Abstract
Sex chromosome dosage (SCD) variation increases risk for neuropsychiatric impairment, which may reflect direct SCD effects on brain organization. Here, we 1) map cumulative X- and Y-chromosome dosage effects on regional cortical thickness (CT) and investigate potential functional implications of these effects using Neurosynth, 2) test if this map is organized by patterns of CT covariance that are evident in health, and 3) characterize SCD effects on CT covariance itself. We modeled SCD effects on CT and CT covariance for 308 equally sized regions of the cortical sheet using structural neuroimaging data from 301 individuals with varying numbers of sex chromosomes (169 euploid, 132 aneuploid). Mounting SCD increased CT in the rostral frontal cortex and decreased CT in the lateral temporal cortex, bilaterally. Regions targeted by SCD were associated with social functioning, language processing, and comprehension. Cortical regions with a similar degree of SCD-sensitivity showed heightened CT covariance in health. Finally, greater SCD also increased covariance among regions similarly affected by SCD. Our study both 1) develops novel methods for comparing typical and disease-related structural covariance networks in the brain and 2) uses these techniques to resolve and identify organizing principles for SCD effects on regional cortical anatomy and anatomical covariance.
Keywords: anatomical coupling, aneuploidy, development, Klinefelter’s, structural magnetic resonance imaging
Introduction
Gene dosage variations—due to duplications or deletions of segments of genomic DNA—represent a major class of genetic risk for cognitive, psychiatric, and neurological impairment (Malhotra and Sebat 2012). The profile of increased risk for different cognitive and behavioral impairments is not the same for all gene dosage disorders (Moreno-De-Luca et al. 2014), suggesting that different gene dosage disorders may show partly dissociable effects across different brain systems. This hypothesis is supported by available neuroimaging reports: gene dosage effects on neuroanatomy are regionally specific (Dennis and Thompson 2013), and the spatial patterning of gene dosage effects on brain anatomy varies depending on the gene sets involved. Such regional specificity remains poorly understood in mechanistic terms, but has been observed in genetic disorders ranging from subchromosomal copy-number variations (Simon et al. 2005; Thompson et al. 2005; Boddaert et al. 2006; Campbell et al. 2006; Chiang et al. 2007; Bearden et al. 2009; Hoeft et al. 2010; Hazlett et al. 2012) to whole chromosome aneuploidies (Hong and Reiss 2014; Lee et al. 2016; Raznahan et al. 2016).
Identifying organizing principles for the spatial targeting of gene dosage effects on brain development would represent a major advance in our biological understanding of this important class of neurogenetic risk. To date, organizing principles for selective anatomical vulnerability of the human brain to neurodevelopmental disorders have only been studied in the context of idiopathic and behaviorally defined conditions such as autism spectrum disorder and schizophrenia (Alexander-Bloch et al. 2014; Wannan et al. 2019). These studies found that spatial patterns of structural brain changes in patient cohorts were partly organized by spatial patterns of structural covariance that were apparent in healthy control populations. Structural covariance is a phenomenon characterized by the tendency of interindividual differences in regional brain morphology to covary with that of other regions (Alexander-Bloch et al. 2013a). Structural covariance patterns in typically developing populations are thought to result from networks of synchronized anatomical change (Raznahan et al. 2011; Alexander-Bloch et al. 2013b) and have been shown to partially reflect underlying white matter tracts (Gong et al. 2012) as well as functional connectivity networks (Alexander-Bloch et al. 2013b; Sotiras et al. 2017).
The fact that observed spatial maps of anatomical change in behaviorally defined neuropsychiatric disorders align with patterns of structural covariance in typical brain development (Alexander-Bloch et al. 2014; Wannan et al. 2019) suggests that a similar phenomenon might explain observed spatial patterns of anatomical change in genetically defined disorders of brain development—although this possibility remains to be empirically assessed. The current study sought to provide a first test of this hypothetical organizing principle—within a family of gene dosage disorders involving X- and/or Y-chromosome aneuploidy. By using sex chromosome aneuploidy (SCA) syndromes to test for evidence of this organizing principle, we also provide new information regarding sex chromosome dosage (SCD) effects on regional cortical anatomy and anatomical covariance. Several considerations motivated our focus on SCA syndromes.
First, SCAs are relatively common neurogenetic disorders with well-replicated effects on regional brain anatomy. Specifically, increases in X- and Y-chromosome dosage have been shown to have convergent effects on cortical thickness (CT) and surface area (SA) within specific frontal, temporal and parietal subregions (Lepage et al. 2014; Raznahan et al. 2016) and on cerebellar subregion volumes (Mankiw et al. 2017) and subcortical structure size and shape (Reardon et al. 2016). Second, SCAs are especially valuable models for studying gene dosage effects on brain organization due to 1) the wide range of naturally occurring SCD variations and 2) the well-established observation that spatial patterns of X- and Y-dosage effects on regional brain anatomy are highly similar to each other (Raznahan et al. 2016; Reardon et al. 2016; Mankiw et al. 2017; Nadig et al. 2018). This spatial concordance in neuroanatomical effects is associated with an overlapping impact of X- and Y-chromosome dosage on cognition and behavior (Lee et al. 2012) and supports parametric analysis of total SCD dosage effects on regional cortical anatomy. Moreover, regions of selective anatomical vulnerability to SCA have shown some spatial correlation with patterns of brain activity for cognitive domains of impairment in SCA (Raznahan et al. 2016). This observation implies that SCA effects on anatomy are partly organized by the topography of functional [and perhaps therefore structural (Alexander-Bloch et al. 2013b)] networks evident in health. Third, there is direct evidence from X-monosomy that altered SCD can modify CT covariance between regions that are themselves altered in CT by X-monosomy (Raznahan et al. 2010). Finally, because SCAs can include addition of one, two, or more sex chromosomes, they provide a unique parametric framework for statistical tests of gene dosage effects on regional brain anatomy and on anatomical covariance between brain regions.
The current study therefore set out to address the following main objectives. First, we directly modeled the impact of total SCD on regional CT using a recently published parcellation that divides the cortex into 308 similarly sized regions that cohere with major gyral boundaries (Romero-Garcia et al. 2012). Foci of peak SCD effects were functionally annotated using the Neurosynth platform for online meta-analysis of functional neuroimaging data (Yarkoni Tal et al. 2011). We next asked if SCD effects on regional CT are spatially embedded within normative patterns of CT covariance that are detectible among typically developing controls. Specifically, we tested if cortical regions that are similarly affected by SCD show stronger structural covariance in health than cortical regions with dissimilar SCD sensitivities. Finally, we characterized SCD effects on CT covariance itself, testing the hypotheses that 1) interregional anatomical covariance would be modified by increasing SCD and 2) pairs of regions with similar SCD effects on CT would show increasing anatomical covariance with mounting SCD. We addressed these goals by harnessing a globally unique structural neuroimaging data set of 301 individuals with diverse SCDs (80 XX, 89 XY, 28 XXX, 58 XXY, 26 XYY, and 20 XXYY).
Methods
Participants
Our study included a total of 301 participants, aged 5–25 years, with varying SCDs: 2 (n = 169: XX, XY), 3 (n = 112: XXX, XXY, XYY), and 4 (n = 20: XXYY). Participant characteristics are detailed in Table 1. Participants with SCA were recruited through advertisements on the National Institutes of Health website and parent support groups. To be included in the study, SCA participants must have had a nonmosaic X/Y aneuploidy and no acquired head injury or neurological condition resulting in gross brain abnormalities. Typically developing participants were singletons enrolled in an ongoing longitudinal study of typical brain development (Giedd et al. 2015). To be included in the study, healthy participants must have never been diagnosed with a mental illness or a condition that impacts the nervous system, taken psychiatric medications, or received mental health treatment or special education services. The participant cohort included in this study has been used in prior studies of SCD effects on several different aspects of brain anatomy (Lin et al. 2015; Raznahan et al. 2016; Reardon et al. 2016b; Fish et al. 2016; Mankiw et al. 2017; Nadig et al. 2018).
Table 1.
Participant characteristics
| Characteristic | XX | XY | XXX | XXY | XYY | XXYY |
|---|---|---|---|---|---|---|
| Sample size | 80 | 89 | 28 | 58 | 26 | 20 |
| Age (years) | ||||||
| Mean | 12.8 | 12.8 | 12.3 | 12.8 | 12.4 | 14.1 |
| SD | 5.07 | 4.61 | 5.68 | 4.93 | 4.91 | 5.45 |
| Range | 5.39-25.13 | 5.24-25.5 | 5.02-24.78 | 5.21-25.97 | 5.71-23.05 | 5.07-22.96 |
| Mean IQ | ||||||
| Full scale* | 115 | 116 | 92 | 96 | 91 | 87 |
| Verbal* | 115 | 113 | 93 | 94 | 89 | 81 |
| Performance* | 111 | 114 | 94 | 99 | 95 | 95 |
| SES | ||||||
| Mean* | 47 | 48 | 41 | 55 | 59 | 46 |
| Tanner stage | ||||||
| 1 | 20 | 24 | 7 | 18 | 9 | 7 |
| 2 | 13 | 14 | 6 | 10 | 4 | 3 |
| 3 | 12 | 14 | 6 | 12 | 7 | 3 |
| 4 | 20 | 17 | 5 | 8 | 4 | 6 |
| 5 | 14 | 17 | 4 | 9 | 2 | 1 |
| Not known | 1 | 3 | 0 | 1 | 0 | 0 |
| Handedness | ||||||
| R | 69 | 79 | 22 | 46 | 21 | 18 |
| M | 5 | 6 | 4 | 7 | 1 | 1 |
| L | 6 | 3 | 2 | 5 | 4 | 1 |
Note: SES, socioeconomic status. SD, standard deviation.
* P < 0.01 for omnibus test of significant variation across groups in core sample.
Neuroimaging
All 301 structural magnetic resonance imaging (sMRI) scans were T1-weighted images with contiguous 1.5 mm axial slices, acquired on the same 1.5 T General Electric SIGNA scanner using a 3D spoiled gradient-recalled echo sequence with the following parameters for image acquisition: echo time, 5 ms; repetition time, 24 ms; flip angle, 45°; acquisition matrix, 256 × 192; number of excitations, 1; and field of view, 24 cm.
Native sMRI scans were submitted to the Montreal Neurological Institute’s CIVET pipeline for automated morphometric analysis (Ad-Dab’bagh et al. 2006). This analysis began with linear transformation, correction of inhomogeneity, and segmentation of each image into white matter, gray matter, and cerebrospinal fluid (CSF) (Zijdenbos et al. 2002). Inner and outer cortical surfaces were then modeled using triangular meshes generated by a constrained Laplacian algorithm (Kim et al. 2005). Finally, CT was measured at ~80 000 vertices across the cortical sheet, which were down-sampled into average CT for each of 308 spatially contiguous patches, approximately equally sized (~5 cm2) regions (Romero-Garcia et al. 2012).
All sMRI scans included in this analysis passed a quality assessment procedure implemented by two independent raters, which consisted of 1) verifying absence of visible motion artifact in the raw scan (Blumenthal et al. 2002; Raznahan et al. 2012; Alexander-Bloch et al. 2016), and 2) verifying absence of visible artifacts in the cortical surfaces extracted by CIVET.
Statistical Analyses
Our analytic pipeline is summarized in Figure 1 and detailed in a step-wise fashion below.
Figure 2.
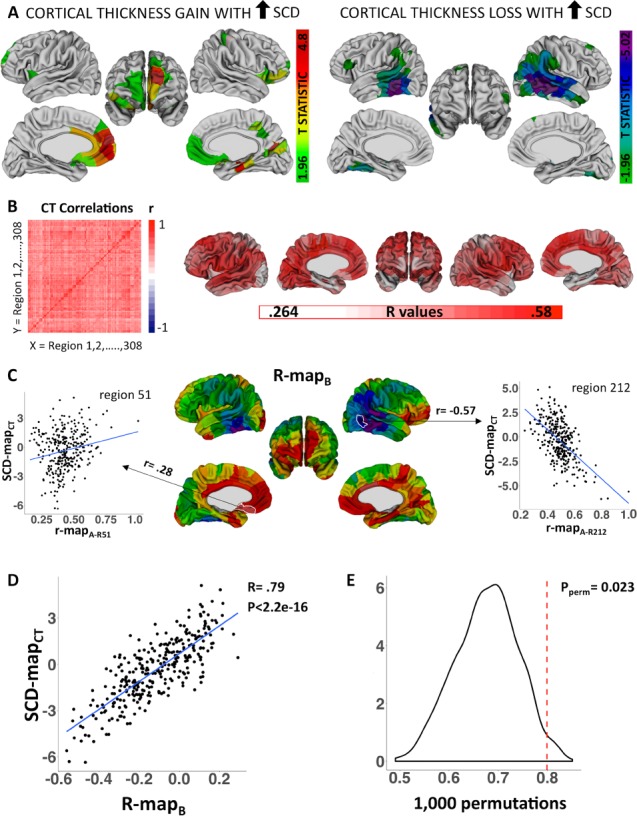
Relating SCD effects on local cortical anatomy to patterns of structural covariance in health. (A) Colored regions are where SCD influences on CT survive FDR correction for multiple comparisons. (B) CT covariance in health: brain-wide CT correlation matrix and visualization of each region’s average CT correlation value. (C) Regional differences in r-mapB values. r-mapB correlation coefficients are projected onto the cortical sheet for visualization. Scatterplots demonstrate that region 51 in the medial orbitofrontal cortex demonstrates stronger CT coupling in health with regions that become thicker with mounting SCD, whereas region 212 in the lateral temporal cortex demonstrates stronger CT coupling in health with regions that become thinner with mounting SCD. (D) Scatterplot demonstrates that regional differences in the alignment between normative CT coupling and CT change in SCD are themselves strongly correlated with regional differences in CT change in SCD. (E) Permutation testing distribution containing 1000 null rB values.
Figure 1.
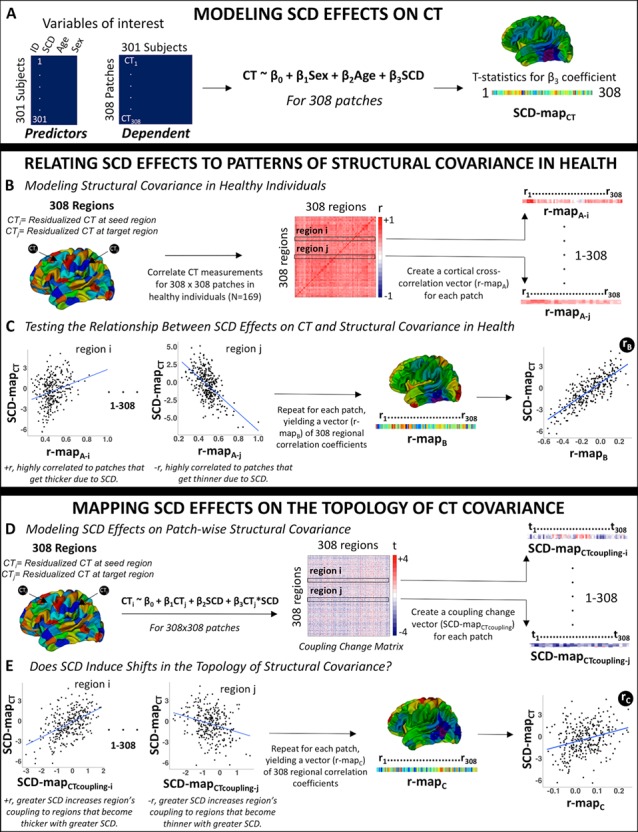
A schematic representation of the analytic pipeline.
Mapping SCD Effects on CT
SCD effects on CT were modeled at each patch for the left and right hemispheres across our entire sample as follows:
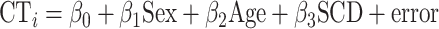 |
(1) |
where CTi is mean thickness across vertices within patch i, sex is a binary variable with females as the reference category, age is a continuous variable centered at mean age of the sample, and SCD is a continuous variable encoding aneuploidic SCD (i.e., 0 for XX and XY|1 for XXX, XXY, and XYY|2 for XXYY). This model was selected after first excluding significant age*SCD and sex*SCD interactions in prediction of regional CT. The exclusion of such interactions indicates that in our dataset, any shifts in CT associate with participant SCD are not significantly modified by participant age or gonadal sex.
Estimation of model (1) at each cortical region above generated a vector of 308 t-values associated with β3 coefficients for the effects of increasing SCD on CT within each cortical region (patch) (“SCD-mapCT”, as illustrated in the schematic shown in Fig. 1A). To define regions of statistically significant SCD effects on CT, P-values associated with regional β3-coefficients from model (1) were corrected for multiple comparisons using False Discovery Rate (FDR) q set at 0.05, and t-statistics for β3-coefficients surviving these corrections were projected onto a reference cortical surface for visualization.
To systematically investigate the functional implications of SCD effects on cortical morphology, we submitted the t-statistic map for SCD effects on CT (SCD-mapCT) to Neurosynth (Yarkoni Tal et al. 2011). Neurosynth is an online platform that extracts brain activation patterns from functional neuroimaging articles and determines which of several thousand prespecified psychological terms is overrepresented in the text of each article (currently Neurosynth includes >14k functional neuroimaging publications). This database can be used to compute association tests between points of interest in the brain and terms (or sets of terms) in the Neurosynth database. We compared the spatial map of SCD effects on CT with the spatial map of association-tests for each of 50 “topics” within the Neurosynth database. These “topics” and their associated brain maps have been previously derived by the creators of Neurosynth by applying latent Dirichlet allocation to a large text collection from neuroimaging literature and can be thought of as underlying components reflecting the conceptual structure of mental functions (Poldrack et al. 2012).
Using Neurosynth (Yarkoni Tal et al. 2011), we identified cognitive and psychological terms that frequently cooccur in the literature with functional activations similar to the observed pattern of SCD effects on cortical morphology (SCD-mapCT). This was achieved by computing cross-cortical spatial correlations between SCD-mapCT values and thresholded “association statistic” maps from Neurosynth for each of 50 “topics” previously derived by a topic mapping dimension reduction of all Neurosynth terms (Poldrack et al. 2012). We identified all topics with an |r| of 0.1 or greater with the SCD-mapCT.
Relating SCD Effects to Patterns of Structural Covariance in Health
Patterns of CT covariance in health were defined using the 169 typically developing participants within our cohort (Fig. 1B). Following well-established protocols for analysis of CT covariance, we regressed age and sex effects out of CT variation at each region using the following linear model:
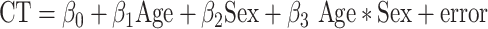 |
(2) |
Note, the age*sex term was included for all vertices to minimize any shared variance between patches that might arise due to subtle differences in age-CT relationships between sexes in each patch.
The regional CT residuals from model (2) were used to calculate a square (308 × 308) symmetric CT covariance (correlation) matrix in health (see schematic Fig. 1B), where Pearson correlation coefficients captured the strength of anatomical coupling between each pair of cortical regions. To test if observed SCD effects on regional CT were organized by this normative anatomical coupling matrix, we correlated each region’s vector of 308 cortical cross-correlations (“r-mapA”) from the CT covariance matrix in health, with the SCD-mapCT vector of 308 region-wise SCD effects (see schematic Fig. 1C). Regions with a positive correlation between their r-mapA and the SCD-mapCT show more positive anatomical coupling in health with regions that become thicker with increasing SCD, whereas regions with a negative correlation show more positive anatomical coupling in health with regions that become thinner with increasing SCD. Repeating this procedure for all cortical regions yielded a vector of 308 regional correlation coefficients (“r-mapB”) measuring the degree of spatial alignment between each region’s cross-cortical CT anatomical coupling in health and the observed map of SCD effects on regional CT (i.e., SCD-mapCT) (Fig. 1C).
To test if the spatial patterning of SCD effects on CT was organized by patterns of CT coupling in health, we quantified the correlation (rB) between r-mapB and SCD-mapCT across all 308 cortical regions (Fig. 1C). A significantly positive rB coefficient would indicate that 1) regions which become thicker with mounting SCD tend to show positive anatomical coupling in health with other regions that become thicker with mounting SCD, whereas 2) regions which become thinner with mounting SCD tend to show positive anatomical coupling in health with other regions that become thinner with mounting SCD. This observation would support the hypothesis that SCD effects on regional CT are organized by normative patterns of CT covariance.
To control for potential inflation of rB by known spatial autocorrelations in CT, we compared the observed value of rB with a null distribution of rB values generated by repeating the above algorithm with 1000 SCD-mapCT vectors derived after permuting scans across study participants (Fig. 2E). For each permutation, this procedure preserves spatial autocorrelations in both SCD-mapCT and r-mapB while removing true SCD effects from SCD-mapCT.
Figure 3.
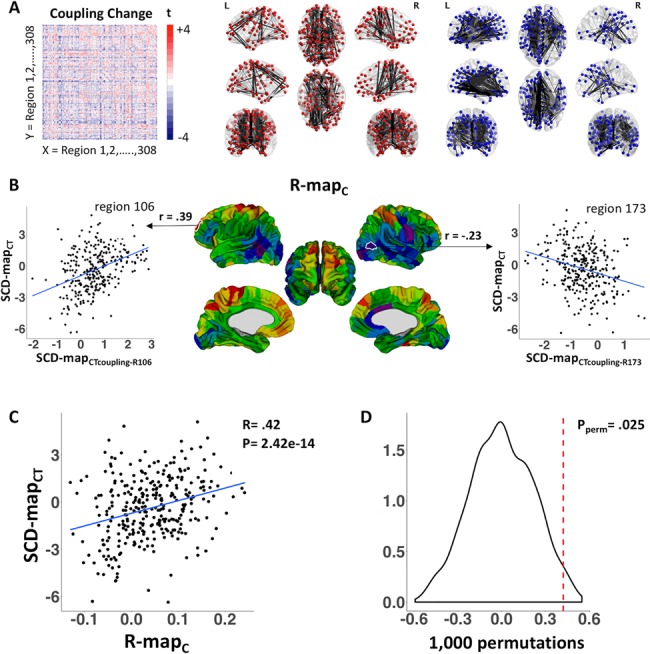
SCD effects on structural covariance. (A) SCD effects on CT coupling between each pair of cortical regions: brain-wide coupling change matrix and visualization of top 5% of positive (red) and negative (blue) edges. (B) Regional differences in r-mapC values. r-mapC correlation coefficients are projected onto the cortical sheet for visualization. Scatterplots demonstrate that region 106 in the rostral frontal cortex demonstrates increased coupling to regions that become thicker with mounting SCD, whereas region 173 in the lateral occipitotemporal junction demonstrates increased coupling to regions that become thinner with mounting SCD. (C) Scatterplot demonstrates that regional differences in the alignment between altered CT coupling and CT change in SCD are themselves strongly correlated with regional differences in CT change in SCD. (D) Permutation testing distribution containing 1000 null rC values.
Mapping SCD Effects on CT Covariance
To examine SCD effects on CT covariance, we first residualized CT at each cortical region for the effects of age and SCD on CT using equation (3) below. This process ensured that any observed SCD effects on CT covariation could not be driven by differences across SCD groups in mean CT.
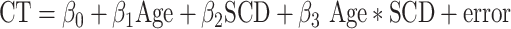 |
(3) |
We then examined SCD effects on the global anatomical coupling of each cortical region using the following linear model:
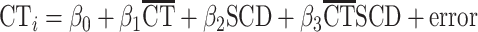 |
(4) |
where the thickness of region i (CTi) is modeled by mean CT across the cortex (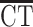 ) and SCD. The β3-coefficient for each cortical region reflects the degree to which its anatomical coupling with mean CT is modified by SCD. The P-values associated with these coefficients were corrected for multiple comparisons across all 308 cortical regions using FDR, q set at 0.05. This analysis tests for SCD effects on the global anatomical coupling of each cortical region, as anatomical coupling with between a region and global mean has been demonstrated to be a reasonable proxy for the average anatomical coupling between a region and every other region (Lerch et al. 2006). However, this procedure does not resolve SCD effects on anatomical coupling between each pair of regions.
) and SCD. The β3-coefficient for each cortical region reflects the degree to which its anatomical coupling with mean CT is modified by SCD. The P-values associated with these coefficients were corrected for multiple comparisons across all 308 cortical regions using FDR, q set at 0.05. This analysis tests for SCD effects on the global anatomical coupling of each cortical region, as anatomical coupling with between a region and global mean has been demonstrated to be a reasonable proxy for the average anatomical coupling between a region and every other region (Lerch et al. 2006). However, this procedure does not resolve SCD effects on anatomical coupling between each pair of regions.
Therefore, potential SCD effects on CT coupling between each pair of cortical regions (i and j), were modeled as follows:
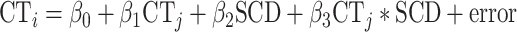 |
(5) |
Estimation of this regression for all unique pairs of cortical regions provided a 308 × 308 matrix of t statistics for the β3 coefficients, which we rendered symmetric by averaging the result for CTi as a function of CTj and CTj as a function of CTi (Fig. 1D). This averaging step was required because model fitting can generate different estimates of β3 in these two cases, although the two are positively correlated (r = 0.49). The resulting “coupling change” matrix represented the effects of SCD on CT coupling between each pair of cortical regions. To test if this effect in turn was related to the effect of SCD on thickness—that is, if the same regions whose thickness was altered by SCD as modeled by equation (1) also manifested SCD-altered interregional anatomical coupling as modeled by equation (5)—we correlated each region’s t-statistics from the coupling change matrix (one row of the coupling change matrix: “SCD-mapCTcoupling” (see schematic Fig. 1D) with the t-statistics from the SCD-mapCT vector (Fig. 1E). Positive values of this correlation coefficient indicated that greater SCD increases a region’s CT coupling with regions that become thicker with greater SCD. In contrast, negative values of this correlation coefficient would indicate that greater SCD increases a region’s CT coupling with regions that become thinner with greater SCD. Repeating this procedure for each region yielded a vector of 308 regional correlation coefficients (“r-mapC”) measuring the degree of spatial alignment between SCD effects on each region’s CT coupling with other regions and the observed map of SCD effects on regional CT (i.e., SCD-mapCT) (see schematic Fig. 1E).
Finally, as a global test of whether the spatial patterning of SCD effects on CT was organized by patterns of SCD effects on CT coupling, we quantified the correlation (rC) between r-mapC and SCD-mapCT across all 308 cortical regions (Fig. 1E). A significantly positive rC coefficient would indicate that 1) regions which become thicker with mounting SCD tend to also become more strongly coupled to other regions that also become thicker with mounting SCD and 2) regions which become thinner with mounting SCD tend to also become more strongly coupled to other regions that also become thinner with mounting SCD. In other words, cortical regions which are brought into “tighter” anatomical coupling with each other also show similar anatomical change with mounting SCD.
Again, to control for potential inflation of rC by known spatial autocorrelations in CT, we compared the observed value of rC with a null distribution of rC values generated by repeating the above algorithm with 1000 SCD-mapCT vectors derived after permuting scans across study participants (Fig. 3D). For each permutation, this procedure preserves spatial autocorrelations in both SCD-mapCT and r-mapC while removing true SCD effects from SCD-mapCT.
Figure 4.

Relating SCD effects on cortical morphology to enriched terms in Neurosynth functional neuroimaging meta-analysis. All Neurosynth Topics (and top three topic terms) with Neurosynth Association Test maps showing an |r| spatial correlation for SCD effects on CT (i.e., SCD-mapCT) > 0.1 in magnitude. The Association Test map for each topic was derived from meta-analysis of functional neuroimaging studies (N-study = number of studies per topic meta-analysis) featuring topic terms. Regions where CT increases with mounting SCD include cortical regions involved in emotion, pain, and inhibitory processing. Conversely, regions where CT decreases with mounting SCD are involved in visual, motor, arithmetic, and attentional processing.
Sensitivity Analyses
Effects of SCD at Different Dosage Ranges
Our primary analytic approach models SCD as an integer numeric variable with three values: 0 (i.e., euploidic controls), 1 (individuals carrying one extra sex chromosome: i.e., XXY, XYY, and XXX groups), and 2 (individuals carrying two extra sex chromosomes: i.e., XXYY). This approach improves statistical power relative to pairwise comparisons between groups with differing SCDs and allows us to develop more broadly applicable methods for modeling the effects of a nonbinary variable on structural covariance in the human brain. However, this analytic approach does assume that SCD effects on regional CT and interregional CT covariance are similar across the two distinct SCD change intervals 0 → 1 and 1 → 2. These two contrasts are necessarily represented by an unequal number of participants given the lower prevalence of sex chromosome tetrasomies versus trisomies (Nielsen and Wohlert 1991). We therefore conducted a series of supplementary analyses to test if SCD effects on CT and CT covariance across the full 0 → 2 range (Fig. 1A and D) were positively correlated to those observed when analysis was restricted to the 0 → 1 and 1 → 2 SCD contrasts. Specifically, we repeated the aforementioned analyses of SCD effects on regional CT (Fig. 1A) and interregional CT covariance (Fig. 1D) in two subsets of our cohort capturing two different SCD contrasts: 1) 0 → 1: that is, euploidic individuals versus participants with a sex chromosome trisomy and 2) 1 → 2: that is, participants with sex chromosome trisomy versus those with sex chromosome tetrasomy. We operationalized the similarity in findings between analyses in these subsets versus our full sample as follows. For SCD effects on CT, we determined the correlation between beta coefficients for SCD effects on CT across all 308 cortical regions in our parcellation scheme. For SCD effects on CT covariance, we determined the correlation between beta coefficients for SCD effects across all 47 278 unique pairwise relationships among the 308 cortical regions in our parcellation.
Alternative Cortical Parcellation
Our primary analyses are all conducted within a previously published cortical parcellation consisting of 308 spatially contiguous and approximately equally sized (~5 cm2) regions (Romero-Garcia et al. 2012). To test for generalization of findings to a different cortical parcellation scheme, we also estimated and compared SCD effects on regional CT and CT covariance using the Desikan-Killany parcellation (68 cortical regions bilaterally based on major gyral landmarks) (Desikan et al. 2006).
Results
SCD Effects on Local Cortical Anatomy
Increasing SCD was associated with a mixed pattern of regionally specific increases and decreases in CT (Fig. 2A). Mounting SCD decreased CT in the lateral temporal cortex and the temporal parietal junction, bilaterally, and in unilateral inferior temporal and anterior temporal cortices. In contrast, SCD increased CT in the rostral frontal cortex, bilaterally, and in unilateral medial parietal, ventromedial temporal, and somatosensory cortex.
Comparison of SCD-mapCT with meta-analyses of functional neuroimaging literature from the Neurosynth platform (methods, (Yarkoni Tal et al. 2011) indicated that regions where CT is increased by mounting SCD include brain systems involved in emotion, pain, and inhibitory processing. Conversely, regions where CT is decreased by mounting SCD are involved in visual, motor, arithmetic, and attentional processing (Fig. 4).
Relating SCD Effects to Patterns of Structural Covariance in Health
In order to relate SCD effects on CT (Fig. 2A) to patterns of CT covariance in health, we next estimated pairwise CT correlations between all cortical regions within our cohort of typically developing individuals (Fig. 2B). Replicating previous work (Lerch et al. 2006), we found that regions with high average cross-cortical CT correlations tended to lie in the association cortices (frontal and temporal), whereas regions with low average cross-cortical CT correlations were localized to primary sensory cortices (Fig. 2B).
The CT correlation matrix in health (Fig. 2B) enabled us to measure how strongly each cortical region’s pattern of CT correlation with other regions followed the spatial distribution of SCD effects on CT. This property varied greatly among cortical regions (Fig. 2C). For example, brain regions in the rostral frontal, medial parietal, ventromedial temporal, and somatosensory cortex tended to show stronger CT coupling in health with cortical regions that become thicker with mounting SCD, whereas the opposite pattern was seen in the lateral temporal cortex and temporal parietal junction (Fig. 2C). Regional differences in this alignment between normative CT coupling and CT change in SCD were themselves strongly correlated with regional differences in CT change (rB = 0.79, P < 2.2e-16, Fig. 2D). Thus, two regions which share a congruent effect of SCD (e.g., both become thicker with greater SCD) tend to show higher CT correlation in health than two regions that are dissimilarly impacted by SCD variation. Permutation testing (see Methods) confirmed that this close relationship between the spatial patterning of CT change with SCD and spatial patterns of CT coupling in health could not be accounted for by shared spatial autocorrelations in SCD-induced CT change and normative CT coupling (P = 0.023, Fig. 2E).
SCD Effects on CT Covariance
Analysis of SCD effects on the coupling between mean global CT and CT of each cortical patch, and correcting for multiple comparisons across patches, did not identify any single cortical region that showed a statistically significant disruption of CT coupling with the rest of the cortical sheet as a consequence of increasing SCD. However, estimating SCD effects on anatomical coupling between each unique pair of cortical regions revealed heterogeneous effects, with mounting SCD tending to increase CT coupling between some region-pairs while decreasing CT coupling between others (Fig. 3A).
The matrix of SCD effects on CT coupling between different cortical regions (Fig. 3A) enabled us to measure how strongly each cortical region’s pattern of SCD-induced CT coupling change followed the spatial distribution of SCD effects on CT (Fig. 3B). Mapping this property across the cortical sheet generated a gradient that was highly similar to the spatial patterning of SCD effects on CT (Fig. 2A)—providing qualitative evidence that SCD effects on CT are organized by SCD effects on CT coupling. For example, in posterior temporal regions, greater SCD was not only associated with CT reduction (Fig. 2A), but also with increased CT coupling with other regions that were thinned by greater SCD (Fig. 3B). Conversely, in rostral frontal regions, greater SCD was not only associated with increased CT (Fig. 2A), but also with increased CT coupling with other regions that were thickened by greater SCD (Fig. 3B).
Quantitatively, we observed a strong positive correlation across the cortical sheet between SCD effects on the interregional CT coupling and SCD effects on regional CT (rC = 0.42, P = 2.422e-14, Fig. 3C). Permutation testing (see Methods) further established that this effect could not be accounted for by shared spatial autocorrelations in SCD-induced CT change and normative CT coupling (P = 0.025, Fig. 3D).
Sensitivity Analyses
Sensitivity analyses verified that interregional differences in SCD effects on CT in our primary analysis (Methods schematic: Fig. 1A, Results: Fig. 2A) were highly correlated with those seen for both the 0 → 1 and the 1 → 2 SCD change contrasts (r = 0.96 and r = 0.81, respectively). We further established that SCD effects on interregional CT covariance in our primary analysis (Methods schematic: Fig. 1D, Results: upper triangle values in the matrix shown in Fig. 3A) were moderately correlated across region pairs with those seen for both the 0 → 1 and the 1 → 2 SCD change contrasts (r = 0.6 and r = 0.4 respectively). Taken together, these supplementary analyses indicate that our primary results across the 0 → 2 SCD range show comparable levels of agreement with results from participant subsets focused in 0 → 1 and 1 → 2 SCD ranges.
All three main findings in our original analyze were also upheld using an alternative (Desikan-Killany) parcellation scheme comprising 68 regions bilaterally (Desikan et al. 2006). First, effects of SCD on regional CT were largely similar between our original parcellation (Fig. 2A) and analyses using the Desikan-Killany parcellation (see Supplementary Fig. 1). Both parcellation schemes revealed that mounting SCD is associated with 1) CT reductions in lateral temporal and temporoparietal junction cortex and 2) CT gains in medial prefrontal and medial parietal cortex. Second, analyses using the Desikan parcellation also replicated our original findings regarding the correspondence of SCD effects on CT with 1) patterns of normative CT covariance (308 parcellation findings: Fig. 2D and E | Desikan results: r = 0.88, Pperm < 0.0001) and 2) patterns of SCD effects on CT covariance (308 parcellation findings: Fig. 3C and D|Desikan results: r = 0.4, Pperm = 0.001).
Discussion
By utilizing a rare neuroimaging cohort of youth with diverse SCAs, we characterize total SCD effects on regional cortical anatomy and establish that cortical regions with a similar pattern of SCD-sensitivity show heightened CT covariance in health. Thus, the targeting of SCD effects to specific cortical regions appears to be organized in part by patterns of interregional CT coupling that are apparent in the typically developing brain. Finally, we show that SCD alters the interregional patterns of CT coupling within the cortical sheet, increasing coupling among similarly targeted cortical regions. We discuss each of these main findings in turn below.
SCD Effects on Local Cortical Anatomy
Previous studies in our cohort have separately examined X- and Y-chromosome dosage effects on the human brain, to identify a surprising degree of spatial overlap between the impact of each chromosome’s copy number on neuroanatomy (Raznahan et al. 2016; Reardon et al. 2016; Mankiw et al. 2017). This overlap has been observed for each of several different phenotypes including regional CT (Raznahan et al. 2016) and volume (Lepage et al. 2014) in the cortical sheet, lobular volumes in the cerebellum (Mankiw et al. 2017), and regional SA in subcortical structures (Reardon et al. 2016). Here, by directly modeling total SCD effects on cortical anatomy, we show that mounting SCD increases CT in the rostral frontal cortex and decreases CT in the lateral temporal cortex.
Step-wise relationships between total SCD and cortical anatomy may reflect the role of those few genes that are shared by both chromosomes: 1) pseudoautosomal region (PAR) genes in the recombining regions of sex chromosomes, (Otto et al. 2011) and 2) non-PAR X-Y homolog genes, or “gametologs” (Bellott et al. 2014). To date, systematic analyses of SCD effects on gene expression in human neural tissue are not available. However, a recent study modeling SCD effects on gene expression in human lymphoblastoid cell lines (LCLs) revealed that X–Y gametolog genes show unusually high levels of expression sensitivity to SCD change and are also closely coexpressed with SCD-sensitive autosomal genes with critical cellular functions (Raznahan et al. 2018). Taken together, these findings propose X-Y gametologs as potential mediators of converging X and Y chromosome effects on brain anatomy.
Our findings also elucidate potential anatomical substrates for cognitive and behavioral alterations across SCA syndromes. We find that excess SCD targets brain regions previously linked to language and social functioning, behavioral domains known to be impaired in individuals with SCAs. These qualitative observations can be formalized using Neurosynth, an online platform for large-scale meta-analysis of existing neuroimaging literature (Yarkoni Tal et al. 2011). This analysis indicates that regions targeted by SCD are particularly associated with socio-emotional and attentional processes. Furthermore, regions we identify as being most anatomically vulnerable to SCD appear to partially overlap with regions of known functional alterations in SCAs. For example, individuals with XXY karyotypes demonstrate increased activation in the middle frontal gyrus, including Broca’s area (Brandenburg-Goddard et al. 2014), and reduced activation in the fusiform gyrus and the superior temporal sulcus (van Rijn et al. 2012) during social information processing tasks, regions we found to be altered by SCD. Multimodal neuroimaging studies will be required to systematically assess the degree of overlap between structural and functional brain changes in SCA.
Relating SCD Effects to Patterns of Structural Covariance in Health
We show that the spatial patterning of SCD effects on cortical anatomy is aligned with patterns of structural covariance between distributed brain regions in health. A similar phenomenon has been described in schizophrenia (Alexander-Bloch et al. 2014; Wannan et al. 2019). To our knowledge, our study is the first to demonstrate that this phenomenon operates within neurodevelopmental disorders of known genetic origin. Thus, the elaboration of SCD effects on brain anatomy may be initialized or constrained by normative structural covariance. Potential mechanisms for this interrelationship are suggested by current theories regarding the biological basis of structural covariance within the human brain.
Several potentially interrelated mechanisms have been proposed as biological bases for structural covariance. One interpretation is that covariance between brain regions arises from physical (i.e., white matter tracts) or functional (i.e., synchronous neuronal activation) connectivity, although existing data suggest that white matter tracts and functional coactivation networks show substantial but incomplete overlap with structural covariance networks in human populations (Gong et al. 2012; Alexander-Bloch et al. 2013a, 2013b; Sotiras et al. 2017). One such study comparing white matter (diffusion imaging) tracts with whole brain CT covariance showed that only 30–40% of interregional covariance occurs between regions that are connected by white matter tracts (Gong et al. 2012). With regard to functional connectivity, Sotiras et al. (2017) observed significant overlap between covariance networks and certain functional networks, with more functional and structural correspondence in low-order networks (motor, sensory, visual, and limbic functional networks) than higher-order networks (frontoparietal control and default mode networks) (Sotiras et al. 2017). Inter-regional structural covariance may also reflect synchronized developmental change among cortical regions. Indeed, preliminary longitudinal neuroimaging studies support this hypothesis, showing that brain regions with strong structural covariance also demonstrate coordinated rates of change in CT during development (Alexander-Bloch et al. 2013b; Raznahan et al. 2011; Khundrakpam et al. 2019), and that brain-wide structural covariance networks are more similar to networks of synchronized anatomical change than to functional connectivity networks (Alexander-Bloch et al. 2013b). Taken together, these findings indicate that structural covariance likely arises from coordinated neurodevelopment between brain regions which share white matter and/or functional connections, which in turn further influence covariance between the regions. Our findings that neurodevelopmental disorders target normative structural covariance networks support the coordinated neurodevelopment interpretation of structural covariance in the human brain, indicating a coordinated disruption in which regions that are similarly vulnerable to SCD become derailed together.
SCD Effects on CT Covariance
Our final set of analyses indicates that SCD effects on cortical anatomy are not only organized by patterns of structural covariance in health, but also operate to disrupt structural covariance itself. Specifically, greater SCD increases covariance among regions that are similarly affected by aneuploidy. To detect this effect, we developed a novel methodology for analysis of nonbinary factors on structural covariance in the human brain. Traditional analyses involve identifying group effects on global structural covariance or on seed-based structural covariance maps, but do not allow for analysis of group effects on interregional (or “edge-level”) measures of structural covariance. Our methodology allows for such analyses and could be generalized to assess shifts in interregional anatomical covariance within the human brain as a function of any ordered or continuous variable.
Our findings of altered structural covariance as a function of excess X- and Y-chromosome dosage indicate that patterns of structural covariance in the brain are sensitive to disease-related genetic variation. This clinical observation aligns with prior quantitative genetic neuroimaging studies in health, which indicate that genetic factors account for a large proportion of anatomical covariance within the human brain (Schmitt et al. 2009; Alexander-Bloch et al. 2017; Schmitt et al. 2017). Furthermore, the heritability of anatomical covariance between different cortical regions has been shown to recapitulate the degree of transcriptomic similarity between brain regions in health (Alexander-Bloch et al. 2017). Thus, SCD effects on anatomical covariance could potentially reflect tightening of similarity in transcriptional programs between cortical regions that are similarly impacted anatomically by SCD variation.
Limitations and Future Considerations
Our findings should be considered in light of certain study limitations. First, our dataset is cross-sectional, and thus we cannot adequately test how our findings may change as a function of age, highlighting the need for a longitudinal follow-up of our cross-sectional findings. Second, the structural brain scans used in this study were acquired on a 1.5 T magnet, and it will be important to repeat analyses when higher field strength datasets have been gathered across patient groups representing a wider range of SCDs. Third, our study focuses on a single morphological variable (CT); however, given the distinct biological origins of other measures of cortical morphometry (volume, SA, and folding), future work could focus on conducting analogous analyses incorporating these other measures. Fourth, our study focuses on the cortical sheet. Future studies examining the relationship between SCD and subcortical covariance will be an important step. Finally, our study provides an analytic workflow for quantifying and comparing the effects of a continuous integer variable (SCD in our case), on CT and CT covariance topography across the cortical mantle. An important goal for future work will be developing methods for modeling how a continuous variable influences topologies of anatomical covariance within the brain. Such work could yield estimates of a continuous variable’s effect on node-wise and edge-wise metrics from graph-theoretical representations of anatomical covariance matrices.
Conclusions
Notwithstanding these limitations, our study implements a novel approach for analysis of group effects on structural covariance in the human brain to identify a number of organizing principles for SCD effects on cortical anatomy. Specifically, we show that SCD effects on cortical anatomy are not only organized by patterns of structural covariance in health, but also operate to disrupt structural covariance itself, with greater SCD increasing covariance among regions that are similarly affected by aneuploidy. These organizing principles may also govern effects of other gene dosage variations on brain development—potentially providing a window into the selective vulnerability of brain and behavior to this important category of genetic risk.
Funding
Intramural Research Program of the National Institute of Mental Health (Clinical trial reg. no. NCT00001246, clinicaltrials.gov; NIH Annual Report Number, ZIA MH002794-13; Protocol number: 89-M-0006).
Notes
The authors would like to thank the patients and their families for their participation in this study. Conflicts of interest: None declared.
Supplementary Material
References
- Ad-Dab’bagh Y., Lyttelton O., Muehlboeck J. S., Lepage C., Einarson D., Mok K., & Evans A. C.. 2006, June. The CIVET image-processing environment: a fully automated comprehensive pipeline for anatomical neuroimaging research In Proceedings of the 12th annual meeting of the organization for human brain mapping (Vol. 2266). Florence, Italy. [Google Scholar]
- Alexander-Bloch A, Clasen L, Stockman M, Ronan L, Lalonde F, Giedd J, Raznahan A. 2016. Subtle in-scanner motion biases automated measurement of brain anatomy from in vivo MRI. Hum Brain Mapp. 37(7):2385–2397. doi: 10.1002/hbm.23180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A, Giedd JN, Bullmore E. 2013a. Imaging structural co-variance between human brain regions. Nat Rev Neurosci. 14(5):322–336. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch A, Raznahan A, Bullmore E, Giedd J. 2013b. The convergence of maturational change and structural covariance in human cortical networks. J Neurosci. 33(7):2889–2899. doi: 10.1523/JNEUROSCI.3554-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch AF, Mathias SR, Fox PT, Olvera RL, Göring HHH, Duggirala R, Curran JE, Blangero J, Glahn DC. 2017. Human cortical thickness organized into genetically-determined communities across spatial resolutions. Cerebral Cortex. 29(1), 106–118. doi: 10.1093/cercor/bhx309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander-Bloch AF, Reiss PT, Rapoport J, McAdams H, Giedd JN, Bullmore ET, Gogtay N. 2014. Abnormal cortical growth in schizophrenia targets normative modules of synchronized development. Biol Psychiatry. 76(6):438–446. doi: 10.1016/j.biopsych.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearden CE, Erp TGM, Dutton RA, Lee AD, Simon TJ, Cannon TD, Emanuel BS, McDonald-McGinn D, Zackai EH, Thompson PM. 2009. Alterations in midline cortical thickness and gyrification patterns mapped in children with 22q11.2 deletions. Cereb Cortex. 19(1):115–126. doi: 10.1093/cercor/bhn064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho T-J, Koutseva N, Zaghlul S, Graves T, Rock S et al. 2014. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 508(7497):494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal JD, Zijdenbos A, Molloy E, Giedd JN. 2002. Motion artifact in magnetic resonance imaging: implications for automated analysis. NeuroImage. 16(1):89–92. [DOI] [PubMed] [Google Scholar]
- Boddaert N, Mochel F, Meresse I, Seidenwurm D, Cachia A, Brunelle F, Lyonnet S, Zilbovicius M. 2006. Parieto-occipital grey matter abnormalities in children with Williams syndrome. NeuroImage. 30(3):721–725. doi: 10.1016/j.neuroimage.2005.10.051. [DOI] [PubMed] [Google Scholar]
- Brandenburg-Goddard MN, van Rijn S, Rombouts SARB, Veer IM, Swaab H. 2014. A comparison of neural correlates underlying social cognition in Klinefelter syndrome and autism. Soc Cogn Affect Neurosci. 9(12):1926–1933. doi: 10.1093/scan/nst190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LE, Daly E, Toal F, Stevens A, Azuma R, Catani M, Ng V, van Amelsvoort T, Chitnis X, Cutter W et al. 2006. Brain and behaviour in children with 22q11.2 deletion syndrome: a volumetric and voxel-based morphometry MRI study. Brain. 129(5):1218–1228. doi: 10.1093/brain/awl066. [DOI] [PubMed] [Google Scholar]
- Chiang M-C, Reiss AL, Lee AD, Bellugi U, Galaburda AM, Korenberg JR, Mills DL, Toga AW, Thompson PM. 2007. 3D pattern of brain abnormalities in williams syndrome visualized using tensor-based morphometry. NeuroImage. 36(4):1096–1109. doi: 10.1016/j.neuroimage.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EL, Thompson PM. 2013. Typical and atypical brain development: a review of neuroimaging studies. Dialogues Clin Neurosci. 15(3):359–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT et al. 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 31(3):968–980. [DOI] [PubMed] [Google Scholar]
- Fish AM, Cachia A, Fischer C, Mankiw C, Reardon PK, Clasen LS, Blumenthal JD, Greenstein D, Giedd JN, Mangin J-F, et al. 2016Influences of brain size, sex, and sex chromosome complement on the architecture of human cortical folding. Cerebral Cortex. 1991: 27(12), 5557–5567. doi: 10.1093/cercor/bhw323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Raznahan A, Alexander-Bloch A, Schmitt E, Gogtay N, Rapoport JL. 2015. Child psychiatry branch of the National Institute of Mental Health longitudinal structural magnetic resonance imaging study of human brain development. Neuropsychopharmacology. 40(1):43–49. doi: 10.1038/npp.2014.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, He Y, Chen ZJ, Evans AC. 2012. Convergence and divergence of thickness correlations with diffusion connections across the human cerebral cortex. NeuroImage. 59(2):1239–1248. doi: 10.1016/j.neuroimage.2011.08.017. [DOI] [PubMed] [Google Scholar]
- Hazlett HC, Poe MD, Lightbody AA, Styner M, MacFall JR, Reiss AL, Piven J. 2012. Trajectories of early brain volume development in fragile X and autism RH: trajectory of brain volume in fragile X. J Am Acad Child Adolesc Psychiatry. 51(9):921–933. doi: 10.1016/j.jaac.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft F, Carter JC, Lightbody AA, Cody Hazlett H, Piven J, Reiss AL. 2010. Region-specific alterations in brain development in one- to three-year-old boys with fragile X syndrome. Proc Natl Acad Sci U S A. 107(20):9335–9339. doi: 10.1073/pnas.1002762107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong DS, Reiss AL. 2014. Cognitive and neurological aspects of sex chromosome aneuploidies. Lancet Neurol. 13(3):306–318. doi: 10.1016/S1474-4422(13)70302-8. [DOI] [PubMed] [Google Scholar]
- Khundrakpam BS, Lewis JD, Jeon S, Kostopoulos P, Itturia Medina Y, Chouinard-Decorte F, Evans AC. 2019. Exploring individual brain variability during development based on patterns of maturational coupling of cortical thickness: a longitudinal MRI study. Cereb Cortex. 29(1):178–188. doi: 10.1093/cercor/bhx317. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Singh V., Lee J. K., Lerch J., Ad-Dab’bagh Y., MacDonald D. & Evans A. C.. 2005. Automated 3-D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification. Neuroimage, 27(1), 210–221. [DOI] [PubMed] [Google Scholar]
- Lee NR, Adeyemi EI, Lin A, Clasen LS, Lalonde FM, Condon E, Driver DI, Shaw P, Gogtay N, Raznahan A et al. 2016. Dissociations in cortical morphometry in youth with down syndrome: evidence for reduced surface area but increased thickness. Cereb Cortex. 26(7):2982–2990. doi: 10.1093/cercor/bhv107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NR, Wallace GL, Adeyemi EI, Lopez KC, Blumenthal JD, Clasen LS, Giedd JN. 2012. Dosage effects of X and Y chromosomes on language and social functioning in children with supernumerary sex chromosome aneuploidies: implications for idiopathic language impairment and autism spectrum disorders. J Child Psychol Psychiatry. 53(10):1072–1081. doi: 10.1111/j.1469-7610.2012.02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage J-F, Hong DS, Raman M, Marzelli M, Roeltgen DP, Lai S, Ross J, Reiss AL. 2014. Brain morphology in children with 47,XYY syndrome: a voxel-and surface-based morphometric study. Genes Brain Behav. 13(2):127–134. doi: 10.1111/gbb.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch JP, Worsley K, Shaw WP, Greenstein DK, Lenroot RK, Giedd J, Evans AC. 2006. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. NeuroImage. 31(3):993–1003. doi: 10.1016/j.neuroimage.2006.01.042. [DOI] [PubMed] [Google Scholar]
- Lin A, Clasen L, Lee NR, Wallace GL, Lalonde F, Blumenthal J, Giedd JN, Raznahan A. 2015. Mapping the stability of human brain asymmetry across five sex-chromosome aneuploidies. J Neurosci. 35(1):140–145. doi: 10.1523/JNEUROSCI.3489-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Sebat J. 2012. CNVs: harbinger of a rare variant revolution in psychiatric genetics. Cell. 148(6):1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankiw C, Park MTM, Reardon PK, Fish AM, Clasen LS, Greenstein D, Giedd JN, Blumenthal JD, Lerch JP, Chakravarty MM et al. 2017. Allometric analysis detects brain size-independent effects of sex and sex chromosome complement on human cerebellar organization. J Neurosci. 37(21):5221–5231. doi: 10.1523/JNEUROSCI.2158-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca D, Moreno-De-Luca A, Cubells JF, Sanders SJ. 2014. Cross-disorder comparison of four neuropsychiatric CNV loci. Curr Genet Med Rep. 2(3):151–161. doi: 10.1007/s40142-014-0045-7. [DOI] [Google Scholar]
- Nadig A, Reardon PK, Seidlitz J, McDermott CL, Blumenthal JD, Clasen LS, Lalonde F, Lerch JP, Chakravarty MM, Raznahan A. 2018. Carriage of supernumerary sex chromosomes decreases the volume and alters the shape of limbic structures. eNeuro. 5(5). doi: 10.1523/ENEURO.0265-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Wohlert M. 1991. Chromosome abnormalities found among 34,910 newborn children: results from a 13-year incidence study in Arhus, Denmark. Hum Genet. 87(1):81–83. doi: 10.1007/bf01213097. [DOI] [PubMed] [Google Scholar]
- Otto SP, Pannell JR, Peichel CL, Ashman T-L, Charlesworth D, Chippindale AK, Delph LF, Guerrero RF, Scarpino SV, McAllister BF. 2011. About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 27(9):358–367. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Poldrack RA, Mumford JA, Schonberg T, Kalar D, Barman B, Yarkoni T. 2012. Discovering relations between mind, brain, and mental disorders using topic mapping. PLoS Comput Biol. 8(10):e1002707. doi: 10.1371/journal.pcbi.1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Cutter W, LaLonde F, Robertson D, Daly E, Conway GS, Skuse DH, Ross J, Giedd JN, Murphy DDGM. 2010. Cortical anatomy in human X monosomy. NeuroImage. 49(4):2915–2923. doi: 10.1016/j.neuroimage.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Lee NR, Greenstein D, Wallace GL, Blumenthal JD, Clasen LS, Giedd JN. 2016. Globally divergent but locally convergent X- and Y-chromosome influences on cortical development. Cereb Cortex. 26(1):70–79. doi: 10.1093/cercor/bhu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A., Parikshak N.N., Chandran V., Blumenthal J.D., Clasen L.S., Alexander-Bloch A.F., Zinn A.R., Wangsa D., Wise J., Murphy D.G.M., Bolton P.F., Ried T., Ross J., Giedd J.N., Geschwind D.H.. 2018. Sex-chromosome dosage effects on gene expression in humans. Proc. Natl. Acad. Sci. U. S. A. 115, 7398–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Lenroot R, Thurm A, Gozzi M, Hanley A, Spence SJ, Swedo SE, Giedd JN. 2012. Mapping cortical anatomy in preschool aged children with autism using surface-based morphometry. NeuroImage Clin. 2:111–119. doi: 10.1016/j.nicl.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Lerch JP, Lee N, Greenstein D, Wallace GL, Stockman M, Clasen L, Shaw PW, Giedd JN. 2011. Patterns of coordinated anatomical change in human cortical development: a longitudinal neuroimaging study of maturational coupling. Neuron. 72(5):873–884. doi: 10.1016/j.neuron.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon PK, Clasen L, Giedd JN, Blumenthal J, Lerch JP, Chakravarty MM, Raznahan A. 2016. An allometric analysis of sex and sex chromosome dosage effects on subcortical anatomy in humans. J Neurosci. 36(8):2438–2448. doi: 10.1523/JNEUROSCI.3195-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijn S, Swaab H, Baas D, de Haan E, Kahn RS, Aleman A. 2012. Neural systems for social cognition in Klinefelter syndrome (47,XXY): evidence from fMRI. Soc Cogn Affect Neurosci. 7(6):689–697. doi: 10.1093/scan/nsr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Garcia R, Atienza M, Clemmensen LH, Cantero JL. 2012. Effects of network resolution on topological properties of human neocortex. NeuroImage. 59(4):3522–3532. doi: 10.1016/j.neuroimage.2011.10.086. [DOI] [PubMed] [Google Scholar]
- Schmitt JE, Giedd JN, Raznahan A, Neale MC. 2017. The genetic contributions to maturational coupling in the human cerebrum: a longitudinal pediatric twin imaging study. Cerebral Cortex, 28(9),3184–3191. doi: 10.1093/cercor/bhx190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Lenroot R, Ordaz SE, Wallace GL, Lerch JP, Evans AC, Prom EC, Kendler KS, Neale MC, Giedd JN. 2009. Variance decomposition of MRI-based covariance maps using genetically-informative samples and structural equation Modeling. NeuroImage. 47(1):56–64. doi: 10.1016/j.neuroimage.2008.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon TJ, Ding L, Bish JP, McDonald-McGinn DM, Zackai EH, Gee J. 2005. Volumetric, connective, and morphologic changes in the brains of children with chromosome 22q11.2 deletion syndrome: an integrative study. NeuroImage. 25(1):169–180. doi: 10.1016/j.neuroimage.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Sotiras A, Toledo JB, Gur RE, Gur RC, Satterthwaite TD, Davatzikos C. 2017. Patterns of coordinated cortical remodeling during adolescence and their associations with functional specialization and evolutionary expansion. Proc Natl Acad Sci U S A. 114(13):3527–3532. doi: 10.1073/pnas.1620928114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Lee AD, Dutton RA, Geaga JA, Hayashi KM, Eckert MA, Bellugi U, Galaburda AM, Korenberg JR, Mills DL et al. 2005. Abnormal cortical complexity and thickness profiles mapped in williams syndrome. J Neurosci. 25(16):4146–4158. doi: 10.1523/JNEUROSCI.0165-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannan CMJ, Cropley VL, Chakravarty MM, Bousman C, Ganella EP, Bruggemann JM, Weickert TW, Weickert CS, Everall I, McGorry P et al. 2019. Evidence for network-based cortical thickness reductions in schizophrenia. Am J Psychiatry. 176(7):552–563. doi: 10.1176/appi.ajp.2019.18040380. [DOI] [PubMed] [Google Scholar]
- Yarkoni Tal PRA, Nichols TE, Essen DCV, Wager TD. 2011. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 8(8):665–670. doi: 10.1038/nmeth.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijdenbos AP, Forghani R, Evans AC. 2002. Automatic “pipeline” analysis of 3-D MRI data for clinical trials: application to multiple sclerosis. IEEE Trans Med Imaging. 21(10):1280–1291. doi: 10.1109/TMI.2002.806283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


