Abstract
Background
Gastrointestinal stromal tumors (GIST) are the most common mesenchymal neoplasms in the gastrointestinal (GI) tract. The mutation of C-KIT is considered to be the crucial step in the tumorigenesis. Targeted therapies are being developed focusing these mutations. Various exon mutations of GIST responded in varied patterns to this targeted therapy. This study was carried out to evaluate the C-KIT exon 11, exon 9 and BRAF V600E mutations among GIST specimens.
Methods
This retrospective study was carried out among 20 DNA extracted specimens from paraffin blocks of GIST received in our tertiary teaching institution for a period of three years. DNA sequencing was carried out for mutational analyses on C-KIT exon 9, C-KIT exon 11 and BRAF V600E genes using Sanger sequencing.
Results
Histologically, majority of the tumors had spindle cell morphology. About 19 cases were positive for CD117. The analysis of type of mutations showed that three cases carried Exon 11 and three cases carried Exon 9 mutations. BRAF V600E mutation was seen in one case.
Conclusion
It is essential to conduct molecular studies on GISTs in order to get a clear understanding of the pathogenesis and behavior pattern. This will also help in designing targeted therapies and assessing recurrence. With the advent of rapidly evolving personalized therapy, the evaluation of genetic mutations is essential for diagnosis and prognostic value.
Keywords: c-kitmutations, braf v600e mutations, dna sequencing, gastrointestinal stromal tumors
Introduction
Gastrointestinal stromal tumours (GISTs) are the most common mesenchymal neoplasms in the gastrointestinal (GI) tract. They account for < 1% of all gastrointestinal tumours and 5% of all sarcomas. GISTs were initially thought to be rare. However, because of an increased ability to reliably diagnose them, their incidence is now estimated to be around 5000 new cases per year in the United States. No supportive data is available from India in this regard.
The spectrum of GIST varies based on the clinical presentation, location, histology and prognostic outcomes. Although GIST can occur throughout the GI tract, it is increasingly common in the stomach and small bowel, compared to colon, rectum and esophagus. In certain situations, an extra gastrointestinal site involvement is also encountered (EGIST).
The cellular origin of gastrointestinal stromal tumours is the pluripotent mesenchymal stem cell programmed to differentiate into Interstitial Cells of Cajal (ICC) [1]. GISTs show many morphologic, immunohistochemical and molecular features common with ICC [2].
The clinical importance of GIST is its characteristic molecular feature of gain of function mutations in the c-kit proto-oncogenes. This mutation is considered to be the crucial step in the tumorigenesis of GISTs and are found in the smallest sub-centimeter of GIST [3]. The c-KIT mutations are scattered along hotspots including exon 9, exon 11, exon 13 and exon 17 of the c-kit gene. Approximately 67% of the KIT mutations involve exon 11, while 10% involve exon 9 and 1% each in exon 13 and 17. However, rare KIT mutations involve exon 8, 12, 14 and 18. Further molecular analyses showed molecular changes in Platelet Derived Growth Factor Receptor Alpha (PDGFRA), B-Raf (BRAFV600E), Succinate Dehydrogenase (SDH) subunits A, B, C, D and Neurofibromatosis type 1 (NF1) genes.
Also, 12-25% of the GISTs lack c-KIT or PDGFRA mutations; these have been identified as wild type GIST. BRAFV600E mutations have been identified in these wild type GIST and account for 3% of all GISTs [4]. The mutations in GISTs showed varied clinical manifestations and prognostic outcomes.
The clinical significance of c-KIT mutations was the development of targeted therapies at molecular level. The initial drug discovered for the treatment of GISTs was imatinib mesylate, a tyrosine kinase inhibitor. This drug acts by competing for the ATP binding site on the target kinase, thereby inhibiting tyrosine kinase and reducing cellular proliferation [5]. With further knowledge in the GISTs, many tyrosine kinase inhibitors were discovered and Sunitinib was found to be suitable for patients with C-Kit mutations with exon 9 involvement. This warranted the need for evaluating the KIT mutations at the exon level, so as to achieve targeted pharmacotherapy.
Although several studies are available on mutational analysis of GISTs, very few studies have been documented in India. In our study, a mutational analysis of C- KIT exon 9, exon 11 and BRAFV600E mutations were carried out in the Indian population. This will prove useful in designing therapies to target specific mutations and provide better therapeutic outcomes.
Materials and methods
This retrospective study was carried out on immunohistochemically proven GIST specimens using DNA extracted from paraffin blocks of all the cases received in the Department of Pathology at our tertiary teaching institution from October 2013 to October 2016. A total of 20 proven cases of GIST were taken up for this study. Permission from the Institutional Ethics Committee was obtained prior to commencing the study(IECNO:CSPMED/15/OCT/25/56).
A structured proforma was used to obtain clinical and demographic information of the patients from the medical records. The formalin fixed paraffin embedded tissues were taken up as 4-5 micron sections and stained with hematoxylin and eosin and graded accordingly. One representative block was selected and immunohistochemically stained for c-KIT. The antibody used was mouse monoclonal antibody (CD117) procured from BioGenex Laboratories Inc.
The steps carried out for mutational analyses on c-KIT exon 9, c-KIT exon 11 and BRAF V600E genes include DNA isolation, assessment of DNA quality and quantity using Hybrid Reader (nanodrop), polymerase chain reaction using forward (F) and reverse(R) primers [Table 1], agarose gel electrophoresis and cycle sequencing.
Table 1. Forward and Reverse Primers used in PCR for C-KIT Exon 11, Exon 9 and BRAF V600E.
| Gene | Primer sequence | BP |
| c-KIT Exon 11(F) | GTGCTCTAATGACTGAGAC | 19 |
| c-KIT Exon 11(R) | TACCCAAAAAGGTGACATGG | 20 |
| c-KIT Exon 9(F) | CTAGAGTAAGCCAGGGCTTTTGTT | 24 |
| c-KIT Exon 9(R) | CCTAAACATCCCCTTAAATTGGATT | 25 |
| BRAF(F) | TCATAATGCTTGCTCTGATAGG | 22 |
| BRAF(R) | GGCCAAAAATTTAATCAGTGG | 21 |
Following this, data analysis was carried out using the Mutation Taster Software. Majority of the participants belonged to the age group of 50-59 years (40%) and were males.The sequencing was analyzed for the type of mutation (in-frame deletions, substitutions, duplications), the nucleotide change, whether the mutation was a heterozygous or a homozygous mutation and whether this mutation was disease causing or not.
Results
All cases of histologically and immunohistochemically proven GISTs diagnosed in the Department of Pathology of our tertiary teaching institution over a period of three years were included in the study. A total of 20 cases were identified.
The age of study participants ranged from 35 to 68 years with 65% cases in men. Stomach was the most common site of the tumor (50%) and about 35% of the tumors were of the size of 5-9 centimeters. Grossly, the tumours were submucosal, circumscribed and unencapsulated. On sectioning, the cut surface varied in colour from grey white to grey brown, solid with few having cystic areas and some tumours had areas of necrosis (Figure 1).
Figure 1. Gastric GIST with a Fleshy appearance and Central Degeneration.
Microscopically, 15 tumours were classified histologically as spindle cell while four cases were classified as mixed type comprising of both spindle cell(Figure 2) and epitheloid (Figure 3) morphology. Epitheloid variant was found in one case only. The staging of the tumors revealed that 50% of the tumors belonged to pT4 stage, while 40% of them belonged to pT3 stage. Majority of the tumors were high risk (45%) followed by intermediate risk (30%).
Figure 2. Microphotograph of Spindle Cell GIST in a Fascicular Pattern (H&E 40x).
Figure 3. : Microphotograph of Epitheloid GIST with a Nested Growth Pattern (H&E 100x).
Immunohistochemistry for CD 117 was done in 20 cases. In 19 cases, strong and diffuse cytoplasmic positivity for CD 117 was seen (Figure 4). One case was negative for CD 117. However, this case showed a positive immunohistochemical staining for Discovered on GIST-1 (DOG1) and PDGFRA.
Figure 4. Microphotograph of C-KIT Immunoreactivity at high power (H&E 200x).
C-Kit mutation analysis
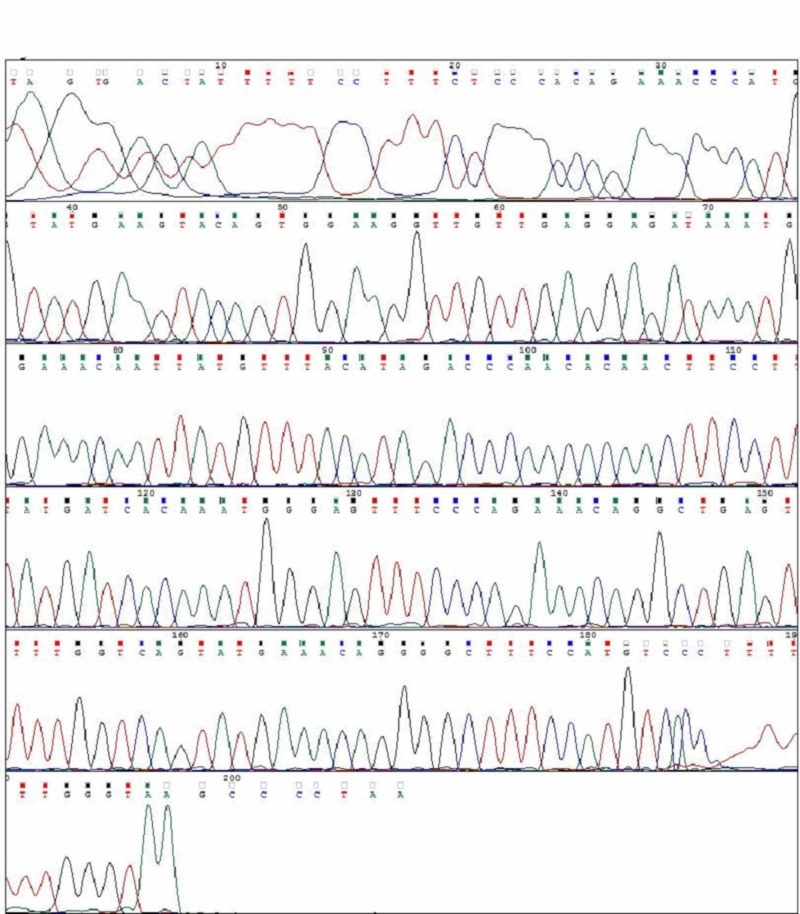
Of the 20 cases, three cases (A1, A2, A3) had mutations in c-KIT exon 11. All the three mutations were a single nucleotide substitution with heterozygous mutations. Their nucleotide change and amino acid sequence were analyzed and using mutation taster software, we predicted whether the change is disease causing or not (Figure 5-7; Table 2). The change in amino acid in case number A1 was from tryptophan at position 557 to stop codon (c.1670G>A), in case number A2 the change in amino acid was from lysine at position 558 to glutamic acid (c.1672A>G) and in case A3 there was no amino acid change but the nucleotide change identified was c.1680 T>G. The rest of the 17 tumours had a normal exon 11 sequence (Figure 8). Correlation with clinico-pathological findings shows that exon 11 substitution mutations occurred in the age group 58-61 years. They were present in different locations namely stomach, duodenum and jejunum. Histologically, all the three cases had a spindle cell morphology. The three tumours were classified as intermediate risk.
Figure 5. Case A1 KIT Exon 11 Sangers Sequence Change.
Figure 7. Case A3 KIT Exon 11 Sangers Sequence Change.
Table 2. Exon 11 KIT Mutations.
| CASE NUMBER | AGE | SEX | SITE | MORPHOLOGY | RISK STRATIFICATION | GENE | EXON | MUTATION STATUS | NUCLEOTIDE CHANGE | AMINO ACID CHANGE | PREDICTION (MUTATION TASTER) |
| A1 | 58yrs | Male | Duodenum | Spindle cell | Intermediate risk | KIT | 11 | Heterozygous | TGG-TAG (c.1670G>A) | Try557Stop codon | Disease causing |
| A2 | 60yrs | Female | Stomach | Spindle cell | Intermediate risk | KIT | 11 | Heterozygous | AAG-GAG (c.1672A>G) | Lys558Glu | Disease causing |
| A3 | 61yrs | Male | Jejunum | Spindle cell | Intermediate risk | KIT | 11 | Heterozygous | GTT-GTG (c.1680 T>G) | Val-Val (no amino acid change) | Disease causing |
Figure 8. Exon 11 Normal Sangers Sequence In Non Mutated Cases .
Figure 6. Case A2 KIT Exon 11 Sangers Sequence Change.
Three cases (A4, A5, A6) had mutations in c-KIT exon 9. All the three cases were heterozygous mutations with single nucleotide substitution. Their nucleotide change and amino acid sequence were analyzed and using mutation taster software, we predicted whether the change is disease causing or not (Figure 9-11; Table 3). The change in amino acid in case number A4 was from alanine at position 507 to proline (c.1519G>C). In A5 and A6 there was no change in the aminoacid but nucleotide change was identified as c.1521A>T. Rest of the 17 tumours had normal exon 9 sequence (Figure 12). Correlation with clinicopathological findings shows that Exon 9 mutations occurred in the age group 45 -60 years and the tumours occurred in different locations namely stomach, jejunum and ileum but predominantly in the small intestine. Histologically, two cases had spindle cell morphology and one case had a mixed pattern of epitheloid and spindle cell morphology. Two tumours were classified as intermediate risk and one tumour as high risk.
Figure 9. Case A4 KIT Exon 9 Sangers Sequence Change.
Figure 11. Case A6 KIT Exon 9 Sangers Sequence Change.
Table 3. Exon 9 KIT Mutations.
| CASE NUMBER | AGE | SEX | SITE | MORPHOLOGY | RISK STRATIFICATION | GENE | EXON | MUTATION STATUS | NUCLEOTIDE CHANGE | AMINO ACID CHANGE | PREDICTION (MUTATION TASTER) |
| A4 | 52yrs | Female | Jejunum | Spindle cell | Intermediate risk | KIT | 9 | Heterozygous | GCA-CCA (c.1519G>c) | Ala507Pro | Disease causing |
| A5 | 60yrs | Female | Ileum | Mixed | High risk | KIT | 9 | Heterozygous | GCA-GCT (c.1521A>T) | Ala-Ala (no amino acid change) | Disease causing |
| A6 | 45yrs | Male | Stomach | Spindle cell | Intermediate risk | KIT | 9 | Heterozygous | GCA-GCT (c.1521A>T) | Ala-Ala (no amino acid change) | Disease causing |
Figure 12. : Exon 9 Normal Sangers Sequence in Non Mutated Cases .
Figure 10. Case A5 KIT Exon 9 Sangers Sequence Change.
BRAF V600E mutation analysis
Among the 20 cases, 19 cases had normal BRAF sequence (Figure 13) and only one tumour (Case number A7) had BRAF V600E mutational sequence (Figure 14, Table 4). A single nucleotide substitution was identified (c.1799T>G) from Valine to glycine at position 600 (c.1799T>G). The clinicopathological correlation revealed that the single case was identified in a 58 years old male patient and the tumour was located in the jejunum. The size of the tumour was 15cm. Histologically, the tumour was composed of a mixed pattern of epitheloid and spindle cell morphology and had a mitotic rate of 12/10HPF. The tumour was classified as high risk.
Figure 13. Normal BRAF V600E Sangers Sequence in Non Mutated Cases.
Figure 14. Case A7 BRAF V600E Sangers Sequence Change.
Table 4. BRAFV600E Mutation.
| CASE NUMBER | AGE | SEX | SITE | MORPHOLOGY | RISK STRATIFICATION | GENE | EXON | MUTATION STATUS | NUCLEOTIDE CHANGE | AMINO ACID CHANGE | PREDICTION (MUTATION TASTER) |
| A7 | 58yrs | Male | Jejunum | Mixed | High risk | BRAF | 15 | Heterozygous | GTG-GGG (c.1799T>G) | Val(600)Gly | Disease causing |
Discussion
The mechanistic basis of GIST is based on multiple genetic alterations. Abnormalities in C-KIT (exon 9, 11, 13 and 17), PDGFRA, BRAFV600E, SDH (A,B,C) and NF1 genes have been implicated in the pathogenesis of GIST. The present study was carried out to evaluate the nature of expression of c-KIT exon 9, 11 and BRAF V600E genetic mutations and their alterations at the molecular level. The mean age of presentation was 54 years with maximum cases seen between 50-59 years (40%). Similar findings were seen in a study done by Bhalgami et al. [6]. Moreover, the peak incidence of GIST was seen among the men (65%) compared to the women (35%) in the present study. This finding was comparable with a study done by DeMatteo et al. [7]. The most common site of presentation of the tumor in the GI tract was stomach (50%) followed by jejunum and ileum. This was in accordance with a study done by Miettinen et al. and Gomez et al. [8,9].
The most common histological type of the GIST was spindle cell type, which was majorly seen in the small intestine. Mixed type was equally present in stomach and small intestine. Among the spindle cell type, the growth pattern commonly seen was fascicular. Epithelioid type was seen in the stomach tumors. Large sized tumors (>10 cm) showed increased mitotic rate. This finding was in accordance with studies done by Vij et al. and Roggen et al. [10,11]. In the present study, 45% of the tumors were categorized as high risk of which majority of the tumors occurred in the stomach. The risk stratification was carried out in accordance to the studies done by Miettinen et al. [12,13]. Moreover, 95% of the specimens morphologically designated as GIST stained positively for CD 117. Based on a study done by Medeiros et al., it is acceptable that all GISTs are not positive for CD117 as these negative tumors may harbor other mutations in other genes such as PDGFRA [14].
The biological mechanism of GIST in the knowledge of c-KIT mutations was first unraveled by Hirota et al. in 1998 [15]. Similar observations were later seen in studies by Rubin et al. and Heinrich et al., where activating mutations on the tyrosine kinase receptor were considered to be crucial in the pathogenesis of GIST [16,17]. In their studies, Colucci et al., Vosseller et al. and Kapur et al. found that signaling pathways activated by KIT oncoproteins modify crucial aspects of the GIST cell phenotype. The GIST signaling pathways differ from those in the non-neoplastic cells, and depending on the location and nature of the mutation, the exact signaling mechanism also varies between GIST tumors. Critical clinicopathological and biological parameters including GIST apoptotic activity, mitotic activity, metastatic capability and imatinib response are all influenced by c-KIT structure and sequence [18-20].
In a study of 100 cases by Pai et al. in the Indian population, exon 11 mutations were identified in 57% of cases. In-frame deletions in 35 cases, 11 substitutions cases, nine cases of double mutations, one case each of insertion and duplication. The most common mutation was tryptophan557_lysine558 del (c.1669_1674delTGGAAG) in 13 cases. The substitution mutations were Val559Asp, Val560Asp, Val559Ala, Val560Gly, Thr574Ile and Leu576Pro; among these nine were homozygous and two were heterozygous [21]. This was not in concordance with our study as we reported different set of missense mutations. In the present study, c-KIT exon 11 mutations were identified in three cases of GISTs. All the three cases had single nucleotide substitution and the missense mutations identified were Trp557Stop (c.1670G>A), Lys558Glu (c.1672A>G), Val-Val (c.1680T>G). All the three cases were heterozygous mutations.
In our study the three C-KIT exon 11 mutations occurred in the stomach, duodenum and jejunum. This was in concordance with a study by Lasota et al. where the exon 11 mutations were seen in different sites including gastric, small intestinal and rectal tumours [22].
In their study of 87 patients, Daniels et al. found in-frame deletions, single nucleotide substitutions and duplications in C-KIT exon 11. In their study all the deletions were classified as high risk, whereas substitution and duplication mutations were classified as intermediate risk [23]. This was in accordance with our study where all the three C-KIT exon 11 mutations were classified as intermediate risk.
In a study by Pai et al., out of 100 cases 10 cases were identified to harbor exon 9 mutations. All the 10 cases had duplications in Alanine to tyrosine at codons 502 -503. Nine tumours were present in the small intestine and one in the retroperitoneum. All the tumours had a spindle cell morphology [21]. Lux et al. and Lasota et al. in their studies found heterozygous exon 9 mutations in GISTs. All the tumours were localized to the small intestine. The most common morphology identified was spindle cell morphology. The mutation seen in these cases were all duplications. The most common duplications were alanine at position 502 and tyrosine at position 503. These mutations activate the receptor via a ligand independent oligomerization [24,25]. These studies were not in concordance with our study where there were single nucleotide substitutions rather than deletions. In our study, the mutations identified were Ala507Pro (c.1519G>C) and two cases had no amino acid change with nucleotide substitution Ala -Ala (c.1521A>T). The site of occurrence of tumour was small intestine (two cases) and stomach (one case). The most common histology was spindle cell type and one mixed type. The reported exon 9 mutations in the present work clearly differs from the prototypical c-KIT exon 9 mutated GIST.
A single case of GIST in stomach with mixed morphology was studied by Grabellus et al., wherein a c-KIT exon 9 single nucleotide substitution Ser476Ile (c.1472G>T) was observed [26]. This patient had a progression free survival of 15 months, since the median progression free survival of usual c-KIT exon 9 mutations is less than 1 year under imatinib standard therapy. The mutation identified was similar to our study. However, the nucleotide substitution was not similar.
BRAF V600E mutations in GISTs were first demonstrated by Agaram et al. in a series of wild type GISTs. BRAF mutations in their study occurred in the small intestine [4]. Our study was in concordance with this study where the site of tumour was in the jejunum. Hostein et al. detected BRAF mutations in 13% of the wild-type GISTs and they identified that the mutation can manifest in any histological pattern including spindle cell, epitheloid and mixed morphology of spindle cell and epitheloid type [27]. This was in concordance with our study where the tumors had a mixed morphology. Agaram et al. demonstrated that these BRAF mutated GISTs are usually classified as high risk tumours. This was also in concordance with our study where the tumour was classified as high risk [4].
Patil et al. demonstrated that mutations in BRAF on exon 15 were a DNA base substitution of thymine for adenine (T to A) that converts valine to glutamic acid of amino acid residue 600 (BRAF V600E) [28]. In the present study, heterozygous mutations were identified in nucleotidec.1799 T>G at exon 15 where there was a substitution of valine to glycine at position 600 (Val600Gly). Though the exon 15 position of mutation (Val600) was in concordance with our study, the nucleotide change was not similar.
In studies by Falchook et al. and Agaram et al., in the therapy for BRAF mutant GISTs, they found that the patient did not respond to a normal dose of imatinib and also required higher doses of imatinib or sorafinib therapy [29,4]. However at present our patient is doing well with the standard doses of imatinib.
Hence, in view of varied clinical outcomes, molecular studies of GISTs is very essential for understanding the pathogenesis, the behavior pattern, designing the spectrum of targeted therapies and assessing recurrence. Primary diagnosis of GISTs is based on histopathology and immunohistochemistry. However, with rapidly evolving personalized therapy, evaluation of mutation will become essential to assess prognosis and design targeted therapies. This study provides an interesting and limited insight in identifying the molecular changes in GISTs at the exonic level using DNA based sanger sequencing.
Conclusions
Molecular pathways play a significant role in determining the pathogenesis and progression of GIST. The c-KIT exon 9 and 11 mutation together constituted 30% of the mutations in our limited series and majority of them were either categorized as intermediate or high risk. One case with BRAFV600E mutation was identified in high risk category. Therefore, with the evolution of personalized therapies for malignancies, assessment of mutations and molecular studies become increasingly valid and essential for both diagnosis and prognosis.
The content published in Cureus is the result of clinical experience and/or research by independent individuals or organizations. Cureus is not responsible for the scientific accuracy or reliability of data or conclusions published herein. All content published within Cureus is intended only for educational, research and reference purposes. Additionally, articles published within Cureus should not be deemed a suitable substitute for the advice of a qualified health care professional. Do not disregard or avoid professional medical advice due to content published within Cureus.
The authors have declared that no competing interests exist.
Human Ethics
Consent was obtained by all participants in this study. Sri Ramachandra Institute of Higher Education and Research issued approval IECNO:CSPMED/15/OCT/25/56
Animal Ethics
Animal subjects: All authors have confirmed that this study did not involve animal subjects or tissue.
References
- 1.Gastrointestinal stromal tumor: advances in diagnosis and management. Patil DT, Rubin BP. https://www.ncbi.nlm.nih.gov/pubmed/21970485. Arch Pathol Lab Med. 2011;135:1298–1310. doi: 10.5858/arpa.2011-0022-RA. [DOI] [PubMed] [Google Scholar]
- 2.Interstitial cells of Cajal as pacemakers in the gastrointestinal tract. Sanders KM, Koh SD, Ward SM. https://doi.org/10.1146/annurev.physiol.68.040504.094718. Annu Rev Physiol. 2006;68:306–343. doi: 10.1146/annurev.physiol.68.040504.094718. [DOI] [PubMed] [Google Scholar]
- 3.KIT mutations are common in incidental gastrointestinal stromal tumors one centimeter or less in size. Corless CL, McGreevet L, Haley A, Town A, Heinrich MC. https://doi.org/10.1016/S0002-9440(10)61103-0. Am J Pathol. 2002 160:1567–1572. doi: 10.1016/S0002-9440(10)61103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novel V600E BRAF mutations in imantinib naïve and imantinib-resistant gastrointestinal stromal tumors. Agaram NP, Wong GC, Guo T, Maki RG, Singer S, DeMatteo RP. https://www.ncbi.nlm.nih.gov/pubmed/18615679. Genes Chromosomes Cancer. 2008;47:853–859. doi: 10.1002/gcc.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. Demetri GD, Von Mehren M, Blanke CD, et al. https://www.ncbi.nlm.nih.gov/pubmed/12181401. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- 6.Clinicopathological study of 113 gastrointestinal stromal tumors. Bhalgami R, Manish K, Patil P, Mehta S, Mohandas KM. https://www.ncbi.nlm.nih.gov/pubmed/23224791. Indian J Gastroenterol. 2013;32:22–27. doi: 10.1007/s12664-012-0273-2. [DOI] [PubMed] [Google Scholar]
- 7.Tumor mitotic rate, size and location independently predict recurrence after resection of primary gastrointestinal stromal tumor (GIST) DeMatteo RP, Gold JS, Saran L, et al. https://www.ncbi.nlm.nih.gov/pubmed/18076015. Cancer. 2008;112:608–615. doi: 10.1002/cncr.23199. [DOI] [PubMed] [Google Scholar]
- 8.Gastrointestinal stromal tumors: pathology and prognosis at different sites. Miettinen M, Lasota J. https://doi.org/10.1053/j.semdp.2006.09.001. Semin Diagn Pathol. 2006;23:70–83. doi: 10.1053/j.semdp.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Molecular alterations of KIT and PDGFRA in GISTs: evaluation of a Portuguese series. Gomes AL, Gouveia A. https://www.ncbi.nlm.nih.gov/pubmed/17827398. J Clin Pathol. 2008;61:203–208. doi: 10.1136/jcp.2007.047043. [DOI] [PubMed] [Google Scholar]
- 10.Gastrointestinal stromal tumors: a clinicopathological and immunohistochemical study of 121 cases. Vij M, Agarwal V, Kumar A, Pandey R. https://www.ncbi.nlm.nih.gov/pubmed/21221881. Indian J Gastroenterol. 2010;29:231–236. doi: 10.1007/s12664-010-0079-z. [DOI] [PubMed] [Google Scholar]
- 11.The histopathological differential diagnosis of gastrointestinal stromal tumors. Graadt van Roggen JF, van Velthuysen MLF. https://www.ncbi.nlm.nih.gov/pubmed/11215292. J Clin Pathol. 2001;54:96–103. doi: 10.1136/jcp.54.2.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gastrointestinal stromal tumors of the stomach a clinicopathologic, immunohistochemical and molecular genetic study of 1765 cases with long term follow up. Miettinen M, Sobin LH, Lasota J. https://www.ncbi.nlm.nih.gov/pubmed/15613856. Am J Surg Pathol. 2005;29:52–68. doi: 10.1097/01.pas.0000146010.92933.de. [DOI] [PubMed] [Google Scholar]
- 13.Gastrointestinal stromal tumors of the jejunum and ileum: a clinicopathologic, immunohistochemical and molecular genetic study of 906 cases before Imantinib with long term follow up. Miettinen M, Makhlouf H, Sobin LH, Lasota J. https://www.ncbi.nlm.nih.gov/pubmed/16625094. Am J Surg Pathol. 2006;30:477–489. doi: 10.1097/00000478-200604000-00008. [DOI] [PubMed] [Google Scholar]
- 14.KIT-negative gastrointestinal stromal tumors: proof of concept and therapeutic implications. Medeiros F, Corless CL, Duensing A, et al. https://www.ncbi.nlm.nih.gov/pubmed/15223958. Am J Surg Pathol. 2004;28:889–894. doi: 10.1097/00000478-200407000-00007. [DOI] [PubMed] [Google Scholar]
- 15.Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Hirota S, Isozaki K, Moriyama Y, et al. http://10.1126/science.279.5350.577. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 16.KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R. https://www.ncbi.nlm.nih.gov/pubmed/11719439. Cancer Res. 2001;61:8118–8121. [PubMed] [Google Scholar]
- 17.Kinase mutations and Imantinib response in patients with metastatic gastrointestinal stromal tumor. Heinrich MC, Corless CL, Demetri GD, et al. https://www.ncbi.nlm.nih.gov/pubmed/14645423. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 18.The receptor tyrosine kinase c-kit provides a critical signal for survival, expansion and maturation of mouse natural killer cells. Colucci F Di Santo JP. https://www.ncbi.nlm.nih.gov/pubmed/10648413. Blood. 2000;95:984–991. [PubMed] [Google Scholar]
- 19.C-KIT receptor signaling through its phosphatidylinositide-3’-kinase-binding site and protein kinase C: Role in mast cell enhancement of degranulation, adhesion and membrane ruffling. Vosseller K, Stella G, Yee NS, Besmer P. https://www.ncbi.nlm.nih.gov/pubmed/9168474. Mol Biol Cell. 1997;8:909–922. doi: 10.1091/mbc.8.5.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A novel mechanism of cooperation between C-KIT and erythropoietin receptor. Stem cell factor induces the expression of STAT5 and erythropoietin receptor resulting in efficient proliferation and survival by erythropoietin. Kapur R, Zhang L. https://www.ncbi.nlm.nih.gov/pubmed/11042182. J Biol Chem. 2001;276:1099–1106. doi: 10.1074/jbc.M007442200. [DOI] [PubMed] [Google Scholar]
- 21.Unraveling the spectrum of KIT mutations in gastrointestinal stromal tumors: An Indian Tertiary Cancer Center Experience. Pai T, Bal M, Shetty O, et al. http://10.4103/sajc.sajc_275_16. South Asian J Cancer. 2017;6:113–117. doi: 10.4103/sajc.sajc_275_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutations in Exon 11 of c-Kit occur preferentially in malignant versus benign gastrointestinal stromal tumors and do not occur in leiomyomas or leiomyosarcomas. Lasota J, Jasinki M. https://www.ncbi.nlm.nih.gov/pubmed/9916918. Am J Pathol. 1999;154:53–60. doi: 10.1016/S0002-9440(10)65250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spectrum of KIT/PDGFRA/BRAF mutations and phosphatidylinositol-3-kinase pathway gene alterations in gastrointestinal stromal tumors (GIST) Daniels M, Lurkin I, Pauli R, et al. https://www.ncbi.nlm.nih.gov/pubmed/21906875. Cancer Lett. 2011;312:43–54. doi: 10.1016/j.canlet.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 24.KIT extracellular and kinase domain mutations in gastrointestinal stromal tumors. Lux ML, Rubin BP, Biase TL, et al. https://www.ncbi.nlm.nih.gov/pubmed/10702394. Am J Pathol. 2000;156:791–795. doi: 10.1016/S0002-9440(10)64946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mutations in exons 9 and 13 of KIT gene are rare events in gastrointestinal stromal. Lasota J, Wozniak A, Sarlomo-Rikala M, et al. https://www.ncbi.nlm.nih.gov/pubmed/11021812. Am J Pathol . 2000;157:1091–1095. doi: 10.1016/S0002-9440(10)64623-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A novel primary KIT exon 9 single nucleotide substitution c.1427G>T (p.Ser476lle) in a gastrointestinal sromal tumor. Grabellus F, Sheu SY, Bauer S, Speich N, Schmid KW, Worm K. https://www.ncbi.nlm.nih.gov/pubmed/20922406. Virchows Arch. 2010;457:623–625. doi: 10.1007/s00428-010-0987-4. [DOI] [PubMed] [Google Scholar]
- 27.BRAF mutation status in gastrointestinal stromal tumors. Hostein I, Faur N, Primois C, et al. https://www.ncbi.nlm.nih.gov/pubmed/20023270. Am J Clin Pathol. 2010;133:141–148. doi: 10.1309/AJCPPCKGA2QGBJ1R. [DOI] [PubMed] [Google Scholar]
- 28.Utility of BRAF V600E mutation specific immunohistochemistry in detecting BRAF V600E mutated gastrointestinal stromal tumors. Patil DT, Ma S, Konishi M, et al. https://www.ncbi.nlm.nih.gov/pubmed/26486743. Am J Clin Pathol. 2015;144:782–789. doi: 10.1309/AJCPRK3V2EIIUPQZ. [DOI] [PubMed] [Google Scholar]
- 29.BRAF mutant gastrointestinal stromal tumor: first report of regression with BRAF inhibitor dabragenib (GSK2118436) and whole exomic sequencing for analysis of acquired resistance. Falchook GS, Trent JC, Heinrich MC, et al. https://doi.org/10.18632/oncotarget.864. Oncotarget. 2013;4:310–315. doi: 10.18632/oncotarget.864. [DOI] [PMC free article] [PubMed] [Google Scholar]