Abstract
Chromosome segregation is a crucial stage of the cell cycle. In general, proteins involved in this process are DNA-binding proteins, and in most bacteria, ParA and ParB are the main players; however, some bacteria manage this process by employing other proteins, such as condensins. The dynamic interaction between ParA and ParB drives movement and exerts positioning of the chromosomal origin of replication (oriC) within the cell. In addition, both ParA and ParB were shown to interact with the other proteins, including those involved in cell division or cell elongation. The significance of these interactions for the progression of the cell cycle is currently under investigation. Remarkably, DNA binding by ParA and ParB as well as their interactions with protein partners conceivably may be modulated by intra- and extracellular conditions. This notion provokes the question of whether chromosome segregation can be regarded as a regulatory stage of the cell cycle. To address this question, we discuss how environmental conditions affect chromosome segregation and how segregation proteins influence other cell cycle processes.
Keywords: chromosome segregation, ParA, ParB, segrosome, cell division, cell elongation
Introduction
Bacteria must adjust their cell cycle to their environmental conditions. Unfavorable conditions such as starvation, oxidative, or osmotic stress alter the energetic state of the cell and trigger the stress signaling molecules (sigma factors, response regulators, signaling nucleotides) (Hengge, 2009; Gottesman, 2019; Latoscha et al., 2019; McLean et al., 2019). As the result, cells modify transcription, increase the generation time, completely inhibit cell division or sometimes form spores or enter dormancy (Errington, 2003; Jones et al., 2013; Heinrich et al., 2015; Desai and Kenney, 2019). While the primary cell cycle checkpoints are the initiation of replication and onset of cell division, these processes must be tightly coordinated with chromosome segregation (for recent reviews, see: Dewachter et al., 2018; Marczynski et al., 2019; Reyes-Lamothe and Sherratt, 2019; Burby and Simmons, 2020). Thus, the chromosome segregation process may link critical stages of the cell cycle.
The role of segregation proteins is to control the positioning of chromosomal (or plasmid) DNA during cell division. Importantly, in most bacterial cells, chromosome segregation begins soon after the initiation of chromosome replication and must be completed before the termination of cell division (Dewachter et al., 2018; Reyes-Lamothe and Sherratt, 2019). During chromosome replication, segregation proteins position newly duplicated chromosomal origin of replication (oriC) regions and ensure proper chromosome organization. Interestingly, in a number of bacterial species, positioning of the oriC and the pattern of chromosome organization may be modified in response to altered environmental conditions, such as limited nutrients (Wang et al., 2013; Badrinarayanan et al., 2015). Perfect examples of this modification are the profound changes in chromosome compaction observed during starvation-induced sporulation of Bacillus subtilis and Streptomyces spp. (Errington, 2001; Flärdh and Buttner, 2009; Jakimowicz and van Wezel, 2012). While in vegetatively growing B. subtilis, the oriC is shifted away from the cell pole, during the formation of endospores, which begins with asymmetric cell division, the oriC is anchored at the poles (Wang et al., 2014). However, even in non-sporulating bacteria, chromosome organization patterns may be altered depending on culture conditions; for example, in Escherichia coli, the chromosome arrangement changes from one in which the oriC adopts a mid-cell position in fast-growing cultures to a longitudinal in slow-growing cells in minimal media (Kleckner et al., 2014; Badrinarayanan et al., 2015). Chromosome topology is controlled by a set of proteins, predominantly topoisomerases and nucleoid-associated proteins (NAPs), whose activities were shown to be influenced by environmental and physiological factors (temperature, pH, salt concentration) (Dorman and Dorman, 2016; Dame et al., 2019). However, the mechanisms by which chromosome arrangement and segregation are adjusted to physiological state of bacterial cell only begun to emerge.
In this review, we discuss how chromosome segregation may be influenced by environmental stress, particularly nutrients limitation, and induced by this factor change of physiological conditions. Furthermore, we explore how changes in segregation protein activity may allow cells to adjust to particular conditions. To address these issues, we focus on the interactions of segregation proteins with DNA and the crosstalk between segregation proteins and their partners.
The Functions of Chromosome Segregation Proteins
Chromosome segregation in bacteria is governed by a set of proteins, among which ParA and ParB are key players. ParA and ParB were first identified as plasmid segregation proteins and further studies revealed that homologs of these proteins control positioning of the chromosomal oriC region (Lee et al., 2003; Mierzejewska and Jagura-Burdzy, 2012; Funnell, 2016). However, ParA and ParB are not fully widespread and chromosome segregation in some bacterial species (i.e., some γ-proteobacteria, including E. coli, which lack parAB genes) exploits the activities of other proteins, such as condensins (Nolivos and Sherratt, 2014; Dewachter et al., 2018). Condensins, which compact the bulk of chromosomal DNA, also play an auxiliary function in ParAB-dependent chromosome segregation (Graumann et al., 1998; Moriya et al., 1998; Wang et al., 2015). Segregation of the terminus-proximal region (ter) usually requires the activities of additional proteins, such as the DNA translocase FtsK or a type II topoisomerase specialized in DNA decatenation (TopoIV) (Kato et al., 1990; Yu et al., 1998; Dewachter et al., 2018) (Figure 1).
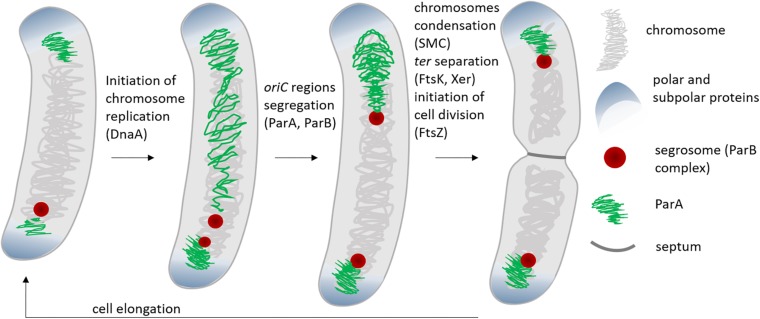
FIGURE 1.
Stages of cell cycle and chromosome segregation in bacteria with polar or subpolar oriC localization (e.g., C. crescentus, V. cholerae, M. smegmatis, M. xanthus). The scheme shows the stage of the cell cycle when activity of DnaA – chromosome replication initiator, ParA, ParB, SMC, and FtsK – proteins involved in chromosome segregation, as well as FtsZ- cell division initiator is required.
The Role of ParA and ParB in oriC Segregation
The ParA and ParB proteins are components of the tripartite segregation system, which also involves parS sequences bound by ParB (parABS system) (Schumacher, 2008; Badrinarayanan et al., 2015). From 1 to 20 parS sequences, depending on the bacterial species, may be scattered over the oriC-proximal chromosomal region, which encompasses a range from 10 kb (in Caulobacter crescentus) to 200 kb in Streptomyces coelicolor, or even up to 650 kb in Pseudomonas aeruginosa (Jakimowicz et al., 2002; Livny et al., 2007; Tran et al., 2018; Kawalek et al., 2020). Upon interaction with ParB, the parS sequence-rich region engages in the formation of a large nucleoprotein complex named the segrosome (Funnell, 2014; Oliva, 2016). parS sites are often referred to as centromeric sites since they mark the chromosomal region that segregates first. Interestingly, in all studied bacterial species that use the ParABS system, ParB binding to DNA was reported to be maintained during majority of the cell cycle. In fact, the initiation of chromosome replication can be detected as the duplication of ParB complexes and the segrosomes mark the positions of the oriC regions throughout the whole cell cycle (Ptacin et al., 2010; Schofield et al., 2010; Shebelut et al., 2010; Harms et al., 2013; Trojanowski et al., 2015; Kois-Ostrowska et al., 2016) (Figure 1).
The organization of the ParB complex is still under investigation, but the studies to date have revealed that its architecture seems to be adjusted for the requirements of each particular bacterial species. Conserved structural features of ParB include the DNA-binding HTH motif in the central part of the polypeptide chain and two conserved sequences named ParB boxes in proximity to the N-terminus (Leonard et al., 2004; Schumacher and Funnell, 2005; Schumacher, 2007, 2008). While, the ParB boxes were shown to be required for segrosome formation, the N-terminal ParB domain is critical for interactions with ParA and C-terminal domain facilitates non-specific DNA binding (Autret et al., 2001; Schumacher and Funnell, 2005; Fisher et al., 2017). Non-specific interactions with DNA allow ParB to spread away from parS sites (Murray et al., 2006; Breier and Grossman, 2007; Kusiak et al., 2011; Chen et al., 2015; Taylor et al., 2015; Song et al., 2017). Moreover, ParB complex assembly was shown to require the bridging of protein molecules bound to spatially distant parS sites (Graham et al., 2014; Song et al., 2017; Soh et al., 2019). This bridging is mediated by dimerization of the arginine-rich ParB box II and was recently shown to be modulated by CTP binding in this region (Osorio-Valeriano et al., 2019; Soh et al., 2019). Notably, while in all bacterial species that use the parABS system, ParB specifically binds parS sequences, the affinity and specificity of ParB toward parS sequences vary among bacteria, resulting in differences in ParB spreading (Jalal et al., 2019). These differences, as well as variations in the number and distribution of parS sites, are reflected in the species-specific architecture of the ParB complex. Nevertheless, the primary role of the ParB complex is to organize the oriC-proximal region of the chromosome to facilitate its movement; the ParB complex thus performs a critical step in chromosome segregation.
The driving force for the chromosomal ParB complex is provided by a P-loop ATPase - ParA. Over the last decade, the hypothesized ParA mechanism of action has changed from filament formation to the generation of a dynamic concentration gradient (Ptacin et al., 2010; Lim et al., 2014; Vecchiarelli et al., 2014; Le Gall et al., 2016). Pivotal for the gradient-based model is non-specific DNA binding by ATP-bound ParA dimers (Leonard et al., 2005; Hester and Lutkenhaus, 2007). Interaction with segrosomes stimulates ParA ATPase activity, while ATP hydrolysis triggers dimer dissociation and protein release from DNA. This generates a ParA-depleted zone in proximity to the ParB complex. Due to their high affinity for DNA-bound ParA dimers, segrosomes move away from the depletion zone toward higher ParA concentration. The directionality of the ParA gradient and ParB movement was suggested to be enhanced by interactions between ParA and the polar or subpolar proteins: TipN and PopZ in C. crescentus, the bactofilin complex in Myxococcus xanthus, HubP in Vibrio cholerae and DivIVA in Mycobacterium smegmatis (Bowman et al., 2008; Ebersbach et al., 2008; Yamaichi et al., 2012; Ginda et al., 2013; Lin et al., 2017). These interactions are critical for proper oriC subcellular localization (Bowman et al., 2008; Ebersbach et al., 2008; Yamaichi et al., 2012; Lin et al., 2017; Pióro et al., 2019). Thus, highly genus- or species-specific interactions play a role in the spatial coordination of chromosome segregation with other cell cycle processes and presumably adjust the segregation machinery to the requirements of the life cycle of a particular bacterium.
The Additional Roles of ParAB Proteins
In addition to their main function in moving the oriC region, in B. subtilis, C. crescentus, and Streptococcus pneumoniae, segrosomes were demonstrated to serve as the loading platform for the condensin complex, which is composed of SMC and the accessory proteins ScpA and ScpB (Sullivan et al., 2009; Wang et al., 2015; Tran et al., 2017). Large, rod-shaped, coiled-coil SMC proteins form dimers due to interactions within the hinge region and ATP-binding head domains. ATP hydrolysis and DNA binding induce conformational changes in the dimer that allow DNA loop extrusion, providing the basis for DNA compaction (Nolivos and Sherratt, 2014; Ganji et al., 2018; Baxter et al., 2019; Marko et al., 2019). Since binding and ATP hydrolysis are crucial for condensin activity, the efficiency of chromosome compaction induced by SMC proteins is presumably dependent on ATP levels in the cell. Importantly, SMC protein loading requires ParB bridging activity (Graham et al., 2014; Wilhelm et al., 2015). Upon loading in proximity to oriC, condensins spread along the chromosome, inducing its overall compaction and longitudinal arrangement.
Finally, an additional function of the ParB complex is its cooperation with proteins engaged in cell division regulation, such as MipZ in C. crescentus and Rhodobacter sphaeroides and PldP as well as FtsZ in Corynebacterium glutamicum (Donovan et al., 2010; Dubarry et al., 2019). Recent studies also shown that ParB also cooperate with NOC and both proteins are required to prevent cell division over nucleoid in B. subtilis (Hajduk et al., 2019) (Table 1).
TABLE 1.
Interaction between proteins engaged in chromosome segregation and their protein partners.
| Microorganism | Segregation protein | Polar or subpolar protein | Replication protein | Chromosome organization protein | Cell division protein | Other cell cycle-involved protein |
| B. subtilis | Soj | DnaA (Murray and Errington, 2008) | ||||
| Spo0J | SMC (Gruber and Errington, 2009) | |||||
| C. crescentus | ParA | TipN, PopZ (Ptacin et al., 2010; Schofield et al., 2010) | ||||
| ParB | PopZ (Bowman et al., 2008; Ebersbach et al., 2008) | MipZ (Thanbichler and Shapiro, 2006) | ||||
| C. glutamicum | ParB | DivIVA (Donovan et al., 2012) | FtsZ, PldP (Donovan et al., 2010) | |||
| M. smegmatis | ParA | DivIVA (Ginda et al., 2013) | DNA glycosylase (Huang and He, 2012) | |||
| M. xanthus | ParA | Bactofilin-PadC (Lin et al., 2017) | ||||
| R. sphaeroides | ParB | MipZ (Dubarry et al., 2019) | ||||
| S. coelicolor | ParA | Scy (Ditkowski et al., 2013) | ParJ (Ditkowski et al., 2010) | |||
| ParB | TopA (Szafran et al., 2013) | |||||
| S. pneumoniae | ParB | SMC (Minnen et al., 2011) | CpsD (Nourikyan et al., 2015) | |||
| V. cholerae | ParAI | HubP (Yamaichi et al., 2012) |
The variety of roles played by segregation proteins in the cell cycles of various bacterial species manifests in the plethora of phenotypes resulting from parAB deletion. While parAB genes were demonstrated to be essential in C. crescentus and M. xanthus, in a number of other bacterial species, including B. subtilis, P. aeruginosa, M. smegmatis, and C. glutamicum, elimination of ParA or ParB leads to chromosome segregation aberrations and mispositioning of the oriC region, eventually resulting in the formation of from 1 to 30% anucleate cells (recently comprehensively reviewed by Kawalek et al., 2020). In some bacteria (V. cholerae and B. subtilis), parB (but not parA) deletion increases the genomic content being manifested as elevated number of oriCs (Lee et al., 2003; Kadoya et al., 2011). Interestingly, parAB deletion may also lead to aberrations in the cell length (in P. aeruginosa, M. smegmatis, C. glutamicum) resulting from septum mispositioning or growth dysregulation (Donovan et al., 2013; Ginda et al., 2013). In some bacteria, parAB deletion results in more pleiotropic phenotypes, such as altered motility in P. aeruginosa, increased transformation competence in S. pneumoniae, reduced resistance to γ-radiation in Deinococcus radiodurans, and inhibited sporulation in B. subtilis (Errington, 2003; Lasocki et al., 2007; Bartosik et al., 2009; Charaka and Misra, 2012; Attaiech et al., 2015). Similarly, elimination of condensins has a bacterial species-dependent impact on chromosome organization. In E. coli and B. subtilis, the deletion of the genes encoding condensins results in a severe growth phenotype and chromosome segregation defects, while their deletion in other bacteria (P. aeruginosa, M. smegmatis, S. coelicolor) leads to a mild phenotype (reviewed by Nolivos and Sherratt, 2014). These observations reinforce the idea that segregation proteins are involved in multiple and varied cellular processes.
Other Proteins Involved in Chromosome Segregation
Interestingly, not all bacterial species employ the ParA and ParB proteins to segregate chromosomes. While the coccoid S. pneumoniae possesses a ParB homolog, it lacks ParA. In these bacteria, ParB cooperates with SMC proteins in chromosome segregation (Minnen et al., 2011). Moreover, some γ-proteobacteria, including E. coli, do not possess any ParA or ParB homologs. In contrast to ParAB-driven segregation, in E. coli, the segregation of newly replicated oriC regions is delayed by their cohesion. Cohesion is controlled by TopoIV, a type II topoisomerase, and SeqA, a protein involved in the regulation of replication initiation (Lu et al., 1994; Joshi et al., 2013; Dewachter et al., 2018). Interestingly, in response to DNA damage-induced stress, the SMC homolog RecN contributes to cohesion control (Vickridge et al., 2017). Moreover, in E. coli, additional ori domain-organizing factors were shown to contribute to positioning of the oriC region. These factors include the cis-acting sites and maoS bound by the characteristic of E. coli MaoP protein as well as migS sites (Yamaichi and Niki, 2004; Valens et al., 2016; Dame et al., 2019). Finally, in E. coli, MukB, a structural SMC homolog strongly contributes to chromosome segregation (Hiraga et al., 1991; Yamazoe et al., 1999). Interestingly, in E. coli, in contrast to SMC in C. crescentus and B. subtilis, MukB does not cause chromosomal arms to adopt a longitudinal arrangement. MukB cooperates with TopoIV and the ter domain-organizing protein MatP (Nolivos et al., 2016; Lioy et al., 2018). MatP-dependent ter organization is a characteristic and unique feature of enterobacteria (Mercier et al., 2008). Whether there is any evolutionary advantage to abandoning the parABS system and adopting the another chromosome arrangement in enterobacteria has not yet been addressed. However, it is tempting to conclude that an elaborate life cycle and/or cell shape (e.g., C. crescentus) demand more complex chromosome segregation machinery.
As the last step of chromosome segregation, the separation of the duplicated chromosome ter regions is the final, critical checkpoint in this process. Interestingly, segregation of the ter regions was observed to be delayed in a number of bacterial species (Thiel et al., 2012). The segregation of ter regions requires the activity of accessory proteins, among which the chromosome translocase FtsK is the most widespread (Massey et al., 2006; Stouf et al., 2013; Crozat et al., 2014). The translocase family also includes the SpoIIIE protein, which is responsible for packaging of the chromosome into the small space of the forespore during B. subtilis sporulation, and TraB homologs involved in the conjugal transfer of DNA (Vogelmann et al., 2011; Thoma and Muth, 2015). As part of the divisiome, FtsK is associated with the newly formed septum via its N-terminal domain, while its C-terminal domain is involved in ATP hydrolysis-dependent DNA translocation as well as recombinase activation (Löwe et al., 2008; Grainge, 2013; Keller et al., 2016). FtsK activity is thus accompanied by DNA decatenation and recombination carried out by TopoIV and XerCD recombinase, respectively (El Sayyed et al., 2016). Replication and segregation of the ter region are tightly coordinated with Z-ring dynamics and the progression of cell division (Espéli et al., 2012; Adams et al., 2014).
Although the main players in the chromosome segregation have been identified, the mechanisms by which the activities of segregation proteins are regulated remain largely unexplored. Nevertheless, the emerging picture is that the process of the chromosome segregation is adjusted to the cell physiological state. Chromosome segregation may be regulated by modulation of the interaction between segregation proteins and DNA, nucleotide binding or other posttranslational protein modifications. Since the activities of various segregation proteins (ParA, SMC/MukB, FtsK, TopoIV) are dependent on the ATP hydrolysis, the overall energetic load of the cell, as manifested by its ATP level, should be considered as an important factor that modifies the efficiency of the segregation process. Importantly, the activities of the segregation proteins may be modulated due to their interactions with protein partners. Subsequently, these interacting partners may alter not only the efficiency of chromosome segregation but also other cell cycle parameters due to their engagement in cell division or cell elongation.
The ParB Complex—Regulation of Its Formation and Its Impact on Chromosome Dynamics
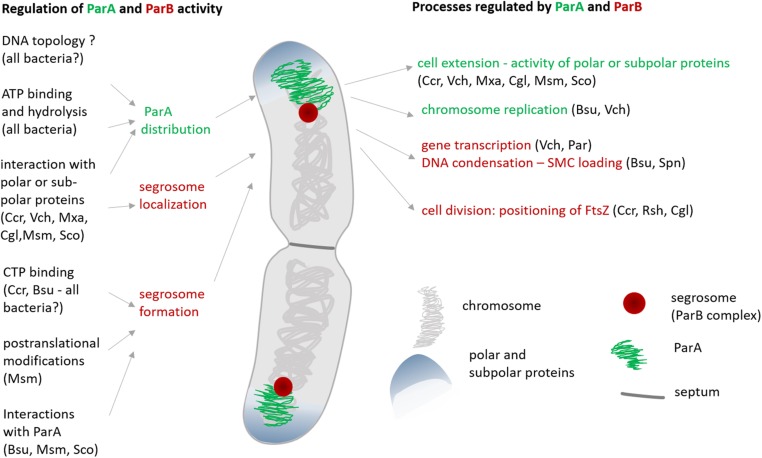
To fulfill their functions, segregation proteins must interact with DNA; hence, the modification of their DNA affinity is critical for the regulation of their activity. Studies in various model bacteria have reported the modification of segrosome formation by the interaction of ParB with CTP or ParA (Breier and Grossman, 2007; Ginda et al., 2013; Baek et al., 2014; Donczew et al., 2016; Osorio-Valeriano et al., 2019; Soh et al., 2019) (Figure 2). Moreover, transcriptional regulation and posttranscriptional modifications of segregation proteins have been described.
FIGURE 2.
The regulation of ParA and ParB activity and the impact of these proteins on cellular processes other than oriC segregation. The abbreviation indicate the species in which particular interactions or influence on activity have been observed: Cgl, C. glutamicum; Cre, C. crescentus; Bsu, B. subtilis; Msm, M. smegmatis; Mxu, M. xanthus; Pae, P. aeruginosa; Rsh, R. sphaeroides; Sco, S. coelicolor; Spe, S. pneumoniae; Vch, V. cholerae; ParA-related regulation is shown in green, ParB-related regulation is shown in red, question mark indicates the connection that has been suggested but has not yet been experimentally confirmed.
Transcriptional Regulation of Segregation Genes
Formation of protein complexes may be controlled by transcriptional regulation. In C. crescentus, this mode of regulation applies to primary cell cycle coordinators, such as the replication initiator DnaA and the cell division protein FtsZ (Laub et al., 2000; Frandi and Collier, 2019). The transcriptional regulation exerted by master regulators such as CtrA and GcrA allows the functional differentiation of two daughter cells, which is characteristic of C. crescentus (Kirkpatrick and Viollier, 2012; Tsokos and Laub, 2012). While the stalked cell is capable of undertaking a new round of chromosome replication, in the swarmer cell, chromosome replication and cell division are inhibited. In C. crescentus, the transcription of genes encoding proteins involved in chromosome topology maintenance (topoisomerases and NAPs) was shown to be developmentally controlled, but the cell cycle-dependent transcriptional regulation of genes encoding segregation proteins has not been reported (Holtzendorff et al., 2004). Fluctuations in ParA and ParB levels between cell divisions cannot be excluded; however, there is no evidence of such regulatory mechanisms in any studied unicellular bacterium. Interestingly, the autoregulation of parAB was demonstrated in case of plasmid segregation proteins (Kwong et al., 2001). The only report showing the life-cycle associated transcriptional induction of chromosomal parAB genes comes from S. coelicolor, a mycelial bacteria that undergo sporulation in response to stress, particularly nutrients limitation. Streptomyces sporulation involves the conversion of multigenomic sporogenic hyphae into chains of unigenomic spores (Flärdh and Buttner, 2009). While during mycelial vegetative growth, cell divisions are rare and not accompanied by chromosome segregation, sporulation requires the synchronous segregation of multiple chromosomal copies synchronized with multiple cell divisions (Jakimowicz and van Wezel, 2012). In S. coelicolor and S. venezuelae, parAB genes, similarly as ftsZ, are controlled by cell cycle regulators associated with the onset of sporulation (WhiA and WhiB) (Flärdh et al., 2000; Jakimowicz et al., 2006; Bush et al., 2013, 2016). Their upregulation allows the induction of parAB and ftsZ required for the formation of numerous ParB complexes and Z-rings, respectively, along sporogenic hyphal cells (Schwedock et al., 1997; Kim et al., 2000; Jakimowicz et al., 2005, 2007). The unprecedented transcriptional upregulation of parAB in Streptomyces fulfils the demands of their unique cell cycle and unusual mode of the chromosome segregation.
The Regulatory Role of Nucleotide Binding and Posttranslational Modifications in ParB
Little is known about the regulation of segrosome architecture; however, CTP binding by ParB was recently demonstrated to modulate interactions during ParB complex formation (Osorio-Valeriano et al., 2019; Soh et al., 2019). CTP is specifically bound within the ParB box II motif, which was previously shown to contribute to the long-range interactions (Graham et al., 2014). ParS binding by ParB induces CTP hydrolysis and increases the protein affinity toward the parS sequence, presumably leading to the complex rearrangement (Osorio-Valeriano et al., 2019; Soh et al., 2019). While the binding of CTP to ParB was demonstrated in B. subtilis and M. xanthus, the CTP interaction interface is highly conserved among ParB homologs, suggesting that this feature is preserved. Moreover, CTP binding by plasmid-encoded ParB proteins (F plasmid ParBF and P1 prophage ParBP1) was demonstrated reinforcing the significance of nucleotide binding for complex formation (Soh et al., 2019). Since CTP is primarily involved in biosynthesis of nucleic acids and phospholipids, rather than the storage and transfer of energy, its intracellular level may reflect the cell capacity to replicate its DNA. Moreover, CTP biosynthesis is tightly regulated, and its level changes in a growth phase-dependent manner (Meng and Switzer, 2001; Jørgensen et al., 2003; Walker et al., 2004). Interestingly, in E. coli, one of NAP (Fis) was shown to be transcriptionally controlled by CTP, indicating the impact of this nucleotide on chromosome topology (Walker et al., 2004). Thus, the dependence of segrosome formation on subcellular CTP levels may influence functional segregation complex formation under unfavorable conditions, such as nutrients limitation, and serve as a link between cell physiology and chromosome segregation.
Furthermore, circumstantial evidence suggests that factors dependent on the cell physiological state, other than nucleotide levels, may also affect segrosome formation. Posttranslational modifications, especially phosphorylation, are well described mechanism to fine-tune the activity of the various proteins in response to changes of environmental conditions, e.g., nutrients availability (Stock et al., 1989; Bernal et al., 2014; Carabetta and Cristea, 2017; Janczarek et al., 2018). While number of nucleoid associated proteins (NAPs) including HU-like proteins were shown to be phosphorylated in B. subtilis and M. tuberculosis and plethora of DNA organizing proteins were suggested to be target for phosphorylation, the evidence for phosphorylation of chromosome segregation proteins is limited (Gupta et al., 2014; Garcia-Garcia et al., 2016). In mycobacteria, ParB was reported to be influenced by phosphorylation, which modified protein affinity toward DNA and abolished its interaction with ParA (Baronian et al., 2015). ParB was shown to be phosphorylated in vitro by several eukaryotic-like Ser/Thr protein kinases whose main role is cell adaptation to changing environmental conditions (e.g., nutrient accessibility). Phosphorylation may potentially mediate fluctuations in ParB activity in relation to cell conditions; however, there is no experimental evidence of such a regulatory mechanism. Among the other posttranslational modifications, acetylation was also shown to influence the activity of numerous DNA associated proteins including topoisomerase I (in E. coli), DNA repair protein Ku and NAP HU (in M. smegmatis) (Zhou et al., 2015, 2017; Ghosh et al., 2016; Anand et al., 2017; Carabetta and Cristea, 2017), however, there are no reports of acetylation influencing directly the activity of chromosome segregation proteins ParA or ParB.
The Influence of ParA on Segresome Assembly
In some bacterial species assembly of the ParB complex was shown to be influenced by ParA. ChIP analyses of B. subtilis, V. cholerae, S. venezuelae, and M. smegmatis indicated that elimination of ParA decreased ParB binding to at least some parS sites (Breier and Grossman, 2007; Ginda et al., 2013; Baek et al., 2014; Donczew et al., 2016). Additionally, in P. aeruginosa, the impact of the ParB complex on chromosome structure was shown to be dependent on ParA (Bartosik et al., 2014). This could be explained by the lower level of ParB in parA deletion strains (detected in B. subtilis and P. aeruginosa but not in S. venezuelae and M. smegmatis) but may also suggest that ParA promotes ParB complex rearrangement. The latter idea is reinforced by the observed influence of ParA on ParB binding to DNA in vitro (Jakimowicz et al., 2007). Because ATP hydrolysis is critical for the activity of ParA, changes in intracellular ATP levels, which reflect fluctuations in the cell energetic state, may impact segrosome formation. In fact, exposure of M. smegmatis cells to stressful conditions modified ParA localization (Ginda et al., 2013; Pióro et al., 2019). Finally, it should be considered that environmental conditions such as increased temperature, pH or osmotic stress influence the chromosome topology (Dorman and Dorman, 2016; Qin et al., 2019), and changes of chromosome topology affect the binding of numerous DNA-interacting proteins including SMC and NAPs (Gruber and Errington, 2009; Dorman and Dorman, 2016; Tran et al., 2017; Qin et al., 2019). Consequently, the activities of segregation proteins may also be easily modified by changes in chromosome topology induced by environmental stress (Figure 2).
Segrosome Impact on Chromosome Topology and Gene Expression
While segrosome assembly may be adjusted in response to environmental clues, its architecture has a profound impact on chromosome structure. In S. coelicolor, segrosomes recruit topoisomerase I, which is required to resolve topological problems and proceed with chromosome segregation (Szafran et al., 2013). Alteration of the ParB complex architecture by changing the parS site position was shown to diminish C. crescentus fitness (Tran et al., 2018). Interestingly, in B. subtilis and S. pneumoniae, changes in the positions of parS sites resulted in the redistribution of ParB but had little effect on chromosome segregation and culture growth (Broedersz et al., 2014; Attaiech et al., 2015; Wang et al., 2015). However, the function of the segrosome in recruiting SMC proteins was affected by abolished ParB bridging (Gruber and Errington, 2009; Minnen et al., 2011; Graham et al., 2014; Wilhelm et al., 2015). Thus, the abovementioned studies indicate that changes in segrosome architecture influence the efficiency of chromosome compaction.
The large nucleoprotein complex formed by ParB is bound to influence chromosome topology and consequently gene expression (Figure 2). The first observation of segregation complex influence on gene expression was made for plasmid ParB proteins (Lynch and Wang, 1995; Rodionov et al., 1999). Considering the interspecies differences in segrosome organization, the impact of this complex on chromosome topology may be diverse. Indeed, in S. pneumoniae, formation of the ParB complex affects the activities of only adjacent genes, particularly the com operon (located 5 kb from parS sites), which encodes proteins involved in competence. This observation explains the increased competence of a S. pneumoniae parB deletion strain. Similar to S. pneumoniae, in V. cholerae, the binding of ParBI to 3 parS sites, results in limited ParB spreading and affects the transcription of only some of the genes in the region bound by ParB (3 of 20 genes) (Baek et al., 2014). Moreover, the transcription of several genes outside of the region bound by ParB is modified in the parAB deletion strain. In contrast to S. pneumoniae and V. cholerae, in P. aeruginosa, ParA and ParB elimination and their overexpression has been shown to affect transcription globally, influencing the expression of genes encoding stress response proteins and putative transcriptional regulators (Bartosik et al., 2014). This phenomenon was explained by P. aeruginosa ParB binding non-restricted to consensus parS sites and ability of this protein to interact with short parS-like motifs (Kawalek et al., 2018). This low DNA-binding specificity of ParB suggests its role in the general organization of DNA, similar to the role of NAPs. Surprisingly, in contrast to the abovementioned bacteria, in B. subtilis, the influence of the ParB complex on gene expression could not be detected (Breier and Grossman, 2007). Although preliminary studies suggested the involvement of parAB in the regulation of sporulation, this phenomenon was later shown to be independent of transcriptional regulation but was explained by the regulation of DnaA activity by ParA (see below). However, other studies reported that deletion of parAB in B. subtilis activated the SOS response by inducing a recA and the gene encoding the cell division inhibitor YneA (Bohorquez et al., 2018). Thus, the influence of the segrosome on the transcription of at least some genes is a common feature of the ParB complex.
Coordination of the Cell Cycle—The Role of Segregation Protein Interactions
Segregation proteins interact with not only DNA and each other but also with proteins engaged in the key cell cycle processes. The ParA–DnaA interaction links chromosome segregation with chromosome replication, the interactions of ParA with polar proteins impact cell elongation, and the ParB–MipZ interaction controls the cell division (Marczynski et al., 2019) (Figure 2). ParA and ParB interaction partners may contribute to the chromosome segregation process; on the other hand, their activity may be controlled by ParA and/or ParB. Importantly, the DNA binding of segregation proteins modulates their availability to the partner-proteins interactions (Murray and Errington, 2008; Schofield et al., 2010; Pióro et al., 2019). Interestingly, most interactions with ParA and ParB are specific to bacterial genera, although some are more widespread and have been detected in various bacterial species.
Interactions Between Segregation and Polar or Subpolar Proteins
The interactions between segregation proteins and polar and subpolar proteins result in the specific localization of oriCs in C. crescentus, V. cholerae, M. xanthus, M. smegmatis, and S. coelicolor, all of which anchor the oriC region at their poles or subapically (Bowman et al., 2008; Ebersbach et al., 2008; Yamaichi et al., 2012; Ginda et al., 2013; Kois-Ostrowska et al., 2016; Lin et al., 2017; Pióro et al., 2019). During the asymmetric cell division of C. crescentus, the unidirectional chromosome segregation must be precisely controlled. The interaction between the ParB protein and the polarity factor PopZ positions the oriC region at the old pole before chromosome replication. PopZ is a small acidic protein that oligomerizes to form a mesh-like structure. In addition to its role in the oriC anchoring, its primary role is to recruit factors involved in stalk morphogenesis (Bowman et al., 2008; Ebersbach et al., 2008). Soon after the initiation of chromosome replication, one of the duplicated segrosomes is moved from the old cell pole to the new pole, and PopZ is simultaneously redistributed to form bipolar foci (Bowman et al., 2008; Ebersbach et al., 2008). Interestingly, PopZ also interacts with ParA monomers released from DNA upon ParA interaction with the ParB complex. Thus, PopZ was suggested to be involved in nucleotide exchange and the regeneration of ParA-ATP bound dimer and restoring its DNA-binding activity. Interestingly, the role of PopZ in regulating ParA activity is partially synergistic with the function of another C. crescentus ParA interaction partner – the coiled-coil TipN protein. TipN is mainly localized at the new pole, and the ParA–TipN interaction is critical for the ParA distribution and the directionality of segrosome movement (Lam et al., 2006; Ptacin et al., 2010). Importantly, ParA was shown to influence the function of PopZ; accumulating at the new pole ParA recruits PopZ, generating a nucleation site that initiates PopZ polymerization (Laloux and Jacobs-Wagner, 2013). It should be noted that formation of PopZ–ParA complex is dependent on availability of ParA released from nucleoid, most often by ongoing chromosome segregation (Laloux and Jacobs-Wagner, 2013). Since PopZ recruits the cell cycle regulator CtrA and its associated kinase as well as CtrA-targeting protease ClpXP (Joshi et al., 2018), the ParA control of PopZ localization possibly indicates the coordination of chromosome segregation with the global cell cycle regulation.
Similar as in C. crescentus, polar anchoring of oriC region and unidirectional chromosome segregation was described for V. cholerae chromosome I (the larger of the two V. cholerae chromosomes). In V. cholerae, ParAI (the ParA protein that governs the segregation of chromosome I) interacts with the polar localized protein HubP, and deletion of hubP abolished polarly oriCI positioning (Yamaichi et al., 2012). HubP also interacts with chemotactic machinery and flagellar proteins; moreover, HubP was also identified to interact with two other ATPases, ParC and FlhG (Yamaichi et al., 2012). Interestingly, in Shewanella oneidensis HubP homolog was also shown to be involved in chromosome segregation. Moreover, the identified in these bacteria interaction between HubP and PdeB, phosphodiesterase that controls c-di-GMP level in the cell, may indicate the potential link between cyclic nucleotide signaling and chromosome segregation (Rossmann et al., 2019). An interesting example of a bacterium in which the oriC is not localized at the poles but rather exhibits subpolar localization is M. xanthus. In this bacterium, the positioning of oriC is exerted by the ParA interaction with PadC, which in turn binds the bactofilin scaffold stretching from the poles (Lin et al., 2017). Only the monomeric form of ParA is recruited to the bactofilin–PadC complex, which is reminiscent of the ParA–PopZ interaction in C. crescentus (Lin et al., 2017). However, in case of V. cholerae and M. xanthus, there is no evidence that ParA influences the activity or localization of its interaction partners.
In the apically extending cells of actinobacteria segregation proteins also interact with polar protein complexes. The main component of the polar complex in these bacteria is the coiled-coil tropomyosin-like protein DivIVA, which recruits peptidoglycan-synthesizing machinery to the poles (Kang et al., 2008; Letek et al., 2008; Flärdh, 2010; Hammond et al., 2019). In M. smegmatis, DivIVA directly interacts with ParA (Ginda et al., 2013). The inhibition of this interaction was shown not only to decrease the efficiency of the chromosome segregation, but also it visibly increased the cell elongation rate, indicating ParA influence on DivIVA activity (Pióro et al., 2019). Considering that the recruitment of ParA to DivIVA was proved to compete with ParA–DNA interaction, the release of ParA from DNA upon interaction with ParB complex or, conceivably, due to changes of DNA topology, may have the impact on cell elongation rate. Markedly, DivIVA in mycobacteria is phosphorylated by the PknA kinase, the activity of which is regulated by the PknB kinase, and both PknA and PknB are essential Ser/Thr protein kinases that control growth rate and morphology (Kang et al., 2005; Jani et al., 2010; Lee et al., 2014). In response to the extracellular signals, these kinases phosphorylate regulators of central carbon metabolism and proteins involved in the stress response, transport and cell wall synthesis. It was shown that growth phase dependent DivIVA phosphorylation status regulates the rate of peptidoglycan synthesis (Jani et al., 2010). Whether the phosphorylation status of DivIVA influences its interaction with ParA, linking environmental conditions with the segregation of chromosomes, remains to be elucidated.
Unlike M. smegmatis, in S. coelicolor, which also belongs to actinobacteria, the interaction between ParA and DivIVA was not detected, but ParA was found to directly interact with the Scy protein – the other component of a polar protein complex named the polarisome, which also includes DivIVA. During S. coelicolor vegetative growth, this interaction is responsible for anchoring of the apical chromosome at the tips of multigenomic hyphal cells. Importantly, the deletion of parA affected the rate of hyphal growth, which was explained by ParA-dependent modulation of Scy activity (Ditkowski et al., 2013; Donczew et al., 2016). Developmentally controlled ParA accumulation during sporulation leads to polarisome disassembly and inhibits hyphal elongation (Ditkowski et al., 2013). Interestingly in S. coelicolor ParA was shown to interact with the other segregation protein ParJ, however, the contribution of this protein to the segregation process is not fully understood (Ditkowski et al., 2010). Unlike in above described actinobacteria, in closely related C. glutamicum ParB directly interacts with DivIVA and this interaction positions oriC at the cell pole. The observation that deletion of parB results in impaired cell extension indicates that ParB–DivIVA interaction may impact DivIVA activity (Donovan et al., 2010, 2012). Thus, in actinobacteria, the interactions of segregation proteins with polar complexes not only contribute to chromosome segregation but also regulate cell elongation.
Interestingly, DivIVA is also involved in anchoring the oriC region during B. subtilis sporulation, though not through its direct interaction with ParAB homologs; DivIVA instead interacts with a complex containing MinD and MinJ (Kloosterman et al., 2016). Moreover, the additional RacA protein, which also specifically binds the oriC-proximal part of the chromosome, contributes to oriC anchoring in B. subtilis (Ben-Yehuda et al., 2005; Schumacher et al., 2016).
Interactions Between Segregation and Replication Proteins
In a number of bacterial species, ParA was also shown to be involved in the regulation of the chromosome replication. The direct interaction of ParA with DnaA was first described in B. subtilis, in which ParA and ParB homologs were originally identified as regulators of sporulation and called Soj and Spo0J, respectively. The elimination of Spo0J was found to inhibit sporulation and that inhibition may be counteracted by deletion of the gene encoding Soj (suppressor of Spo0J, a homolog of ParA) (Quisel et al., 1999). Later studies showed that this effect is indirect and results from Soj-dependent regulation of DnaA, which subsequently negatively regulates transcription of sporulation genes (Murray and Errington, 2008; Scholefield et al., 2011, 2012). Monomeric Soj directly interacts with DnaA and reduce its interaction with DNA inhibiting its oligomerization. In the absence of Spo0J, the DNA-bound Soj dimer is more stable, and the level of monomeric Soj available to interact with DnaA is decreased; therefore, DnaA replication activity is elevated. Thus, the function of Soj as a DnaA inhibitor depends on interaction between segregation protein and DNA. Since DNA binding by ParA homologs may be influenced by intracellular ATP level, Soj likely links the changes of the cell physiological state and environmental conditions that have the impact on cell energetic state with the DnaA replication (Murray and Errington, 2008; Scholefield et al., 2011, 2012). Additionally, in V. cholerae, DnaA interacts with ParA as well as ParB, while in D. radiodurans (another bacterium with a multipartite genome: two chromosomes and a megaplasmid), the DnaA protein interacts with ParB (Kadoya et al., 2011; Maurya et al., 2019). The involvement of the segregation proteins in replication regulation explains the increased number of oriC resulting from parB deletion.
Interactions Between Segregation and Cell Division Proteins
Chromosome segregation proteins also interact with the cell division proteins. In the α-proteobacteria C. crescentus and R. sphaeroides, the interaction between the ParB protein and MipZ (a ParA homolog) was detected (Thanbichler and Shapiro, 2006; Dubarry et al., 2019). MipZ is an inhibitor of FtsZ polymerization that exhibits dynamic localization characteristic of the ParA family of ATPases. In C. crescentus, MipZ forms a cloud-like structure with the lowest MipZ concentration in the middle of the cell, which restricts Z-ring formation to the cell center (Thanbichler and Shapiro, 2006). The localization of MipZ in R. sphaeroides is different; MipZ is situated mainly at the cell poles but also at the mid-cell position. In both C. crescentus and R. sphaeroides, the localization of MipZ depends on ParB, but unlike ParA, MipZ dimers are recruited and stabilized by ParB (Thanbichler and Shapiro, 2006; Dubarry et al., 2019). Importantly, in C. crescentus, the transcription level of mipZ changes during cell cycle progression and in response to environmental cues (e.g., nitrogen starvation) (Collier, 2019). In C. glutamicum, ParB also interacts with PldP – a ParA homolog involved in the regulation of cell division (Donovan et al., 2010). Moreover, in these bacteria, the direct interaction between ParB and FtsZ—a cell division initiator—was shown (Donovan et al., 2010). These interactions presumably account for the observed influence of parAB deletion on septum placement (Donovan and Bramkamp, 2014).
Interestingly, in S. pneumoniae (which lacks the ParA component of the parABS system), ParB interacts with CpsD, which is homologous to ParA tyrosine (BY) kinase and localizes at the site of cell division (Bender and Yother, 2001). BY-kinases are autokinases that regulate polymerization and the export of capsular polysaccharides. Inhibition of CpsD phosphorylation delayed chromosome segregation, while increased CpsD phosphorylation enhanced ParB mobility. The interaction between ParB and CpsD may coordinate chromosome segregation with capsular formation and the cell division (Nourikyan et al., 2015). Recent studies identified another ParB interacting partner in S. pneumoniae—the RocS protein. RocS is required for chromosome segregation but also interacts with FtsZ and CspD (Mercy et al., 2019). The above examples show that chromosome segregation and cell division are coupled due to the interactions of segregation proteins.
Some of the interactions of segregation proteins were shown to be critical under stress conditions. These include discovered in M. smegmatis interaction between ParA and 3-methyladenine DNA glycosylase, a protein mainly involved in DNA repair (Huang and He, 2012). This interaction stimulates the ATPase activity of ParA and regulates cell growth and morphology independent of DNA glycosylase activity. In B. subtilis, ParAB was shown to cooperate with another segregation protein, WhiA, which was suggested to maintain DNA integrity (Bohorquez et al., 2018). Interestingly, double parAB/whiA deletion was lethal and could be explained by the blockade of cell division.
Concluding Remarks
Studies of last two decades have shed light on chromosome segregation, revealing the concerted actions of segregation proteins, dissecting the mechanisms of their activities and describing their interactions. However, evidence that the chromosome segregation process is adjusted to environmental conditions has only started to emerge. Environmental stress factors, such as nutrients limitation, modify cell physiology and require adjustment of the cell cycle process. The possible pathways that can be used to coordinate the cell cycle with stress response are those based on intracellular nucleotide levels and chromosome topology. In particular, the finding that the ParB–CTP interaction is a critical factor for segrosome formation opens a new avenue for the exploration of chromosome segregation regulation. Finally, the impact of polar proteins (TipN, PopZ, HubP, or DivIVA) on the activities of segregation proteins under unfavorable conditions also remains to be further investigated to identify the links between changes in cell physiology and chromosome segregation.
Furthermore, recent studies have indicated the impact of chromosome segregation proteins on other cell cycle processes. Interestingly, due to their involvement in highly species-specific interactions (including both DNA interactions during segrosome formation and protein–protein interactions), the involvement of segregation proteins in coordination of the cell cycle is diverse and species dependent. Common regulatory pathways (identified in at least two unrelated organisms) include the regulation of gene transcription, chromosome replication, and the regulation of the cell elongation and division (Marczynski et al., 2019). Notably, the availability of ParA to interact with their protein partners (DnaA, PopZ, DivIVA) depend on the ParA binding to chromosome. Since this interaction is plausibly influenced by environmental factors, it may serve as the important regulatory circuit. However, further studies are required to fully understand the complex regulatory networks behind the identified connections and the impact of external factors on the global coordination of cell cycle processes.
Author Contributions
MP and DJ wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Jolanta Zakrzewska-Czerwiñska, Marcin Szafran, and Joanna Hołówka for critical reading and comments on the manuscript.
Footnotes
Funding. The authors acknowledge financial support from the National Science Center, Poland (OPUS grant 2017/27/B/NZ1/00823).
References
- Adams D. W., Wu L. J., Errington J. (2014). Cell cycle regulation by the bacterial nucleoid. Curr. Opin. Microbiol. 22C 94–101. 10.1016/j.mib.2014.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand C., Garg R., Ghosh S., Nagaraja V. (2017). A Sir2 family protein Rv1151c deacetylates HU to alter its DNA binding mode in Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 493 1204–1209. 10.1016/j.bbrc.2017.09.087 [DOI] [PubMed] [Google Scholar]
- Attaiech L., Minnen A., Kjos M., Gruber S., Veening J. W. (2015). The ParB-parS chromosome segregation system modulates competence development in Streptococcus pneumoniae. mBio 6:e00662-15. 10.1128/mBio.00662-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autret S., Nair R., Errington J. (2001). Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis?: evidence for separate domains involved in DNA binding and interactions with Soj protein. Mol. Microbiol. 41 743–755. 10.1046/j.1365-2958.2001.02551.x [DOI] [PubMed] [Google Scholar]
- Badrinarayanan A., Le T. B. K., Laub M. T. (2015). Bacterial Chromosome Organization and Segregation. Annu. Rev Cell Dev. Biol. 31 171–199. 10.1146/annurev-cellbio-100814-125211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek J. H., Rajagopala S. V., Chattoraj D. K. (2014). Chromosome segregation proteins of Vibrio cholerae as transcription regulators. mBio 5:e01061-14. 10.1128/mBio.01061-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baronian G., Ginda K., Berry L., Cohen-Gonsaud M., Zakrzewska-Czerwiñska J., Jakimowicz D., et al. (2015). Phosphorylation of Mycobacterium tuberculosis ParB participates in regulating the ParABS chromosome segregation system. PLoS One 10:e0119907. 10.1371/journal.pone.0119907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosik A., Mierzejewska J., Thomas C. M., Jagura-Burdzy G. (2009). ParB deficiency in Pseudomonas aeruginosa destabilizes the partner protein ParA and affects a variety of physiological parameters. Microbiology 155 1080–1092. 10.1099/mic.0.024661-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosik A. A., Glabski K., Jecz P., Mikulska S., Fogtman A., Koblowska M., et al. (2014). Transcriptional profiling of para and ParB mutants in actively dividing cells of an opportunistic human pathogen Pseudomonas aeruginosa. PLoS One 9:e87276. 10.1371/journal.pone.0087276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter J., Oliver A. W., Schalbetter S. A. (2019). Are SMC complexes loop extruding factors? Linking theory with fact. Bioessays 41:1800182. 10.1002/bies.201800182 [DOI] [PubMed] [Google Scholar]
- Bender M. H., Yother J. (2001). CpsB is a modulator of capsule-associated tyrosine kinase activity in Streptococcus pneumoniae. J. Biol. Chem. 276 47966–47974. 10.1074/jbc.M105448200 [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S., Fujita M., Liu X. S., Gorbatyuk B., Skoko D., Yan J., et al. (2005). Defining a centromere-like element in Bacillus subtilis by identifying the binding sites for the chromosome-anchoring protein RacA. Mol. Cell 17 773–782. 10.1016/j.molcel.2005.02.023 [DOI] [PubMed] [Google Scholar]
- Bernal V., Castaño-Cerezo S., Gallego-Jara J., Écija-Conesa A., de Diego T., Iborra J. L., et al. (2014). Regulation of bacterial physiology by lysine acetylation of proteins. N. Biotechnol. 31 586–595. 10.1016/j.nbt.2014.03.002 [DOI] [PubMed] [Google Scholar]
- Bohorquez L. C., Surdova K., Jonker M. J., Hamoen L. W. (2018). The conserved DNA binding protein WhiA influences chromosome. J. Bacteriol. 200:e00633-17. 10.1128/JB.00633-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman G. R., Comolli L. R., Zhu J., Eckart M., Koenig M., Downing K. H., et al. (2008). A polymeric protein anchors the chromosomal Origin/ParB complex at a bacterial cell pole. Cell 134 945–955. 10.1016/j.cell.2008.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier A. M., Grossman A. D. (2007). Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 64 703–718. 10.1111/j.1365-2958.2007.05690.x [DOI] [PubMed] [Google Scholar]
- Broedersz C. P., Wang X., Meir Y., Loparo J. J., Rudner D. Z., Wingreen N. S. (2014). Condensation and localization of the partitioning protein ParB on the bacterial chromosome. Proc. Natl. Acad. Sci. U.S.A. 111 8809–8814. 10.1073/pnas.1402529111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burby P. E., Simmons L. A. (2020). Regulation of cell division in bacteria by monitoring genome integrity and DNA replication status. J. Bacteriol. 202:e00408-19. 10.1128/jb.00408-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M. J., Bibb M. J., Chandra G., Findlay K. C., Buttner M. J. (2013). Genes required for aerial growth, cell division, and chromosome segregation are targets of whia before sporulation in Streptomyces venezuelae. mBio 4:e00684-13. 10.1128/mBio.00684-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush M. J., Chandra G., Bibb M. J., Findlay K. C., Buttner M. J. (2016). Genome-wide chromatin immunoprecipitation sequencing analysis shows that WhiB is a transcription factor that cocontrols its regulon with WhiA to initiate developmental cell division in Streptomyces. mBio 7:e00523-16. 10.1128/mBio.00523-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabetta V. J., Cristea I. M. (2017). Regulation, function, and detection of protein acetylation in bacteria. J. Bacteriol. 199:e00107-17. 10.1128/JB.00107-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charaka V. K., Misra H. S. (2012). Functional characterization of the role of the chromosome I partitioning system in genome segregation in Deinococcus radiodurans. J. Bacteriol. 194 5739–5748. 10.1128/JB.00610-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.-W., Lin M.-H. H., Chu C.-H. H., Hsu C.-E. E., Sun Y.-J. J. (2015). Insights into ParB spreading from the complex structure of Spo0J and parS. Proc. Natl. Acad. Sci. U.S.A. 112 6613–6618. 10.1073/pnas.1421927112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J. (2019). Cell division control in Caulobacter crescentus. Biochim. Biophys. Acta Gene Regul. Mech. 1862 685–690. 10.1016/j.bbagrm.2018.04.005 [DOI] [PubMed] [Google Scholar]
- Crozat E., Rousseau P., Fournes F., Cornet F. (2014). The FtsK Family of DNA translocases finds the ends of circles. J. Mol. Microbiol. Biotechnol. 24 396–408. 10.1159/000369213 [DOI] [PubMed] [Google Scholar]
- Dame R. T., Rashid F.-Z. M., Grainger D. C. (2019). Chromosome organization in bacteria: mechanistic insights into genome structure and function. Nat. Rev. Genet. 12 1–16. 10.1038/s41576-019-0185-4 [DOI] [PubMed] [Google Scholar]
- Desai S. K., Kenney L. J. (2019). Switching lifestyles is an in vivo adaptive strategy of bacterial pathogens. Front. Cell. Infect. Microbiol. 9:421. 10.3389/fcimb.2019.00421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewachter L., Verstraeten N., Fauvart M., Michiels J. (2018). An integrative view of cell cycle control in Escherichia coli. FEMS Microbiol. Rev. 42 116–136. 10.1093/femsre/fuy005 [DOI] [PubMed] [Google Scholar]
- Ditkowski B., Holmes N., Rydzak J., Donczew M., Bezulska M., Ginda K., et al. (2013). Dynamic interplay of ParA with the polarity protein, Scy, coordinates the growth with chromosome segregation in Streptomyces coelicolor. Open Biol. 3:130006. 10.1098/rsob.130006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditkowski B., Troc P., Ginda K., Donczew M., Chater K. F. K. F., Zakrzewska-Czerwiñska J., et al. (2010). The actinobacterial signature protein ParJ (SCO1662) regulates ParA polymerization and affects chromosome segregation and cell division during Streptomyces sporulation. Mol. Microbiol. 78 1403–1415. 10.1111/j.1365-2958.2010.07409.x [DOI] [PubMed] [Google Scholar]
- Donczew M., Mackiewicz P., Wrobel A., Flardh K., Zakrzewska-Czerwínska J., Jakimowicz D., et al. (2016). ParA and ParB coordinate chromosome segregation with cell elongation and division during Streptomyces sporulation. Open Biol. 6:150263. 10.1098/rsob.150263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan C., Bramkamp M. (2014). Cell division in Corynebacterineae. Front. Microbiol. 5:132. 10.3389/fmicb.2014.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan C., Schauss A., Krämer R., Bramkamp M. (2013). Chromosome segregation impacts on cell growth and division site selection in Corynebacterium glutamicum. PLoS One 8:e55078. 10.1371/journal.pone.0055078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan C., Schwaiger A., Krämer R., Bramkamp M. (2010). Subcellular localization and characterization of the ParAB system from Corynebacterium glutamicum. J. Bacteriol. 192 3441–3451. 10.1128/JB.00214-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan C., Sieger B., Krämer R., Bramkamp M. (2012). A synthetic Escherichia coli system identifies a conserved origin tethering factor in Actinobacteria. Mol. Microbiol. 84 105–116. 10.1111/j.1365-2958.2012.08011.x [DOI] [PubMed] [Google Scholar]
- Dorman C. J., Dorman M. J. (2016). DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression. Biophys. Rev. 8 89–100. 10.1007/s12551-016-0238-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubarry N., Willis C. R., Ball G., Lesterlin C., Armitage J. P. (2019). In vivo imaging of the segregation of the 2 chromosomes and the cell division proteins of rhodobacter sphaeroides reveals an unexpected role for MipZ. mBio 10:e2515-18. 10.1128/mBio.02515-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G., Briegel A., Jensen G. J., Jacobs-Wagner C. (2008). A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134 956–968. 10.1016/j.cell.2008.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sayyed H., Le Chat L., Lebailly E., Vickridge E., Pages C., Cornet F., et al. (2016). Mapping topoisomerase IV binding and activity sites on the E. coli Genome. PLoS Genet. 12:e1006025. 10.1371/journal.pgen.1006025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. (2001). Septation and chromosome segregation during sporulation in Bacillus subtilis. Curr. Opin. Microbiol. 4 660–666. 10.1016/s1369-5274(01)00266-1 [DOI] [PubMed] [Google Scholar]
- Errington J. (2003). Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1 117–126. 10.1038/nrmicro750 [DOI] [PubMed] [Google Scholar]
- Espéli O., Borne R., Dupaigne P., Thiel A., Gigant E., Mercier R., et al. (2012). A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 31 3198–3211. 10.1038/emboj.2012.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher G. L. M., Pastrana C. L., Higman V. A., Koh A., Taylor J. A., Butterer A., et al. (2017). The structural basis for dynamic DNA binding and bridging interactions which condense the bacterial centromere. eLife 6:e28086. 10.7554/eLife.28086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flärdh K. (2010). Cell polarity and the control of apical growth in Streptomyces. Curr. Opin. Microbiol. 13 758–765. 10.1016/j.mib.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Flärdh K., Buttner M. J. (2009). Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7 36–49. 10.1038/nrmicro1968 [DOI] [PubMed] [Google Scholar]
- Flärdh K., Leibovitz E., Buttner M. J., Chater K. F. (2000). Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Mol. Microbiol. 38 737–749. 10.1046/j.1365-2958.2000.02177.x [DOI] [PubMed] [Google Scholar]
- Frandi A., Collier J. (2019). Multilayered control of chromosome replication in Caulobacter crescentus. Biochem. Soc. Trans. 47 187–196. 10.1042/BST20180460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell B. E. (2014). How to build segregation complexes in bacteria: use bridges. Genes Dev. 28 1140–1142. 10.1101/gad.244517.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell B. E. (2016). ParB partition proteins: complex formation and spreading at bacterial and plasmid centromeres. Front. Mol. Biosci. 3:44. 10.3389/fmolb.2016.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji M., Shaltiel I. A., Bisht S., Kim E., Kalichava A., Haering C. H., et al. (2018). Real-time imaging of DNA loop extrusion by condensin. Science 360 102–105. 10.1126/science.aar7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Garcia T., Poncet S., Derouiche A., Shi L., Mijakovic I., Noirot-Gros M. F. (2016). Role of protein phosphorylation in the regulation of cell cycle and DNA-related processes in bacteria. Front. Microbiol. 7:184. 10.3389/fmicb.2016.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Padmanabhan B., Anand C., Nagaraja V. (2016). Lysine acetylation of the Mycobacterium tuberculosis HU protein modulates its DNA binding and genome organization. Mol. Microbiol. 100 577–588. 10.1111/mmi.13339 [DOI] [PubMed] [Google Scholar]
- Ginda K., Bezulska M., Ziółkiewicz M., Dziadek J., Zakrzewska-Czerwiñska J., Jakimowicz D. (2013). ParA of Mycobacterium smegmatis co-ordinates chromosome segregation with the cell cycle and interacts with the polar growth determinant DivIVA. Mol. Microbiol. 87 998–1012. 10.1111/mmi.12146 [DOI] [PubMed] [Google Scholar]
- Gottesman S. (2019). Trouble is coming: signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 294 11685–11700. 10.1074/jbc.REV119.005593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham T. G. W., Wang X., Song D., Etson C. M., Van Oijen A. M., Rudner D. Z., et al. (2014). ParB spreading requires DNA bridging. Genes Dev. 28 1228–1238. 10.1101/gad.242206.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainge I. (2013). Simple topology: FtsK-directed recombination at the dif site. Biochem. Soc. Trans. 41 595–600. 10.1042/BST20120299 [DOI] [PubMed] [Google Scholar]
- Graumann P. L., Losick R., Strunnikov A. V. (1998). Subcellular localization of Bacillus subtilis SMC, a protein involved in chromosome condensation and segregation. J. Bacteriol. 180 5749–5755. 10.1128/jb.180.21.5749-5755.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S., Errington J. (2009). Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell 137 685–696. 10.1016/j.cell.2009.02.035 [DOI] [PubMed] [Google Scholar]
- Gupta M., Sajid A., Sharma K., Ghosh S., Arora G., Singh R., et al. (2014). HupB, a nucleoid-associated protein of mycobacterium tuberculosis, is modified by serine/threonine protein kinases in vivo. J. Bacteriol. 196 2646–2657. 10.1128/JB.01625-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduk I. V., Mann R., Rodrigues C. D. A., Harry E. J. (2019). The ParB homologs, Spo0J and Noc, together prevent premature midcell Z ring assembly when the early stages of replication are blocked in Bacillus subtilis. Mol. Microbiol. 112 766–784. 10.1111/mmi.14319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond L. R., White M. L., Eswara P. J. (2019). vIVA la DivIVA! J. Bacteriol. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A., Treuner-Lange A., Schumacher D., Søgaard-Andersen L. (2013). Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLoS Genet. 9:e1003802. 10.1371/journal.pgen.1003802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich K., Leslie D. J., Jonas K. (2015). Modulation of bacterial proliferation as a survival strategy. Adv. Appl. Microbiol. 92 127–171. 10.1016/bs.aambs.2015.02.004 [DOI] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7 263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- Hester C. M., Lutkenhaus J. (2007). Soj (ParA) DNA binding is mediated by conserved arginines and is essential for plasmid segregation. Proc. Natl. Acad. Sci. U.S.A. 104 20326–20331. 10.1073/pnas.0705196105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S., Niki H., Imamura R., Ogura T., Yamanaka K., Feng J., et al. (1991). Mutants defective in chromosome partitioning in E. coli. Res. Microbiol. 142 189–194. 10.1016/0923-2508(91)90029-A [DOI] [PubMed] [Google Scholar]
- Holtzendorff J., Hung D., Brende P., Reisenauer A., Viollier P. H., McAdams H. H., et al. (2004). Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science 304 983–987. 10.1126/science.1095191 [DOI] [PubMed] [Google Scholar]
- Huang F., He Z.-G. G. (2012). Characterization of a conserved interaction between DNA glycosylase and ParA in Mycobacterium smegmatis and M. tuberculosis. PLoS One 7:e38276. 10.1371/journal.pone.0038276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D., Chater K., Zakrzewska-Czerwínska J. (2002). The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol. Microbiol. 45 1365–1377. 10.1046/j.1365-2958.2002.03102.x [DOI] [PubMed] [Google Scholar]
- Jakimowicz D., Gust B., Zakrzewska-Czerwiñska J., Chater K. F. K. F., Zakrzewska-Czerwinska J., Chater K. F. K. F. (2005). Developmental-stage-specific assembly of ParB complexes in Streptomyces coelicolor hyphae. J. Bacteriol. 187 3572–3580. 10.1128/JB.187.10.3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D., Mouz S., Zakrzewska-Czerwiñska J., Chater K. F. K. F. (2006). Developmental control of a parAB promoter leads to formation of sporulation-associated ParB complexes in Streptomyces coelicolor. J. Bacteriol. 188 1710–1720. 10.1128/JB.188.5.1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D., van Wezel G. P. G. P. (2012). Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere? Mol. Microbiol. 85 393–404. 10.1111/j.1365-2958.2012.08107.x [DOI] [PubMed] [Google Scholar]
- Jakimowicz D., Zydek P., Kois A., Zakrzewska-Czerwiñska J., Chater K. F. K. F. (2007). Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol. Microbiol. 65 625–641. 10.1111/j.1365-2958.2007.05815.x [DOI] [PubMed] [Google Scholar]
- Jalal A. S. B., Pastrana C. L., Tran N. T., Stevenson C. E., Lawson D. M., Moreno-Herrero F., et al. (2019). Structural and biochemical analyses of Caulobacter crescentus ParB reveal the role of its N- terminal domain in chromosome segregation Adam. bioRxiv [Preprint]. 10.1101/816959 [DOI] [Google Scholar]
- Janczarek M., Vinardell J. M., Lipa P., Karaś M. (2018). Hanks-type serine/threonine protein kinases and phosphatases in bacteria: roles in signaling and adaptation to various environments. Int. J. Mol. Sci. 19:2872. 10.3390/ijms19102872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani C., Eoh H., Lee J. J., Hamasha K., Sahana M. B., Han J.-S., et al. (2010). Regulation of polar peptidoglycan biosynthesis by Wag31 Phosphorylation in Mycobacteria. BMC Microbiol. 10:327. 10.1186/1471-2180-10-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. H., Vail K. M., McMullen L. M. (2013). Filament formation by foodborne bacteria under sublethal stress. Int. J. Food Microbiol. 165 97–110. 10.1016/j.ijfoodmicro.2013.05.001 [DOI] [PubMed] [Google Scholar]
- Jørgensen C. M., Hammer K., Martinussen J. (2003). CTP limitation increases expression of CTP synthase in Lactococcus lactis. J. Bacteriol. 185 6562–6574. 10.1128/JB.185.22.6562-6574.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi K. K., Battle C. M., Chien P. (2018). Polar localization hub protein PopZ restrains adaptor-dependent ClpXP proteolysis in Caulobacter crescentus. J. Bacteriol. 200:e00221-18. 10.1128/JB.00221-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi M. C., Magnan D., Montminy T. P., Lies M., Stepankiw N., Bates D. (2013). Regulation of sister chromosome cohesion by the replication fork tracking protein SeqA. PLoS Genet. 9:e1003673. 10.1371/journal.pgen.1003673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoya R., Baek J. H., Sarker A., Chattoraj D. K. (2011). Participation of chromosome segregation protein ParAI of Vibrio cholerae in chromosome replication. J. Bacteriol. 193 1504–1514. 10.1128/JB.01067-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C., Abbott D. W., Park S. T., Dascher C. C., Cantley L. C., Husson R. N. (2005). The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19 1692–1704. 10.1101/gad.1311105.nism [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.-M. M., Nyayapathy S., Lee J.-Y. Y., Suh J.-W. W., Husson R. N. (2008). Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology 154 725–735. 10.1099/mic.0.2007/0140760 [DOI] [PubMed] [Google Scholar]
- Kato J. I., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H. (1990). New topoisomerase essential for chromosome segregation in E. coli. Cell 63 393–404. 10.1016/0092-8674(90)90172-B [DOI] [PubMed] [Google Scholar]
- Kawalek A., Bartosik A. A., Glabski K., Jagura-Burdzy G. (2018). Pseudomonas aeruginosa partitioning protein ParB acts as a nucleoid-associated protein binding to multiple copies of a parS-related motif. Nucleic Acids Res. 46 4592–4606. 10.1093/nar/gky257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawalek A., Wawrzyniak P., Bartosik A. A., Jagura-Burdzy G. (2020). Rules and exceptions?: the role of chromosomal ParB in DNA segregation and other cellular processes. Microorganisms 8:105. 10.3390/microorganisms8010105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A. N., Xin Y., Boer S., Reinhardt J., Baker R., Arciszewska L. K., et al. (2016). Activation of Xer-recombination at dif: structural basis of the FtsKγ-XerD interaction. Sci. Rep. 6:33357. 10.1038/srep33357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Calcutt M. J., Schmidt F. J., Chater K. F. (2000). Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 182 1313–1320. 10.1128/jb.182.5.1313-1320.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick C. L., Viollier P. H. (2012). Decoding Caulobacter development. FEMS Microbiol. Rev. 36 193–205. 10.1111/j.1574-6976.2011.00309.x [DOI] [PubMed] [Google Scholar]
- Kleckner N., Fisher J. K., Stouf M., White M. A., Bates D., Witz G. (2014). The bacterial nucleoid: nature, dynamics and sister segregation. Curr. Opin. Microbiol. 22 127–137. 10.1016/j.mib.2014.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman T. G., Lenarcic R., Willis C., Roberts D. M., Hamoen L. W. (2016). Complex polar machinery required for proper chromosome segregation in vegetative and sporulating cells of Bacillus subtilis. Mol. Microbiol. 101 333–350. 10.1111/mmi.13393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kois-Ostrowska A., Strzałka A., Lipietta N., Tilley E., Zakrzewska-Czerwiñska J., Herron P. R., et al. (2016). Unique Function of the Bacterial Chromosome Segregation Machinery in Apically Growing Streptomyces - Targeting the Chromosome to New Hyphal Tubes and its Anchorage at the Tips. PLoS Genet. 12:e1006488. 10.1371/journal.pgen.1006488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusiak M., Gapczynska A., Plochocka D., Thomas C. M., Jagura-Burdzy G., Gapczyñska A., et al. (2011). Binding and spreading of ParB on DNA determine its biological function in Pseudomonas aeruginosa. J. Bacteriol. 193 3342–3355. 10.1128/JB.00328-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong S. M., Chew C. Y., Chit L. P. (2001). Molecular analysis of the pRA2 partitioning region: ParB autoregulates parAB transcription and forms a nucleoprotein complex with the plasmid partition site, parS. Mol. Microbiol. 40 621–633. 10.1046/j.1365-2958.2001.02405.x [DOI] [PubMed] [Google Scholar]
- Laloux G., Jacobs-Wagner C. (2013). Spatiotemporal control of PopZ localization through cell cycle-coupled multimerization. J. Cell Biol. 201 827–841. 10.1083/jcb.201303036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H., Schofield W. B., Jacobs-Wagner C. (2006). A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124 1011–1023. 10.1016/j.cell.2005.12.040 [DOI] [PubMed] [Google Scholar]
- Lasocki K., Bartosik A. A., Mierzejewska J., Thomas C. M., Jagura-Burdzy G. (2007). Deletion of the parA (soj) homologue in Pseudomonas aeruginosa causes ParB instability and affects growth rate, chromosome segregation, and motility. J. Bacteriol. 189 5762–5772. 10.1128/JB.00371-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latoscha A., Wörmann M. E., Tschowri N. (2019). Nucleotide second messengers in streptomyces. Microbiology 165 1153–1165. 10.1099/mic.0.000846 [DOI] [PubMed] [Google Scholar]
- Laub M. T., McAdams H. H., Feldblyum T., Fraser C. M., Shapiro L. (2000). Global analysis of the genetic network controlling a bacterial cell cycle. Science 290 2144–2148. 10.1126/science.290.5499.2144 [DOI] [PubMed] [Google Scholar]
- Le Gall A., Cattoni D. I., Guilhas B., Mathieu-Demazière C., Oudjedi L., Fiche J.-B., et al. (2016). Bacterial partition complexes segregate within the volume of the nucleoid. Nat. Commun. 7:12107. 10.1038/ncomms12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. J., Kang C. M., Lee J. H., Park K. S., Jeon J. H., Lee S. H. (2014). Phosphorylation-dependent interaction between a serine/threonine kinase PknA and a putative cell division protein Wag31 in Mycobacterium tuberculosis. New Microbiol. 37 525–533. [PubMed] [Google Scholar]
- Lee P. S., Lin D. C. H., Moriya S., Grossman A. D. (2003). Effects of the chromosome partitioning protein Spo0J (ParB) on oriC positioning and replication initiation in Bacillus subtilis. J. Bacteriol. 185 1326–1337. 10.1128/JB.185.4.1326-1337.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard T. A., Butler P. J., Lowe J. (2005). Bacterial chromosome segregation?: structure and DNA binding of the Soj dimer — a conserved biological switch. EMBO J. 24 270–282. 10.1038/sj.emboj.7600530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard T. A., Butler P. J. G., Löwe J. (2004). Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol. Microbiol. 53 419–432. 10.1111/j.1365-2958.2004.04133.x [DOI] [PubMed] [Google Scholar]
- Letek M., Ordóñez E., Vaquera J., Margolin W., Flärdh K., Mateos L. M., et al. (2008). DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. J. Bacteriol. 190 3283–3292. 10.1128/JB.01934-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H. C., Surovtsev I. V., Beltran B. G., Huang F., Bewersdorf J., Jacobs-Wagner C. (2014). Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. eLife 3:e02758. 10.7554/eLife.02758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Osorio Valeriano M., Harms A., Søgaard-Andersen L., Thanbichler M. (2017). Bactofilin-mediated organization of the ParABS chromosome segregation system in Myxococcus xanthus. Nat. Commun. 8:1817. 10.1038/s41467-017-02015-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy V. S., Cournac A., Koszul R., Mozziconacci J., Espeli O., Boccard F., et al. (2018). Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell 172 1–13. 10.1016/j.cell.2017.12.027 [DOI] [PubMed] [Google Scholar]
- Livny J., Yamaichi Y., Waldor M. K. (2007). Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J. Bacteriol. 189 8693–8703. 10.1128/JB.01239-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J., Ellonen A., Allen M. D., Atkinson C., Sherratt D. J., Grainge I. (2008). Molecular Mechanism of Sequence-Directed DNA Loading and Translocation by FtsK. Mol. Cell 31 498–509. 10.1016/j.molcel.2008.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Campbell J. L., Boye E., Kleckner N. (1994). SeqA: a negative modulator of replication initiation in E. coli. Cell 77 413–426. 10.1016/0092-8674(94)90156-2 [DOI] [PubMed] [Google Scholar]
- Lynch A. S., Wang J. C. (1995). SopB protein-mediated silencing of genes linked to the sopC locus of Escherichia coli F plasmid. Proc. Natl. Acad. Sci. U.S.A. 92 1896–1900. 10.1073/pnas.92.6.1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczynski G. T., Petit K., Patel P. (2019). Crosstalk regulation between bacterial chromosome replication and chromosome partitioning. Front. Microbiol. 10:279. 10.3389/fmicb.2019.00279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko J. F., De Los Rios P., Barducci A., Gruber S. (2019). DNA-segment-capture model for loop extrusion by structural maintenance of chromosome (SMC) protein complexes. Nucleic Acids Res. 47 6956–6972. 10.1093/nar/gkz497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey T. H., Mercogliano C. P., Yates J., Sherratt D. J., Löwe J. (2006). Double-stranded DNA translocation: structure and mechanism of Hexameric FtsK. Mol. Cell 23 457–469. 10.1016/j.molcel.2006.06.019 [DOI] [PubMed] [Google Scholar]
- Maurya G. K., Kota S., Misra H. S. (2019). Characterisation of ParB encoded on multipartite genome in Deinococcus radiodurans and their roles in radioresistance. Microbiol. Res. 223–225 22–32. 10.1016/j.micres.2019.03.005 [DOI] [PubMed] [Google Scholar]
- McLean T. C., Lo R., Tschowri N., Hoskisson P. A., Al Bassam M. M., Hutchings M. I., et al. (2019). Sensing and responding to diverse extracellular signals: an updated analysis of the sensor kinases and response regulators of streptomyces species. Microbiology 165 929–952. 10.1099/mic.0.000817 [DOI] [PubMed] [Google Scholar]
- Meng Q., Switzer R. L. (2001). Regulation of transcription of the Bacillus subtilis pyrG gene, encoding cytidine triphosphate synthetase. J. Bacteriol. 183 5513–5522. 10.1128/JB.183.19.5513-5522.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R., Petit M. A., Schbath S., Robin S., El Karoui M., Boccard F., et al. (2008). The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135 475–485. 10.1016/j.cell.2008.08.031 [DOI] [PubMed] [Google Scholar]
- Mercy C., Ducret A., Slager J., Lavergne J. P., Freton C., Nagarajan S. N., et al. (2019). RocS drives chromosome segregation and nucleoid protection in Streptococcus pneumoniae. Nat. Microbiol. 4 1661–1670. 10.1038/s41564-019-0472-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzejewska J., Jagura-Burdzy G. (2012). Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle. Plasmid 67 1–14. 10.1016/j.plasmid.2011.08.003 [DOI] [PubMed] [Google Scholar]
- Minnen A., Attaiech L., Thon M., Gruber S., Veening J.-W. (2011). SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol. Microbiol. 81 676–688. 10.1111/j.1365-2958.2011.07722.x [DOI] [PubMed] [Google Scholar]
- Moriya S., Tsujikawa E., Hassan A. K. M., Asai K., Kodama T., Ogasawara N. (1998). A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol. 29 179–187. 10.1046/j.1365-2958.1998.00919.x [DOI] [PubMed] [Google Scholar]
- Murray H., Errington J. (2008). Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135 74–84. 10.1016/j.cell.2008.07.044 [DOI] [PubMed] [Google Scholar]
- Murray H., Ferreira H., Errington J. (2006). The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol. Microbiol. 61 1352–1361. 10.1111/j.1365-2958.2006.05316.x [DOI] [PubMed] [Google Scholar]
- Nolivos S., Sherratt D. (2014). The bacterial chromosome: architecture and action of bacterial SMC and SMC-like complexes. FEMS Microbiol. Rev. 38 380–392. 10.1111/1574-6976.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolivos S., Upton A. L., Badrinarayanan A., Muller J., Zawadzka K., Wiktor J., et al. (2016). MatP regulates the coordinated action of topoisomerase IV and MukBEF in chromosome segregation. Nat. Commun. 7:10466. 10.1038/ncomms10466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourikyan J., Kjos M., Mercy C., Cluzel C., Morlot C., Noirot-Gros M. F., et al. (2015). Autophosphorylation of the bacterial tyrosine-kinase CpsD connects capsule synthesis with the cell cycle in Streptococcus pneumoniae. PLoS Genet. 11:e1005518. 10.1371/journal.pgen.1005518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva M. A. (2016). Segrosome complex formation during DNA trafficking in bacterial cell division. Front. Mol. Biosci. 3:51. 10.3389/fmolb.2016.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio-Valeriano M., Altegoer F., Steinchen W., Urban S., Liu Y., Bange G., et al. (2019). ParB-type DNA segregation proteins Are CTP-dependent molecular switches. Cell 179 1512–1524.e15. 10.1016/j.cell.2019.11.015 [DOI] [PubMed] [Google Scholar]
- Pióro M., Małecki T., Portas M., Magierowska I., Trojanowski D., Sherratt D., et al. (2019). Competition between DivIVA and the nucleoid for ParA binding promotes segrosome separation and modulates mycobacterial cell elongation. Mol. Microbiol. 111 204–220. 10.1111/mmi.14149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin J. L., Lee S. F., Garner E. C., Toro E., Eckart M., Comolli L. R., et al. (2010). A spindle-like apparatus guides bacterial chromosome segregation. Nat. Cell Biol. 12 791–798. 10.1038/ncb2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L., Erkelens A. M., Ben Bdira F., Dame R. T. (2019). The architects of bacterial DNA bridges: a structurally and functionally conserved family of proteins. Open Biol. 9:190223. 10.1098/rsob.190223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quisel J. D., Lin D. C., Grossman A. D. (1999). Control of development by altered localization of a transcription factor in B. subtilis. Mol. Cell 4 665–672. 10.1016/s1097-2765(00)80377-9 [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe R., Sherratt D. J. (2019). The bacterial cell cycle, chromosome inheritance and cell growth. Nat. Rev. Microbiol. 17 467–478. 10.1038/s41579-019-0212-7 [DOI] [PubMed] [Google Scholar]
- Rodionov O., Łobocka M., Yarmolinsky M. (1999). Silencing of genes flanking the P1 plasmid centromere. Science 283 546–549. 10.1126/science.283.5401.546 [DOI] [PubMed] [Google Scholar]
- Rossmann F. M., Rick T., Mrusek D., Sprankel L., Dörrich A. K., Leonhard T., et al. (2019). The GGDEF domain of the phosphodiesterase PdeB in Shewanella putrefaciens mediates recruitment by the polar landmark protein HubP. J. Bacteriol. 201 1–14. 10.1128/JB.00534-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield W. B., Lim H. C., Jacobs-Wagner C. (2010). Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 29 3068–3081. 10.1038/emboj.2010.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield G., Errington J., Murray H. (2012). Soj/ParA stalls DNA replication by inhibiting helix formation of the initiator protein DnaA. EMBO J. 31 1542–1555. 10.1038/emboj.2012.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield G., Whiting R., Errington J., Murray H. (2011). Spo0J regulates the oligomeric state of Soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Mol. Microbiol. 79 1089–1100. 10.1111/j.1365-2958.2010.07507.x [DOI] [PubMed] [Google Scholar]
- Schumacher M. (2007). Structural biology of plasmid segregation proteins. Curr. Opin. Struct. Biol. 17 103–109. 10.1016/j.sbi.2006.11.005 [DOI] [PubMed] [Google Scholar]
- Schumacher M. (2008). Structural biology of plasmid partition?: uncovering the molecular mechanisms of DNA segregation. Biochem. J. 412 1–18. 10.1042/BJ20080359 [DOI] [PubMed] [Google Scholar]
- Schumacher M., Funnell B. E. (2005). Structures of ParB bound to DNA reveal mechanism of partition complex formation. Nature 438 516–519. 10.1038/nature04149 [DOI] [PubMed] [Google Scholar]
- Schumacher M. A., Lee J., Zeng W., Duke N. H. (2016). Molecular insights into DNA binding and anchoring by the Bacillus subtilis sporulation kinetochore-like RacA protein. Nucleic Acids Res. 44 5438–5449. 10.1093/nar/gkw248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwedock J., McCormick J. R., Angert E. R., Nodwell J. R., Losick R. (1997). Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 25 847–858. 10.1111/j.1365-2958.1997.mmi507.x [DOI] [PubMed] [Google Scholar]
- Shebelut C. W., Guberman J. M., van Teeffelen S., Yakhnina A. A., Gitai Z., Van Teeffelen S., et al. (2010). Caulobacter chromosome segregation is an ordered multistep process. Proc. Natl. Acad. Sci. U.S.A. 107 14194–14198. 10.1073/pnas.1005274107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh Y.-M., Davidson I. F., Zamuner S., Basquin J., Bock F. P., Taschner M., et al. (2019). Self-organization of parS Centromeres by the ParB CTP Hydrolase. Science 366 1129–1133. 10.1126/science.aay3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Rodrigues K., Graham T. G. W., Loparo J. J. (2017). A network of cis and trans interactions is required for ParB spreading. Nucleic Acids Res. 45 7106–7117. 10.1093/nar/gkx271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. (1989). Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53 450–490. 10.1128/mmbr.53.4.450-490.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouf M., Meile J. C., Cornet F. (2013). FtsK actively segregates sister chromosomes in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 110 11157–11162. 10.1073/pnas.1304080110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N. L., Marquis K. A., Rudner D. Z. (2009). Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell 137 697–707. 10.1016/j.cell.2009.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafran M., Skut P., Ditkowski B., Ginda K., Chandra G., Zakrzewska-Czerwiñska J., et al. (2013). Topoisomerase I (TopA) is recruited to ParB complexes and is required for proper chromosome organization during Streptomyces coelicolor sporulation. J. Bacteriol. 195 4445–4455. 10.1128/JB.00798-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. A., Pastrana C. L., Butterer A., Pernstich C., Gwynn E. J., Sobott F., et al. (2015). Specific and non-specific interactions of ParB with DNA: implications for chromosome segregation. Nucleic Acid Res. 43 719–731. 10.1093/nar/gku1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M., Shapiro L. (2006). MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126 147–162. 10.1016/j.cell.2006.05.038 [DOI] [PubMed] [Google Scholar]
- Thiel A., Valens M., Vallet-Gely I., Espéli O., Boccard F. (2012). Long-range chromosome organization in E. coli: a site-specific system isolates the ter macrodomain. PLoS Genet. 8:e1002672. 10.1371/journal.pgen.1002672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma L., Muth G. (2015). The conjugative DNA-transfer apparatus of Streptomyces. Int. J. Med. Microbiol. 305 224–229. 10.1016/j.ijmm.2014.12.020 [DOI] [PubMed] [Google Scholar]
- Tran N. T., Laub M. T., Le T. B. K. (2017). SMC progressively aligns chromosomal arms in Caulobacter crescentus but is antagonized by convergent transcription. Cell Rep. 20 2057–2071. 10.1016/j.celrep.2017.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran N. T., Stevenson C. E., Som N. F., Thanapipatsiri A., Jalal A. S. B., Le T. B. K. (2018). Permissive zones for the centromere-binding protein ParB on the Caulobacter crescentus chromosome. Nucleic Acids Res. 46 1196–1209. 10.1093/nar/gkx1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski D., Ginda K., Pióro M., Hołówka J., Skut P., Jakimowicz D., et al. (2015). Choreography of the mycobacterium replication machinery during the cell cycle. mBio 6:e02125-14. 10.1128/mBio.02125-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokos C. G., Laub M. T. (2012). Polarity and cell fate asymmetry in Caulobacter crescentus. Curr. Opin. Microbiol. 15 744–750. 10.1016/j.mib.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valens M., Thiel A., Boccard F. (2016). The MaoP/maoS Site-Specific System Organizes the Ori Region of the E. coli Chromosome into a Macrodomain. PLoS Genet. 12:e1006309. 10.1371/journal.pgen.1006309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli A. G., Neuman K. C., Mizuuchi K. (2014). A propagating ATPase gradient drives transport of surface-confined cellular cargo. Proc. Natl. Acad. Sci. U.S.A. 111 4880–4885. 10.1073/pnas.1401025111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickridge E., Planchenault C., Cockram C., Junceda I. G., Espéli O. (2017). Management of E. coli sister chromatid cohesion in response to genotoxic stress. Nat. Commun. 8:14618. 10.1038/ncomms14618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann J., Ammelburg M., Finger C., Guezguez J., Linke D., Flötenmeyer M., et al. (2011). Conjugal plasmid transfer in Streptomyces resembles bacterial chromosome segregation by FtsK/SpoIIIE. EMBO J. 30 2246–2254. 10.1038/emboj.2011.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker K. A., Mallik P., Pratt T. S., Osuna R. (2004). The Escherichia coli fis promoter is regulated by changes in the levels of its transcription initiation nucleotide CTP. J. Biol. Chem. 279 50818–50828. 10.1074/jbc.M406285200 [DOI] [PubMed] [Google Scholar]
- Wang X., Le T. B. K., Lajoie B. R., Dekker J., Laub M. T., Rudner D. Z. (2015). Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 29 1661–1675. 10.1101/gad.265876.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Montero Llopis P., Rudner D. Z. (2013). Organization and segregation of bacterial chromosomes. Nat. Rev. Genet. 14 191–203. 10.1038/nrg3375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Montero Llopis P., Rudner D. Z. (2014). Bacillus subtilis chromosome organization oscillates between two distinct patterns. Proc. Natl. Acad. Sci. U.S.A. 111 12877–12882. 10.1073/pnas.1407461111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm L., Bürmann F., Minnen A., Shin H. C., Toseland C. P., Oh B. H., et al. (2015). SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. eLife 4:e06659. 10.7554/eLife.06659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y., Bruckner R., Ringgaard S., Möll A., Ewen Cameron D., Briegel A., et al. (2012). A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev. 26 2348–2360. 10.1101/gad.199869.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y., Niki H. (2004). migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J. 23 221–233. 10.1038/sj.emboj.7600028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe M., Onogi T., Sunako Y., Niki H., Yamanaka K., Ichimura T., et al. (1999). Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 18 5873–5884. 10.1093/emboj/18.21.5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. C., Weihe E. K., Margolin W. (1998). Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J. Bacteriol. 180 6424–6428. 10.1128/.180.23.6424-6428.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Zhou Y. N., Jin D. J., Tse-Dinh Y. C. (2017). Deacetylation of topoisomerase i is an important physiological function of E. coli CobB. Nucleic Acids Res. 45 5349–5358. 10.1093/nar/gkx250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Chen T., Zhou L., Fleming J., Deng J., Wang X., et al. (2015). Discovery and characterization of Ku acetylation in Mycobacterium smegmatis. FEMS Microbiol. Lett. 362:fnu051. 10.1093/femsle/fnu051 [DOI] [PubMed] [Google Scholar]