2 depletion assay; “extract + H2O2”—untreated extracts after H2O2 depletion assay; “extract + PoxB”—pyruvate depleted extracts before H2O2 depletion assay; “extract + PoxB + H2O2”—pyruvate depleted extracts after H2O2 depletion assay; “H2O2”—H2O2 solution before H2O2 depletion assay). d S. mutans (not encoding spxB) and spxB deletion mutants of various streptococci were tested for their protective capacity with H2O2 protection assays. Paired t-tests (two-tailed) were performed testing each mutant against its respective H2O2 control. Note that the displayed control represents the average of all experiments. The averages and standard deviations of four independent experiments are shown with representative images. e The protective capacity of S. mutans and spxB mutants of various streptococci are shown together with the pyruvate concentrations of their respective extracts. The inhibition zones from Fig. 7D were transformed to “relative protection” for a better comparability (0% = no protection = full inhibition as it appears in the control; 100% = full protection (no inhibition zone visible)). All tested extracts were derived from three independent experiments. Significance levels: *p < 0.05; **p < 0.01; ***p < 0.001 (two-tailed t-test, unpaired).
We also included the wild type of SK36 and its isogenic ccpA mutant in this assay. Low but significant amounts of pyruvate were detected, which increased due to the deletion of ccpA (Fig. 7b).
The protective phenotype is not limited to SK36 and DL1
In addition to S. sanguinis SK36 and S. gordonii DL1, several other spxB mutants were generated (Fig. 7d). All spxB-encoding species exhibited protection against H2O2 mediated killing, whereas the spxB− species S. mutans completely lacked this ability. Consistent with SK36 and DL1, the abundance of excreted pyruvate closely matched the pattern of H2O2 protection in all cases (Fig. 7e). Hence, the protective phenotype (mediated by excreted pyruvate) is not exclusively confined to S. sanguinis SK36 and S. gordonii DL1, rather this is likely a common characteristic of spxB+ streptococci in general.
Discussion
The new phenotype
The release of H2O2 masked a previously unrecognized characteristic of spxB+ streptococci: the ability to actively deplete H2O2 from the surrounding environment by the secretion of pyruvate.
Compared with their respective wild types, SK36 ΔccpA and DL1 ΔccpA demonstrated a decreased growth inhibition capacity against H2O2-susceptible S. mutans, while the release of H2O2 was not affected or even increased (detected by Prussian blue indicator plates, Fig. 1b). Here, we demonstrate that a significantly increased amount of pyruvate released into the environment (by the ccpA mutant) is responsible for the scavenging of H2O2 (demonstrated in the spxB deletion background). Consequently, final H2O2 concentrations dropped to noninhibitory levels for the ccpA deletion mutants (as indicated in Fig. 1a, plain BHI, without Prussian blue indicator), which exhibit increased pyruvate production. Supporting this assumption, the pyruvate release in the ccpA deletion mutant of SK36 was elevated compared with the parental wild-type strain, albeit not significant (Fig. 7b).
A H2O2 dependent self-inhibitory effect could also explain our observation, by an increased amount of H2O2 inhibiting the growth of the ccpA deletion mutant itself and thus resulting in an overall reduction of the final H2O2 concentration. However, since there is no significant increased H2O2 release in DL1 ΔccpA, which could trigger such a self-inhibitory effect, it might only play a minor role under the given conditions.
The role of Dps and Nox
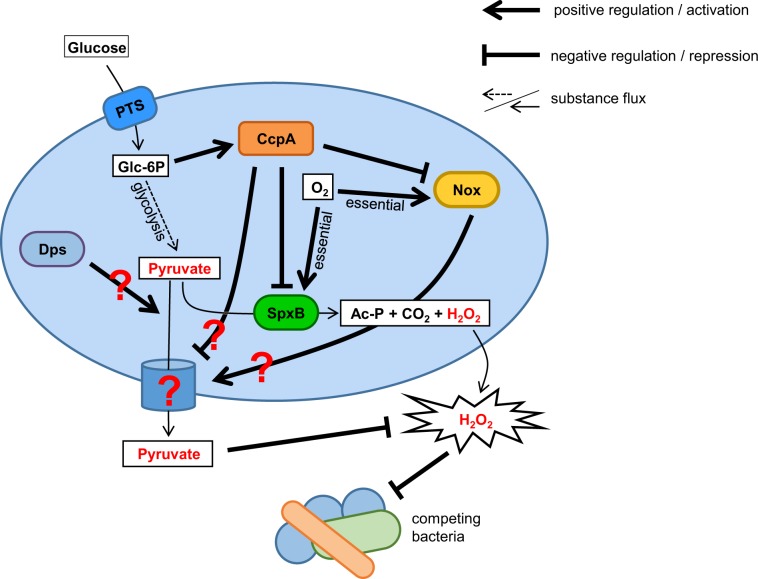
Transposon mutagenesis and targeted gene deletions identified dps and nox as associated with the protective phenotype. Dps is a ferritin-like protein that mediates resistance to H2O2 by chelating iron and binding DNA, thus preventing Fenton-mediated killing mechanisms [14, 17, 43]. Since the protective effect is the result of pyruvate excreted into the environment, the intracellular protein Dps is unlikely to be directly involved in the protection process. Thus, we speculate there is a potential regulatory function for Dps, possibly through its predicted DNA binding ability. Alternatively, the deletion of Dps might increase intracellular iron levels, which in turn decrease the intracellular concentration of pyruvate due to a possible reaction of both compounds [44]. As a result, less pyruvate would be available for secretion as depicted in the overview figure of the here presented regulatory factors and substances influencing H2O2 and pyruvate release (Fig. 8).
Fig. 8. Components modulating pyruvate release.
Glycolysis provides the cell with energy and pyruvate, which can be exported by an unknown mechanism and function as a scavenger for extracellular H2O2. Pyruvate is also the substrate for SpxB and therefore needed for the generation of H2O2, acetyl-phosphate (Ac-P), and CO2. Extracellular glucose can be translocated through the cell membrane by the phosphotransferase system (PTS), which also phosphorylate glucose to glucose-6-phosphate (Glc-6P), which in turn can be metabolized through glycolysis. In addition, Glc-6P functions as a signal molecule for the presence of glucose and initializes catabolite repression through the CcpA pathway. Activated CcpA represses transcription of spxB, nox, and other genes (not shown), and thus influences H2O2 as well as pyruvate release. Nox is an oxygen dependent NADH oxidase, which is also involved in pyruvate release by an unknown mechanism. Dps also alters the release on pyruvate by an unknown pathway. Potential mechanisms are discussed in the text. H2O2 generation via SpxB as well as pyruvate release depends on the presence of oxygen. Ac-P acetyl phosphate, Glc-6P glucose-6-phosphate, PTS phosphotransferase system.
The NADH Oxidase Nox seems to play an essential role in the protective phenotype, as evident from the inability of the nox mutant to confer protection. Interestingly, the secretion of pyruvate as well as the H2O-forming activity of Nox are both oxygen-dependent processes (Fig. 4c) [45, 46]. In addition, nox expression is under the control of CcpA (http://regprecise.lbl.gov [41, 47, 48]), which might explain the CcpA dependency of the protective phenotype. Although the activity of Nox is critical for the protective effect, NADH oxidase is also present in other streptococci like S. mutans (81% identity on amino acid level compared with SK36), which did not exhibit any detectable protective activity or pyruvate secretion. Furthermore, it is presently unclear how Nox stimulates pyruvate secretion. Nox is an oxygen dependent NADH oxidase and therefore involved in the regeneration of NADH to NAD+, important for an undisrupted glycolysis [49] and thus for the generation of pyruvate. The lack of NAD+ would affect growth dramatically, but we did not observe any growth deficiencies comparing oxic and anoxic growth, neither in the wild-type background nor in the spxB deletion mutant. This might be explained by alternative metabolic pathways to regenerate NAD+ via lactate dehydrogenase or formation of ethanol [50]. Thus, pyruvate secretion could also be influenced by the disrupted SpxB pathway. This would also explain why the effect is limited to spxB+ species, since they prefer the more beneficial pathway (one more ATP per pyruvate). Other species are limited to lactate or ethanol formation and might therefore do not secrete pyruvate.
The activity of Nox also affects the intracellular redox potential through the NAD+/NADH-ratio. Therefore, the loss of Nox alters expression of genes involved in energy metabolism, transport, several genes encoding transcription factors, and signaling molecules [51–53]. Thus, we speculate that Nox modulates pyruvate release by affecting activity or expression of other factors that are needed for the release of pyruvate.
Pyruvate release and the newly identified protective effect of spxB positive streptococci
Pyruvate is a critical central intermediate of cellular metabolism and is involved in energy generation, amino acid biosynthesis, and acid tolerance [29, 54–56]. Strikingly, pyruvate is also the substrate for the pyruvate oxidase SpxB and therefore directly involved in the production of H2O2 [29] (Fig. 8). As a potent H2O2 scavenger, pyruvate has been reported to protect neurons and other eukaryotic cells from H2O2 toxicity [57–59]. Recent studies demonstrated that microorganisms also utilize pyruvate to detoxify intracellular H2O2 or can benefit from host derived pyruvate to passively escape the human immune response [60–62]. Here, we describe the first prokaryotic example of an active pyruvate secretion to mitigate the deleterious effects of H2O2. Hence, pyruvate secretion has the potential to extend the streptococcal toolbox for the management of reactive oxygen species stress and might be an important component that aides in direct H2O2 detoxification [14, 17, 37–40, 63].
Our knowledge of the biological relevance of the here described pyruvate secretion phenotype is limited at this time. We could demonstrate that S. sanguinis wild type is secreting detectable amounts of pyruvate under conditions that are highly selective for optimized H2O2 production. This suggests that not all pyruvate is reacting with the H2O2 produced by the wild type, potentially being able to serve as antioxidant for nearby species. However, this speculation requires further validation for example with a genetically controlled release of pyruvate. Efforts are currently underway to identify the respective pyruvate secretion mechanism in oral streptococci to address the biological relevance in the context of the oral biofilm. Of note, naturally occurring spxB mutants were described for the typically spxB+ species S. pneumoniae. The tested mutants were more virulent compared with their SpxB expressing (and H2O2 releasing) counterparts [64–66]. As demonstrated in the present study, the mutation of spxB and thus the reduced release of H2O2 results in an elevated pyruvate release, which in turn could be related to the increased virulence level. Therefore, future investigations targeting the in vivo meaning of the here described effect will have to focus also on naturally occurring spxB mutants, since there are no data available regarding their abundancy.
Pyruvate transport has been characterized as an inwardly directed energy-dependent process [67]. A recent study identified a member of the peptide transporter carbon starvation (CstA) family as a specific pyruvate/H+ symporter in E. coli [68, 69]. Nevertheless, no obvious homologous SK36 proteins could be identified for any of the known pyruvate transporters (CstA, BtsT from E. coli [68, 69], MPC1 and MPC2 known as mitochondrial pyruvate carrier [70], or MctC from Corynebacterium glutamicum [71]). However, another recent study suggests a novel class of transporters as involved in the passive transport of pyruvate in S. mutans via the lrgAB system, which had been previously described as a holin/antiholin system in Bacillus subtilis and Staphylococcus aureus [72–74]. We examined the respective lrgAB orthologs in SK36 (SSA_0842 and SSA_0843) as well as the LrgAB-associated two-component system lytST (SSA_1119 and SSA_1120) [72, 73] and observed no connection to the secretion of pyruvate (data not shown). Hence, it is still unclear how the cells manage to release pyruvate. The fact that no transporter could be identified via transposon mutagenesis suggests that the respective transporter is either essential or there are redundant pyruvate transporters.
Implications for the development and maturation of mixed-species oral biofilms
The ability to release H2O2 is recognized a key mechanism for streptococcal interspecies antagonism during oral biofilm development [11, 20, 21]. Abundant SpxB+ streptococci inhibit susceptible bacteria by the secretion of growth inhibiting amounts of H2O2, which generates highly reactive hydroxyl radicals intracellularly via the Fenton reaction triggering irreparable DNA damage [75]. In SK36 and DL1, spxB expression and H2O2 release are controlled by CcpA [24, 25, 33], the primary carbon catabolite regulator in Gram-positive bacteria. CcpA regulates the cellular response to the availability of carbohydrates [76]. Remarkably, the response to an elevated amount of glucose is different in both species: while SK36 does not alter the release of H2O2 [21, 33], DL1 releases significantly less H2O2 in the presence of high-glucose concentrations [25, 33]. The H2O2 protective phenotype is also under the control of CcpA and follows the same pattern both species. For example, in DL1 both H2O2 release and the protective effect follow classical catabolite repression, while in SK36 only the protective effect is subject to catabolite control whereas spxB expression seems to be under permanent CcpA repression [21, 33]. This observation suggests additional regulatory mechanisms likely exist to modulate these antagonistic systems of H2O2 homeostasis. Remarkably, the H2O2 protective ability seems to be present in most or all spxB+ streptococci, including low-passage clinical isolates as well as the human pathogen S. pneumoniae. Thus, this ability might impact many different microbiome communities outside of the oral cavity, as oral streptococci are frequently found in the nasopharynx and respiratory tract, while spxB+ streptococci (especially S. pneumoniae) are also associated with severe invasive infections [77–81].
Pyruvate secretion and the immune response
Several recent studies have revealed additional functions of H2O2. It has been demonstrated to serve as an effective signal molecule for the immune response and is also as a key player in the genesis of lung carcinoma [82–85]. Hence, the ability of bacteria to modulate the level of external H2O2 could have a much broader impact and might not be limited to interspecies competition among the microbiome. Since the ability to modulate the level of H2O2 seems to be present in spxB+ commensals as well as in some pathogenic streptococci, it will be interesting to determine whether these organisms are potentially able to actively influence the host immune response through the modulation of H2O2 availability.
Limitations of the study
The here described experiments focus on a protective compound being effective against a range of 20–100 mM H2O2. This range was selected to optimally display the protective effect on the tested mutants. Moreover, H2O2 generation and pyruvate release were measured under ideal laboratory conditions (optimal aeration) and do not necessarily mimic the actual physiological environment. H2O2 concentrations under laboratory conditions have been shown to reach up to 1.6 mM in single-species biofilms of S. gordonii [86] depending on growth conditions, time period, and method of measurement. The transient concentration in saliva has been calculated to be around 10 μM based on known concentrations of thiocyanate and hypothiocyanite in saliva [12], but no data is available for actual oral biofilms in situ. Furthermore, in this study we focused on spxB mutants and its derivatives, since wild-type strains as well as isogenic ccpA mutants produce significant amounts of H2O2, which masked the protective effect making it difficult to present. Nevertheless, a significant amount of released pyruvate was detectable for S. sanguinis (Fig. 7b). Therefore, the ecological meaning of active pyruvate secretion regarding H2O2 homeostasis needs further investigation.
Supplementary information
Supplemental Material - Material and Methods
Acknowledgements
The authors would like to acknowledge the following National Institutes of Health grants: R56-DE021726 (JK), R35-DE028252 (JM). The project described was supported, in part by the Oregon State University Research Office. The content is solely the responsibility of the authors and does not necessarily represent the official views of the OSU Mass Spectrometry Center. The authors acknowledge the OSU Mass Spectrometry Center at Oregon State University and specific institutional instrument grants. Orbitrap Fusion Lumos—NIH # 1S10OD020111-01, Waters Ion Mobility ToF Mass Spectrometer—NIH #1S10RR025628-01, Applied Biosystems 4000Qtrap—NIH # 1S10RR022589-01, ABSciex Triple ToF 5600-NIH #1S10RR027878-01.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-0592-8) contains supplementary material, which is available to authorized users.
References
- 1.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jefferson KK. What drives bacteria to produce a biofilm? FEMS Microbiol Lett. 2004;236:163–73.. doi: 10.1016/j.femsle.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Huang R, Li M, Gregory RL. Bacterial interactions in dental biofilm. Virulence. 2011;2:435–44. doi: 10.4161/viru.2.5.16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts FA, Darveau RP. Beneficial bacteria of the periodontium. Periodontology 2000. 2002;30:40–50. doi: 10.1034/j.1600-0757.2002.03004.x. [DOI] [PubMed] [Google Scholar]
- 5.Avila M, Ojcius DM, Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 2009;28:405–11. doi: 10.1089/dna.2009.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asikainen S, Alaluusua S. Bacteriology of dental infections. Eur Heart J. 1993;14:43–50. [PubMed] [Google Scholar]
- 7.Li X, Kolltveit KM, Tronstad L, Olsen I. Systemic diseases caused by oral infection. Clin Microbiol Rev. 2000;13:547–58. doi: 10.1128/cmr.13.4.547-558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosan B, Lamont RJ. Dental plaque formation. Microbes Infect. 2000;2:1599–607.. doi: 10.1016/s1286-4579(00)01316-2. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Ge X, Dou Y, Wang X, Patel JR, Xu P. Identification of hydrogen peroxide production-related genes in Streptococcus sanguinis and their functional relationship with pyruvate oxidase. Microbiology. 2011;157:13–20. doi: 10.1099/mic.0.039669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velsko IM, Chakraborty B, Nascimento MM, Burne RA, Richards VP. Species designations belie phenotypic and genotypic heterogeneity in oral streptococci. mSystems. 2018;3:e00158–18. [DOI] [PMC free article] [PubMed]
- 11.Redanz S, Cheng X, Giacaman RA, Pfeifer CS, Merritt J, Kreth J. Live and let die: hydrogen peroxide production by the commensal flora and its role in maintaining a symbiotic microbiome. Mol Oral Microbiol. 2018;33:337–52. [DOI] [PMC free article] [PubMed]
- 12.Zhu L, Kreth J. The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid Med Cell Longev. 2012;2012:717843. doi: 10.1155/2012/717843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng LY, Itzek A, Chen ZY, Kreth J. Oxygen dependent pyruvate oxidase expression and production in Streptococcus sanguinis. Int J Oral Sci. 2011;3:82–9. doi: 10.4248/IJOS11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujishima K, Kawada-Matsuo M, Oogai Y, Tokuda M, Torii M, Komatsuzawa H. dpr and sod in Streptococcus mutans are involved in coexistence with S. sanguinis, and PerR is associated with resistance to H2O2. Appl Environ Microbiol. 2013;79:1436–43. doi: 10.1128/AEM.03306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pericone CD, Park S, Imlay JA, Weiser JN. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the fenton reaction. J Bacteriol. 2003;185:6815–25. doi: 10.1128/JB.185.23.6815-6825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei Y, Zhang Y, Guenther BD, Kreth J, Herzberg MC. Mechanism of adhesion maintenance by methionine sulphoxide reductase in Streptococcus gordonii. Mol Microbiol. 2011;80:726–38.. doi: 10.1111/j.1365-2958.2011.07603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Itzek A, Kreth J. Comparison of genes required for H2O2 resistance in Streptococcus gordonii and Streptococcus sanguinis. Microbiology. 2014;160:2627–38. doi: 10.1099/mic.0.082156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu P, Alves JM, Kitten T, Brown A, Chen Z, Ozaki LS, et al. Genome of the opportunistic pathogen Streptococcus sanguinis. J Bacteriol. 2007;189:3166–75. doi: 10.1128/JB.01808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelmann S, Hecker M. Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol Lett. 1996;145:63–9. doi: 10.1111/j.1574-6968.1996.tb08557.x. [DOI] [PubMed] [Google Scholar]
- 20.Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol. 2008;190:4632–40. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol. 2005;187:7193–203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemos JA, Quivey RG, Jr, Koo H, Abranches J. Streptococcus mutans: a new Gram-positive paradigm? Microbiology. 2013;159:436–45. doi: 10.1099/mic.0.066134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krzysciak W, Jurczak A, Koscielniak D, Bystrowska B, Skalniak A. The virulence of Streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis. 2014;33:499–515. doi: 10.1007/s10096-013-1993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng L, Chen Z, Itzek A, Ashby M, Kreth J. Catabolite control protein A controls hydrogen peroxide production and cell death in Streptococcus sanguinis. J Bacteriol. 2011;193:516–26. doi: 10.1128/JB.01131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng L, Itzek A, Chen Z, Kreth J. Environmental influences on competitive hydrogen peroxide production in Streptococcus gordonii. Appl Environ Microbiol. 2011;77:4318–28. doi: 10.1128/AEM.00309-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev. 2015;39:649–69. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed: NY: Cold Spring Harbor Laboratory, Cold Spring Harbor; 1989.
- 28.Zhu L, Zhang Y, Fan J, Herzberg MC, Kreth J. Characterization of competence and biofilm development of a Streptococcus sanguinis endocarditis isolate. Mol Oral Microbiol. 2011;26:117–26.. doi: 10.1111/j.2041-1014.2010.00602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng X, Redanz S, Cullin N, Zhou X, Xu X, Joshi V, et al. Plasticity of the pyruvate node modulates hydrogen peroxide production and acid tolerance in multiple oral streptococci. Appl Environ Microbiol. 2018;84:e01697–17. [DOI] [PMC free article] [PubMed]
- 30.Redanz S, Standar K, Podbielski A, Kreikemeyer B. Heterologous expression of sahH reveals that biofilm formation is autoinducer-2-independent in Streptococcus sanguinis but is associated with an intact activated methionine cycle. J Biol Chem. 2012;287:36111–22. doi: 10.1074/jbc.M112.379230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnsborg O, Blomquist T, Kilian, M, Håvarstein, L.S. Biologically active peptides in streptococci. In: Molecular biology of streptococci. Hakenbeck R, Chhatwal S, editors. Norwich: Horizon Scientific Press. 2007:p. 42–7.
- 32.Saito M, Seki M, Iida K, Nakayama H, Yoshida S. A novel agar medium to detect hydrogen peroxide-producing bacteria based on the prussian blue-forming reaction. Microbiol Immunol. 2007;51:889–92.. doi: 10.1111/j.1348-0421.2007.tb03971.x. [DOI] [PubMed] [Google Scholar]
- 33.Redanz S, Masilamani R, Cullin N, Giacaman RA, Merritt J, Kreth J. Distinct regulatory role of carbon catabolite protein A (CcpA) in oral StreptococcalspxB expression. J Bacteriol. 2018;200:e00619–17. [DOI] [PMC free article] [PubMed]
- 34.van Opijnen T, Lazinski DW, Camilli A. Genome-wide fitness and genetic interactions determined by Tn-seq, a high-throughput massively parallel sequencing method for microorganisms. Curr Protoc Microbiol. 2015;36:1E 3 1–24. doi: 10.1002/9780471729259.mc01e03s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan G, Gao Y, Shi M, Zhang X, He S, Chen Z, et al. SiteFinding-PCR: a simple and efficient PCR method for chromosome walking. Nucleic Acids Res. 2005;33:e122. doi: 10.1093/nar/gni124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu A, Romero R, Petty HR. A sensitive fluorimetric assay for pyruvate. Anal Biochem. 2010;396:146–51. doi: 10.1016/j.ab.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lushchak VI. Glutathione homeostasis and functions: potential targets for medical interventions. J Amino Acids. 2012;2012:736837. doi: 10.1155/2012/736837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng X, Zhang K, Zhou X, Liu C, Li M, Li Y, et al. Involvement of gshAB in the interspecies competition within oral biofilm. J Dent Res. 2013;92:819–24. doi: 10.1177/0022034513498598. [DOI] [PubMed] [Google Scholar]
- 39.Owens RA, Hartman PE. Export of glutathione by some widely used Salmonella typhimurium and Escherichia coli strains. J Bacteriol. 1986;168:109–14. doi: 10.1128/jb.168.1.109-114.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Tong H, Dong X. PerR-regulated manganese ion uptake contributes to oxidative stress defense in an oral streptococcus. Appl Environ Microbiol. 2014;80:2351–9. doi: 10.1128/AEM.00064-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, et al. RegPrecise 3.0 - a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genom. 2013;14:745. doi: 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu P, Ge X, Chen L, Wang X, Dou Y, Xu JZ, et al. Genome-wide essential gene identification in Streptococcus sanguinis. Sci Rep. 2011;1:125. doi: 10.1038/srep00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calhoun LN, Kwon YM. Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J Appl Microbiol. 2011;110:375–86.. doi: 10.1111/j.1365-2672.2010.04890.x. [DOI] [PubMed] [Google Scholar]
- 44.Muchowska KB, Varma SJ, Moran J. Synthesis and breakdown of universal metabolic precursors promoted by iron. Nature. 2019;569:104–7. doi: 10.1038/s41586-019-1151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge X, Yu Y, Zhang M, Chen L, Chen W, Elrami F, et al. Involvement of NADH oxidase in competition and endocarditis virulence in Streptococcus sanguinis. Infect Immun. 2016;84:1470–7. doi: 10.1128/IAI.01203-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carvalho SM, Farshchi Andisi V, Gradstedt H, Neef J, Kuipers OP, Neves AR, et al. Pyruvate oxidase influences the sugar utilization pattern and capsule production in Streptococcus pneumoniae. PloS one. 2013;8:e68277. doi: 10.1371/journal.pone.0068277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novichkov PS, Brettin TS, Novichkova ES, Dehal PS, Arkin AP, Dubchak I, et al. RegPrecise web services interface: programmatic access to the transcriptional regulatory interactions in bacteria reconstructed by comparative genomics. Nucleic Acids Res. 2012;40:W604–8. [DOI] [PMC free article] [PubMed]
- 48.Novichkov PS, Laikova ON, Novichkova ES, Gelfand MS, Arkin AP, Dubchak I, et al. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 2010;38:D111–8. doi: 10.1093/nar/gkp894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higuchi M, Yamamoto Y, Poole LB, Shimada M, Sato Y, Takahashi N, et al. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J Bacteriol. 1999;181:5940–7. doi: 10.1128/jb.181.19.5940-5947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carvalho SM, Kuipers OP, Neves AR. Environmental and nutritional factors that affect growth and metabolism of the pneumococcal serotype 2 strain D39 and its nonencapsulated derivative strain R6. PLoS One. 2013;8:e58492. doi: 10.1371/journal.pone.0058492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afzal M, Shafeeq S, Kuipers OP. NADH-Mediated Gene Expression in Streptococcus pneumoniae and Role of Rex as a Transcriptional Repressor of the Rex-Regulon. Front Microbiol. 2018;9:1300. doi: 10.3389/fmicb.2018.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker JL, Derr AM, Karuppaiah K, MacGilvray ME, Kajfasz JK, Faustoferri RC, et al. Streptococcus mutans NADH oxidase lies at the intersection of overlapping regulons controlled by oxygen and NAD+ levels. J Bacteriol. 2014;196:2166–77. doi: 10.1128/JB.01542-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng C, Ren S, Xu J, Zhao X, Shi G, Wu J, et al. Contribution of NADH oxidase to oxidative stress tolerance and virulence of Streptococcus suis serotype 2. Virulence. 2017;8:53–65. doi: 10.1080/21505594.2016.1201256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deibel RH, Niven CF., Jr Pyruvate fermentation by Streptococcus faecalis. J Bacteriol. 1964;88:4–10. doi: 10.1128/jb.88.1.4-10.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su YB, Peng B, Li H, Cheng ZX, Zhang TT, Zhu JX, et al. Pyruvate cycle increases aminoglycoside efficacy and provides respiratory energy in bacteria. Proc Natl Acad Sci USA. 2018;115:E1578–E87. doi: 10.1073/pnas.1714645115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartel T, Eylert E, Schulz C, Petruschka L, Gierok P, Grubmuller S, et al. Characterization of central carbon metabolism of Streptococcus pneumoniae by isotopologue profiling. J Biol Chem. 2012;287:4260–74. doi: 10.1074/jbc.M111.304311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Desagher S, Glowinski J, Premont J. Pyruvate protects neurons against hydrogen peroxide-induced toxicity. J Neurosci. 1997;17:9060–7. doi: 10.1523/JNEUROSCI.17-23-09060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, Perez E, Liu R, Yan LJ, Mallet RT, Yang SH. Pyruvate protects mitochondria from oxidative stress in human neuroblastoma SK-N-SH cells. Brain Res. 2007;1132:1–9. doi: 10.1016/j.brainres.2006.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Long LH, Halliwell B. Artefacts in cell culture: pyruvate as a scavenger of hydrogen peroxide generated by ascorbate or epigallocatechin gallate in cell culture media. Biochem Biophys Res Commun. 2009;388:700–4. doi: 10.1016/j.bbrc.2009.08.069. [DOI] [PubMed] [Google Scholar]
- 60.van Niel EW, Hofvendahl K, Hahn-Hagerdal B. Formation and conversion of oxygen metabolites by Lactococcus lactis subsp. lactis ATCC 19435 under different growth conditions. Appl Environ Microbiol. 2002;68:4350–6. doi: 10.1128/AEM.68.9.4350-4356.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim JG, Park SJ, Sinninghe Damste JS, Schouten S, Rijpstra WI, Jung MY, et al. Hydrogen peroxide detoxification is a key mechanism for growth of ammonia-oxidizing archaea. Proc Natl Acad Sci USA. 2016;113:7888–93. doi: 10.1073/pnas.1605501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Troxell B, Zhang JJ, Bourret TJ, Zeng MY, Blum J, Gherardini F, et al. Pyruvate protects pathogenic spirochetes from H2O2 killing. PloS One. 2014;9:e84625. doi: 10.1371/journal.pone.0084625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mishra S, Imlay J. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys. 2012;525:145–60.. doi: 10.1016/j.abb.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bryant JC, Dabbs RC, Oswalt KL, Brown LR, Rosch JW, Seo KS, et al. Pyruvate oxidase of Streptococcus pneumoniae contributes to pneumolysin release. BMC Microbiol. 2016;16:271. doi: 10.1186/s12866-016-0881-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pericone CD, Overweg K, Hermans PW, Weiser JN. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun. 2000;68:3990–7. doi: 10.1128/iai.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idanpaan-Heikkila I, Rosenow C, et al. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol. 1996;19:803–13. doi: 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 67.Lang VJ, Leystra-Lantz C, Cook RA. Characterization of the specific pyruvate transport system in Escherichia coli K-12. J Bacteriol. 1987;169:380–5. doi: 10.1128/jb.169.1.380-385.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kristoficova I, Vilhena C, Behr S, Jung K. BtsT, a novel and specific pyruvate/H(+) symporter in Escherichia coli. J Bacteriol. 2018;200:e00599–17. [DOI] [PMC free article] [PubMed]
- 69.Hwang S, Choe D, Yoo M, Cho S, Kim SC, Cho S, et al. Peptide transporter CstA imports pyruvate in Escherichia coli K-12. J Bacteriol. 2018;200:e00771–17. [DOI] [PMC free article] [PubMed]
- 70.McCommis KS, Finck BN. Mitochondrial pyruvate transport: a historical perspective and future research directions. Biochem J. 2015;466:443–54. doi: 10.1042/BJ20141171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jolkver E, Emer D, Ballan S, Kramer R, Eikmanns BJ, Marin K. Identification and characterization of a bacterial transport system for the uptake of pyruvate, propionate, and acetate in Corynebacterium glutamicum. J Bacteriol. 2009;191:940–8. doi: 10.1128/JB.01155-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Charbonnier T, Le Coq D, McGovern S, Calabre M, Delumeau O, Aymerich S, et al. Molecular and physiological logics of the pyruvate-induced response of a novel transporter in Bacillus subtilis. mBio. 2017;8:e00976–17. [DOI] [PMC free article] [PubMed]
- 73.Sadykov MR, Bayles KW. The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr Opin Microbiol. 2012;15:211–5. doi: 10.1016/j.mib.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahn SJ, Deep K, Turner ME, Ishkov I, Waters A, Hagen SJ, et al. Characterization of LrgAB as a stationary phase-specific pyruvate uptake system in Streptococcus mutans. BMC Microbiol. 2019;19:223. doi: 10.1186/s12866-019-1600-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winterbourn CC. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol Lett. 1995;82-83:969–74. doi: 10.1016/0378-4274(95)03532-x. [DOI] [PubMed] [Google Scholar]
- 76.Deutscher J. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol. 2008;11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 77.Seppala H, Haanpera M, Al-Juhaish M, Jarvinen H, Jalava J, Huovinen P. Antimicrobial susceptibility patterns and macrolide resistance genes of viridans group streptococci from normal flora. J Antimicrob Chemother. 2003;52:636–44. doi: 10.1093/jac/dkg423. [DOI] [PubMed] [Google Scholar]
- 78.Siegel SJ, Weiser JN. Mechanisms of bacterial colonization of the respiratory tract. Annu Rev Microbiol. 2015;69:425–44. doi: 10.1146/annurev-micro-091014-104209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cahill TJ, Prendergast BD. Infective endocarditis. Lancet. 2016;387:882–93.. doi: 10.1016/S0140-6736(15)00067-7. [DOI] [PubMed] [Google Scholar]
- 80.Liu YT, Lin CF, Lee YL. Streptococcus sanguinis meningitis following endoscopic ligation for oesophageal variceal haemorrhage. J Med Microbiol. 2013;62:794–6. doi: 10.1099/jmm.0.054213-0. [DOI] [PubMed] [Google Scholar]
- 81.O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 82.Wittmann C, Chockley P, Singh SK, Pase L, Lieschke GJ, Grabher C. Hydrogen peroxide in inflammation: messenger, guide, and assassin. Adv Hematol. 2012;2012:541471. doi: 10.1155/2012/541471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gough DR, Cotter TG. Hydrogen peroxide: a Jekyll and Hyde signalling molecule. Cell Death Dis. 2011;2:e213. doi: 10.1038/cddis.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Di Marzo N, Chisci E, Giovannoni R. The role of hydrogen peroxide in redox-dependent signaling: homeostatic and pathological responses in mammalian cells. Cells. 2018;7: E156. [DOI] [PMC free article] [PubMed]
- 85.Upadhyay S, Vaish S, Dhiman M. Hydrogen peroxide-induced oxidative stress and its impact on innate immune responses in lung carcinoma A549 cells. Mol Cell Biochem. 2019;450:135–47. [DOI] [PubMed]
- 86.Liu X, Ramsey MM, Chen X, Koley D, Whiteley M, Bard AJ. Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc Natl Acad Sci USA. 2011;108:2668–73. doi: 10.1073/pnas.1018391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material - Material and Methods