Abstract
Some deep-sea chemosynthetic invertebrates and their symbiotic bacteria can use molecular hydrogen (H2) as their energy source. However, how much the chemosynthetic holobiont (endosymbiont-host association) physiologically depends on H2 oxidation has not yet been determined. Here, we demonstrate that the Campylobacterota endosymbionts of the gastropod Alviniconcha marisindica in the Kairei and Edmond fields (kAlv and eAlv populations, respectively) of the Indian Ocean, utilize H2 in response to their physical and environmental H2 conditions, although the 16S rRNA gene sequence of both the endosymbionts shared 99.6% identity. A thermodynamic calculation using in situ H2 and hydrogen sulfide (H2S) concentrations indicated that chemosynthetic symbiosis could be supported by metabolic energy via H2 oxidation, particularly for the kAlv holobiont. Metabolic activity measurements showed that both the living individuals and the gill tissues consumed H2 and H2S at similar levels. Moreover, a combination of fluorescence in situ hybridization, quantitative transcript analyses, and enzymatic activity measurements showed that the kAlv endosymbiont expressed the genes and enzymes for both H2- and sulfur-oxidations. These results suggest that both H2 and H2S could serve as the primary energy sources for the kAlv holobiont. The eAlv holobiont had the ability to utilize H2, but the gene expression and enzyme activity for hydrogenases were much lower than for sulfur-oxidation enzymes. These results suggest that the energy acquisitions of A. marisindica holobionts are dependent on H2- and sulfur-oxidation in the H2-enriched Kairei field and that the mechanism of dual metabolism is controlled by the in situ H2 concentration.
Subject terms: Biogeochemistry, Biogeochemistry, Biogeochemistry
Introduction
It has been about 40 years since the discovery of deep-sea chemosynthetic animals in a hydrothermal system of the Galapagos rift [1–5]. To date, more than a thousand sites, including hydrothermal vent fields, methane-seep fields, and whale fall sites, have been discovered in the global ocean and their associated chemosynthetic invertebrate communities have been identified [6]. These invertebrates harbor specific lineages of chemosynthetic bacteria in the cells of specific tissues (endosymbiosis) or on their tissues (ectosymbiosis). Most of the chemosynthetic holobionts (host-symbiont associations) are sustained by energy metabolisms of either sulfur- or methane-oxidation (single thiotrophy or methanotrophy) or both (dual thiotrophy and methanotrophy) [6]. This fact raises the question why the deep-sea chemosynthetic holobionts do not adopt H2 oxidation (hydrogenotrophy) as their primary energy metabolism when H2 represents one of the most abundant gas components of hydrothermal systems and most free-living microbial populations adopt hydrogenotrophy as their predominant energy metabolism [7–12].
Petersen et al. showed for the first time that the symbiotic deep-sea mussel, Bathymodiolus puteoserpentis, living in the Logatchev hydrothermal field of the Mid-Atlantic Ridge (MAR) that hosts serpentinization-driven H2-abundant fluids, had the ability to utilize H2 as its chemosynthetic energy source [13]. It was also demonstrated by fluorescent in situ hybridization (FISH) and immunohistochemistry that the sulfur-oxidizing endosymbiont cells in the gills expressed genes for H2 oxidation [13]. Although H2 oxidization was likely an advantageous energy metabolism for chemosynthetic symbiosis in the H2-enriched Logatchev hydrothermal environment, different B. puteoserpentis ecotypes and their symbionts living in other MAR H2-depleted hydrothermal systems also seemed to have hydrogenotrophic ability [13]. Beinart et al. extensively surveyed Alviniconcha spp. holobionts in the Eastern Lau Spreading Center (ELSC) by quantitative molecular approach coupled with geochemical analyses using both gastight fluid sampler and in situ electrochemical instrument [14]. They demonstrated that both the host and symbiont species were genetically distinct in accordance with the geochemical conditions of their habitats, suggesting that the biogeographical distribution of holobionts was determined by the hydrothermal fluid chemistry [14]. Subsequently, based on the newly developed in situ RNA extracting instrument and metatranscriptomic analysis, Sanders et al. also demonstrated that some of the Sulfurimonas endosymbionts in Campylobacterota [15] expressed genes of Ni–Fe-hydrogenase in the northern ELSC vent fields with relatively abundant H2, implying that the specific Alviniconcha holobionts would also utilize H2 as their chemosynthetic energy source [14, 16]. Ikuta et al. [17] also demonstrated that hupL and hupS genes, which encode small and large subunits of hydrogenase, were expressed in the symbiont of Bathymodiolus septemdierum from the Myojin Knoll hydrothermal vent site. In addition, genome-sequence analyses demonstrated that several holobionts of vent-endemic chemosynthetic invertebrates have genes that are potentially involved in H2-oxidization, although the functional and molecular information of these genes is still not known [18–20]. Nevertheless, no deep-sea chemosynthetic holobiont that depends only on hydrogenotrophy has yet been identified, and it is yet unresolved how much the potentially hydrogenotrophic chemosynthetic symbiosis physiologically depends on H2 oxidation (as compared with sulfur and/or methane oxidation) and is regulated by the physical and chemical conditions of the hydrothermal fluids and habitats.
Alviniconcha gastropods dwell widely in many deep-sea hydrothermal systems of the western Pacific and Indian Ocean [21–23]. Each of the Alviniconcha populations harbors a specific lineage of chemosynthetic endosymbiont in the gill [6, 14, 22–25], while the host Alviniconcha and endosymbiont species represent biogeographic diversity in different deep-sea hydrothermal systems [6, 14, 22–25]. Alviniconcha strummeri in the Lau Basin, Alviniconcha hessleri in the Mariana Trough, and Alviniconcha kojimai in the Manus Basin and the Lau Basin harbor predominantly gammaproteobacterial endosymbionts [23]. Alviniconcha marisindica in the Indian Ocean and Alviniconcha boucheti in the Manus Basin and the Lau Basin harbor Campylobacterota endosymbionts [22, 23, 26]. It has been reported that the Campylobacterota endosymbionts of A. boucheti in relatively H2-abundant ELSC deep-sea hydrothermal systems express the genes related with H2 oxidation [16].
In this study, the holobionts of A. marisindica in the Kairei and Edmond hydrothermal fields of Central Indian Ridge (CIR) [27, 28] are investigated. The Kairei and Edmond hydrothermal vent fields show distinct hydrothermal fluid chemistry; the endmember hydrothermal fluids in the Kairei field are highly enriched with H2 (3.7–8.2 mM) [29–32], while the endmember fluid in the Edmond field is relatively H2 depleted (22–250 μM) [29, 30, 32]. It was also demonstrated that the free-living microbial communities in the hydrothermal mixing zones of the Kairei field are mainly sustained by hydrogenotrophic chemolithoautotrophy [12, 33]. The populations of A. marisindica in the Kairei and Edmond hydrothermal fields (kAlv and eAlv) harbor phylogenetically related Campylobacterota endosymbionts [26], which are also closely related to the hydrogenotrophic and thiotrophic Sulfurovum sp. NBC37-1 (97.1% sequence similarity) [34]. Moreover, in Sulfurovum sp. NBC37-1, it was proposed that the activity of the hydrogenase, an enzyme that oxidizes H2, was induced and strongly elevated under the high H2 conditions [35].
On the basis of these observations, we hypothesized that the holobionts of A. marisindica, particularly kAlv, would be able to utilize H2 as a primary energy source. If this hypothesis would be justified, we are next interested in how both Alviniconcha holobionts engaged to oxidize H2 and reduced sulfur compounds in the H2-abundant and -depleted habitats. We carried out multiple analyses including onboard incubation experiments and quantitative molecular and enzymatic analyses to understand the dual energy metabolisms of the holobionts of A. marisindica, and to address how the configuration of dual energy metabolisms are controlled by the environmental conditions of the habitats.
Materials and methods
Sampling sites
A. marisindica individuals and the water samples of representative habitats were collected from the Kairei (25° 19.23′ S, 70° 02.42′ E; 2415–2460 m of water depth) (Fig. 1a) and the Edmond (23° 52.68′ S, 69° 35.80′ E; 3290–3320 m of water depth) (Fig. 1b) hydrothermal vent fields in the CIR using a human-occupied vehicle, Shinkai6500, during the YK05-16 Leg2 (February 2006), YK09-13 Leg2 (November 2009), YK13-03 (March 2013), and YK16-E02 (February 2016) cruises of R/V Yokosuka. The distance between these two hydrothermal fields is 165 km. The history of this study, which explored the Kairei and Edmond fields is summarized in Table S1.
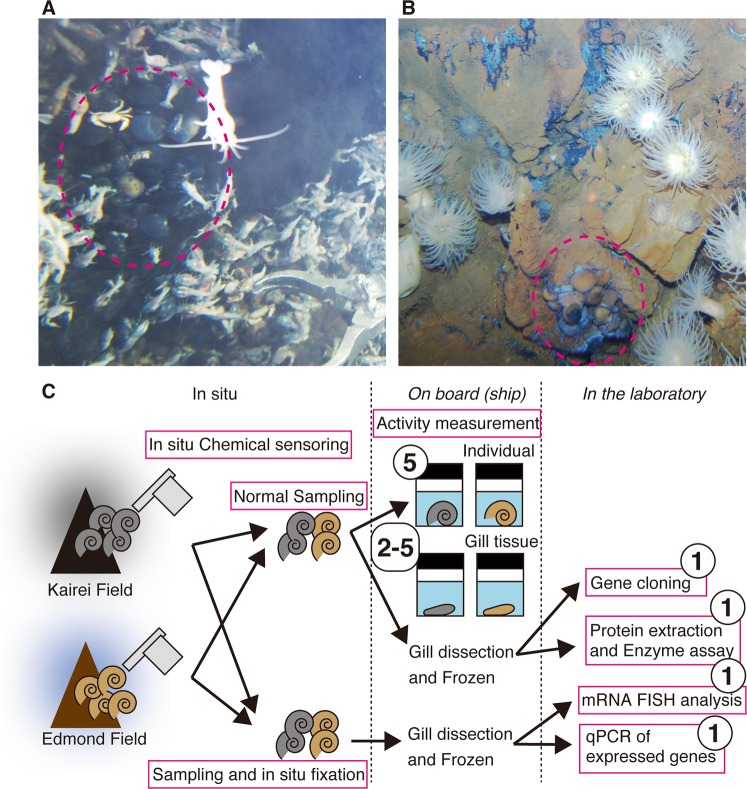
Fig. 1. Photographs of habitats and colonies of two populations of Alviniconcha marisindica in the Central Indian Ridge and analytic and experimental set-ups used in this study.
Magenta circles indicate the typical habitats and colonies of kAlv in the Kairei field (a) and of eAlv in the Edmond field (b). The schematic illustration shows representative experiments and analyses conducted in situ, onboard, and in the laboratory (c). The numbers in circles represent samples used in each experiment.
In situ sensor measurements and water sampling
To simultaneously collect water samples surrounding the gastropod’s colonies (colony water samples) and measure the in situ H2 and H2S concentrations, we assembled a waterflow line with the in situ H2 and H2S sensor system that was equipped with a Shinkai6500, as summarized in Fig. S1 in Supplemental materials. Using this waterflow line, we were able to measure the chemical conditions by sensing with or without sampling water. For the in situ calibration of chemical sensors (under different temperature and pressure conditions), 5 l of chilled seawater that contained approximately 200 μM H2 and 500 μM Na2S (pH 7.5) without dissolved O2, or 50 μM CH4 with saturated dissolved O2, was packed in a 5-L Rontainer (Sekisui Seikei Co Ltd, Osaka, Japan), which is a thin-polyethylene container, and was deployed in the payload of Shinkai6500 to connect the water sampling line before the dive. At the seafloor during the dive, these in situ standard waters were measured by the chemical sensors and were collected with a Hachiren water sampler. After the recovery of in situ standard water samples onboard, dissolved gas extraction from the Hachiren water samples was conducted using previously reported techniques [36] and the exact dissolved H2 and CH4 concentrations of the in situ standard water samples were determined by gas chromatography with helium ionization detector (GC-HID, GL Science, Torrance, CA, USA). The concentration of sulfide in the standard water sample was spectrophotometrically determined using the methylene blue method [37, 38]. The values of chemical sensors in the deep seawater distantly located from the deep-sea hydrothermal fields were used for the zero standard of H2 and H2S.
A phosphorescence dissolved oxygen (DO) sensor (ARO-USB, JFE Advantech Co. Ltd, Kobe, Japan) was used to measure in situ dissolved O2 concentration, in which the analyzed data were recorded. Measurement of DO for the invertebrate colony was carried out by directly placing this instrument on the shells of invertebrates for 5 min.
Sampling and preparation of gastropods
Individuals of A. marisindica (kAlv and eAlv populations) were collected with a suction pump sampler equipped on the Shinkai6500 from representative habitats of the Kairei and Edmond fields, respectively. As summarized in Fig. S2 of the Supplemental materials, some of the gastropod individuals were collected and then were fixed in situ by using RNA later (25 mM sodium citrate, pH 5.2, 10 mM EDTA, 0.7 kg/L ammonium sulfate) [39]. This method (without the in situ homogenization of the gastropod) did not assure appropriate fixation of the endosymbiont’s intracellular RNA by potentially incomplete infiltration of RNA later solution into the tissue, while it was confirmed that the gastropod individuals after this fixation process provided greater abundances of quantification for the 16S rRNA and other functional gene transcripts from the gill endosymbiotic cells than the individuals without the in situ fixation in the quantitative PCR (qPCR) analyses described later. After recovery onboard, most of the individuals were reared in tanks containing fresh surface seawater previously cooled at 4 °C for more than 12 h. Other individuals were dissected and the gill tissues (and some of the mantle tissues) were subsampled and stored at −80 °C and 4 °C. The frozen gill and mantle tissues were used for DNA and protein analyses.
To measure H2- and sulfide-consumption activity, both living A. marisindica individuals and dissected whole gill tissues (from individuals representing 3.5–4.0 cm shell width and 5.0–6.0 cm shell length, and within 12 h of onboard recovery) were washed twice with chilled surface seawater (previously passed through a 0.2 μm sterile filter) and incubated with substrates of either H2 or sulfide. In addition, the gill tissues dissected from the in situ fixed individuals were preserved into RNAprotect Bacteria Reagent (Qiagen, Valencia, CA, USA) and then stored at −80 °C for the subsequent transcript and ISH analyses. The procedure of this study is illustrated in Fig. 1c.
Measurement of H2- and sulfide-consumption
Total 29 of living Alviniconcha individuals were acclimated in cold surface seawater at 4 °C for 12 h after recovery on the ship, then a whole individual or dissected whole gill tissue was placed in a sterile 250-ml glass bottle with 200 ml filtrated seawater and the glass bottles were sealed with a butyl rubber stopper under atmosphere. For the H2-consumption experiments, 5 ml of H2 was added to the headspace in a bottle (at a final soluble H2 concentration of ~100 μM) and the bottle including a living individual or gill tissue was incubated at 5 °C. At 8–12 h intervals, 1 ml of seawater was extracted from the bottle and was then injected into a 20 ml vacuumed vial. After the addition of 30 ml of helium (He) gas into the vial, 400 μl of gas phase was measured using a GC-HID. At the same time, we prepared glass bottles without individuals or gill tissue and measured the background consumption as the controls. The initial velocity of H2 decrease was calculated from differences between the bottles with and without Alviniconcha individuals. The total experiment time was set to be 40 h.
Sulfide-consumption experiments were carried out according to a method described previously [40] with minor modifications. Four milliliters of neutralized Na2S solution (10 mM) was added to a bottle (at a final sulfide concentration of ~200 μM) and the bottle, including a living individual or a gill tissue, was incubated at 5 °C. At 1-h intervals, 0.5 ml of seawater was extracted from the bottle and the sulfide concentration was measured by methylene blue reduction at an absorbance of 668 nm [37, 38]. To calculate activity, we used control bottles, which contained neither individuals nor gill tissue. This control bottle contained the same concentration of Na2S as the bottle including a living individual or gill tissue. Total experiment time was set to be 4 h. The initial sulfide concentration in this experiment was designed based on the in situ H2S concentrations determined by the sensor.
Cloning and phylogenetic analysis of 16S rRNA, H2- and sulfur-oxidizing enzyme genes of gill endosymbionts and cytochrome c oxidoreductase I gene of host’s mitochondria
DNA extraction from 0.4 g frozen A. marisindica gill or mantle tissue was carried out using an Ultraclean Soil DNA Isolation Kit (MO BIO Laboratories, Solana Beach, CA, USA). The 16S rRNA, H2-uptaking hydrogenases (hydAB), sulfide-quinone-reductase (sqr), and sulfur-oxidizing enzyme complex subunit B and C (soxB and soxC) genes of Campylobacterota endosymbionts were amplified with the extracted DNA from the gill tissue by PCR using LA Taq with GC buffer (Takara Bio, Otsu, Japan). In these PCRs, oligonucleotide primers were used from previously described degenerated primers (for the amplification of 16S rRNA gene [41] and sqr gene [42]) or were designed based on our whole-genome sequencing results (for the amplification of hydA1B1, hydA2B2, soxB and C genes, gyrB and gap; unpublished data). The cytochrome c oxidoreductase I (mtCOI) gene from host-mitochondrial DNA was also amplified using the DNA extracted from mantle tissue by PCR with EX taq DNA polymerase (Takara Bio). All the oligonucleotide primers and PCR conditions used in this study are summarized in Tables S2 and S3. The amplicons were purified via agarose gel electrophoresis and a MiniElute Gel Extraction Kit (Qiagen) and ligated into the pCR2.1 vector in a TA cloning kit (Invitrogen, Carlsbad, CA, USA). A clone library of the amplicon of each gene was constructed using Escherichia coli INVαF′ (Invitrogen) with the ligation mixture. Eight of cloned fragments for each gene were directly sequenced using Sanger et al.’s method by using Applied Biosystem 3730xl Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA) [43]. After confirmation by direct sequencing, the plasmids containing the correct amplicons were extracted and used for the subsequent full-length sequence and phylogenetic analyses. The determined 16S rRNA gene sequence was analyzed using the ARB software package [44]. The tree of 16S rRNA gene was constructed using the neighbor-joining method of the ARB software package. Phylogenetic analyses of hydB, sqr, soxB, and mtCOI genes were performed using the neighbor-joining method in the MEGA5 software package [45].
RNA extraction and quantification of 16S rRNA and transcripts of hydABs, sqr, and soxBC genes
RNA extraction and the subsequent reverse transcription (RT) were carried out according to the method previously described [46] with some minor modifications. RNA extracts from frozen gill tissues (0.233 g for kAlv and 1.297 g for eAlv) were subjected to in situ fixation using an RNA PowerSoil Total RNA Isolation Kit (MO BIO Laboratories). The extracted RNA (500 ng) was treated with 1 μl of 100 U/ml DNaseI (Invitrogen) overnight at room temperature. Then, RT was carried out to synthesize cDNA using a Superscript III (Invitrogen) with random hexamers according to the kit manual. The synthesized cDNA assemblage was subjected to PCR to amplify 16S rRNA, hydABs, sqr soxB, soxC, gyrB, and gap genes. The primers and amplification conditions of RT-PCR are summarized in Tables S2 and S3. The amplified fragments were confirmed by cloning into the PCR2.1 vector and then sequenced according to the method described for cloning of the functional genes.
All of the qPCR analyses against the synthesized cDNA assemblages were performed with a 7500 Real Time PCR system and StepOnePlus Real Time PCR system (Applied Biosystems, Foster City, CA, USA). The primer sets and conditions of the quantitative RT-PCR are listed in Tables S2 and S3, respectively. In addition, genes encoding DNA gyrase subunit B (gyrB) and glyceraldehyde 3-phosphate dehydrogenase (gap) were used in qPCR as internal controls, which were cloned from the endosymbionts by the method as described in cloning of functional genes. Standard curves were established by plasmids described in cloning of the functional genes (Fig. S8 in the Supplement materials). These standard curves spanned eight orders of magnitude and were designed to allow the addition of 100–10−7 μg/ml of the constructed plasmid as 1 μl of template in each PCR. Ten-microliter reaction was used with a final concentration of 1× TB Green premix EX taq, 1× ROX (Takara Bio). The PCR cycling conditions were summarized in Table S3 in the Supplemental materials. To confirm whether target and/or nontarget fragments were amplified, dissociation program was added to final steps of every PCR programs and electrophoresis was carried out against the amplicons after qPCR. Data were analyzed by the system software of the instruments and then were converted to gene copies. The coefficients of determination (R2) were 0.9765–0.9991.
FISH analyses of 16S rRNA and transcripts of hydAB and soxB
RNA probes for FISH were prepared as described previously [47, 48]. To synthesize RNA probes, amplifications of 16S rRNA, hydABs, and soxB genes were conducted by PCR using primers, which were designed based on the sequence data from Tables S2 to S3 and using the constructed plasmids as templates; then the amplicons were ligated into a pTA2 vector (Toyobo, Osaka Japan). Cloned genes were sequenced by direct sequencing of the fragments [43]. Using 200 ng of the DNA fragments as templates, which were amplified from the plasmids containing each of the gene fragments, antisense and sense RNA probes were synthesized with T3 and T7 RNA polymerase, respectively, using digoxigenin (DIG) or fluorescein RNA labeling mix (F. Hoffmann-La Roche Ltd, Basel, Switzerland).
In situ hybridization was carried out as described previously [47, 48], with the following modifications. The gill tissues excised from the in situ fixed individuals were re-fixed in Davidson’s solution (10% glycerol, 40% formaldehyde, 30% ethanol, and 30% filtered artificial seawater) at −30 °C for 24 h and at 4 °C for 16 h, followed by stepwise dehydration in an ethanol series. The fixed gill was embedded in paraffin and sliced into 4-μm transverse sections. Hybridization in the presence of both DIG- and fluorescein-labeled probes and the following washing procedures were performed at 60 °C. After two-color fluorescence staining using a TSA plus cyanine 3/fluorescein kit (PerkinElmer, MA, USA), all sections were mounted in a Vectashield (Vector Laboratories, CA, USA) and covered with a coverslip. Images were taken using a Keyence (Osaka, Japan) BZ-9000 microscope, and micrographs were processed with Adobe Photoshop CS6 (Adobe Systems, CA, USA). All of the fluorescent micrographs were obtained using the same settings.
Activity measurement of hydrogenase and sulfur-oxidizing enzymes in crude extracts of gill
Dissected gill tissue (1.8 g each) was homogenized in liquid nitrogen in a mortar and pestle and then suspended in 8 ml of extraction buffer containing 100 mM Tris–HCl and 1 mM dithiothreitol (pH 8.0). After addition of 1.0 g of glass beads (0.710–1.18 mm diameter) into the slurries, cells were thoroughly disrupted by vortex mixing. After centrifugation at 7500 × g for 30 min at 4 °C, the supernatants were recovered into glass vials and used for the enzyme assays as crude extracts of gill tissues. These procedures were carried out under a 95% N2 and 5% H2 atmosphere in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI, USA).
The specific activities of hydrogenase and Sox enzymes were determined according to the following method, previously described by Yamamoto et al. [35]. All measurements were taken in an anaerobic environment. When we measured hydrogenase activity in the crude extract, 10 μl of the crude extract was added to 1 ml of the reaction mixture (100 mM MOPS, pH 7.0, 5 mM methylene blue, 1 atm of 100% H2) that was preincubated at assay temperature (25, 45, or 65) for 5 min, and the reaction was monitored at the same temperature following the decrease in absorbance at 665 nm. Instead of 10 μl of the crude extract, we used 10 μl of the extraction buffer without any enzyme was added to the reaction mixture as a control. Specific activity of hydrogenase was calculated from the absorption coefficient of methylene blue, 74.0 cm2 μmol−1. One unit was defined as the amount of enzyme that reduced 1 μmol of electron acceptor per min. When we measured Sox activity in crude extract, 10 μl of the crude extract was added to 1 ml of the reaction solution (100 mM HEPES, pH 8.0, 35 μM horse heart cytochrome c (Sigma, St. Louis, Mo., USA), 10 mM sodium thiosulfate) that was also preincubated at assay temperature (25, 45, or 65 °C) for 5 min and the reaction was monitored at the same temperature following the decrease in absorbance at 550 nm. Instead of 10 μl of the crude extract, we used 10 μl of the extraction buffer without any enzyme was added to the reaction mixture as a control. Specific activity of Sox enzyme was calculated from the absorption coefficient of cytochrome c, 27.8 cm2 μmol−1. One unit was defined as the amount of enzyme that reduced 1 μmol of electron acceptor per min.
Measuring of sulfide:quinone reductase (Sqr) enzyme activity in crude extract was carried out by the modified method described by Wakai et al. [49]. Assay solution (250 μl) containing 50 mM BisTris, pH 7.0, 20 mM glucose, 3 mM DUQ (decylubiquinone, SIGMA) and 10 μl of the crude extract was flashed with N2 gas, in which one unit of glucose oxidase and ten units of catalase was added to establish anoxic conditions. Instead of 10 μl of the crude extract, we used 10 μl of the extraction buffer without any enzyme was added to the reaction mixture as a control. This prepared assay solution was preincubated at 30 °C for 5 min, then the reaction was initiated by addition of 100 μM of Na2S solution and monitored at 30 °C following the decrease in absorbance at 275 nm. Specific activity of Sqr enzyme was calculated from the absorption coefficient of ubiquinone, 12.5 cm2 μmol−1. One unit was defined as the amount of enzyme that reduced 1 μmol of ubiquinone per min.
Protein concentrations in the crude extracts were measured using the bicinchoninic acid method according to the manufacturer’s manual (BCA Protein Assay Reagent Kit, Pierce, Rockford, IL, USA).
Nucleotide sequence accession numbers
All the sequences obtained in this study were deposited into DDBJ (DNA data bank of Japan) database under accession numbers AB610411–AB610419 and LC474371–LC474378.
Results and discussion
Phylogenetic relationship of Alviniconcha marisindica holobionts in the Kairei and the Edmond fields
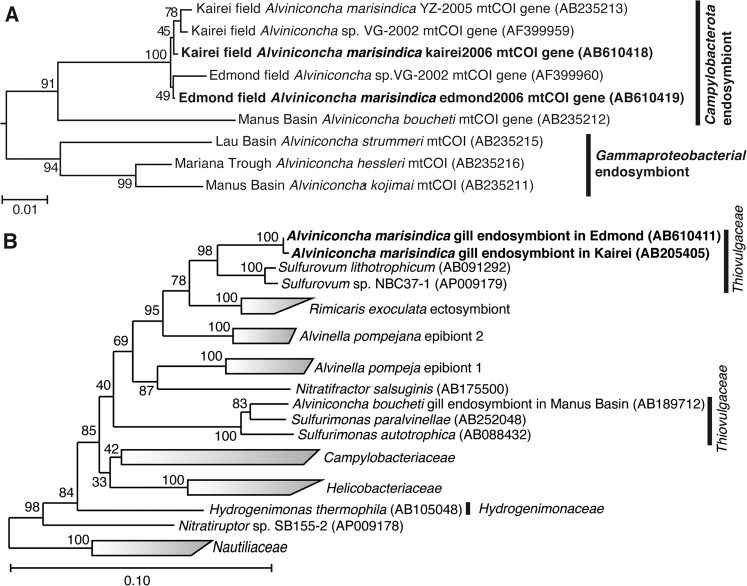
We cloned and sequenced partial mitochondrial cytochrome c reductase (mtCOI) genes from individuals of both A. marisindica populations. The sequence of the mtCOI gene of kAlv showed 99.4% sequence identity with that of eAlv, indicating that both populations of A. marisindica taxonomically belong to the same species. The phylogenetic tree of the mtCOI gene, including the data obtained from the previous studies [50], also demonstrated that kAlv and eAlv were very closely related (Fig. 2a).
Fig. 2. Phylogenetic trees based on the sequences of mitochondrial cytochrome c reductase (mtCOI) gene from hosts and 16S rRNA gene from endosymbionts in two populations of Alviniconcha marisindica.
The phylogenetic trees were constructed by the neighbor-joining method using MEGA5 software [45] based on the mtCOI gene sequence (544 base position) (a) and using the ARB software package [44] based on 16S rRNA gene sequences (1402 base position) (b). The mtCOI gene sequences obtained from A. marisindica individuals in this study have been indicated in bold. The mtCOI genes of Ifremeria nautilei from North Fiji Basin (AB235214) and Manus Basin (AB235217) [22] were used as an outgroup for this tree. The 16S rRNA gene sequences obtained from A. marisindica individuals in this study have been indicated in bold. The 16S rRNA gene of Aquifex aeolicus (AJ309733) was used as an outgroup of this tree.
The 16S rRNA genes of endosymbionts of both kAlv and eAlv were cloned and sequenced. The 16S rRNA gene sequences of the kAlv and eAlv endosymbionts determined in this study were identical to the previously reported sequences of them [26] and none of the sequences potentially derived from minor symbiontic bacteria was obtained in the previous and present studies. The sequence identity between the kAlv and eAlv endosymbionts was 99.6%, indicating that the endosymbionts of kAlv and eAlv are taxonomically the same species. The both endosymbionts belong to the genus Sulfurovum within the class Campylobacterota and the most closely related cultivated strain is Sulfurovum sp. NBC37-1, whose genome sequence has previously been determined [19] (Fig. 2b).
These results showed that it may be interpreted that all the molecular and functional properties of both populations in this study are derived from a taxonomically identical holobiont system, the same species of endosymbiont and host animal, although further genome-sequence analyses of both endosymbionts and hosts will clarify the genomic differences between the holobionts of kAlv and eAlv in the future.
Physical and chemical conditions and chemolithotrophic metabolic potentials of in situ habitats of Alviniconcha marisindica in the Kairei and the Edmond fields
During the seafloor observation at the Kairei and Edmond hydrothermal fields, we found several A. marisindica colonies (Fig. 1).
In the Kairei field, the largest colony of A. marisindica (kAlv) was found in the diffusing fluid flow zones at the foot of the Monju chimney site and consisted of several hundred or over a thousand individuals with Chrysomallon squamiferum behind the large population of Rimicaris kairei (shrimp) (Fig. 1a) [26, 28–30, 51, 52]. When the temperature of A. marisindica colony water (i.e., the seawater in and around the colony of the A. marisindica population) was measured by a temperature probe equipped with the top of water sampler inlet during the water sampling and sensing (at the positions attaching the shells of individuals), the temperature ranged from 11.9 to 60.0 °C (average 19.3 °C) (Table 1).
Table 1.
Physical and chemical conditions of habitats of Alviniconcha marisindica populations (kAlv and eAlv populations) and Chrysomallon squamiferum in the Kairei and Edmond hydrothermal fields.
| Sampling site | Temp. range [average] | DO range [average] | H2 range [average] | H2S range [average] |
|---|---|---|---|---|
| (°C) | (μM) | (μM) | (μM) | |
| Kairei field | ||||
| Reference bottom seawater |
1.7–1.8 [1.8] |
207 | <0.03 | <0.5 |
| A. marisindica (kAlv) colony at the Monju chimney |
11.9–60.0 [19.3] |
104–166 [144] |
20.1–44.6 [34.1] in situ |
137–211 [185] in situ |
|
11.8–20.2 [16.5] onboard |
<1.5 onboard |
|||
| C. squamiferum colony at the Monju chimney |
8.4–13.5 [12.6] |
133–187 [158] |
8.00–8.60 [8.40] in situ |
23.5–29.6 [25.1] in situ |
|
6.37–10.2 [8.16] onboard |
<1.5 onboard |
|||
| Edmond field | ||||
| Reference bottom seawater |
1.8–1.9 [1.9] |
208 | <0.03 | <0.5 |
| A. marisindica (eAlv) colony at the Giant shrimp chimney |
6.5–38.1 [26.8] |
147–202 [184] |
0.39–0.45 [0.42] in situ |
109–127 [120] in situ |
|
0.70–4.52 [2.52] onboard |
19–26 [23.3] onboard |
|||
To determine the chemical conditions within the kAlv colony, we introduced in situ electrochemical sensors for H2 and H2S concentrations, together with a DO sensor. The H2 concentration of kAlv colony water was found to be 20.1–40.6 μM (average 34.1 μM) (Table 1). The H2S concentration of water in the kAlv colony was found to be 137–211 μM (average 185 μM) (Table 1).
In the Edmond field, we discovered many small colonies (10–20 individuals) of A. marisindica (eAlv) along the cracks with diffusing fluid flows at the foot of the enormous hydrothermal mound (giant shrimp chimney) (Fig. 1b). The temperature of the water in several representative colonies ranged from 6.5 to 38.1 °C (average 26.8 °C) (Table 1). The H2 concentration in water at the eAlv habitats ranged from 0.39 to 0.45 μM (average 0.42 μM) (Table 1), indicating that eAlv inhabits water with much lower H2 concentrations that kAlv. In contrast, the H2S concentration in the eAlv habitats ranged from 109 to 127 μM (average 120 μM), and the DO concentration was found to be 147–202 μM (average 184 μM) (Table 1). In contrast to the H2 concentration, both the kAlv and eAlv would colonize under similar concentrations of H2S and DO.
To determine the H2 and H2S concentrations in both kAlv and eAlv waters, we also used the water samples recovered onboard (Table 1). However, the H2 and/or H2S concentrations in the recovered water samples were strongly reduced probably due to microbial consumption and chemical oxidation during the recovery (Table 1).
Using these physical and chemical properties of the in situ habitats and the bimodal mixing model between each of the endmember hydrothermal fluids and ambient seawater previously described [12], we calculated potential metabolic energy (J/kg water) of H2 oxidization with O2 reduction (aerobic hydrogenotrophy) and H2S oxidization with O2 reduction (aerobic thiotrophy) for free living and/or symbiotic microbial populations in the kAlv and eAlv colonies (Table 2). The calculated energy yield of aerobic hydrogenotrophy (4.4–9.4 J/kg water) using the in situ data of the kAlv colony was considerably lower than the yield based on the mixing model calculation of the kAlv colony (24.1–41.1 J/kg water) (Table 2). In contrast, the energy yield of aerobic thiotrophy in the kAlv colony was similar between the calculations based on the in situ data (39.2–65.0 J/kg water) and the mixing model (31.8–37.9 J/kg water) (Table 2). This was because the measured in situ H2 concentrations were lower than the H2 concentrations simply expected from the bimodal mixing between the endmember fluid and the seawater. Since the diffusing fluids (even high-temperature fluids) had already experienced subseafloor-microbial activities and physical–chemical processes prior to the seafloor discharge [33, 53], the H2 concentrations of the seafloor diffusing fluids were strongly reduced from the initial concentrations during the seafloor mixing process. Even under the in situ H2 concentrations, however, aerobic hydrogenotrophy is thermodynamically energy yielding and is a competitive metabolism with aerobic thiotrophy for the free living and symbiotic microbes in the kAlv population (Table 2).
Table 2.
Potential metabolic energy yield estimated from the measured in situ physical and chemical data and the bimodal mixing model between each of the endmember hydrothermal fluids and ambient seawater.
| Calculated temperature range (°C) | H2-trophy colony water (J/kg) | H2S-trophy colony water (J/kg) | |
|---|---|---|---|
| Kairei field | |||
| A. marisindica (kAlv) colony | |||
| Using the measured in situ data | 11.9–60.0 | 4.4–9.4 | 39.2–65.0 |
| Using the bimodal mixing model | 12–60 | 24.1–41.1 | 31.8–37.9 |
| C. squamiferum colony | |||
| Using the measured in situ data | 8.4–13.5 | 1.8–2.0 | 18.2–23.1 |
| Using the bimodal mixing model | 8–13 | 14.3–26.4 | 37.7–38.4 |
| Edmond field | |||
| A. marisindica (eAlv) colony | |||
| Using the measured in situ data | 6.5–38.1 | 0.08–0.1 | 56.1–79.5 |
| Using the bimodal mixing model | 6–40 | 0.3–2.5 | 33.7–36.2 |
Similarly, the calculated energy yield of aerobic hydrogenotrophy (0.08–0.1 J/kg water) using the in situ data of the eAlv colonies was found to be considerably lower than the yield (0.3–2.5 J/kg water) based on the mixing model calculation of the eAlv colony, while the energy yield of aerobic thiotrophy was similar between the calculations based on the in situ data (56.1–79.5 J/kg water) and the mixing model (33.7–36.2 J/kg water) (Table 2). The available energy potentials of aerobic hydrogenotrophy in the eAlv colonies were much lower than those in the kAlv colony. Thus, aerobic hydrogenotrophy would be highly disadvantageous for the microbial populations of eAlv colonies, including eAlv endosymbionts, compared with aerobic thiotrophy from the thermodynamic aspect. These thermodynamic estimations suggest that hydrogenotrophic energy metabolism may be the viable option for the kAlv endosymbiont but not eAlv endosymbiont, together with the thiotrophic metabolism.
H2- and sulfide-consumption experiments
Onboard H2- and sulfide-consumption experiments were conducted using both living A. marisindica individuals and dissected whole gill tissues within 12 h of onboard recovery. During the incubation of living kAlv individuals with a supply of H2, the dissolved H2 concentration constantly decreased and the average H2-consumption rate was calculated to be 15.7 ± 8.23 μmol/h/individual (Table 3). In the case of incubations with dissected kAlv gill tissues, the H2-consuming rate was evident but lower than for the living individuals (the average rate was 0.59 ± 0.45 μmol/h/gill) (Table 3). These results clearly indicate that the holobiont of kAlv consumes H2 in the habitats. On the other hand, during the incubations with a supply of sulfide, the average sulfide-consuming rates of kAlv individuals and dissected gill tissues were 20.3 ± 6.4 μmol/h/individual and 2.78 ± 0.92 μmol/h/gill, respectively (Table 3). These rates were slightly higher than the H2-consuming rates.
Table 3.
H2 and sulfide-consuming rates of living individuals and dissected gill tissues of Alviniconcha marisindica populations.
| Specimen | H2 consumption | H2S consumption |
|---|---|---|
| kAlv | ||
| Living individual | (n = 3) | (n = 3) |
| (μmol/h/individual) | 15.7 ± 8.23 | 20.3 ± 6.4 |
| (μmol/h/g of individual) | 0.44 ± 0.23 | 0.56 ± 0.18 |
| Gill tissue | (n = 5) | (n = 5) |
| (μmol/h/gill) | 0.59 ± 0.45 | 2.78 ± 0.92 |
| (μmol/h/g of gill tissue) | 0.11 ± 0.086 | 0.53 ± 0.18 |
| eAlv | ||
| Living individual | (n = 3) | (n = 3) |
| (μmol/h/individual) | 6.0 ± 1.6 | 9.8 ± 0.35 |
| (μmol/h/g of individual) | 0.16 ± 0.042 | 0.26 ± 0.0093 |
| Gill tissue | (n = 5) | (n = 2) |
| (μmol/h/gill) | 0.11 ± 0.21 | 2.1 ± 0.47 |
| (μmol/h/g of gill tissue) | 0.024 ± 0.046 | 0.46 ± 0.10 |
The numbers in parentheses show the specimen numbers of the living individuals or gill tissues used for independent incubations.
When the living individuals and dissected gill tissues of eAlv were incubated with H2 and sulfide supplies, the average H2- and sulfide-consuming rates were found to be 6.0 ± 1.6 μmol/h/individual for H2 and 9.8 ± 0.35 μmol/h/individual for sulfide and 0.11 ± 0.21 μmol/h/gill for H2 and 2.1 ± 0.47 μmol/h/gill for sulfide (Table 3). The eAlv individuals showed one-third of the H2-consuming and a half of the sulfide-consuming rates of the kAlv individuals, although the dissected gill tissues of both populations showed similar H2- and sulfide-consuming rates (Table 3). It should be noted that experimental H2 concentration in the eAlv individuals and gill tissue (~100 μM) was much higher than in the natural habitat (0.4 μM, Table 1) and the experimental H2S concentration (~200 μM) was similar to the natural habitat (120 μM, Table 1). Thus, such a high concentration of H2 in the incubation would induce possible hydrogenase expression and activity of the eAlv endosymbionts during incubation, as observed in the previous study of Campylobacterota [35]. However, these results suggest that not only the kAlv holobiont but also the eAlv entity would have the metabolic potential to utilize both H2 and H2S in their natural habitat.
The considerably lower H2- and sulfide-consumption rates of the dissected gill tissues compared with the living individuals were observed in both populations of A. marisindica. One explanation for this may be the potentially limited supply of electron acceptors (for the most part O2 but possibly nitrate also) for H2- and sulfide-oxidation in endosymbiont cells in the dissected gill tissue due to the lack of blood ventilation. Without the blood ventilation of the living individual, electron acceptors may not be sufficiently provided to the intracellularly-immobilized bacterial cells. In addition, the lack of blood ventilation may cause a serious deficiency of endosymbiont viability and metabolic activity in the absence of certain physiological factors from the host. Although we thoroughly washed the shell and body surfaces of the living individuals prior to the experiments, we cannot exclude the inevitable contribution of H2- and/or sulfur-oxidizing microbial populations that adhere to the shell and body surface or in the other tissues to the H2- and sulfide-consumption of the living individual.
Molecular analyses of hydABs, sqr, and soxBC genes of endosymbionts
It is known that H2 oxidation is catalyzed only by a membrane-bound [Ni–Fe]-hydrogenase in the Campylobacterota [10, 34]. Acquired electrons from H2 are transferred to the terminal electron-accepting enzymes, such as polysulfide reductase, cytochrome c oxidase, or denitrification enzymes, potentially via ferredoxin [35]. In the draft genome sequence of the endosymbionts of kAlv, two paralogous hydrogenase genes (hydA1B1 and hydA2B2) were found (unpublished data). The identity in amino acid sequence between both hydrogenases was 49% in HydA and 47% in HydB. On the other hand, sulfur oxidation is likely catalyzed by the Sox multi-enzyme system in Sulfurovum sp. NBC37-1, which is taxonomically closely related to the endosymbiont of A. marisindica [35, 39]. The Sox multi-enzyme system performs the conversion of sulfur compounds to sulfate. Acquired electrons are transferred to cytochrome c oxidase or denitrification enzymes potentially through cytochrome c [35]. The draft genome sequence of the kAlv endosymbiont demonstrated that genes encoding Sox proteins were distributed in two gene clusters (unpublished data). One consisted of soxC-cyc-soxY-soxZ genes, and the other contained soxX-soxY-soxZ-soxA-soxB genes. It is also known that Sqr is involved in the conversion of sulfide to polysulfide in many of the Campylobacterota [35, 54]. During this conversion, electrons are received by quinone and transported to the terminal enzymes. We found a sqr gene encoding type IV sulfide:quinone reductase in the draft genome sequence of the kAlv endosymbiont (unpublished data). These enzymes seem to be basic components for H2- and sulfur-oxidizing energy metabolisms of the endosymbionts of A. marisindica. Therefore, to evaluate the expression level of the transcripts of these functional genes related to hydrogenotrophic and thiotrophic metabolisms, we investigated hydA1B1, hydA2B2, soxC, soxB, and sqr genes from both endosymbionts.
The functional genes from both the endosymbionts of kAlv and eAlv of H2- and sulfur-oxidizing energy metabolisms such as hydABs, sqr, and soxBC genes were successfully amplified using degenerated primers [41, 42] or primers based on the genome sequence from the endosymbiont of kAlv, except for the hydA2B2 gene. We also attempted to amplify the hydA2B2 gene using several primers inferred from the kAlv endosymbiont genome but could not obtain the gene from eAlv endosymbiont (Fig. S3). Thus, we tentatively conclude that the eAlv endosymbiont either has nonhomologous hydA2B2 genes or lacks the hydA2B2 gene. The hydA1B1, sqr, soxB, and soxC gene sequences from both endosymbionts were determined. The amino acid sequence identities of hydA1B1, sqr, soxB, and soxC between the kAlv and eAlv endosymbionts were 99.8% (1 amino acid difference per 149 amino acids on HydB), 99.5% (2 amino acid differences per 247 amino acids), 99.9% (1 amino acid difference per 257 amino acids), and 100% (0 amino acid differences per 231 amino acids), respectively.
In addition to the 16S rRNA gene sequence comparison, these results reinforce the idea that both endosymbionts are equipped with almost identical gene repertoires for H2- and sulfur-oxidizing energy metabolisms, except for the hydrogenase paralogs. In other words, the different metabolic behaviors between the holobionts of kAlv and eAlv are derived primarily from regulation at the transcription, expression, and modification levels, responding to the environmental conditions rather than from the genome and genetic structures. Phylogenetic analysis also showed that these genes from the endosymbionts are closely related with those of Sulfurovum species, sharing more than 97% 16S rRNA identity with the endosymbionts of both kAlv and eAlv (Figs. S4–S7). Based on the phylogenetic tree of HydB, it was suggested that the H2-oxidization of Campylobacterota could be attributed to the products of hydA1B1 genes (Fig. S3), and we expected that the hydA2B2 gene products were not responsible for the H2-oxidization of the kAlv holobiont. However, as the functions of the hydA2B2 gene products are unknown, we conducted analyses using both hydA1B1 and hydA2B2 genes.
In addition, to normalize the comparative expression abundance of the functional genes between both kAlv and eAlv endosymbionts, we used gyrB and gap genes, which encode the DNA gyrase B subunit and glyceraldehyde 3-phosphate dehydrogenase, respectively. These genes are widely used as internal controls for the functional gene expression of bacteria and humans [55–58]. The amplified fragments of gyrB (695 bp) and gap (620 bp) shared 99.6% (three-bases mismatch) and 99.8% (one-base mismatch), respectively, with the endosymbionts.
Estimation of cellular expression of functional gene transcripts in endosymbionts
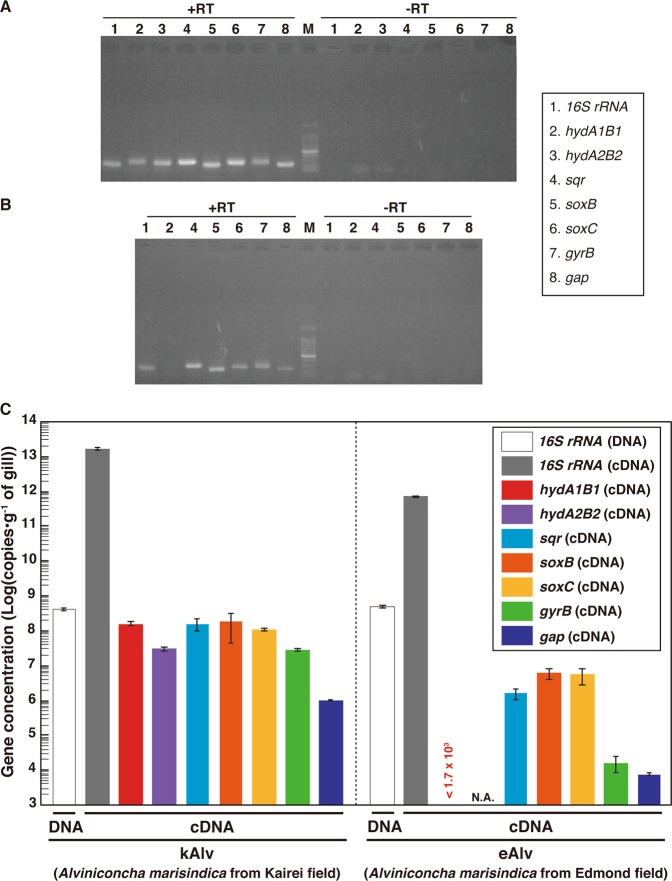
To evaluate the cellular expression of the functional genes involved in the H2- and sulfur-oxidizing energy metabolisms, we conducted synthesis of cDNAs derived from the transcripts of 16S rRNA, hydABs, sqr, soxB, soxC and reference genes in the endosymbionts from individuals of both kAlv and eAlv. Total RNA assemblages were extracted from the gill tissues of both populations and RT of the total RNAs were conducted. In these procedures, we confirmed that all the double-stranded DNA was completely removed before the RT step by DNaseI digestion (Fig. 3a, b). Then, RT-PCR was performed with the synthesized cDNA assemblages using the primer sets specific to each of the target genes of 16S rRNA, hydAB, sqr, soxB, and soxC genes. The agarose gel electrophoresis of the RT-PCR products demonstrated that the amplifications of 16S rRNA, sqr, and soxB cDNAs were observed in both the kAlv and eAlv gills while the amplification of the hydA1B1 gene was observed only in the kAlv gill under the experimental conditions (Fig. 3a, b).
Fig. 3. Quantitative analyses of 16S rRNA and transcripts of hydABs, sqr, soxB, soxC, gyrB, and gap genes by RT-PCR.
a Agarose gel electrophoresis of PCR products for 16S rRNA, hydABs, sqr, soxBC, gyrB, and gap genes with or without RT of total RNA extracts from kAlv gills. b Agarose gel electrophoresis of PCR products for 16S rRNA, hydA1B1, sqr, soxBC, gyrB, and gap genes with or without RT of total RNA extracts from eAlv gills. c Comparative abundances of 16S rRNA gene in the DNA extracts from kAlv and eAlv gills by quantitative PCR analysis and of hydA1B1, hydA2B2, sqr, soxB, gyrB, and gap gene transcripts in the cDNA assemblages from kAlv and eAlv gills by quantitative RT-PCR analyses. Quantity of transcript (<1.7 × 103 copies/g) of hydA1B1 gene in eAlv holobiont was lower than the detection limit by qPCR analyses. NA indicates not analyzed because the hydA2B2 gene was not detected by PCR in the eAlv endosymbiont.
To further estimate the expression levels of functional genes, we performed qPCR analyses. We confirmed the sensitivity and confidence of quantification in our method by using the clones constructed in the cloning step (Fig. S8). First, we conducted qPCR analysis for the 16S rRNA gene of the endosymbiont using the total DNA extracts from the kAlv and eAlv gills. The estimated quantity of 16S rRNA was 4.5 × 108 and 5.4 × 108 copies/g for kAlv and eAlv, respectively (Fig. 3c). This result indicated that the endosymbiont cell number in 1 g of gill tissue was similar between the kAlv and eAlv individuals. However, when qPCR analysis of endosymbiont 16S rRNA was conducted using the cDNA assemblages from the kAlv and eAlv gills, it was found that the estimated number of 16S rRNA was 22 times higher in the kAlv (1.71 × 1013 copies/g) than in the eAlv (7.66 × 1011 copies/g). This result suggested that the protein synthesis activity of the kAlv endosymbiont was much higher than that of the eAlv endosymbiont. In response to the different abundances of transcript 16S rRNA, we performed qPCR analyses of the gyrB and gap gene transcripts, which were used as references for the expression levels of constitutional genes. The results confirmed that the abundances of gyrB and gap transcripts in the kAlv endosymbiont (3.11 × 107 and 9.94 × 105 copies/g, respectively) were 1870 times greater in gyrB and 130 times greater in gap than those in the eAlv endosymbiont (1.66 × 104 and 7.73 × 103 copies/g, respectively). These results indicate that the expression abundances of constitutional genes related with cellular replication, translation, and central metabolism in the endosymbionts are greater in the kAlv than in the eAlv.
In a similar manner, the RT-qPCR indicated that all functional genes analyzed in this study were more abundantly expressed in the kAlv than in the eAlv (Fig. 3c). These results strongly suggest that the kAlv endosymbiont would be functionally more active than the eAlv endosymbiont. The largest difference of expression between the kAlv and eAlv gills was found in the hydA1B1 gene. The number of hydA1B1 gene transcripts for the kAlv was estimated to be 1.60 × 108 copies/g, while the number for eAlv was below the detection limit (<1.7 × 103 copies/g) (Fig. 3c). However, the estimated abundances of sqr, soxB, and soxC gene transcripts in the kAlv gill were 1.61 × 108, 2.00 × 108, and 1.18 × 108 copies/g, respectively, which were 20–100 times greater than that of the eAlv gill (1.74 × 106, 6.81 × 106, and 6.04 × 106 copies/g for the sqr, soxB, and soxC gene transcripts, respectively) (Fig. 3c). In comparison with various functional genes in the endosymbionts, these results demonstrated that the functional genes for H2- and sulfur-oxidizing energy metabolisms were expressed at similar levels in the kAlv gill, while the functional genes for sulfur-oxidizing energy metabolism were more abundantly expressed than for H2 oxidation in the eAlv gill. In addition, the expression abundance ratios of hydrogenase and sulfur-oxidizing genes against the gap gene were 47.1 (for hydA1B1), 47.5 (for sqr), 59.0 (for soxB), and 34.9 (for soxC) in the kAlv endosymbiont, and <0.021 (hydA1B1), 20.0 (sqr), 78.0 (soxB), and 69.2 (soxC) in the eAlv endosymbiont. These values were similar between the kAlv and eAlv endosymbionts except for the expression of hydA1B1 gene, suggesting the possible functional relevance between energy and carbon metabolisms such as glycogenesis and glycolysis.
Although the hydA2B2 gene was only found in the kAlv endosymbiont, the estimated abundance of the transcribed gene in the gill tissue was 3.26 × 107 copies/g, 4.9 times lower than that of hydA1B1 gene (Fig. 3c). It seems likely, therefore, that the H2 oxidation of the endosymbiont is primarily catalyzed by products of the hydA1B1 gene, although the function of hydA2B2 is still uncertain.
In the kAlv endosymbiont, the results of functional gene expressions seem to be consistent with the results of the H2- and sulfide-consumption experiments of the living individuals. The kAlv holobiont showed higher H2- and sulfide-consuming activities than the eAlv holobiont (Table 3) and showed a similar level of activity for both H2- and sulfide-consumption (Table 3). In contrast, although the eAlv holobiont showed slightly lower H2-consuming activity than sulfide-consuming activity (Table 3), the expression of hydA1B1 genes in the eAlv endosymbiont was below detection limit (<1.7 × 103 copies/g gill) (Fig. 3c). As discussed above, this difference between the onboard consuming experiment and the in situ transcript analysis would be derived from the possible experimental induction of hydrogenase expression and activity by the relatively high H2 concentration used for the onboard consuming experiment of eAlv holobiont. It seems likely therefore that the dual energy metabolism configuration of A. marisindica holobionts may be regulated by the expression of related functional genes responding to the physical and environmental H2 conditions of their habitats.
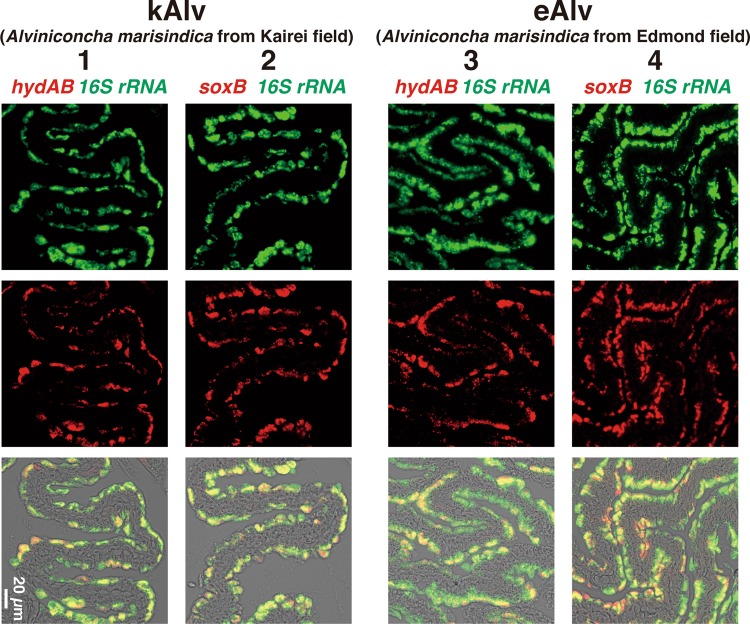
FISH analyses of hydA1B1 and soxB gene transcripts in gill tissue
To identify the localization of the functional gene transcripts in the gills of both populations, we designed probes to target the hydA1B1 and soxB gene transcripts that were likely involved in the H2- and sulfur-oxidation metabolisms, respectively, and then conducted FISH analysis using the individuals fixed in situ. Consistent with the distribution pattern of endosymbionts shown in the previous study of Alviniconcha [59], signals for the 16S rRNA of Campylobacterota endosymbionts in both populations were detected throughout the apical side of the gill epithelium (Fig. 4). Similarly, signals for the hydA1B1 and soxB gene transcripts were detected at the apical side of the gill epithelium in both the kAlv and eAlv, and mostly overlapped with the signals for the 16S rRNA (Fig. 4). When a sense probe was used as a control, no positive signal was observed (Fig. S9 in Supplemental materials). These results verified that both hydA1B1 and soxB expression occurred not only in the kAlv endosymbiont cells but also the eAlv ones. The FISH results seem to contradict with the qPCR results indicating that the abundance of hydA1B1 transcript in the eAlv endosymbiont was estimated as below detection limit (<1.7 × 103 copies/g gill) and was much lower than that of the kAlv endosymbiont (Fig. 3c). Although other interpretations cannot be excluded, a possible explanation is that the given incubation time of catalyzed FISH in this study may have resulted in excess amplification of initial signals derived from the probes bound with the target functional gene transcripts and may not properly represent the cellular abundances of hydA1B1 and soxB gene transcripts in the kAlv and eAlv endosymbionts. However, it is evident that the hydA1B1 and soxB genes are co-expressed in the identical Campylobacterota endosymbiont cells in the Alviniconcha gill tissues.
Fig. 4. Fluorescent micrographs of gill sections of two populations of Alviniconcha marisindica by FISH analyses targeting 16S rRNA and functional gene transcripts.
Each column shows the same section image taken by different excitation wavelengths of light. Left two columns (1 and 2) and right two columns (3 and 4) show the results of kAlv and eAlv gills, respectively. Green signals in the top panels are derived from 16S rRNA, and red signals in the middle panels are from hydAB gene transcripts using antisense probe (columns 1 and 3) and soxB gene transcripts using antisense probe (columns 2 and 4). The bottom panels show the synthesized fluorescence of 16S rRNA and each functional gene transcript.
Enzymatic activity analysis in crude gill extract
To estimate the enzymatic basis of H2- and sulfur-oxidizing energy metabolisms in the holobionts of kAlv and eAlv, we determined specific activities of hydrogenase and Sox enzymes in the crude extracts of gill tissues from both kAlv and eAlv individuals. We also attempted to determine the specific activity of Sqr in the gill crude extracts from both populations, but it was difficult to detect the Sqr activity due to the high concentration of various types of proteins in the crude extracts, which interfered with the spectroscopic measurement of the reduced form of quinone.
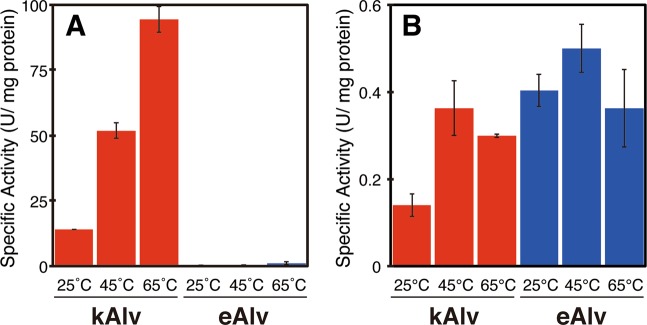
The specific activity of hydrogenase in the kAlv gill crude extract was found to be 14.1, 51.8, and 94.4 U/g at 25 °C, 45 °C, and 65 °C, respectively (Fig. 5a). In contrast, the specific activity of hydrogenase in the eAlv gill crude extract was 0.025, 0.16, and 1.22 U/g at 25 °C, 45 °C and 65 °C, respectively. It was interesting that the hydrogenase activities of A. marisindica endosymbionts were thermophilic despite the average in situ temperatures of their habitats (20–30 °C) (Table 3). The comparison of hydrogenase activities in the gills clearly indicated that the kAlv holobiont had much greater activity (>100 times) than the eAlv holobiont. The difference of hydrogenase activity between the kAlv and eAlv holobionts was consistent with the difference in hydA1B1 gene expression level between them (Figs. 3 and 5a). Although the living kAlv individuals or their dissected gills showed only three to six times higher H2-consumption activity than living eAlv individuals or their dissected gills under the H2-supplemented incubation condition (Table 3), the gill tissues of freshly sampled individuals from the habitats showed drastic differences in expression and activity of the H2-oxidizing enzyme between the kAlv and eAlv holobionts. The differences observed in H2-consuming experiments using individuals or gill may be due to the initial hydrogenase abundance in the endosymbiont shown in this enzyme assay using crude extract.
Fig. 5. Specific activities of hydrogenase and Sox enzymes in crude extracts of gills.
The activities of both enzymes were measured at 25 °C, 45 °C, and 65 °C. Red and blue bars indicate the specific activities of hydrogenase (a) and Sox enzymes (b) in crude extracts of gills from kAlv and eAlv, respectively.
On the other hand, the specific activities of Sox enzymes in the kAlv gill crude extracts were 0.140, 0.362, and 0.299 U/g at 25 °C, 45 °C, and 65 °C, respectively, while those in the eAlv gill crude extract were found to be 0.404, 0.500, and 0.362 U/g at 25 °C, 45 °C, and 65 °C, respectively (Fig. 5b). The Sox enzymes from both the kAlv and the eAlv gills showed the highest specific activity at 45 °C, despite the highest activity of hydrogenase observed at 65 °C. This result may represent the substantial enzymatic property of Sox enzymes of endosymbionts, while it is probably due to an analytical artifact because the cytochrome c used in the Sox enzyme activity assay is obtained from bovine serum (Sigma) and thus becomes unstable at 65 °C. The comparison of Sox enzyme activities in the gills indicates that both the kAlv and the eAlv holobionts had similar activities. This result slightly contradicts the comparative results of sulfide-consumption activities of living individuals and expression abundance of soxB gene between the kAlv and eAlv holobionts, in which the sulfide-consumption activity of living individuals and the soxB gene expression were greater in the kAlv holobiont than in the eAlv holobiont (Table 3 and Fig. 3b).
Since the specific activities of hydrogenase and Sox enzymes are usually proportional to the abundance of enzymes in the extract solutions and are substantially coupled with the net energy yield, the difference seen in the enzyme activities in the gills may be derived from the different cellular expression of hydrogenase and Sox enzymes potentially responding to the environmental H2 concentrations in their habitats.
Conclusive remarks and future perspectives
In this study, the chemosynthetic holobionts of two populations of A. marisindica in the CIR deep-sea hydrothermal fields were investigated to justify the hypothesized occurrence of dual energy metabolisms (H2- and sulfur-oxidation). Based on the phylogenetic analysis of representative genes from the endosymbionts and hosts, the two populations (kAlv and eAlv) had approximately identical holobiont systems that consisted of the same endosymbiotic and host species (Fig. 2a, b). The polyphasic physiological and molecular analyses clearly demonstrated that the holobionts of the two populations had metabolic potentials of both H2- and sulfur-oxidation (Fig. 3 and Table 3) but showed different configurations and operations of the dual energy metabolisms. The results strongly suggest that the kAlv holobiont adopts H2 oxidation as its primary energy metabolism as much as, or possibly more than, sulfur oxidation (Figs. 3c and 5a), while the eAlv holobiont was, for the most part, dependent on sulfur oxidation for chemosynthetic production (Figs. 3c and 5a). In the previous studies of Alviniconcha spp. in the ESLC [14, 16], it was suggested that the energy metabolisms of holobionts were controlled by the niche separation of different genetic and metabolic holobiont types (different couples of host and symbiont species) responding to the physical and environmental H2 conditions of the habitats. However, in the CIR A. marisindica, it is shown that the energy metabolisms of holobionts are regulated by the expression and function of dual energy metabolic genes and enzymes in the same holobiont type (the same couple of host and symbiont species). However, due to the limited opportunities for, and numbers of, dive observations, onboard experiments, and samples, statistical verification of different expression and function patterns in dual energy metabolisms was not fully conducted. In addition, as shown in the previous study of mussels in the MAR deep-sea hydrothermal fields [13], the carbon fixation and transport of holobionts associated with energy metabolisms should be clarified. These experiments will be our foci in future studies.
What would drive different configurations and operations of dual energy metabolisms in the taxonomically near-identical holobiont type of A. marisindica? At present, it seems very likely that the in situ H2 concentration of the habitat serves as one of the key factors. The kAlv holobiont dwells in habitats where the H2 concentration is around 100-fold higher than that of the eAlv holobiont (Table 1). If the environmental abundance of H2 can induce the relatively rapid change of configuration and operation of dual energy metabolisms, the hypothetical habitat exchange experiment between the kAlv and eAlv holobionts would result in the transformation of hydrogeno- and thiotrophic eAlv and thiotrophic kAlv holobionts from the native phenotypes. If other substantial factor(s) other than H2 (e.g., strict genotype selection and acquisition of endosymbiont at the initial holobiont formation and intergenerations of host-symbiont genetic interaction responding to the environmental conditions) can control the configuration and operation of dual energy metabolisms, the habitat exchange experiment would not affect the expression of new phenotypes. Not only habitat exchange experiments but also onboard rearing experiments under different H2 conditions will provide important insights into understanding the energy and carbon metabolisms of chemosynthetic symbioses in deep-sea invertebrates. For onboard rearing experiments in the future, the effect of in situ hydrostatic pressure may be considered, although it was revealed that the in situ hydrostatic pressure did not affect the specific activities of energy metabolisms in the case of the symbiotic microbial community of Shinkaia crosnieri (squat lobster) [39].
The H2-depdendent chemosynthetic symbiosis of deep-sea invertebrates was first demonstrated in a deep-sea mussel, B. puteoserpentis, living in the Logatchev hydrothermal field of MAR by Petersen et al. [13]. Based on the FISH and immunohistochemistry analyses and the radio-isotope-labeled bicarbonate tracer experiments, Petersen et al. [13] pointed out that hydrogenotrophy may serve as primary energy metabolism competitively with thiotrophy in the chemosynthetic holobiont. In addition, it has been reported that some chemosynthetic holobionts may use H2 as an energy source because the genes encoding hydrogenase, and even the expression of the genes, were found in the symbiont genomes and transcriptomes [14, 16–20]. However, it has been also shown that these holobionts have molecular and physiological potentials of other energy metabolisms (sulfur and/or methane oxidation). Actually, Mitchell et al., demonstrated that the endosymbionts of tubeworm, Riftia pachyptila, which possessed genes for hydrogenases did not utilized hydrogen but utilized sulfide as a major electron donor [60]. Thus, the physiological demand and metabolic response to the target energy source in the chemosynthetic holobiont should be addressed at multiple levels, including the environmental conditions of habitats, the transcription level of related genes, the expression level of related enzymes and the physiological level of individuals. In this context, we provide multiple lines of evidence that a deep-sea gastropod, A. marisindica, and its endosymbiont in the Kairei field utilize H2 as the primary energy source as well as H2S (sulfide).
Supplementary information
Acknowledgements
We are very grateful to the crew of R/V Yokosuka and the operation team members of HOV Shinkai6500 for their great support in our research. We benefited greatly from the cooperation of on-board scientists that joined in related cruises. We would also like to thank Mr Masaki Saito (JAMSTEC) for his technical support. Part of this study was supported by a Grant-in-Aid for Scientific Research (no. 23780098) from the Japan Society for the Promotion of Science.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-0605-7) contains supplementary material, which is available to authorized users.
References
- 1.Lonsdale PA. A deep-sea hydrothermal site on a strike-slip fault. Nature. 1979;281:531–34. [Google Scholar]
- 2.Jones ML. Riftia pachyptila Jones: observations on the vestimentiferan worm from the Galapagos Rift. Science. 1981;213:333–36. doi: 10.1126/science.213.4505.333. [DOI] [PubMed] [Google Scholar]
- 3.Felbeck H. Chemoautotrophic potential of the hydrothermal vent tube worm, Riftia pachyptila Jones (Vestimentifera) Science. 1981;213:336–38. doi: 10.1126/science.213.4505.336. [DOI] [PubMed] [Google Scholar]
- 4.Rau GH. Hydrothermal vent clam and tube worm 13C/12C: further evidence of nonphotosynthetic food sources. Science. 1981;213:338–40. doi: 10.1126/science.213.4505.338. [DOI] [PubMed] [Google Scholar]
- 5.Cavanaugh CL, Gardiner SL, Jones ML, Jannasch HW, Waterbury JB. Prokaryotic cells in the hydrothermal vent tube worm Riftia pachyptila Jones: possible chemoautotrophic symbionts. Science. 1981;213:340–42. doi: 10.1126/science.213.4505.340. [DOI] [PubMed] [Google Scholar]
- 6.Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol. 2008;6:725–40. doi: 10.1038/nrmicro1992. [DOI] [PubMed] [Google Scholar]
- 7.Campbell BJ, Jeanthon C, Kostka JE, Luther GW, III, Cary SC. Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl Environ Microbiol. 2001;67:4566–72. doi: 10.1128/AEM.67.10.4566-4572.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alain K, Querellou J, Lesongeur F, Pignet P, Crassous P, Raguenes G, et al. Cambon-Bonavita MA. Caminibacter hydrogeniphilus gen. nov., sp. nov., a novel thermophilic, hydrogen-oxidizing bacterium isolated from an East Pacific Rise hydrothermal vent. Int J Syst Evol Microbiol. 2002;52:1317–23. doi: 10.1099/00207713-52-4-1317. [DOI] [PubMed] [Google Scholar]
- 9.Miroshnichenko ML, Kostrikina NA, L’Haridon S, Jeanthon C, Hippe H, Stackebrandt E, et al. Nautilia lithotrophica gen. nov., sp. nov., a thermophilic sulfur-reducing ε-proteobacterium isolated from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2002;52:1299–304. doi: 10.1099/00207713-52-4-1299. [DOI] [PubMed] [Google Scholar]
- 10.Takai K, Campbell BJ, Cary SC, Suzuki M, Oida H, Nunoura T, et al. Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl Environ Microbiol. 2005;71:7310–20. doi: 10.1128/AEM.71.11.7310-7320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takai K, Nakamura K. Compositional, physiological and metabolic variability in microbial communities associated with geochemically diverse, deep-sea hydrothermal vent fluids. In: Barton L, Mendl M, Loy A, editors. Geomicrobiology: molecular & environmental perspective. NY: Springer; 2010. p. 251–83.
- 12.Nakamura K, Takai K. Theoretical constraints of physical and chemical properties of hydrothermal fluids on variations in chemolithotrophic microbial communities in seafloor hydrothermal. Prog Earth Planet Sci. 2014 doi: 10.1186/2197-4284-1-5. [DOI] [Google Scholar]
- 13.Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, et al. Hydrogen is an energy source for hydrothermal vent symbioses. Nature. 2011;476:176–80. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- 14.Beinart RA, Sanders JG, Faure B, Sylva SP, Lee RW, Becker EL, et al. Evidence for the role of endosymbionts in regional-scale habitat partitioning by hydrothermal vent symbioses. Proc Natl Acad Sci USA. 2012;109:E3241–50. doi: 10.1073/pnas.1202690109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, et al. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.) Front Microbiol. 2017;8:4962–19. doi: 10.3389/fmicb.2017.00682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders JG, Beinart RA, Stewart FJ, Delong EF, Girguis PR. Metatranscriptomics reveal differences in in situ energy and nitrogen metabolism among hydrothermal vent snail symbionts. ISME J. 2013;7:1556–67. doi: 10.1038/ismej.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikuta T, Takaki Y, Nagai Y, Shimamura S, Tsuda M, Kawagucci S, et al. Heterogeneous composition of key metabolic gene clusters in a vent mussel symbiont population. ISME J. 2016;10:990–1001. doi: 10.1038/ismej.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markert S, Arndt C, Felbeck H, Becher D, Sievert SM, Hügler M, et al. Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila. Science. 2007;315:247–50. doi: 10.1126/science.1132913. [DOI] [PubMed] [Google Scholar]
- 19.Nakagawa S, Shimamura S, Takaki Y, Suzuki Y, Murakami S, Watanabe T, et al. Allying with armored snails: the complete genome of gammaproteobacterial endosymbiont. ISME J. 2014;8:40–51. doi: 10.1038/ismej.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Liles MR, Halanych KM. Endosymbiont genomes yield clues of tubeworm success. ISME J. 2018;12:2785–95. doi: 10.1038/s41396-018-0220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warén A, Bouchet P. Gastropoda and monoplacophora from hydrothermal vents and seeps; new taxa and records. Veliger. 2001;44:116–231. [Google Scholar]
- 22.Suzuki Y, Kojima S, Sasaki T, Suzuki M, Utsumi T, Watanabe H, et al. Host-symbiont relationships in hydrothermal vent gastropods of the genus Alviniconcha from the Southwest Pacific. Appl Environ Microbiol. 2006;72:1388–93. doi: 10.1128/AEM.72.2.1388-1393.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson SB, Warén A, Tunnicliffe V, Dover CV, Wheat CG, Schultz TF, et al. Molecular taxonomy and naming of five cryptic species of Alviniconcha snails (Gastropoda: Abyssochrysoidea) from hydrothermal vents. Syst Biodivers. 2014;13:278–95. [Google Scholar]
- 24.Stein JL, Cary SC, Hessler RR, Ohta S, Vetter RD, Childress JJ, et al. Chemoautotrophic symbiosis in a hydrothermal vent gastropod. Biol Bull. 1988;174:373–8. [Google Scholar]
- 25.Suzuki Y, Sasaki T, Suzuki M, Tsuchida S, Nealson KH, Horikoshi K. Molecular phylogenetic and isotopic evidence of two lineages of chemoautotrophic endosymbionts distinct at the subdivision level harbored in one host-animal type: the genus Alviniconcha (Gastropoda: Provannidae) FEMS Microbiol Ecol. 2005;249:105–12. doi: 10.1016/j.femsle.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki Y, Sasaki T, Suzuki M, Nogi Y, Miwa T, Takai K, et al. Novel chemoautotrophic endosymbiosis between a member of the Epsilonproteobacteria and the hydrothermal-vent gastropod Alviniconcha marisindica (Gastropoda: Provannidae) from the Indian Ocean. Appl Environ Microbiol. 2005;71:5440–50. doi: 10.1128/AEM.71.9.5440-5450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashimoto J, Ohta S, Gamo T, Chiba H, Yamaguchi T, Tsuchida S, et al. First hydrothermal vent communities from the Indian Ocean discovered. Zool Sci. 2001;18:717–21. [Google Scholar]
- 28.Van Dover CL, Humphris SE, Fornari D, Cavanaugh CM, Collier R, Goffredi SK, et al. Biogeography and ecological setting of Indian Ocean hydrothermal vents. Science. 2001;294:818–23. doi: 10.1126/science.1064574. [DOI] [PubMed] [Google Scholar]
- 29.Gallant RM, Von Damm KL. Geochemical controls on hydrothermal fluids from the Kairei and Edmond vent fields, 23°-25°S, Central Indian Ridge. Geochem Geophys Geosyst. 2006;7:Q06018. [Google Scholar]
- 30.Kumagai H, Nakamura K, Toki T, Morishita T, Okino K, Ishibashi J, et al. Geological background of the Kairei and Edmond hydrothermal fields along the Central Indian Ridge: implications of their vent fluids’ distinct chemistry. Geofluid. 2008;8:239–51. [Google Scholar]
- 31.Nakamura K, Morishita T, Bach W, Klein F, Hara K, Okino K, et al. Serpentinized troctolites exposed near the Kairei hydrothermal field, Central Indian Ridge: insights into the origin of the Kairei hydrothermal fluid supporting a unique microbial ecosystem. Earth Planet Sci Lett. 2009;193:371–9. [Google Scholar]
- 32.Miyazaki J, Makabe A, Matsui Y, Ebina N, Tsutsumi S, Ishibashi J, et al. WHATS-3: an improved flow-through multi-bottle fluid sampler for deep-sea geofluid research. Front Earth Sci. 2017;5:202–13. [Google Scholar]
- 33.Takai K, Gamo T, Tsunogai U, Nakayama N, Hirayama H, Nealson KH, et al. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles. 2004;8:269–82. doi: 10.1007/s00792-004-0386-3. [DOI] [PubMed] [Google Scholar]
- 34.Nakagawa S, Takaki Y, Shimamura S, Reysenbach AL, Takai K, Horikoshi K. Deep-sea vent ε-proteobacterial genomes provide insights into emergence of pathogens. Proc Natl Acad Sci USA. 2007;104:12146–50. doi: 10.1073/pnas.0700687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamamoto M, Nakagawa S, Shimamura S, Takai K, Horikoshi K. Molecular characterization of inorganic sulfur-compound metabolism in the deep-sea epsilonproteobacterium Sulfurovum sp. NBC37-1. Environ Microbiol. 2010;12:1144–53. doi: 10.1111/j.1462-2920.2010.02155.x. [DOI] [PubMed] [Google Scholar]
- 36.Kawagucci S, Miyazaki J, Noguchi T, Okamura K, Shibuya T, Watsuji T, et al. Fluid chemistry in the Solitaire and Dodo hydrothermal fields of the Central Indian Ridge. Geofluids. 2016;16:988–1005. [Google Scholar]
- 37.Fogo JK, Chemistry MPA. Spectrophotometric determination of hydrogen sulfide. Anal Chem. 1949;21:732–4. [Google Scholar]
- 38.Cline J. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–8. [Google Scholar]
- 39.Watsuji T, Yamamoto A, Takaki Y, Ueda K, Kawagucci S, Takai K. Diversity and methane oxidation of active epibiotic methanotrophs on live Shinkaia crosnieri. ISME J. 2014;8:1020–31. doi: 10.1038/ismej.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watsuji T, Motoki K, Hada E, Nagai Y, Takaki Y, Yamamoto A, et al. Compositional and functional shifts in the epibiotic bacterial community of Shinkaia crosnieri Baba & Williams (a squat lobster from hydrothermal vents) during methane-fed rearing. Microbes Environ. 2018;33:348–56. doi: 10.1264/jsme2.ME18072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–9. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pham VH, Yong JJ, Park SJ, Yoon DN, Chung WH, Rhee SK. Molecular analysis of the diversity of the sulfide:quinone reductase (sqr) gene in sediment environments. Environ Microbiol. 2008;154:3112–21. doi: 10.1099/mic.0.2008/018580-0. [DOI] [PubMed] [Google Scholar]
- 43.Sanger F, Nicklen EF, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–7. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyazaki J, Higa R, Toki T, Ashi J, Tsunogai U, Nunoura T, et al. Molecular characterization of potential nitrogen fixation by anaerobic methane-oxidizing archaea in the methane seep sediments at the number 8 Kumano Knoll in the Kumano Basin, offshore of Japan. Appl Environ Microbiol. 2009;75:7153–62. doi: 10.1128/AEM.01184-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hongo Y, Ikuta T, Takaki Y, Shimamura S, Shigenobu S, Maruyama T, et al. Expression of genes involved in the uptake of inorganic carbon in the gill of a deep-sea vesicomyid clam harboring intracellular thioautotrophic bacteria. Gene. 2016;585:228–40. doi: 10.1016/j.gene.2016.03.033. [DOI] [PubMed] [Google Scholar]
- 48.Takishita K, Takaki Y, Chikaraishi Y, Ikuta T, Ozawa G, Yoshida T, et al. Genomic evidence that methanotrophic endosymbionts likely provide deep-sea Bathymodiolus mussels with a sterol intermediate in cholesterol biosynthesis. Genome Biol Evol. 2017;9:1148–60. doi: 10.1093/gbe/evx082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wakai S, Kikumoto M, Kanao T, Kamimura K. Involvement of sulfide:quinone oxidoreductase in sulfur oxidation of an acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans NASF-1. Biosci Biotechnol Biochem. 2004;68:2519–28. doi: 10.1271/bbb.68.2519. [DOI] [PubMed] [Google Scholar]
- 50.Beedessee G, Watanabe H, Ogura T, Nemoto S, Yahagi T, Nakagawa S, et al. High connectivity of animal populations in deep-sea hydrothermal vent fields in the Central Indian Ridge relevant to its geological setting. PLoS One. 2013;8:e81570. doi: 10.1371/journal.pone.0081570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warén A, Bengtson S, Goffredi SK, Van Dover CL. A hot-vent gastropod with iron sulfide dermal sclerites. Science. 2003;302:1007–7. doi: 10.1126/science.1087696. [DOI] [PubMed] [Google Scholar]
- 52.Chen C, Linse K, Copley JT, Rogers AD. The “scaly-foot gastropod”: a new genus and species of hydrothermal vent-endemic gastropod (Neomphalina: Peltospiridae) from the Indian Ocean. J Molluscan Stud. 2015;81:322–34.
- 53.Takai K, Nunoura T, Ishibashi J, Lupton J, Suzuki R, Hamasaki H, et al. Variability in the microbial communities and hydrothermal fluid chemistry at the newly-discovered Mariner hydrothermal field, southern Lau Basin. J Geophys Res. 2008;113:G02031. [Google Scholar]
- 54.Yamamoto M, Takai K. Sulfur metabolisms in Epsilon- and Gamma-proteobacteria in deep-Sea hydrothermal fields. Front Microbiol. 2011;2. 10.3389/fmicb.2011.00192. [DOI] [PMC free article] [PubMed]
- 55.Cusick KD, Fitzgerald LA, Cockrell AL, Biffinger JC. Selection and evaluation of reference genes for reverse transcription-quantitative PCR expression studies in a thermophilic bacterium grown under different culture conditions. PLoS One. 2015;10:e0131015. doi: 10.1371/journal.pone.0131015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao W, Li Y, Gao P, Sun Z, Sun T, Zhang H. Validation of reference genes for real-time quantitative PCR studies in gene expression levels of Lactobacillus casei Zhang. J Ind Microbiol Biotechnol. 2010;38:1279–86. [DOI] [PubMed]
- 57.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beinart RA, Nyholm SV, Dubilier N, Girguis PR. Intracellular oceanospirillales inhabit the gills of the hydrothermal vent snail Alviniconcha with chemosynthetic, γ-proteobacterial symbionts. Environ Microbiol Rep. 2014;6:656–64. doi: 10.1111/1758-2229.12183. [DOI] [PubMed] [Google Scholar]
- 60.Mitchell JH, Leonard JM, Delaney J, Girguis PR, Scott KM. Hydrogen does not appear to be a major electron donor for symbiosis with the deep-sea hydrothermal vent tubeworm Riftia pachyptila. Appl Environ Microbiol. 2019;86:e01522–19. doi: 10.1128/AEM.01552-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.