Abstract
Porphyromonas gingivalis, like other bacteria belonging to the phylum Bacteroidetes, synthesizes sphingolipids (SLs). However, their exact roles in microbial physiology and their potential role in mediating interactions with their eukaryotic host are unclear. Our working hypothesis for this study was that synthesis of SLs (host-like lipids) affords a mechanism that allows P. gingivalis to persist in homeostasis with its host. In a previous study, we deleted a gene (PG1780 in strain W83) predicted to encode a serine palmitoyl transferase (SPT)—the enzyme that catalyzes the first conserved step in the synthesis of SLs—and we determined that the mutant was unable to synthesize SLs. Here, we characterized the SPT enzyme encoded by PG1780, analyzed the impact of SPT deletion on P. gingivalis gene expression (RNA-Seq analysis), and began to define the impact of SL synthesis on its interactions with host cells. Enzymatic analysis verified that the protein encoded by PG1780 is indeed an SPT. RNA-Seq analysis determined that a lack of SL synthesis results in differential expression of extracytoplasmic function sigma factors, components of the type IX secretion system (T9SS), and CRISPR and cas genes. Our data demonstrate that when human THP1 macrophage-like cells were challenged with the wild type (W83) and the SL-null mutant (W83 ΔSPT), the SL-null strain elicited a robust inflammatory response (elevated IL-1β, IL-6, IL-10, IL-8, RANTES, and TNFα) while the response to the parent strain W83 was negligible. Interestingly, we also discovered that SLs produced by P. gingivalis can be delivered to host cells independent of cell-to-cell contact. Overall, our results support our working hypothesis that synthesis of SLs by P. gingivalis is central to its ability to manipulate the host inflammatory response, and they demonstrate the integral importance of SLs in the physiology of P. gingivalis.
Keywords: homeostasis, Bacteroidetes, inflammation, dihydroceramides, membrane microdomains, sigma factors
Introduction
Sphingolipids (SLs) are a class of amphipathic lipids containing a long-chain amino alcohol backbone (also called a sphingoid base) attached via an amide linkage to a fatty acyl chain. The first committed step in the generation of SLs is the condensation of an amino acid, often serine, and palmitoyl CoA to form sphinganine by the enzyme serine palmitoyl transferase (SPT; Merrill and Carman 2015; Harrison et al. 2018). SLs play a prominent role in numerous eukaryotic cellular processes, including inflammation, cell migration, adhesion, growth, and apoptosis (Hannun and Obeid 2008, 2018; Maceyka and Spiegel 2014; Merrill and Carman 2015), and they have been linked to a growing number of inborn genetic diseases (Dunn et al. 2019).
While SL synthesis is ubiquitous in eukaryotes, it is rare in prokaryotes. Intriguingly, a variety of bacteria belonging to the phylum Bacteroidetes that persist in the oral microbiome, including Porphyromonas gingivalis, Tannerella forsythia, and Prevotella intermedia, are proficient in SL synthesis (Olsen and Jantzen 2001). Although SLs produced by these bacteria are highly similar to the host SLs, these lipids are distinct in their head groups and an isomethyl branch in the long chain base and ceramide component (Harrison et al. 2018.). Practically, these chemical distinctions are highly significant since they have been used to detect and distinguish bacterially derived SLs (Nichols et al. 2004; Brown et al. 2019). In particular, the SLs produced by oral anaerobes, includingP. gingivalis, have been shown to permeate host tissues (Nichols 1998; Nichols and Rojanasomsith 2006; Nichols et al. 2011), and the types of SLs were found to be distinct in healthy versus diseased tissues (Nichols and Rojanasomsith 2006; Nichols et al. 2011). Given that P. gingivalis is strongly implicated in the etiology of periodontal disease (Lamont and Jenkinson 1998; Socransky et al. 1998; Byrne et al. 2009; Darveau 2010), understanding the impact of SLs on the physiology of this bacterium as well as defining their impact on the host as purified lipids has been investigated (Moye et al. 2016; Olsen and Nichols 2018). Purified SLs derived from P. gingivalis induce a number of changes in the physiology of eukaryotic cells in vitro (Olsen and Nichols 2018), and often these effects are observed only for SLs bearing a particular headgroup. For example, phosphoglycerol dihydroceramides induce the RANKL-dependent pathway of osteoclastogenesis in osteoclasts (Kanzaki et al. 2017), initiate apoptosis in endothelial cells (Zahlten et al. 2007), and increase the generation of prostaglandin E2 by gingival fibroblasts (Nichols et al. 2004). In model systems of disease, phosphoethanolamine dihydroceramides induced inflammation in a murine model of experimental autoimmune encephalomyelitis (Nichols et al. 2009). Thus, SLs synthesized by P. gingivalis profoundly affect a variety of eukaryotic signaling pathways in a highly cell- and lipid-specific manner and may form a link to systemic conditions.
While there are few reports describing the role of bacterially derived SLs in bacterial physiology or membrane structure and function, the data indicate that they may function in similar ways as in eukaryotic cells (Heaver et al. 2018). Studies with Bacteroides fragilis have demonstrated the formation of SL-dependent membrane microdomains, similar to eukaryotic lipid rafts, and that SLs are essential for mounting a stress response and long-term survival, suggesting that SLs play a role in regulating gene expression (An et al. 2011). We recently demonstrated that SLs are essential for P. gingivalis survival under oxidative stress. Also, we determined that select SLs are present in outer membrane vesicles, thereby identifying a potential mechanism of SL secretion (Moye et al. 2016). Here, we define the enzyme kinetics of the SPT produced by P. gingivalis and describe a working model where SLs regulate gene expression via ECF sigma factors. Furthermore, we show a hyperinflammatory response of macrophage-like cells when cultured with the SL null mutant. Interestingly, we also discovered that P. gingivalis can deliver its SLs to host cells in a contact-independent manner. Overall, our studies exemplify the integral importance of SLs in the physiology of P. gingivalis and provide new evidence supporting the concept that, like other members of the Bacteroidetes, synthesis of SLs by P. gingivalis is likely central to its ability to manipulate the host inflammatory response.
Methods
See Appendix for details.
Purification and Characterization of SPT Enzyme
The PG1780 gene (strain W83) was cloned into expression plasmids that contained a C-terminal stop codon in lieu of a tag (PgSPT), a C-terminal 10-histidine tag (pEBSRCTEVC10HIS), or an N-terminal 6-histidine tag (pEHISTEV). Constructs were transformed into Escherichia coli BL21 (DE3)–competent cells. PgSPT was purified either by nickel affinity column chromatography (His-tagged) or by HiTrap anion exchange chromatography (nontagged), followed by gel filtration chromatography. Purification was monitored by SDS-PAGE and size characterized by LC-ESI-MS (liquid chromatography electrospray ionization mass spectrometry). Dissociation constants (Kd) were determined by ultraviolet-visible absorbance spectrophotometry. Kinetic experiments were performed with a 5,5′-dithiobis-2-nitrobenzoic acid (DTNB) assay, and resultant products were measured with MALDI-TOF-MS (matrix assisted laser desorption ionization-time of flight mass spectrometry).
RNA-Seq Analysis
P. gingivalis strain W83 was used in this study with the matching SPT mutant (W83 ΔPG1780), which was generated and characterized previously (Moye et al. 2016). RNA was extracted from cells grown anaerobically in TSBHK to an OD600 of 1.0; then, the quality was assessed, and sequencing was performed and analyzed as previously described (Moradali et al. 2019; Moye et al. 2019).
Host Cell Cytokine/Chemokine Profiling
The human cell line THP-1 was maintained in RPMI-1640 + 10% FBS and differentiated for 48 h into macrophage-like cells with 100nM phorbol 12-myristate 13-acetate (PMA), and 5 × 105 cells were seeded into 24-well tissue culture plates. Parent or ΔSPT mutant P. gingivalis (cultured as stated earlier) was added to THP-1 cells (multiplicity of infection, 100). Following 2 h, 6 h, and 24 h of incubation, cell culture supernatant fluids were collected, and cytokine and chemokine levels were determined by Milliplex Multiplex Assays with a Luminex 200 system. THP-1 cell viability was assessed by MTT assay.
P. gingivalis SL Labeling and Tracking
SL labeling was performed as previously described for Bacteroides thetaiotaomicron (Johnson et al. 2019) with slight modifications. Briefly, P. gingivalis strains were cultured for 24 h in rich medium and transferred into chemically defined medium (Vermilyea et al. 2019) supplemented with chemically modified palmitic acid containing an alkyne (PAA). PMA-differentiated THP-1 cells were placed in the bottom wells of 24-well plates containing sterile glass coverslips. Sterile 0.4-µm-pore Transwell inserts were placed into the wells of the cell culture dishes, and 1 × 109 bacteria were then placed in the upper chamber. After 24 h of culture at 37 °C + 5% CO2, coverslips were removed, washed, and then click labeled with an azide 488 fluorophore per manufacturer specifications. Coverslips were mounted onto slides with a DAPI-containing medium and imaged by fluorescence microscopy. W83 parent and SPT mutant + PAA were click labeled directly to validate PAA incorporation and labeling only in the parent.
Results
SPT Sequence Comparisons
All bacterial SPTs are members of the pyridoxal-5′phosphate (PLP)–dependent α-oxoamine synthase family, catalyzing Claisen-like condensation reactions between acyl-CoA substrates and amino acid to form different α-oxoamine products (Harrison et al. 2018). In the case of SPT, this would be ketodihydrosphingosine (KDS). The average amino acid sequence similarity across the α-oxoamine synthase enzymes is ~30% to 35%, depending on different functions (see Appendix Table 1 and Appendix Fig. 1A). The amino acid sequence alignment among Sphingomonas paucimobilis SPT (SpSPT, Q93UV0; Yard et al. 2007), B. fragilis SPT (BfSPT, Q5LCK4), and P. gingivalis SPT (PgSPT, W1R7E5) shows high sequence homology, with conservation of key residues involved in PLP binding and catalysis. Moreover, gut human microbial BfSPT shares the highest amino acid sequence identity (76%) with PgSPT.
Expression and Purification of Recombinant PgSPT
Recombinant PgSPT was prepared in a manner similar to that described for SpSPT (Yard et al. 2007; Raman et al. 2009). Briefly, the PgSPT gene (PG1780 from strain W83) was cloned and expressed in E. coli from plasmid pET-28a/PgSPT with a 6His-affinity tag at the C-terminus. A combined HisTrap column and size-exclusive chromatography (Sephadex HR S200; GE Healthcare) approach was used to isolate the dimeric PLP-bound holoform of the enzyme, and 10% glycerol was added to avoid PgSPT precipitation. The purity of the protein was assessed by SDS-PAGE (Appendix Fig. 1B).
Spectroscopic Properties of C′ Terminal-Tagged PgSPT
The ultraviolet-visible spectrum of a PLP-dependent enzyme such as SPT usually shows 2 absorption maxima at 335 nm and 425 nm, due to the properties of the 2 forms of the internal aldimine PLP Schiff base: enolimine and ketoenamine. In contrast to the SpSPT enzyme, the ultraviolet-visible spectrum of PgSPT displays an absorbance maximum at 425 nm, suggesting that the PLP cofactor was present predominantly as the ketoenamine form (Fig. 1A). By analyzing the change in the absorbance at 425 nm with varying changes in the concentration of L-serine, the dissociation constant (KdSer) was determined to be 5.46 ± .60 mM (Fig. 1B). This value is approximately 5 times weaker than that determined for SpSPT (KdSer = 1.1 mM; Raman et al. 2009).
Figure 1.
Characterization of recombinant Porphyromonas gingivalis serine palmitoyl transferase (SPT). (A) Absorption ultraviolet-visible spectrum of PLP-dependent (pyridoxal-5′phosphate) P. gingivalis SPT. Upon addition of L-serine, the enzyme (20 μM) converts from the internal aldimine to the external aldimine form, performed in 20mM potassium phosphate, 250mM NaCl, pH 7.5, at 25 °C. Solid line (0mM L-serine) or dashed lines in the presence of 0.1mM to 100mM L-serine. (B) Analysis of L-serine binding to C-terminal PgSPT by monitoring the change in absorbance at 425 nm. (C) Michaelis-Menten kinetic analysis of SPT with substrates L-serine (0.1 to 100 mM) and palmitoyl-CoA (250 μM) with 1μM enzyme, 100mM HEPES, pH 7.0, 250mM NaCl, and 0.2mM DTNB and measured spectrophotometrically at 412 nm. (D) The concentration of L-serine (20 mM) with different palmitoyl-CoA concentrations (1 to 1000 μM). All data are plotted as mean readings ± 2-SD error bars.
C′ Terminal PgSPT Activity and Kinetics
To find the optimal conditions for PgSPT activity, the enzyme was initially tested in buffers of different pH, and the highest reaction rate was observed in 100mM HEPES at pH 7.0. Here we used a convenient coupled assay that uses DTNB reagent that reacts with the CoASH product. The resulting TNB thiolate anion absorbs strongly at 412 nm (εmax = 14,150 M-1cm-1; Raman et al. 2009). The enzyme was analyzed with both substrates, L-serine and palmitoyl-CoA, to obtain the kinetic parameters, and the Michaelis-Menten plot for C′ terminal His-tagged PgSPT (Fig. 1C, D) showed that the enzyme bound L-serine and palmitoyl-CoA, with Km values of 0.52 ± .06 mM and 84 ± 11.7 μM, respectively. The enzyme turned over with a kcat of 43.5 ± 0.4 × 10–3 s−1 and an efficiency (kcat/Km) of 84.6 M−1s−1 for L-serine and 524 M−1s−1 for pimeloyl-CoA. This compares to similar values determined for SpSPT with respect to substrate binding but with the PgSPT turning over much slower.
Identification of KDS Formation by PgSPT
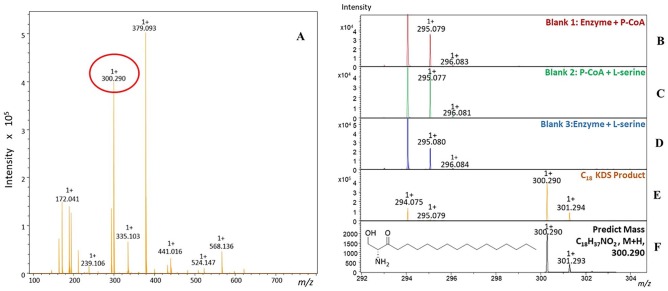
Since the kinetic assay is indirect and measures only CoASH release, we confirmed that PgSPT catalyzed conversion of L-serine and palmitoyl-CoA to the product KDS. For this, we used MALDI-TOF MS analysis of the PgSPT assay to detect the formation of the molecular ion related to the product C18:0 KDS [C18H37NO2, M+H]+ (m/z 300.290; Fig. 2A). A series of controls (Fig. 2B–D) confirmed that the KDS was formed only in the presence of the enzyme and both substrates.
Figure 2.
MALDI-ToF (matrix assisted laser desorption ionization-time of flight) mass spectra analysis of the PgSPT (Porphyromonas gingivalis serine palmitoyl transferase) reaction between L-serine and palmitoyl-CoA. Each assay, which contained 1μM enzyme, 100mM HEPES, pH 7.0, 250mM NaCl, 0.2mM 5,5′-dithiobis-2-nitrobenzoic acid, and 20mM L-serine or 250μM palmitoyl-CoA, was added dependent on samples. All reaction samples were eluted with 100% acetonitrile (ACN) by C4 zip-tip and mixed with α-Cyano-4-hydroxycinnamic acid matrix dissolved in 50% ACN within 0.25% trifluoroacetic acid. The spectrum was analyzed on positive ion mode in triplicates. (A) Observation of the product ketodihydrosphingosine (KDS) with m/z = 300.290 during a sweep of masses (m/z = 100 to 800 amu). (B–D) Negative controls. (E) Full assay with PgSPT, L-serine, and P-CoA with a mass range of m/z = 292 to 304. (F) Theoretical mass spectrum based on the KDS formula (M+H)+.
RNA-Seq Analysis
The rigid structural characteristic of SLs serves an important functional role in eukaryotic cells by condensing around signaling proteins in the cell membrane and forming densely packed regions of the membrane known as lipid rafts. These puncta of closely associated lipids and proteins are thought to increase the efficiency of cellular signaling pathways by bringing signaling proteins into close proximity. This information led us to hypothesize that a SL-null mutant may possess a defect in gene expression. Transcriptomic analysis of the SL-null strain in comparison with the parent strain identified 120 genes that were differentially expressed (≥2-fold, q value <0.01). Of the 120 genes, the expression of 61 genes was lower while expression of 59 genes was higher. Most notably, 3 extracytoplasmic function (ECF) sigma factors were found to be differentially expressed: 1 gene (PG0985) was 3.2-fold lower while the other 2 (PG0162 and PG0214) were expressed 2.2- and 6.1-fold higher, respectively. In addition, the data show that all of the genes harbored in 2 distinct loci encoding CRISPR-associated genes (PG1981-PG1989 and PG2013-PG2020) were lower, while genes encoding type IX secretion structural and cargo proteins were among the most overexpressed. As expected, numerous genes encoding hypothetical proteins were differentially expressed (23 reduced and 21 overexpressed; Tables 1 and 2).
Table 1.
Genes Expressed at Lower Levels in the SPT Mutant versus the Parent Strain W83.
| Name | Gene ID | Product | Q Value | Fold Change |
|---|---|---|---|---|
| SPT | PG1780 | Serine palmitoyltransferase | 0 | 0.01 |
| Proteolysis and amino acid metabolism | ||||
| pepD-2 | PG0537 | Aminoacyl-histidine dipeptidase | 1.30E-216 | 0.17 |
| pruA | PG1269 | Delta-1-pyrroline-5-carboxylate dehydrogenase | 2.01E-35 | 0.38 |
| — | PG1270 | PLP-dependent aminotransferase | 3.38E-35 | 0.37 |
| — | PG1271 | Acetylornithine aminotransferase | 2.92E-13 | 0.38 |
| Transposon | ||||
| — | PG0549 | ISPg1, transposase | 4.72E-19 | 0.45 |
| — | PG0872 | Mobilizable transposon, Xis protein | 1.20E-09 | 0.50 |
| — | PG1480 | Conjugative transposon protein TraI | 1.68E-07 | 0.50 |
| — | PG1482 | Conjugative transposon protein TraF | 1.62E-13 | 0.33 |
| — | PG1483 | Conjugative transposon protein TraE | 6.11E-11 | 0.50 |
| Hypothetical | ||||
| — | PG0354 | Hypothetical protein | 1.26E-07 | 0.50 |
| — | PG0554 | Hypothetical protein | 7.25E-13 | 0.46 |
| — | PG0609 | Hypothetical protein | 6.13E-15 | 0.50 |
| — | PG0617 | Hypothetical protein | 7.14E-14 | 0.41 |
| — | PG0727 | Hypothetical protein | 7.89E-57 | 0.28 |
| — | PG0835 | Hypothetical protein | 2.58E-26 | 0.33 |
| — | PG0914 | Hypothetical protein | 1.54E-29 | 0.40 |
| — | PG0986 | Hypothetical protein | 4.38E-24 | 0.37 |
| — | PG0987 | Hypothetical protein | 1.42E-125 | 0.21 |
| — | PG1229 | Hypothetical protein | 1.68E-09 | 0.50 |
| — | PG1268 | Hypothetical protein | 1.54E-53 | 0.32 |
| — | PG1494 | Hypothetical protein | 5.30E-05 | 0.50 |
| — | PG1508 | Hypothetical protein | 0.003359 | 0.35 |
| — | PG1510 | Hypothetical protein | 1.57E-21 | 0.40 |
| — | PG1511 | Hypothetical protein | 2.39E-21 | 0.37 |
| — | PG1512 | Hypothetical protein | 2.21E-18 | 0.36 |
| — | PG1516 | Hypothetical protein | 2.53E-06 | 0.48 |
| — | PG1547 | Hypothetical protein | 7.02E-06 | 0.50 |
| — | PG1549 | Hypothetical protein | 4.25E-17 | 0.33 |
| — | PG1795 | Hypothetical protein | 0.005 | 0.38 |
| — | PG1798 | Hypothetical protein | 1.79E-12 | 0.44 |
| — | PG1871 | Hypothetical protein | 7.38E-05 | 0.33 |
| — | PG1908 | Hypothetical protein | 2.23E-04 | 0.44 |
| CRISPR loci | ||||
| cas2-1 | PG1981 | CRISPR-associated Cas2 family protein | 2.67E-11 | 0.42 |
| — | PG1982 | CRISPR-associated Cas1 family protein | 2.27E-12 | 0.44 |
| — | PG1983 | CRISPR-associated Cmr5 family protein | 2.25E-06 | 0.50 |
| — | PG1984 | Hypothetical protein | 1.27E-19 | 0.33 |
| — | PG1985 | CRISPR-associated Cmr4 family protein | 1.13E-20 | 0.42 |
| — | PG1986 | CRISPR-associated Cmr3 family protein | 2.10E-32 | 0.35 |
| — | PG1987 | CRISPR-associated Csm1 family protein | 4.80E-30 | 0.29 |
| — | PG1988 | Hypothetical protein | 2.31E-40 | 0.24 |
| — | PG1989 | Hypothetical protein | 6.29E-65 | 0.27 |
| cas2-2 | PG2013 | CRISPR-associated Cas2 family protein | 2.73E-12 | 0.47 |
| cas1 | PG2014 | CRISPR-associated Cas1 family protein | 5.31E-31 | 0.39 |
| cas4 | PG2015 | CRISPR-associated Cas4 family protein | 1.92E-26 | 0.40 |
| cas3 | PG2016 | CRISPR-associated helicase Cas3 | 5.50E-09 | 0.33 |
| — | PG2017 | Hypothetical protein | 4.44E-14 | 0.33 |
| — | PG2018 | Hypothetical protein | 3.10E-11 | 0.33 |
| — | PG2019 | Hypothetical protein | 6.56E-22 | 0.31 |
| PG2020 | CRISPR-associated Cas5e family protein | ? | 0.39 | |
| Redox homeostasis | ||||
| — | PG0616 | Thioredoxin | 0.003 | 0.38 |
| Cell wall | ||||
| — | PG0726 | Putative lipoprotein, s-layer | 8.42E-08 | 0.25 |
| Transcription | ||||
| — | PG0985 | ECF subfamily RNA polymerase sigma factor | 3.41E-68 | 0.29 |
| — | PG1535 | Transcriptional regulator | 1.88E-10 | 0.50 |
| Metabolism | ||||
| hprA | PG1190 | Glycerate dehydrogenase | 8.69E-12 | 0.49 |
| — | PG1504 | NAD dependent protein | 0.009 | 0.33 |
| — | PG1509 | HAD superfamily hydrolase | 3.63E-24 | 0.34 |
| — | PG1514 | Glycerol dehydrogenase | 3.16E-11 | 0.44 |
| — | PG1515 | Ribulose bisphosphate carboxylase-like protein | 1.01E-14 | 0.45 |
| Biosynthesis of cofactors | ||||
| — | PG1505 | Radical SAM domain-containing protein | 3.28E-15 | 0.32 |
SPT, serine palmitoyl transferase.
Table 2.
Genes Expressed at Higher Levels in the SPT Mutant versus the Parent Strain W83.
| Name | Gene ID | Product | Q Value | Fold Change |
|---|---|---|---|---|
| Type IX secretion system | ||||
| — | PG0027 | Hypothetical protein | 1.81E-28 | 2.83 |
| porP | PG0287 | Hypothetical protein porP | 3.33E-56 | 2.58 |
| porK | PG0288 | Putative lipoprotein porK | 1.24E-32 | 2.65 |
| porL | PG0289 | Hypothetical protein porL | 1.14E-40 | 2.56 |
| porM | PG0290 | Hypothetical protein porM | 8.78E-20 | 2.21 |
| porN | PG0291 | Hypothetical protein porN | 2.53E-29 | 2.58 |
| porT | PG0751 | porT protein | 8.54E-30 | 2.00 |
| sov | PG0809 | Hypothetical protein | 3.61E-10 | 2.10 |
| PG0810 | Hypothetical protein | 4.20E-55 | 2.60 | |
| tpr | PG1055 | Thiol protease | 0 | 8.00 |
| — | PG1947 | Hypothetical protein | 1.48E-17 | 2.00 |
| TapA | PG2100 | TapA | 0 | 10.88 |
| TapB | PG2101 | TapB | 0 | 20.33 |
| TapC | PG2102 | TapC | 0 | 20.33 |
| Hypothetical and other | ||||
| ispF | PG0028 | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | 1.19E-28 | 2.20 |
| — | PG0161 | Hypothetical protein | 1.58E-134 | 3.34 |
| — | PG0216 | Hypothetical protein | 1.49E-182 | 4.80 |
| — | PG0217 | Hypothetical protein | 1.57E-298 | 4.69 |
| — | PG0218 | Hypothetical protein | 0 | 5.00 |
| — | PG0241 | Putative lipoprotein | 2.60E-05 | 2.07 |
| — | PG0297 | Hypothetical protein | 1.52E-18 | 2.00 |
| — | PG0323 | Hypothetical protein | 2.96E-36 | 2.31 |
| — | PG0419 | Hypothetical protein | 7.47E-20 | 2.27 |
| — | PG0606 | Hypothetical protein | 2.13E-29 | 2.25 |
| — | PG0607 | Hypothetical protein | 3.11E-13 | 2.44 |
| clpB | PG1118 | clpB protein | 9.12E-23 | 2.28 |
| — | PG1374 | Hypothetical protein | 6.87E-16 | 2.51 |
| — | PG1527 | Hypothetical protein | 1.39E-19 | 2.00 |
| — | PG1571 | Metallo-beta-lactamase superfamily protein | 5.44E-10 | 2.00 |
| — | PG1625 | Hypothetical protein | 1.62E-11 | 2.17 |
| — | PG1626 | Hypothetical protein | 3.75E-17 | 2.32 |
| — | PG1634 | Hypothetical protein | 3.84E-20 | 2.16 |
| — | PG1662 | Hypothetical protein | 7.73E-27 | 2.12 |
| — | PG1682 | Glycosyl transferase | 8.04E-54 | 2.44 |
| — | PG1683 | Hypothetical protein | 2.03E-26 | 2.10 |
| — | PG1684 | Hypothetical protein | 5.65E-42 | 3.20 |
| udk | PG1781 | Uridine kinase | 2.11E-30 | 2.30 |
| — | PG1835 | Putative lipoprotein | 3.05E-10 | 2.12 |
| aroA | PG1944 | 3-phosphoshikimate 1-carboxyvinyltransferase | 1.40E-20 | 2.04 |
| — | PG1945 | Hypothetical protein | 3.76E-34 | 2.31 |
| — | PG1967 | Hypothetical protein | 2.07E-34 | 2.33 |
| — | PG2103 | Hypothetical protein | 1.68E-37 | 2.30 |
| Transport | ||||
| — | PG0064 | CzcA family heavy metal efflux protein | 3.76E-42 | 2.29 |
| — | PG0280 | ABC transporter permease | 1.97E-11 | 2.00 |
| — | PG0281 | ABC transporter permease | 2.05E-14 | 2.00 |
| — | PG0282 | ABC transporter ATP-binding protein | 4.46E-13 | 3.00 |
| — | PG0680 | RND family efflux transporter MFP subunit | 3.77E-07 | 2.00 |
| — | PG1010 | ABC transporter ATP-binding protein | 5.74E-33 | 2.00 |
| — | PG1117 | MATE efflux family protein | 1.10E-07 | 2.00 |
| — | PG1176 | ABC transporter ATP-binding protein | 0.001661967 | 2.00 |
| — | PG1663 | ABC transporter ATP-binding protein | 6.34E-34 | 2.05 |
| — | PG1664 | ABC transporter permease | 2.07E-33 | 2.11 |
| — | PG1665 | ABC transporter permease | 3.06E-24 | 2.00 |
| — | PG1946 | ABC transporter | 1.56E-25 | 2.25 |
| Transcription | ||||
| — | PG0162 | ECF subfamily RNA polymerase sigma factor | 4.84E-10 | 2.10 |
| — | PG0214 | ECF subfamily RNA polymerase sigma factor | 0 | 5.87 |
| — | PG0215 | Putative anti-sigma factor | 1.25E-199 | 4.46 |
| — | PG1007 | GntR family transcriptional regulator | 9.11E-24 | 2.10 |
SPT, serine palmitoyl transferase.
Synthesis of SLs by P. gingivalis Limits the Host Capacity to Mount a Robust Proinflammatory Response
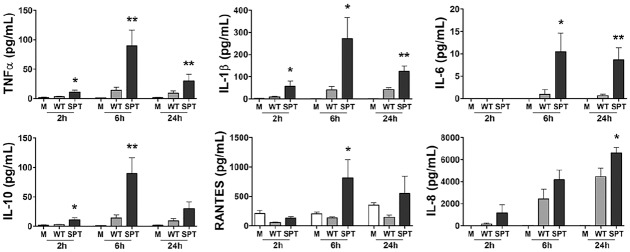
To examine the contribution of SL synthesis to the host inflammatory response, we cultured macrophage-like THP-1 cells with P. gingivalis wild type (WT) and the corresponding SPT mutant for up to 24 h and measured cytokine and chemokine levels. THP-1 is a transformed cell line of human origin. It is a frequently used model cell for investigating macrophage function, a cell that is central to periodontal disease. Our data show that THP-1 cells cultured with the SPT mutant produced a robust immune response that was not observed from cells cultured with the WT (Fig. 3). Even as rapidly as 2 h after initiation of coculture, significant increases in the levels of TNF-α, IL-1β, and IL-10 were measured from the cultures infected with the SL-null mutant as compared with those elicited by parent W83 (P < 0.05 for all by t test). By 6 h, the signature of elevated inflammation initiated by the SPT mutant accelerated, with the addition of a significant increase in IL-6 and RANTES also observed (Fig. 3). The trend of lower cytokine and chemokine production in response to the WT remained evident at 24 h of coculture but trended lower than observed at 6 h. No significant differences in THP-1 cell viability was observed between cells cultured with SPT mutant and WT per the MTT assay (P > 0.05 by analysis of variance; Appendix Table 2). These findings support our hypothesis that in the context of live bacteria, synthesis of SLs limits and/or suppresses the host capacity to mount a robust proinflammatory response to this organism.
Figure 3.
The inability of Porphyromonas gingivalis to synthesize sphingolipids (SLs) leads to an enhanced cytokine and chemokine response. PMA-treated human macrophage–like THP-1 cells were directly cultured with P. gingivalis W83 (wild type [WT]; gray bars) or the P. gingivalis W83 SL-null mutant (serine palmitoyl transferase [SPT]; black bars) at a multiplicity of infection of 100. Cell culture supernatant fluids were collected at 2, 6, and 24 h of coculture, and the levels of TNFα, IL-1β, IL-6, IL-10, RANTES, and IL-8 were measured by multiplex immunoassay. Medium alone (M; white bars) served as unchallenged control. Data are presented as mean ± SEM (n = 8 independent experiments). *P < 0.05 and **P < 0.01 versus wild type (WT) P. gingivalis with unpaired t tests. PMA, phorbol 12-myristate 13-acetate.
Transfer of SLs from P. gingivalis to THP-1 Cells in a Transwell System
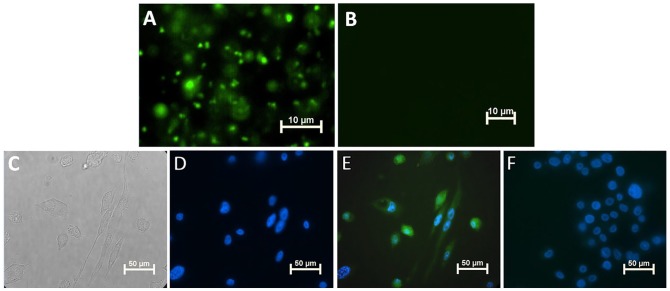
Last, we assessed whether SLs could be transferred from P. gingivalis to THP-1 macrophages. By employing a 0.4-µm-pore Transwell system, metabolically labeled P. gingivalis (grown in the presence of PAA to allow for specific click labeling of SLs with a fluorophore) were placed in the upper well of the Transwell, with THP-1 cells placed in the lower well. After 24 h of Transwell coculture, click chemistry verified that P. gingivalis SLs were transferred to THP-1 cells without physical contact (Fig. 4).
Figure 4.
Sphingolipids transfer from Porphyromonas gingivalis to THP-1 cells in a Transwell system. (A) Epifluorescent image of wild-type W83 bacterial cells shows detection of palmitic acid alkyne (PAA) when bacteria were grown with addition of PAA (green–azide Fluor 488) by click chemistry. (B) As expected, the serine palmitoyl transferase–null mutant did not incorporate PAA. (C) Bright field image of THP-1 cells on cover slip in the lower well of a Transwell system after 24-h coculture with strain W83. (D) Epifluorescent image of the same THP-1 cells shows DAPI (blue) staining of nucleus, and (E) THP-1 cells incorporated the P. gingivalis alkyne-tagged sphingolipids (green) that were transferred from W83 constrained to the upper well of the Transwell system. (F) Click labeling of THP-1 cells cocultured with W83 grown in medium without PAA via the Transwell system; no green–azide Fluor 488 was detected.
Discussion
P. gingivalis can be present in subgingival plaque even during periodontal health (Griffen et al. 1998), suggesting that the host does not always respond to this bacterium as a pathogen. While other members of the phylum Bacteroidetes, in particular members of the genus Bacteroides, are viewed as symbiotic or pathobionts, this framework of a symbiotic relationship with the host is not typically applied to P. gingivalis. Our view of P. gingivalis as a pathobiont (Cugini et al. 2013) led us to consider its unusual ability to synthesize lipids almost identical to its host as a strategy to evade host immune activation.
To evaluate function, gene PG1780, encoding a predicted SPT, was cloned, and the recombinant protein was isolated, characterized, and confirmed as an SPT (PgSPT) by determining the kinetics of the reaction with the canonical substrates L-serine and palmitoyl-CoA. Formation of the KDS product was confirmed by MALDI-TOF-MS analysis. This allowed a comparison with another well-characterized bacterial SpSPT from S. paucimobilis (Harrison et al. 2019). The PgSPT bound both substrates with a similar affinity to SpSPT, but in contrast to this isoform, PgSPT displayed much slower kinetics. The molecular details of these differences may be revealed by a comparative x-ray structural analysis, and to that end, crystal trials of PgSPT enzyme are underway. Once the protein structure is known, a comparative evolutionary study of the microbial SPTs will be carried out to explore the species-specific features of the bacterial and eukaryotic SPTs (Harrison et al. 2018; Heaver et al. 2018).
Lipid microdomains are known to position proteins associated with signal transduction, membrane trafficking (protein secretion systems), and regulation of metabolism (protease complexes) in close proximity (Bramkamp and Lopez 2015; Lopez 2015). It is tempting to speculate that a subset of the SLs may support T9SS machinery, given their known function in protein secretion systems. Furthermore, our RNA-Seq analysis indicates that SLs may indeed stabilize certain proteins involved in signal transduction, in particular sequestration of antisigma factors. Antisigma factors are known to be localized to the inner membrane, where they bind their target ECF sigma factors, preventing transcription. Our working model is that, when SLs are not produced, the targets are overexpressed because the ECF sigma factors are free to interact with target promoters. Some T9SS genes have been shown to be regulated via ECF sigma factors, and we identified 14 T9SS genes that are expressed at higher levels in the SPT mutant, some as much as 20-fold. Importantly, the genes encoding gingipains were not differentially expressed in the mutant. Our prior studies showed that the SL-null mutant actually demonstrated elevated secreted gingipain activity, not less, suggesting that the higher levels of cytokines are not due to a lack of gingipain activity. That being said, since these proteases are proficient at degrading cytokines, studies are ongoing to further evaluate a link between SL synthesis and secreted gingipain activity.
Our cell infection modeling shows that SL synthesis leads to a reduced inflammatory response, suggesting that synthesis supports homeostasis. This discovery in some ways contradicts published results. Prior studies with purified P. gingivalis SLs point to TLR-2-inducing activity (Nichols et al. 2009), stimulation of cellular inflammatory responses (Nichols et al. 2001), and driving of apoptosis (Zahlten et al. 2007). Yet, our findings parallel studies on SL function in other members of the Bacteroidetes phylum that strongly support a role for SLs in immune suppression (An et al. 2011; An et al. 2014; Heaver et al. 2018; Brown et al. 2019). Specifically, one study reported an inverse relationship between SL synthesis by Bacteroides and inflammatory bowel disease, indicating that bacterial SLs can serve as key factors that mechanistically promote intestinal homeostasis (Brown et al. 2019). As gingival tissues from periodontally healthy and diseased individuals contain SLs and yet the SL types are distinct, our working model has been that, not only does SL synthesis play a central role in membrane trafficking in P. gingivalis, but the secreted SLs may also directly influence host cell function. Our in vitro findings agree with clinical findings that P. gingivalis releases and/or secretes its SLs; moreover, our findings support that P. gingivalis SLs are transferred to host cells. This later discovery is particularly compelling, as transfer of SLs from bacteria to host suggests an intriguing interplay, which may serve an important role by which host and microbe interact and which in turn may control oral inflammation, as shown for B. thetaiotaomicron in the gut (Johnson et al. 2019). Last, our results show that the absence of SLs elicited high levels of proinflammatory cytokines, as well as IL-10, a highly expressed anti-inflammatory cytokine. Our findings of the presence of pro- and anti-inflammatory cytokines occurring concurrently is not fully understood; however, these results are consistent with clinical profiles observed in inflamed periodontal tissues. The ultimate outcome of this unusual inflammatory pattern requires further evaluation.
In summary, P. gingivalis is often described as a master manipulator of the immune response (Hajishengallis and Lamont 2014), primarily due to its ability to degrade immunoglobulins, complement, and cytokines via its repertoire of secreted proteases (Hajishengallis and Lambris 2012). We posit that SL synthesis is another mechanism of control. Future studies testing these findings in the context of periodontal disease may identify novel approaches to control SL production by P. gingivalis and thus shift the balance of inflammation elicited by the subgingival biofilm to a more homeostatic state.
Author Contributions
F.G. Rocha, Z.D. Moye, F.C. Gibson III, M.E. Davey, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; G. Ottenberg, contributed to data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; P. Tang, D.J. Campopiano, contributed to data acquisition, analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034520908784 for Porphyromonas gingivalis Sphingolipid Synthesis Limits the Host Inflammatory Response by F.G. Rocha, Z.D. Moye, G. Ottenberg, P. Tang, D.J. Campopiano, F.C. Gibson and M.E. Davey in Journal of Dental Research
Acknowledgments
The authors thank the School of Chemistry, University of Edinburgh, and the Edinburgh Global Research Scholarship for PhD studentship funding (P.T.). The authors also thank Dr. Peter Harrison, Dr. Bohdan Mykhaylyk, Dr. Jo Simpson, and Dr. Van Kelly for helpful discussion on SPT, as well as all members of the Davey laboratory for many helpful discussions on SLs.
Footnotes
A supplemental appendix to this article is available online.
The research was supported by the National Institute of Craniofacial Research (R01DE019117 and R01DE24580 to M.E.D.; T90 DE021990 to Z.D.M. and F.G.R.; R90 DE22530 to F.G.R.) as well as start-up funds (to F.C.G. and M.E.D.).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- An D, Na C, Bielawski J, Hannun YA, Kasper DL. 2011. Membrane sphingolipids as essential molecular signals for Bacteroides survival in the intestine. Proc Natl Acad Sci U S A. 108 Suppl 1:4666–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. 2014. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 156(1–2):123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramkamp M, Lopez D. 2015. Exploring the existence of lipid rafts in bacteria. Microbiol Mol Biol Rev. 79(1):81–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EM, Ke X, Hitchcock D, Jeanfavre S, Avila-Pacheco J, Nakata T, Arthur TD, Fornelos N, Heim C, Franzosa EA, et al. 2019. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe. 25(5):668–680e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SJ, Dashper SG, Darby IB, Adams GG, Hoffmann B, Reynolds EC. 2009. Progression of chronic periodontitis can be predicted by the levels of Porphyromonas gingivalis and Treponema denticola in subgingival plaque. Oral Microbiol Immunol. 24(6):469–477. [DOI] [PubMed] [Google Scholar]
- Cugini C, Klepac-Ceraj V, Rackaityte E, Riggs JE, Davey ME. 2013. Porphyromonas gingivalis: keeping the pathos out of the biont. J Oral Microbiol. 5:19804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 8(7):481–490. [DOI] [PubMed] [Google Scholar]
- Dunn TM, Tifft CJ, Proia RL. 2019. A perilous path: the inborn errors of sphingolipid metabolism. J Lipid Res. 60(3):475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen AL, Becker MR, Lyons SR, Moeschberger ML, Leys EJ. 1998. Prevalence of Porphyromonas gingivalis and periodontal health status.J Clin Microbiol. 36(11):3239–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lambris JD. 2012. Complement and dysbiosis in periodontal disease. Immunobiology. 217(11):1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. 2014. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol. 44(2):328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 9(2):139–150. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. 2018. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol. 19(3):175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Dunn TM, Campopiano DJ. 2018. Sphingolipid biosynthesis in man and microbes. Nat Prod Rep. 35(9):921–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Gable K, Somashekarappa N, Kelly V, Clarke DJ, Naismith JH, Dunn TM, Campopiano DJ. 2019. Use of isotopically labeled substrates reveals kinetic differences between human and bacterial serine palmitoyltransferase. J Lipid Res. 60(5):953–962. Erratum in: J Lipid Res. 2019;60(8):1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaver SL, Johnson EL, Ley RE. 2018. Sphingolipids in host-microbial interactions. Curr Opin Microbiol. 43:92–99. [DOI] [PubMed] [Google Scholar]
- Johnson EL, Heaver SL, Waters JL, Kim BI, Bretin A, Goodman AL, Gewirtz AT, Worgall TS, Ley RE. 2019. Sphingolipid production by gut Bacteroidetes regulates glucose homeostasis. bioRxiv. doi: 10.1101/632877 [DOI] [Google Scholar]
- Kanzaki H, Movila A, Kayal R, Napimoga MH, Egashira K, Dewhirst F, Sasaki H, Howait M, Al-Dharrab A, Mira A, et al. 2017. Phosphoglycerol dihydroceramide, a distinctive ceramide produced by Porphyromonas gingivalis, promotes RANKL-induced osteoclastogenesis by acting on non-muscle myosin II-A (Myh9), an osteoclast cell fusion regulatory factor. Biochim Biophys Acta Mol Cell Biol Lipids. 1862(5):452–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 62(4):1244–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D. 2015. Molecular composition of functional microdomains in bacterial membranes. Chem Phys Lipids. 192:3–11. [DOI] [PubMed] [Google Scholar]
- Maceyka M, Spiegel S. 2014. Sphingolipid metabolites in inflammatory disease. Nature. 510(7503):58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill AH, Jr, Carman GM. 2015. Introduction to thematic minireview series: novel bioactive sphingolipids. J Biol Chem. 290(25):15362–15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradali MF, Ghods S, Angelini TE, Davey ME. 2019. Amino acids as wetting agents: surface translocation by Porphyromonas gingivalis. ISME J. 13(6):1560–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye ZD, Gormley CM, Davey ME. 2019. Galactose impacts the size and intracellular composition of the asaccharolytic oral pathobiont Porphyromonas gingivalis. Appl Environ Microbiol. 85(4):e02268-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moye ZD, Valiuskyte K, Dewhirst FE, Nichols FC, Davey ME. 2016. Synthesis of sphingolipids impacts survival of Porphyromonas gingivalis and the presentation of surface polysaccharides. Front Microbiol. 7:1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols FC. 1998. Novel ceramides recovered from Porphyromonas gingivalis: relationship to adult periodontitis. J Lipid Res. 39(12):2360–2372. [PubMed] [Google Scholar]
- Nichols FC, Housley WJ, O’Conor CA, Manning T, Wu S, Clark RB. 2009. Unique lipids from a common human bacterium represent a new class of toll-like receptor 2 ligands capable of enhancing autoimmunity. Am J Pathol. 175(6):2430–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols FC, Levinbook H, Shnaydman M, Goldschmidt J. 2001. Prostaglandin e2 secretion from gingival fibroblasts treated with interleukin-1beta: effects of lipid extracts from Porphyromonas gingivalis or calculus. J Periodontal Res. 36(3):142–152. [DOI] [PubMed] [Google Scholar]
- Nichols FC, Riep B, Mun J, Morton MD, Bojarski MT, Dewhirst FE, Smith MB. 2004. Structures and biological activity of phosphorylated dihydroceramides of Porphyromonas gingivalis. J Lipid Res. 45(12):2317–2330. [DOI] [PubMed] [Google Scholar]
- Nichols FC, Rojanasomsith K. 2006. Porphyromonas gingivalis lipids and diseased dental tissues. Oral Microbiol Immunol. 21(2):84–92. [DOI] [PubMed] [Google Scholar]
- Nichols FC, Yao X, Bajrami B, Downes J, Finegold SM, Knee E, Gallagher JJ, Housley WJ, Clark RB. 2011. Phosphorylated dihydroceramides from common human bacteria are recovered in human tissues. PLoS One. 6(2):e16771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen I, Jantzen E. 2001. Sphingolipids in bacteria and fungi. Anaerobe. 7(2):103–112. [Google Scholar]
- Olsen I, Nichols FC. 2018. Are sphingolipids and serine dipeptide lipids underestimated virulence factors of Porphyromonas gingivalis? Infect Immun. 86(7):e00035-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman MC, Johnson KA, Yard BA, Lowther J, Carter LG, Naismith JH, Campopiano DJ. 2009. The external aldimine form of serine palmitoyltransferase: structural, kinetic, and spectroscopic analysis of the wild-type enzyme and HSAN1 mutant mimics. J Biol Chem. 284(25):17328–17339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. 1998. Microbial complexes in subgingival plaque. J Clin Periodontol. 25(2):134–144. [DOI] [PubMed] [Google Scholar]
- Vermilyea DM, Ottenberg GK, Davey ME. 2019. Citrullination mediated by PPAD constrains biofilm formation in P. gingivalis strain 381. NPJ Biofilms Microbiomes. 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yard BA, Carter LG, Johnson KA, Overton IM, Dorward M, Liu H, McMahon SA, Oke M, Puech D, Barton GJ, et al. 2007. The structure of serine palmitoyltransferase; gateway to sphingolipid biosynthesis. J Mol Biol. 370(5):870–886. [DOI] [PubMed] [Google Scholar]
- Zahlten J, Riep B, Nichols FC, Walter C, Schmeck B, Bernimoulin JP, Hippenstiel S. 2007. Porphyromonas gingivalis dihydroceramides induce apoptosis in endothelial cells. J Dent Res. 86(7):635–640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034520908784 for Porphyromonas gingivalis Sphingolipid Synthesis Limits the Host Inflammatory Response by F.G. Rocha, Z.D. Moye, G. Ottenberg, P. Tang, D.J. Campopiano, F.C. Gibson and M.E. Davey in Journal of Dental Research