Abstract
Cell differentiation and proliferation require Hedgehog (HH) signaling, and aberrant HH signaling causes birth defects or cancers. In this signaling pathway, the N-terminally palmitoylated and C-terminally cholesterylated HH ligand is secreted into the extracellular space with help of Dispatched-1 (DISP1) and Scube2 proteins. Patched-1 (PTCH1) protein releases its inhibition of the oncoprotein Smoothened (SMO) after binding the HH ligand, triggering downstream signaling events. In this review, we will discuss the recent structural and biochemical studies on four major components in HH pathway: HH ligand, DISP1, PTCH1, and SMO. This research provides mechanistic insights into how HH signaling is generated and transduced from the cell surface into the intercellular space and will aid in facilitating treatment of HH related diseases.
Keywords: Hedgehog, Patched, Smoothened, Dispatched, Sterol, Signal transduction
Physiology of the HH signaling pathway
The Hedgehog (HH) signaling pathway plays a vital role in developmental patterning of animal embryogenesis and regeneration of adult tissues [1–4], and the HH ligand functions in a paracrine signaling (see Glossary) manner to regulate the HH signaling pathway [5]. Importantly, abnormal activation of HH signaling is implicated in many cancers, including basal cell carcinoma (BCC) and medulloblastoma (MB) [6]. Because of the complexity of the HH signaling pathway, the molecular mechanism of this pathway remains largely unknown and controversial. Therefore, studies of the proteins involved in the HH signaling pathway will reveal the molecular mechanism of animal development and provide insight into cancer treatment and regenerative medicine. Here, we provide a brief overview of the HH signaling pathway and delve into deeper discussions about specific steps in the pathway, including HH ligand secretion and reception, Patched-1 (PTCH1) inhibition, and Smoothened (SMO) activation, primarily focusing on structural studies of proteins in the pathway.
Overview of the HH signaling pathway
Although the HH signaling pathway is conserved in metazoan cells, the HH family contains different numbers of homologues owing to evolutionary divergence [7]. For example, while in fruit fly there is only one HH ligand, there are three HH members in mammals: Sonic Hedgehog (SHH), Desert Hedgehog (DHH), and Indian Hedgehog (IHH). All three mammalian HH ligands share high sequence homology and function as initial ligands that trigger HH signaling.
In the first step of the signaling pathway, the HH precursor undergoes autocatalytic cholesterol-dependent processing in the endoplasmic reticulum (ER) to release the HH N-terminal signaling domain (HH-N) that is covalently coupled to cholesterol at the carboxyl terminus [8, 9] (Figure 1A). Next, a membrane-bound O-acyltransferase, named Skinny HH in flies [10, 11], or HH acyltransferase (HHAT) in vertebrates [12], transfers the thiol group of palmitoyl-CoA to the N-terminal cysteine of HH-N. In the final step of generating extracellular mature HH, on the cell surface, a membrane protein called Dispatched-1 (DISP1) facilitates the release of the doubly lipidated HH-N (referred to as “native HH-N” in this review) into the extracellular space [13].
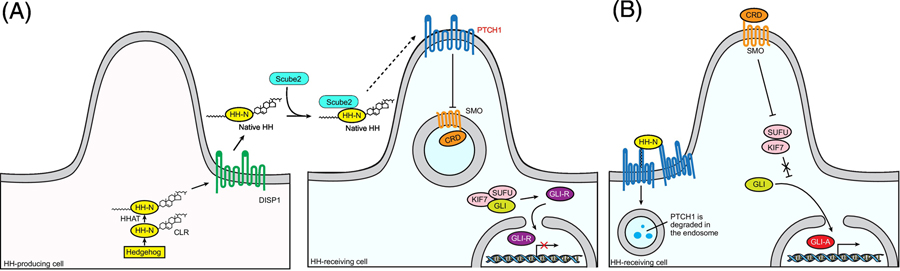
Figure 1. Key proteins and important processes in vertebrate HH signaling pathway.
(A) The production and trafficking of HH protein. The precursor of HH is auto-cleaved and HH-N is covalently modified by a cholesterol at the C-terminus; then HH-N is further palmitoylated by HHAT on ER membrane. After these processes, DISP1, a membrane protein located on the cell surface, facilitates the release of HH-N into the extracellular space from the producing cells. Scube2, an extracellular protein assists in the trafficking of HH-N in the extracellular space. In the HH receiving cells, before binding HH-N, the receptor PTCH1 localizes to the cilia and suppress the downstream protein SMO, keeping the GLI transcription factors in an inactive form (GLI-R). (B) The HH signal transduction from PTCH1 to SMO. The HH binding to PTCH1 relieves the inhibition of PTCH1 to SMO, causing SMO to relocate to the cilia. The activated SMO can promote the maturation of GLI to its active form (GLI-A). GLI-A enters the nuclei to turn on the HH signal.
In the extracellular space, Scube proteins serve as chaperones to bind the native HH ligand and assist in its trafficking from the producing cells to the receiving cells [14, 15] (Figure 1A). This trafficking is also regulated by several membrane anchored proteins, such as negatively charged heparan sulfate proteoglycans (HSPGs) [16] and HH co-receptors like CAM-related/down-regulated by oncogenes (CDO), Brother of CDO (BOC), Growth arrest-specific 1 (Gas1) and Hedgehog interacting protein (HHIP) [17–20]. Particularly, CDO, BOC, and Gas1 have been shown to facilitate HH binding to PTCH1, a plasma membrane protein that localizes to the primary cilia of vertebrates and the cell surface in Drosophila in the absence of HH [21] to suppress HH signaling [22, 23].
There are two ptch genes (ptch1 and ptch2) in humans [24]; PTCH1 consists of 1447 residues and includes twelve transmembrane helices (TMs), three ~30kD soluble domains, two extracellular domains (ECD), and one C-terminal domain (CTD)) [25, 26], and acts as the primary receptor for HH ligands, recognized by the ECDs [27, 28]. PTCH2 shares a conserved TM domain and two ECDs with PTCH1, but lacks the CTD [29, 30], and there is a possibility that PTCH2 binds HH proteins in different cellular environments to regulate HH signaling [31].
After binding HH, PTCH1 is inhibited and forms oligomers, which are further moved out of the cilia and degraded in the endosome [32]. It also releases its inhibition of SMO, a Frizzled-Class (Class-F) G protein–coupled receptor (GPCR), which relocates to the cilia (Figure 1B). This activates the Glioma-associated oncogene (GLI) transcription factors to up-regulate target genes [4, 33].
Notably, TMs 2–6 of PTCH1 and DISP1 form sterol-sensing domains (SSDs) that share sequence homology with other membrane proteins in cholesterol metabolism and signaling, including Niemann-Pick disease, type C1 (NPC1) protein, Niemann-Pick disease, type C1-like 1 (NPC1L1) protein, HMG-CoA reductase, and SREBP cleavage-activating protein [25, 34, 35]. The TM portions of NPC1, PTCH1, and DISP1, including the SSD, are topologically similar to prokaryotic Resistance-nodulation-division (RND) multidrug transporters [36, 37]. Mutations in the SSDs have been shown to interfere with the functions of these proteins. For example, a D584N mutation in the Drosophila PTCH-SSD abolishes HH signaling without interrupting HH binding [38].
More importantly, PTCH1 and SMO are associated with human diseases. PTCH1 is a tumor suppressor associated with BCC, MB, primitive neuroectodermal tumors [39], and holoprosencephaly-7, a structural anomaly of the brain [40]. SMO is an oncoprotein and the target of many anti-cancer drugs [41, 42]. Notably, a SMO inhibitor named Vismodegib has been approved for the treatment of BCC [43].
The release and trafficking of HH ligand from the producing cells to the receiving cells
There are three disp genes and three scube genes in humans, and the DISP1 and Scube2 proteins are well studied among their homologues. DISP1 contains 1524 amino acids including 12 TMs, two ECDs, and N-terminal and C-terminal cytosolic flexible regions (Figure 2, top). Previous work showed that a proprotein convertase named Furin can cleave the ECD-I of DISP1 triggering HH-N release, that this cleavage is required for the activation of DISP1 in vivo, and that mutations of the cleavage site (R279A/E280A) prevent the release of HH-N [44]. Interestingly, a variant of DISP1 named “NNN” [45] that includes three Asp to Asn mutations in the TM domain can bind HH-N with a higher affinity than wild type; however, this variant cannot release HH-N [15]. This finding implies that the TM domains of DISP1 may affect the binding of HH-N allosterically, or that the modifications of HH-N may facilitate its release by binding the TM domains of DISP1.
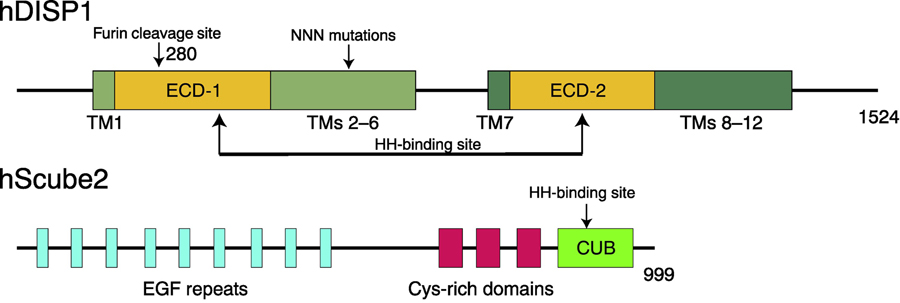
Figure 2. Secondary Structures of human DISP1 (hDISP1) and human Scube2 (hScube2).
DISP1 shares a similar structure with PTCH1, consisting of 12 TMs, two ECDs, and N-terminal and C-terminal cytosolic flexible regions. The ECDs are involved in HH-N binding (indicated by arrows). A proprotein convertase named Furin can cleave the ECD-I (cleavage site is labeled) of DISP1 triggering HH-N release. TMs 2–6 form an SSD and mutations in the SSD affect the release of HH-N. Scube2 protein works in the extracellular space and consists of 9 EGF-like repeats, three CRDs, and one CUB domain, which is essential for HH-N binding.
Recently, Cannac et al reported cryo-electron microscopy (EM) structures of Drosophila melanogaster DISP at 3.2 Å resolution and its complex with unmodified HH ligand at 4.7 Å resolution [46]. The TM domains of DISP show a similar arrangement to the RND transporters, PTCH1, and NPC1. However, unlike NPC1, the ECDs of DISP spread apart, revealing an open conformation and creating a large bowl-shaped cavity that was shown to accommodate the HH ligand. Notably, a loop of ECD-I between residues E90 and S247 is not resolved in the structures. This unstructured loop may serve as a molecular gate for receiving and releasing the HH ligand. This observation is consistent with the finding that a loop of DISP1 ECD-I has to be cleaved for its maturation [44].
Scube2 consists of nine EGF-like repeats, three Cys-rich domains, and one CUB domain that is required for binding the cholesterol modification of HH-N [14, 15] (Figure 2, bottom); deletion of the CUB domain disrupts this binding and prevents the movement of HH-N in the extracellular space [15]. It is a mystery how the cholesterol modification of HH protein recognizes Scube2, since the structure of Scube2 is still not available. It is also unknown whether Scube2 can complex with DISP1 and HH-N to receive the HH-N from DISP1. Importantly, the cholesterol modification of HH-N is required for the release of HH-N mediated by DISP1 and Scube2 [14, 15] (Figure 1A), but beyond the examples provided, the underlying mechanism remains a mystery. Further structural investigations of DISP1–native HH-N and Scube2–native HH-N complexes will elucidate the necessity of the cholesterol modification in HH-N release and trafficking.
The recognition of HH ligand by PTCH1
PTCH1 and N-terminal palmitoylation of HH-N
N-terminal palmitoylation has been shown to be indispensable for HH signaling in both vertebrates and Drosophila by differentiation assays [47–49] and GLI-dependent HH signaling assays [15, 32], and the embryonic development in both Drosophila and mice can be interfered with by abolishing the palmitoylation of HH-N [10, 11, 49–52]. The indicated assays showed that fatty acylated SHH-N is far more active than unacylated SHH-N. Moreover, inhibitors of HHAT that block the palmitoylation of SHH-N prevent HH signaling [53, 54] (Figure 1A). Particularly, palmitoylated SHH peptide (first 22 amino acids of SHH-N) binds PTCH1 to partially stimulate GLI-dependent HH signaling [32]. Additionally, our previous pull-down assay showed that native SHH-N can bind PTCH1 variant (see below) with a higher affinity than His-tagged unmodified SHH-N. This finding implies that the modifications of SHH-N may be involved in its interaction with PTCH1 [55]. Those studies demonstrate the physiological importance of the palmitate moiety in HH signal transduction.
HH-N interfaces with PTCH1
Additionally, SHH-N has been used for biochemical and cell biology experiments to validate the interaction between PTCH1 or HH co-receptors and SHH-N. Previous structural studies showed that the calcium-mediated interface of HH-N can bind to HH co-receptors, and a specific antibody named 5E1 could block HH signaling [56] (Figure 3A). The biochemical assays also provided evidence that PTCH1 can bind to the calcium-mediated interface of HH-N, while a point mutation, R153E, on the calcium-mediated interface of SHH-N decreases GLI-dependent HH signaling [57]. Particularly, CDO, a HH co-receptor, can bind SHH-N and this binding can be competitively inhibited by the addition of PTCH1 in vitro [58]. These studies provide strong molecular evidence that PTCH1 recognizes the calcium-mediated interface of SHH-N. In contrast, in vivo studies revealed that co-receptors can form a hetero-trimeric complex with HH-N and PTCH1 to promote either embryonic development or cell proliferation [19, 20].
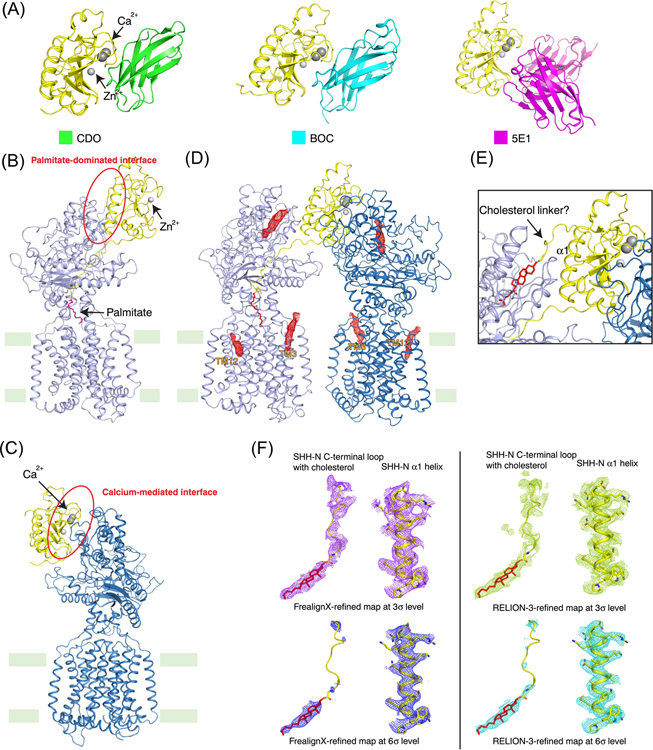
Figure 3. Structures of PTCH1–HH complexes.
(A) Structures of HH with CDO (PDB ID: 3D1M), BOC (PDB ID: 3N1M) and 5E1 antibody (PDB ID: 3MXW). CDO and BOC are HH-N co-receptors that up-regulate HH signaling; 5E1 down-regulates HH signaling. They bind HH-N via the calcium-mediated interface. (B) Structure of PTCH1 with native HH-N (PDB ID: 6OEV). At 1:1 molar ratio, the palmitate moiety of native HH-N inserts in to the ECDs of PTCH1, forming the palmitate-dominated interface. (C) Structure of PTCH1 with His-tagged HH-N (PDB ID: 6DMY). His-tagged HH-N lacks the palmitate modification and binds PTCH1 through the calcium-mediated interface in presence of Ca2+. (D) Structure of 2:1 PTCH-1 and native HH-N complex (PDB ID: 6E1H). PTCH1 and native HH-N were incubated at 2:1 molar ratio with 1 mM Ca2+. In this structure, native HH-N employs both interfaces to bind two PTCH1 molecules. The sterol-like molecules that are observed in the cryo-EM map shown in red mesh. They are located in both TMs and ECDs. (E) The potential insertion of cholesterol modification of HH-N into PTCH1 (PDB ID: 6RVD). The cholesterol modification of native HH-N may insert into the ECD-I of PTCH1, but further structural and functional data are needed to prove this hypothesis. (F) SHH-N C-terminal loop and cholesterol. The cryo-EM maps of the linker region between HH-N and its cholesterol modification that were refined by FrealignX or RELION-3 at different signal levels. Although the overall EM maps are almost the same, densities of the linker region are quite different due to refinements by different software. This suggests the densities at this region may not be suitable for building the structural model. The palmitate is shown in magenta sticks. Cholesterol is shown in red sticks. The calcium and zinc ions were shown in gray balls.
A major barrier to investigating the interaction between HH-N and PTCH1 is that full-length human PTCH1 behaves poorly in detergent solution. However, cell biology and GLI-dependent HH signaling assays showed that deletion of the cytosolic C-terminus and loop between TM6 and TM7 can retain this PTCH1 variant in cilia with similar signal activity as the wild type protein [57], and this construct (named PTCH1* here) is well-folded and homogenous after purification with detergent. The PTCH1* protein sample, which is suitable for structural investigation, has been used by two groups [55, 59], and has been shown to share similar topology with NPC1 protein [55, 59–62]. The cryo-EM structure of PTCH1* in complex with native SHH-N at a 1:1 molar ratio reveals an unexpected interface [55] (Figure 3B). Specifically, the ECD-I of PTCH1 engages the helix 1 of SHH-N (called “palmitate-dominated interface” here) instead of its calcium-mediated interface. Notably, the N-terminus of SHH-N with its palmitate moiety inserts into a cavity between the two ECDs. This is consistent with pull-down assays showing that the modification of SHH-N is involved in complex assembly. Mutagenesis on each interface of PTCH1 and SHH-N validated the physiological importance of the interfaces in cells [55].
Why is this interface different from the one previously predicted? Differences in the experimental set-up may explain these disparate findings. Although His-tagged SHH-N can bind PTCH1 with less affinity than native SHH-N, Ca2+ in the solution or medium can enhance the binding between His-tagged SHH-N and PTCH1 [55]. The further pull-down assay showed that 5E1 can form the complex with native SHH-N and PTCH1; however, this antibody can disrupt the complex of PTCH1 and His-tagged SHH-N at a 1 mM Ca2+ concentration [55]. This finding suggests that the native SHH-N employs its palmitate-dominated interface to bind PTCH1, while SHH-N lacking palmitate deviates from the palmitate-dominated binding mode by interacting with PTCH1 through the calcium-mediated interface. This is consistent with the observations on the structure of PTCH1 complexed with the unmodified SHH-N [61, 62] (Figure 3C).
An additional cryo-EM structure of PTCH1*–SHH-N complex at a 2:1 molar ratio in presence of 1mM Ca2+ demonstrated that one SHH-N molecule can engage both epitopes to concomitantly bind two PTCH1 receptors in an asymmetric arrangement, and that both HH surfaces are necessary for the complex [63] (Figure 3D). Functional assays using PTCH1 or SHH-N mutants that disrupted the individual interfaces demonstrated that simultaneous engagement of both interfaces between SHH-N and PTCH1 are essential for signaling in cells [63].
PTCH1 and cholesterol modification of HH-N
Remarkably, several sterol-like molecules were observed in the cryo-EM map of PTCH1* [55, 59, 61–63] (Figure 3D); this observation is consistent with the prediction that PTCH1 works as a transporter and regulates SMO catalytically by transporting sterols. One sterol-like molecule was in the SSD of PTCH1*, implying the SSD may serve as part of a tunnel for sterol transport (see below), and an in vitro binding assay showed that binding between unmodified HH and PTCH1 may depend on the sterol-like molecule in ECD-I [61].
The cryo-EM structure of a different PTCH1 variant (with a shorter C-terminus but with the loops between TM6 and TM7) in complex with the palmitoylated SHH-N showed the 2:1 complex dimerized to 4:2 stoichiometric ratio complex through an asymmetric paradigm [64]. The intracellular domains of PTCH1 promoted the formation of the 4:2 complex; however, the interaction details of these domains were not resolved owing to the low local resolution. From the previously reported cryo-EM map of native SHH-N and PTCH1*, Qian et al interpreted that the cholesterol modification of SHH-N inserts into the ECD-I of PTCH1 [64]. Later, Rudolf et al reported the crystal structure of PTCH1 ECD with a nanobody in the presence of Cholesteryl Hemisuccinate (CHS). Based on the reassessment of the previously reported 2:1 PTCH1-SHH-N complex, they too claimed that the cholesterol of SHH-N inserts into ECD-I [65] (Figure 3E). They also provided functional evidence that SHH-N with the cholesterol modification can upregulate GLI1 mRNA when compared with non-modified SHH-N or a SHH-N with a five amino acid linker truncation (residues 189–193) between the cholesterol modification and the globular portion of SHH-N [65]. It would be interesting to simultaneously compare GLI1 mRNA levels after treating cells with native SHH-N, palmitoylated SHH-N, cholesterylated SHH-N, and unmodified SHH-N individually to compare the contributions of different modifications of HH-N to the signaling activity in one assay.
Though the cryo-EM map of the 2:1 PTCH1*–SHH-N complex is not clear enough to build the linker between SHH-N and the sterol-like molecule in the ECD-I (Figure 3F), two works increased the signal level of the map in order to trace the linker between the cholesterol modification and globular portion of SHH-N [64, 65]. However, as a result, the noise of the map was amplified as well. That said, it is apparent that the maps of the linker area are not identical when the same cryo-EM data set is processed by different software, though the overall maps are almost the same (Figure 3F). This means the densities that show up in the linker area are not well-defined. This point should be considered, and requires further investigations as to whether cholesterol modification is involved in the interaction between PTCH1 and SHH-N. It is possible that cholesterol can insert into the ECD-I in vitro since hydrophobic cholesterol may not be exposed in solution. The binding between PTCH1 and cholesterol modification of SHH-N is still lacking physiological evidence.
The putative mechanism of palmitoylated HH mediated signaling
PTCH1 catalytically controls the activity of SMO without direct interaction [66]; PTCH1 may transport some small molecules, changing their concentrations in the cell membrane. These small molecules may stimulate or inhibit SMO, further regulating HH signaling. Since PTCH1 shares a very similar topology with the putative LDL-derived cholesterol transporter NPC1, it has been predicted that PTCH1 serves as a sterol transporter to regulate the HH signal [16]. Structural analysis revealed a putative tunnel through PTCH1 extending to the SSD [59, 61, 63]. Based on structural observations, HH-N binds to PTCH1 by its palmitate moiety, disrupting the tunnel of PTCH1 and leading to an accumulation of accessible cholesterol and other sterols, generating a partial HH signal as a result (Figure 3B and 4A). Consistent with that, a palmitoylated SHH-N peptide can induce the HH signal, although this signal is lower than that from palmitoylated SHH-N addition to cells [32]. Introducing mutations on the palmitate-binding site of PTCH1 mimics SHH-N palmitate insertion, abolishing the inhibitory ability of PTCH1 to the signaling pathway, suggesting that the palmitate can abolish the activity of PTCH1 by blocking its tunnel [59, 63]. Recently, the structure of human NPC1 with its inhibitor itraconazole has been reported [67]. It shows that itraconazole binds to the center of NPC1 intermolecular channel, as well as to the palmitate of HH, blocking a similar site in the homologous tunnel of PTCH1 and implying a conserved mechanism for sterol transport in these proteins.
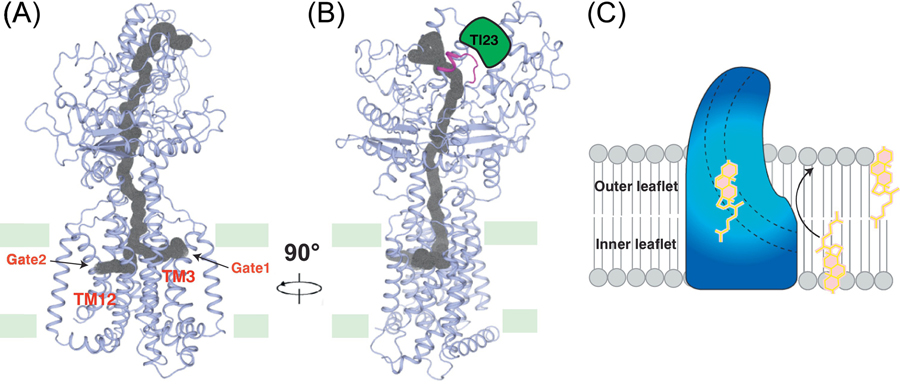
Figure 4. The putative sterol transport tunnel in PTCH1.
(A) and (B) The putative sterol tunnel in PTCH1. A tunnel across PTCH1 was predicted based on protein structure. PTCH1 may transport sterols through this tunnel. Nanobody Tl23 squeezes the tunnel and may inhibit the activity of PTCH1. The tunnel is indicated in gray and the two gates in the transmembrane domain are indicated. Tl23 (green) binding site is colored in magenta. (C) Model of PTCH1-mediated sterol transport. PTCH1 may transport the sterol from the inner leaflet to outer leaflet of the cell membrane.
More specifically, a triad of charged residues in TM4 and TM10 were shown to be essential for PTCH1 transporter activity, probably by conducting Na+ flow [68]. Indeed, a mouse PTCH1 mutant could not suppress HH signaling in the Ptch1−/− cell line [59]. Remarkably, a recent study showed a nanobody named Tl23 can increase the HH signal through binding to PTCH1 without the addition of SHH-N [69]. The cryo-EM structure of PTCH1-Tl23 demonstrates that Tl23 binds ECD-I to squeeze the putative tunnel of PTCH1 preventing sterol transport for triggering the HH signal (Figure 4B).
How does PTCH1 change the distribution of sterol on the cilia membrane? Zhang et al used a set of sensors derived from the cholesterol-binding domain of Perfringolysin O to measure cholesterol concentration in the membrane [59]. The results showed that PTCH1 can transport cholesterol from the inner to the outer leaflet of plasma membrane (Figure 4C). This discovery is consistent with the result from an in vitro assay that demonstrates that SMO activity can be regulated by cholesterol levels of the plasma membrane [68], and structural and functional studies that have shown there is a sterol binding site in SMO-7TMs [70, 71] (also see the discussion below). Interestingly, the transport direction of PTCH1 (from inner leaflet to the lumen) [59] is opposite its structural homolog NPC1, which is responsible for transporting cholesterol from the lumen to the membrane [72].
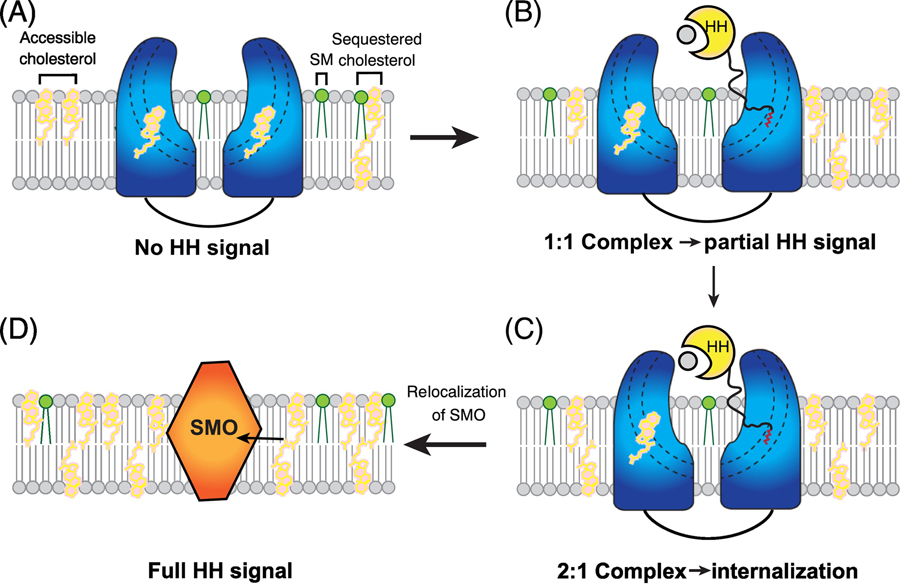
There are three pools of cholesterol on the cell surface: accessible cholesterol (free cholesterol), sphingomyelin (SM)-sequestered cholesterol, and essential cholesterol [73]. The free cholesterol can be used for triggering the HH signal. PTCH1 can transport the sterols preventing their accumulations on the cilia. After the palmitate-dominated interface binding to PTCH1, the calcium-mediated interface of the HH ligand can be recognized by the other PTCH1 molecule, and this interaction generates the 2:1 complex, triggering an endocytosis of this complex [32] to create a PTCH1-free cilia environment for a future accumulation of the accessible cholesterol and/or other sterols and allowing the SMO to relocate to the cilia [63] (Figure 5). Notably, depletion of sphingomyelin by myriocin might increase accessible cholesterol thereby enhancing GLI1 mRNA level in cells [74].
Figure 5. Model of HH signaling generation.
In the absence of HH protein, PTCH1 locates on the cilia membrane and transports sterols out of the membrane to keep a low sterol concentration in cilia. After the palmitate-dominated interface of HH-N binds to PTCH1, the bound PTCH1 is inhibited. Then, the calcium-mediated interface of the HH-N can bind to the other PTCH1 molecule. These interactions generate the 2:1 complex, triggering an endocytosis of this complex to create a PTCH1-free cilia environment for the future accumulation of free cholesterol and other sterols. Then, SMO relocates to the cilia membrane to obtain the sterols for activating the HH signaling. Cholesterol and sphingomyelin (SM) are shown in pink and green sticks. PTCH1, HH and SMO are colored in blue, yellow and orange, respectively.
Signal transduction from the cell surface into intercellular space
The mechanism of how the HH protein triggers GLI activation via the PTCH1–SMO system is still unclear. As a member of the Class-F GPCR family, SMO includes 7-TMs and a Cysteine-Rich Domain (CRD) at the amino terminus [75] (Figure 1). Previous studies showed cholesterol, or its derivatives such as 20(S)-hydroxycholesterol, can bind the CRD of SMO on the cell surface to directly stimulate the HH signal [76–80]. Importantly, the CRD of SMO has been shown to be covalently modified by cholesterol at residue D95 [81]. The 7-TMs of SMO contain a ligand-binding pocket that can bind agonistic or antagonistic ligands [41, 42, 82, 83] (Figure 5A). Moreover, glycosylated SMO has been verified to induce a non-canonical signal through Gαi, suggesting post-translational modification of SMO can impact the HH signal transduction [84].
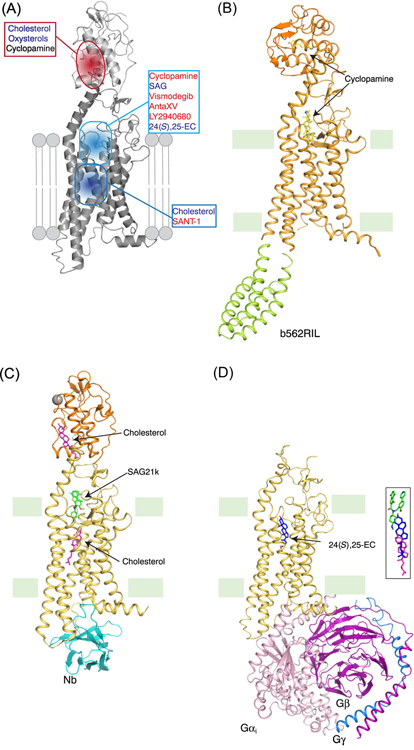
Several previous studies showed that cholesterol can serve as an endogenous ligand to activate SMO [78, 79]. The crystal structures of cholesterol-bound Xenopus laevis SMO (xSMO) and cyclopamine-bound xSMO, with a C-terminal deletion and a replacement of intracellular loop 3 (ICL3) with thermostabilized apocytochrome b562RIL, showed that SMO adopts an active conformation with its CRD rotated considerably compared to other published crystal structures of SMO [82] (Figure 5B). These two structures reveal that cyclopamine binds in both the TM domain and the CRD, while cholesterol is only observed in the CRD. A recently published crystal structure of mouse SMO (mSMO) bound to the agonist SAG21k and an intracellular binding nanobody revealed a sterol-binding site under the SAG21k-binding site in the 7-TMs, suggesting that the inactivation of PTCH1 by HH allows a sterol to access the 7-TMs of SMO from the inner leaflet of the cell membrane [71] (Figure 5C). Another work presents a potential cholesterol binding site between TM2 and TM3 in the outer leaflet side of the cell surface [85]. This site may serve as a gate for cholesterol entry to the 7-TMs of SMO. Additionally, the CRD of active mouse SMO adopts a similar conformation as that of inactive human SMO (hSMO) but not xSMO (Figure 5B) [71]. Since the structure of active mSMO was obtained with synthetic agonist SAG21k, it remains unknown whether the conformation of mSMO with a 7-TMs bound sterol can retain an active conformation.
SMO can activate Gi-family protein, which inhibits cAMP production, thereby activating GLI [86–88]. Pertussis toxin, which prevents the interaction between Gαi and active GPCRs, weakens the activation of GLI [87]. Another study shows that 24,25-epoxycholesterol (24,25-EC), which was identified from the purified PTCH1 by mass spectrometry, can directly stimulate HH signaling in cells and trigger SMO to bind heterotrimeric Gi protein in vitro [70]. This finding is consistent with a recent discovery that 24,25-EC is the most enriched oxysterol in sea urchin embryo cilia, and 24(S),25-EC can activate HH signaling in cells [89]. The cryo-EM structure of hSMO also revealed an activation mechanism of SMO and a putative sterol-binding site in the 7-TMs, which is consistent with the observation from the crystal structure of mSMO [71] (Figure 5D). The CRD in the structure was not determined, implying that the CRD might be flexible or adopt multiple conformations in the active state to release its repression of the 7-TMs. Remarkably, the Gi protein in the complex exhibits a different arrangement from that of Class-A GPCR–Gi complex. This work provides structural evidence that SMO is a functional GPCR protein.
Previous studies by different groups also showed that the inhibition of oxysterol or cholesterol synthase can block HH signaling in cells [89–91] and that signaling can be rescued by different oxysterols. These discoveries reveal that either cholesterol or distinctive oxysterols can be associated with SMO as an endogenous ligand to regulate the signal. 24(S),25-EC serves as a sterol representative to reveal how sterol can trigger SMO activation without other agonists. Additionally, cell signaling assays were performed by adding cholesterol into the medium and measuring GLI activity after 24–48 hours. It is also possible that the cholesterol could be oxidized to convert to an oxysterol for signal activation, even if F7-cholesterol (a fluorinated sterol that prevents oxidation on the 25C, 26C and 27 C-position) and 25F-cholesterol (a fluorinated sterol that prevents oxidation on the 25C position) can trigger the HH signal directly [78].
Although the structures of SMO with different ligands have been reported, further investigations are needed to determine whether the CRD or 7-TMs of SMO serve as an initial binding site for the endogenous sterol ligand, how the CRD represses the activation of SMO, and how this repression can be released owing to the ligand binding. Furthermore, how does the endogenous ligand access the 7-TMs? Through the membrane or by delivery via the CRD? Which other proteins or co-factors facilitate SMO trafficking from the intercellular space to the cilia and how is SMO trafficking regulated? The answers will give us a better understanding of cilia biology and help with pharmacological design of SMO inhibitors.
An additional question is how does SMO transduce the signal to GLI? Previous studies showed that the suppressor of fused (SUFU) represses GLI transcription factor activation, and active SMO releases this inhibition [92, 93]. The C-terminal tail of SMO can be phosphorylated by several kinases (protein kinase A (PKA), casein kinase 1 (CK1) and G-protein-coupled receptor kinase 2 (GRK2) in Drosophila; CK1 and GRK2 in mammals), and this phosphorylation further promotes GLI activation [94–96]. Interestingly, HH stimulates the formation of SMO-kinase complexes (SMO–PKA in Drosophila and SMO–CK1 in mammals) to initiate downstream signaling events [97, 98]. Solving the structure of full-length SMO in complex with its kinase may provide insight into how SMO transduces the HH signal through its C-terminal intracellular tail. Recent studies in Drosophila and mammalian cells showed that with HH stimulation the Fused (Fu) family kinases phosphorylate Ci/GLI interfering with its binding to SUFU, resulting in activation of Ci/GLI [99]. How SMO activates mammalian Fu kinase family members remains to be determined. Finally, the SUMO pathway and E3 ubiquitin ligases can also target SMO to regulate its trafficking and HH signaling, [100, 101] but the underlying mechanism awaits further investigation.
Concluding Remarks
The recent structural and functional studies have provided molecular insights into the generation and regulation of HH signaling. Particularly, the technological breakthrough of cryo-EM accelerated these discoveries. Structures of DISP and its complex with unmodified HH-N provided a primary understanding of HH-N secretion; 1:1 PTCH1–HH-N and 2:1 PTCH1–HH-N complexes explained the physiological importance of the modifications of HH-N and revealed the inhibition mechanism of PTCH1 by HH-N; crystal and cryo-EM structures of SMO in different conformations shed light on how sterols regulate SMO activity, though these results also posed enigmas for understanding the role of the CRD and the method of sterol binding. Future studies may focus on 1) the structures of DISP, native HH-N, and Scube2 complexes to understand the secretion and trafficking of HH; 2) the mechanism of SMO regulation by sterols, including the role of the CRD, the sterol binding sites, and the trafficking to the cell surface; 3) the link between SMO and GLI to explain how the HH signal is transduced to the downstream components of the pathway. Structural studies on the proteins in the HH pathway will guide the development of therapies against HH related cancers, such as the design of antibodies to block the interfaces between HH-N and PTCH1; and small molecules to compete the sterol binding sites in SMO. In addition, studies on PTCH1 and SMO proteins will also bring light to the mechanism of sterol transport or metabolism by SSD and signaling transduction by Class-F GPCRs.
Figure 6. Structures of SMO with distinct ligands.
(A) Multiple ligand-binding sites in SMO. As a member of class-F GPCR, SMO has multiple ligand binding sites in 7-TMs and one additional site in the CRD. Recent structural studies have revealed the binding details of numerous ligands including both agonists and antagonists. (B) Structure of xSMO with cyclopamine (PDB ID: 6D32). Compared with hSMO, the CRD of this xSMO structure undergoes a dramatic rotation, representing a possible active state of the CRD. (C) Structure of mSMO with SAG21k, cholesterol and a nanobody (Nb) (PDB ID: 6O3C). The mSMO is in an active conformation and the cholesterol molecule binds in the lower site of 7-TMs. The conformation of the CRD is similar with the inactive hSMO. (D) The Structure of hSMO with heterotrimeric Gi in the presence of 24(S),25-EC (PDB ID: 6OT0). The hSMO is in an active conformation and the 24(S),25-EC binding site is higher than that of cholesterol in mSMO structure. The CRD density in this cryo-EM structure is not observed. The right panel shows the position comparison of the ligands in panel (C) and (D). The ligands are shown in sticks with different colors as in C and D.
Outstanding Questions.
How do DISP1 and Scube2 facilitate the secretion and diffusion of HH ligand?
How do HH co-receptors work with PTCH1 to upregulate or downregulate the HH signal?
Does PTCH1 have transporter activity in vitro? The in vitro study of PTCH1 will facilitate the investigations on the substrate(s), energy source and transport direction of PTCH1.
What is the endogenous ligand(s) of SMO and what is the initial binding site of the ligand(s)?
How is the subcellular localization of SMO regulated?
How is the HH signal transduced from SMO to GLI?
Highlights:
The Hedgehog (HH) signaling pathway is essential for the development of animals and is involved in many human cancers.
The secreted ligand HH binds its receptor Patched-1 (PTCH1) and releases the inhibition of Smoothened (SMO) by PTCH1; PTCH1 regulates SMO catalytically via sterols.
How HH recognizes and inhibits PTCH1, how SMO is regulated, and how HH signal is transduced into cell remain unknown and controversial.
Recent structural studies on HH–PTCH1 complexes and SMO in different conformations reveal the mechanism of PTCH1 recognition and inhibition by HH, and provide insights into the mechanism of SMO activation.
Acknowledgement
We apologize to our colleagues whose works have not be mentioned owing to space limitations. We thank E. Coutavas, L. Friedberg, J. Jiang and P. Schmiege for their time and efforts on the manuscript preparation. This work was supported by NIH grants R01 GM135343 and P01 HL020948. X.Q. is the recipient of DDBrown Fellow of the Life Sciences Research Foundation. X.L. is a Damon Runyon-Rachleff Innovator supported by the Damon Runyon Cancer Research Foundation (DRR-53-19) and a Rita C. and William P. Clements Jr. Scholar in Biomedical Research at UT Southwestern Medical Center.
Glossary:
- CAM-related/down-regulated by oncogenes (CDO) and Brother of CDO (BOC)
co-receptors of HH on cell surface that can complex with HH via the calcium-mediated interface and upregulate HH signaling.
- Dispatched-1 (DISP1)
a membrane protein that facilitates the secretion of native HH from the producing cells. DISP is a structural homolog of PTCH, including 12 transmembrane helices and two extracellular domains.
- Glioma-associated oncogene (GLI)
a zinc finger transcription factor that mediates transcriptional responses to HH signaling. In mammals there are three GLI homologs that are homologs of Drosophila Ci (Cubitus interruptus).
- Growth arrest-specific 1 (Gas1)
a HH binding protein in mammals without a homolog in Drosophila. It locates on cell surface and acts as a co-receptor to regulate HH signaling.
- G protein–coupled receptor (GPCR)
a class of seven-transmembrane helices proteins that activate heterotrimeric G proteins.
- Hedgehog acyltransferase (HHAT)
also called skinny hedgehog homology in humans. The enzyme catalyzes the N-terminal palmitoylation of SHH.
- Hedgehog interacting protein (HHIP)
a negative regulator of HH pathway that is upregulated by HH signaling and presumed to function in negative feedback by sequestering HH ligand away from Patched.
- Hedgehog N-terminal signaling domain (HH-N)
the ~20kD secreted ligand of the Hedgehog signaling pathway, palmitoylated at its N terminus and cholesterol modified at its C terminus.
- Niemann-Pick disease, type C1 (NPC1) protein
NPC1 protein transports LDL-derived cholesterol from the lysosome lumen to the membrane. Loss-of-function mutations of NPC1 protein cause an accumulation of cholesterol in lysosomes leading to Niemann-Pick disease, type C.
- Niemann-Pick disease, type C1-like 1 (NPC1L1) protein
a cell surface protein. Shares a similar topology with NPC1 protein and plays an essential role in intestinal cholesterol absorption.
- Paracrine signaling
a mode of signaling where the sending cell produces the signal ligand (e.g. HH ligand), but another cell (the receiving cell) binds to and responds to
- Patched-1 (PTCH1)
the HH receptor at the cell surface or in primary cilia, which binds to HH to initiate ligand-dependent signaling. It includes 12 transmembrane helices, two extracellular domains and a cytosol domain at the C-terminus. In the absence of its ligand, it inhibits SMO to turn off the signal.
- Resistance-nodulation-division (RND) multidrug transporters
a family of bacterial efflux pumps located on the cytoplasmic membrane that can transport multiple substrates.
- Smoothened (SMO)
a Frizzled-Class GPCR protein that transmits the HH signal across the membrane, leading to activation of the GLI transcription factors. It is also the target of several HH pathway inhibitors and anti-cancer drugs.
- Sterol-sensing domain (SSD)
a protein domain that consists of five transmembrane helices. It is predicted to bind cholesterol and presents in proteins involved in cholesterol transport, signaling and metabolism.
- Suppressor of fused (SUFU)
a negative regulator of the HH pathway, binds to GLI proteins preventing their entry into the nucleus. It acts as a transcriptional co-repressor; SUFU deletion or inhibition increases HH signaling activity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing financial interests.
References
- 1.Lum L and Beachy PA (2004) The Hedgehog response network: sensors, switches, and routers. Science 304 (5678), 1755–9. [DOI] [PubMed] [Google Scholar]
- 2.Pak E and Segal RA (2016) Hedgehog Signal Transduction: Key Players, Oncogenic Drivers, and Cancer Therapy. Dev Cell 38 (4), 333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandit T and Ogden SK (2017) Contributions of Noncanonical Smoothened Signaling During Embryonic Development. J Dev Biol 5 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang J and Hui CC (2008) Hedgehog signaling in development and cancer. Dev Cell 15 (6), 801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scales SJ and de Sauvage FJ (2009) Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci 30 (6), 303–12. [DOI] [PubMed] [Google Scholar]
- 6.Rimkus TK et al. (2016) Targeting the Sonic Hedgehog Signaling Pathway: Review of Smoothened and GLI Inhibitors. Cancers (Basel) 8 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burglin TR (2008) The Hedgehog protein family. Genome Biol 9 (11), 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JJ et al. (1994) Autoproteolysis in hedgehog protein biogenesis. Science 266 (5190), 1528–37. [DOI] [PubMed] [Google Scholar]
- 9.Porter JA et al. (1996) Cholesterol modification of hedgehog signaling proteins in animal development. Science 274 (5285), 255–9. [DOI] [PubMed] [Google Scholar]
- 10.Amanai K and Jiang J (2001) Distinct roles of Central missing and Dispatched in sending the Hedgehog signal. Development 128 (24), 5119–27. [DOI] [PubMed] [Google Scholar]
- 11.Chamoun Z et al. (2001) Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 293 (5537), 2080–4. [DOI] [PubMed] [Google Scholar]
- 12.Buglino JA and Resh MD (2008) Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J Biol Chem 283 (32), 22076–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burke R et al. (1999) Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 99 (7), 803–15. [DOI] [PubMed] [Google Scholar]
- 14.Creanga A et al. (2012) Scube/You activity mediates release of dually lipid-modified Hedgehog signal in soluble form. Genes Dev 26 (12), 1312–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tukachinsky H et al. (2012) Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep 2 (2), 308–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrov K et al. (2017) Sending and Receiving Hedgehog Signals. Annu Rev Cell Dev Biol 33, 145–168. [DOI] [PubMed] [Google Scholar]
- 17.Okada A et al. (2006) Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature 444 (7117), 369–73. [DOI] [PubMed] [Google Scholar]
- 18.Tenzen T et al. (2006) The cell surface membrane proteins Cdo and Boc are components and targets of the Hedgehog signaling pathway and feedback network in mice. Dev Cell 10 (5), 647–56. [DOI] [PubMed] [Google Scholar]
- 19.Allen BL et al. (2011) Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev Cell 20 (6), 775–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izzi L et al. (2011) Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Dev Cell 20 (6), 788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huangfu D et al. (2003) Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426 (6962), 83–7. [DOI] [PubMed] [Google Scholar]
- 22.Ingham PW et al. (1991) Role of the Drosophila patched gene in positional signalling. Nature 353 (6340), 184–7. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y and Struhl G (1996) Dual roles for patched in sequestering and transducing Hedgehog. Cell 87 (3), 553–63. [DOI] [PubMed] [Google Scholar]
- 24.Ingham PW and McMahon AP (2001) Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15 (23), 3059–87. [DOI] [PubMed] [Google Scholar]
- 25.Kuwabara PE and Labouesse M (2002) The sterol-sensing domain: multiple families, a unique role? Trends Genet 18 (4), 193–201. [DOI] [PubMed] [Google Scholar]
- 26.Johnson RL et al. (1996) Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272 (5268), 1668–71. [DOI] [PubMed] [Google Scholar]
- 27.Marigo V et al. (1996) Biochemical evidence that patched is the Hedgehog receptor. Nature 384 (6605), 176–9. [DOI] [PubMed] [Google Scholar]
- 28.Stone DM et al. (1996) The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384 (6605), 129–34. [DOI] [PubMed] [Google Scholar]
- 29.Cohen MM Jr. (2003) The hedgehog signaling network. Am J Med Genet A 123A (1), 5–28. [DOI] [PubMed] [Google Scholar]
- 30.Motoyama J et al. (1998) Ptch2, a second mouse Patched gene is co-expressed with Sonic hedgehog. Nat Genet 18 (2), 104–6. [DOI] [PubMed] [Google Scholar]
- 31.Rahnama F et al. (2004) Distinct roles of PTCH2 splice variants in Hedgehog signalling. Biochem J 378 (Pt 2), 325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tukachinsky H et al. (2016) Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proc Natl Acad Sci U S A 113 (40), E5866–E5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arensdorf AM et al. (2016) Smoothened Regulation: A Tale of Two Signals. Trends Pharmacol Sci 37 (1), 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldstein JL et al. (2006) Protein sensors for membrane sterols. Cell 124 (1), 35–46. [DOI] [PubMed] [Google Scholar]
- 35.Altmann SW et al. (2004) Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science 303 (5661), 1201–4. [DOI] [PubMed] [Google Scholar]
- 36.Davies JP et al. (2000) Transmembrane molecular pump activity of Niemann-Pick C1 protein. Science 290 (5500), 2295–8. [DOI] [PubMed] [Google Scholar]
- 37.Li X et al. (2016) Structure of human Niemann-Pick C1 protein. Proc Natl Acad Sci U S A 113 (29), 8212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson RL et al. (2002) Distinct consequences of sterol sensor mutations in Drosophila and mouse patched homologs. Dev Biol 242 (2), 224–35. [DOI] [PubMed] [Google Scholar]
- 39.Briscoe J and Therond PP (2013) The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 14 (7), 416–29. [DOI] [PubMed] [Google Scholar]
- 40.Ming JE et al. (2002) Mutations in PATCHED-1, the receptor for SONIC HEDGEHOG, are associated with holoprosencephaly. Hum Genet 110 (4), 297–301. [DOI] [PubMed] [Google Scholar]
- 41.Wang C et al. (2013) Structure of the human smoothened receptor bound to an antitumour agent. Nature 497 (7449), 338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byrne EFX et al. (2016) Structural basis of Smoothened regulation by its extracellular domains. Nature 535 (7613), 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basset-Seguin N et al. (2015) Efficacy of Hedgehog pathway inhibitors in Basal cell carcinoma. Mol Cancer Ther 14 (3), 633–41. [DOI] [PubMed] [Google Scholar]
- 44.Stewart DP et al. (2018) Cleavage activates dispatched for Sonic Hedgehog ligand release. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Y et al. (2002) Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 111 (1), 63–75. [DOI] [PubMed] [Google Scholar]
- 46.Cannac Fe.a. (2019) Cryo-EM structure of the Hedgehog release protein Dispatched. bioRxiv, doi: 10.1101/707513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pepinsky RB et al. (1998) Identification of a palmitic acid-modified form of human Sonic hedgehog. J Biol Chem 273 (22), 14037–45. [DOI] [PubMed] [Google Scholar]
- 48.Williams KP et al. (1999) Functional antagonists of sonic hedgehog reveal the importance of the N terminus for activity. J Cell Sci 112 ( Pt 23), 4405–14. [DOI] [PubMed] [Google Scholar]
- 49.Kohtz JD et al. (2001) N-terminal fatty-acylation of sonic hedgehog enhances the induction of rodent ventral forebrain neurons. Development 128 (12), 2351–63. [DOI] [PubMed] [Google Scholar]
- 50.Chen MH et al. (2004) Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev 18 (6), 641–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dawber RJ et al. (2005) Differential range and activity of various forms of the Hedgehog protein. BMC Dev Biol 5, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JD and Treisman JE (2001) Sightless has homology to transmembrane acyltransferases and is required to generate active Hedgehog protein. Curr Biol 11 (14), 1147–52. [DOI] [PubMed] [Google Scholar]
- 53.Petrova E et al. (2013) Inhibitors of Hedgehog acyltransferase block Sonic Hedgehog signaling. Nat Chem Biol 9 (4), 247–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodgers UR et al. (2016) Characterization of Hedgehog Acyltransferase Inhibitors Identifies a Small Molecule Probe for Hedgehog Signaling by Cancer Cells. ACS Chem Biol 11 (12), 3256–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi X et al. (2018) Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 560 (7716), 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beachy PA et al. (2010) Interactions between Hedgehog proteins and their binding partners come into view. Genes Dev 24 (18), 2001–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fleet A et al. (2016) Activities of the Cytoplasmic Domains of Patched-1 Modulate but Are Not Essential for the Regulation of Canonical Hedgehog Signaling. J Biol Chem 291 (34), 17557–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McLellan JS et al. (2008) The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature 455 (7215), 979–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y et al. (2018) Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched. Cell 175 (5), 1352–1364 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X et al. (2017) 3.3 A structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gong X et al. (2018) Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science 361 (6402). [DOI] [PubMed] [Google Scholar]
- 62.Qi C et al. (2019) Structural basis of sterol recognition by human hedgehog receptor PTCH1. Sci Adv 5 (9), eaaw6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qi X et al. (2018) Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science. [DOI] [PMC free article] [PubMed]
- 64.Qian H et al. (2019) Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm. Nat Commun 10 (1), 2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rudolf AF et al. (2019) The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism. Nat Chem Biol 15 (10), 975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taipale J et al. (2002) Patched acts catalytically to suppress the activity of Smoothened. Nature 418 (6900), 892–7. [DOI] [PubMed] [Google Scholar]
- 67.Long T et al. (2020) Structural basis for itraconazole-mediated NPC1 inhibition. Nat Commun 11 (1), 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Myers BR et al. (2017) Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc Natl Acad Sci U S A 114 (52), E11141–E11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y e.a. (2019) Hedgehog pathway activation through conformational blockade of the Patched sterol conduit. bioRxiv [DOI] [PMC free article] [PubMed]
- 70.Qi X et al. (2019) Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi. Nature 571 (7764), 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deshpande I et al. (2019) Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature 571 (7764), 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pfeffer SR (2019) NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes. J Biol Chem 294 (5), 1706–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Das A et al. (2014) Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kinnebrew M et al. (2019) Cholesterol accessibility at the ciliary membrane controls Hedgehog signaling. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu A and Song BL (2019) The interplay of Patched, Smoothened and cholesterol in Hedgehog signaling. Curr Opin Cell Biol 61, 31–38. [DOI] [PubMed] [Google Scholar]
- 76.Nachtergaele S et al. (2013) Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. Elife 2, e01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nedelcu D et al. (2013) Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat Chem Biol 9 (9), 557–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang P et al. (2016) Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling. Cell 166 (5), 1176–1187 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Luchetti G et al. (2016) Cholesterol activates the G-protein coupled receptor Smoothened to promote Hedgehog signaling. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rana R et al. (2013) Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling. Nat Commun 4, 2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiao X et al. (2017) Cholesterol Modification of Smoothened Is Required for Hedgehog Signaling. Mol Cell 66 (1), 154–162 e10. [DOI] [PubMed] [Google Scholar]
- 82.Huang P et al. (2018) Structural Basis of Smoothened Activation in Hedgehog Signaling. Cell 175 (1), 295–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen B et al. (2016) Posaconazole, a Second-Generation Triazole Antifungal Drug, Inhibits the Hedgehog Signaling Pathway and Progression of Basal Cell Carcinoma. Mol Cancer Ther 15 (5), 866–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marada S et al. (2015) Functional Divergence in the Role of N-Linked Glycosylation in Smoothened Signaling. PLoS Genet 11 (8), e1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hedger G et al. (2019) Cholesterol Interaction Sites on the Transmembrane Domain of the Hedgehog Signal Transducer and Class F G Protein-Coupled Receptor Smoothened. Structure 27 (3), 549–559 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.DeCamp DL et al. (2000) Smoothened activates Galphai-mediated signaling in frog melanophores. J Biol Chem 275 (34), 26322–7. [DOI] [PubMed] [Google Scholar]
- 87.Riobo NA et al. (2006) Activation of heterotrimeric G proteins by Smoothened. Proc Natl Acad Sci U S A 103 (33), 12607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ogden SK et al. (2008) G protein Galphai functions immediately downstream of Smoothened in Hedgehog signalling. Nature 456 (7224), 967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Raleigh DR et al. (2018) Cilia-Associated Oxysterols Activate Smoothened. Mol Cell 72 (2), 316–327 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Corcoran RB and Scott MP (2006) Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc Natl Acad Sci U S A 103 (22), 8408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cooper MK et al. (2003) A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat Genet 33 (4), 508–13. [DOI] [PubMed] [Google Scholar]
- 92.Dunaeva M et al. (2003) Characterization of the physical interaction of Gli proteins with SUFU proteins. J Biol Chem 278 (7), 5116–22. [DOI] [PubMed] [Google Scholar]
- 93.Svard J et al. (2006) Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell 10 (2), 187–97. [DOI] [PubMed] [Google Scholar]
- 94.Li S et al. (2016) Regulation of Smoothened Phosphorylation and High-Level Hedgehog Signaling Activity by a Plasma Membrane Associated Kinase. PLoS Biol 14 (6), e1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao Y et al. (2007) Hedgehog regulates smoothened activity by inducing a conformational switch. Nature 450 (7167), 252–8. [DOI] [PubMed] [Google Scholar]
- 96.Jia J et al. (2004) Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature 432 (7020), 1045–50. [DOI] [PubMed] [Google Scholar]
- 97.Chen Y et al. (2011) Sonic Hedgehog dependent phosphorylation by CK1alpha and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol 9 (6), e1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li S et al. (2014) Hedgehog induces formation of PKA-Smoothened complexes to promote Smoothened phosphorylation and pathway activation. Sci Signal 7 (332), ra62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han Y et al. (2019) Phosphorylation of Ci/Gli by Fused Family Kinases Promotes Hedgehog Signaling. Dev Cell 50 (5), 610–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li S et al. (2012) Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. PLoS Biol 10 (1), e1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li S et al. (2018) Hedgehog reciprocally controls trafficking of Smo and Ptc through the Smurf family of E3 ubiquitin ligases. Sci Signal 11 (516). [DOI] [PMC free article] [PubMed] [Google Scholar]