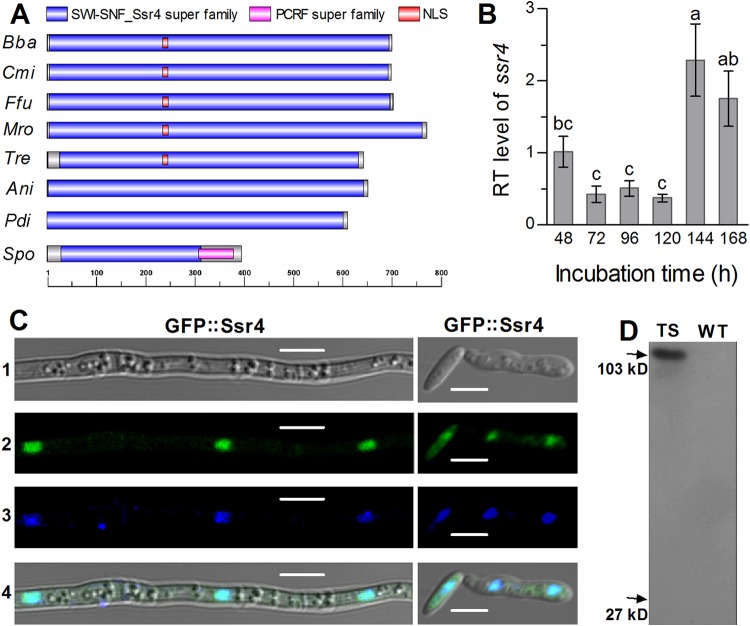
FIG 1.
Properties of Ssr4 in B. bassiana. (A) Structural comparison of Ssr4 orthologs found in B. bassiana (Bba), Cordyceps militaris (Cmi), Fusarium fugikuroi (Ffu), Metarhizium roberstii (Mro), Trichoderma reesei (Tre), Aspergillus nidulans (Ani), Penicillium digitatum (Pdi), and S. pombe (Spo). The main domains and nuclear localization signal (NLS) were predicted at http://smart.embl-heidelberg.de and http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi, respectively. (B) Relative transcript (RT) level of ssr4 in the SDAY culture of a WT strain during a 7-day incubation at the optimal regimen with respect to a standard at the end of 48 h. Values are means ± standard deviations (SD) (error bars) from three cDNA samples. Different lowercase letters denote significant differences (Tukey’s HSD test, P < 0.05). (C) LSCM images for subcellular localization of GFP::Ssr4 fusion protein in the hyphal cells collected from 3-day-old SDBY culture and stained with DAPI. Panels 1 to 4 show bright, expressed, stained, and merged views of the same field, respectively. Note that the expressed signal (green) overlapped well with the stained signal (blue) in nuclei. Bars, 5 μm. (D) Western blots for GFP (27 kDa) and GFP::Ssr4 (102.9 kDa) in the protein extracts isolated from the 3-day-old SDBY cultures of transgenic strain (TS) versus WT (control) and probed with anti-GFP antibodies.