Abstract
The plasticity-related protein Synaptopodin (SP) has been implicated in neuronal plasticity. SP is targeted to dendritic spines and the axon initial segment, where it organizes the endoplasmic reticulum (ER) into the spine apparatus and the cisternal organelle, respectively. Here, we report an inducible third localization of SP in the somata of activated granule cell ensembles in mouse dentate gyrus. Using immunofluorescence and fluorescence in situ hybridization, we observed a subpopulation of mature granule cells (~1–2%) exhibiting perinuclear SP protein and a strong somatic SP mRNA signal. Double immunofluorescence labeling for Arc demonstrated that ~ 75% of these somatic SP-positive cells are also Arc-positive. Placement of mice into a novel environment caused a rapid (~2–4 h) induction of Arc, SP mRNA, and SP protein in exploration-induced granule cell ensembles. Lesion experiments showed that this induction requires input from the entorhinal cortex. Somatic SP colocalized with α-Actinin2, a known binding partner of SP. Finally, ultrastructural analysis revealed SP immunoprecipitate on dense plates linking cytoplasmic and perinuclear ER cisterns; these structures were absent in granule cells of SP-deficient mice. Our data implicate SP in the formation of contextual representations in the dentate gyrus and the behaviorally induced reorganization of cytoplasmic and perinuclear ER.
Keywords: Arc, endoplasmic reticulum, entorhinal cortex lesion, hippocampus, learning and memory
Introduction
The dentate gyrus is the entry gate to the hippocampus relaying spatial and episodic information from the entorhinal cortex to Ammon’s horn (van Strien et al. 2009). It functions as a filter separating known information patterns from novel ones (e.g., Schmidt et al. 2012). At the cellular level, young granule cells mediate pattern separation, whereas older granule cells facilitate pattern completion (Nakashiba et al. 2012).
The ensemble of granule cells activated in a specific environmental context can be identified using immediate early genes as markers, for example, activity-regulated cytoskeleton-associated protein (Arc) (Guzowski et al. 2001; Pinaud et al. 2001; Guzowski 2002; Temple et al. 2003; Chawla et al. 2005; Ramirez-Amaya et al. 2005; Ramirez-Amaya et al. 2013). These immediate early genes are expressed by only ~ 1–2% of granule cells, in line with dentate sparse coding (Diamantaki et al. 2016; Severa et al. 2017). Placing an animal into a novel environment activates another ensemble and transiently doubles the number of activated granule cells (Guzowski et al. 2001; Pinaud et al. 2001; Guzowski 2002; Chawla et al. 2005). Importantly, granule cell ensembles are necessary and sufficient for the formation and recall of episodic memory traces (Liu et al. 2012, 2014; Ramirez et al. 2013) and their valence can be modified by experience (Redondo et al. 2014). The mechanisms and molecular players involved in their formation and plasticity are incompletely understood, however.
The plasticity-related protein Synaptopodin (SP) is found in kidney podocytes and telencephalic neurons (Mundel et al. 1997). It is tightly associated with the spine apparatus of dendritic spines (Deller et al. 2000) and the cisternal organelle of the axon initial segment (AIS; Bas Orth et al. 2007). Both organelles consist of stacks of endoplasmic reticulum (ER) and require SP for their formation (Deller et al. 2003; Bas Orth et al. 2007). Using plasticity protocols (Yamazaki et al. 2001; Fukazawa et al. 2003) as well as loss- (Deller et al. 2003; Jedlicka et al. 2009; Zhang et al. 2013) and gain-of-function (Vlachos et al. 2013) strategies, a role in Hebbian (Jedlicka and Deller 2017) and homeostatic (Vlachos et al. 2013) synaptic plasticity was established. Mechanistically, SP influences spine head expansion (Okubo-Suzuki et al. 2008; Vlachos et al. 2009; Zhang et al. 2013), the recruitment of AMPA receptors (Vlachos et al. 2009), and Ca2+-transients in spines (Korkotian and Segal 2011; Korkotian et al. 2014). Thus, SP is an essential component of the machinery executing changes in synaptic strength (Jedlicka and Deller 2017) and a major plasticity-related protein.
In this study, we provide first evidence that the plasticity-related protein SP is rapidly induced in activated dentate granule cell ensembles. Surprisingly, SP accumulates in the somatic compartment of activated granule cells, where it is not normally abundant. Ultrastructural analysis revealed SP-positive dense plates linking perinuclear and somatic ER cisterns, thus changing the geometry of the ER labyrinth. We propose that this alteration could affect ER-dependent signaling from synapse to the nucleus and the ability of granule cells to express plasticity.
Materials and Methods
Animals
Adult male mice (C57BL6/J, Janvier) and SP-deficient mice (Deller et al., 2003; C57BL6/J background; bred at the animal facility in Frankfurt) were used for experimental analysis. Animal care and experimental procedures were performed in agreement with the German law on the use of laboratory animals (animal welfare act; TierSchG). Animal welfare was supervised and approved by the Institutional Animal Welfare Officer.
Tissue Preparation
Mice were deeply anesthetized with an overdose of pentobarbital (500 mg/kg body weight) and transcardially perfused with 0.9% sodium chloride (NaCl) followed by 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS, pH 7.4) for in situ hybridization and immunofluorescence staining or using 4% PFA and 0.5% glutaraldehyde (Polysciences) in 0.1 M cacodylate buffer (pH 7.4) for electron microscopy analysis. For immunohistochemistry, brains were postfixed in 4% PFA overnight (o/n) at 4 °C, whereas for in situ hybridization, brains were postfixed for 4 h in 4% PFA followed by 20% sucrose at 4 °C o/n. For electron microscopy, brains were postfixed in 4% PFA and 0.5% glutaraldehyde in 0.1 M cacodylate buffer at 4 °C o/n. Frontal sections (40 μm) of dorsal hippocampus were cut with a vibratome (VT 1000S, Leica) for immunohistochemistry or using a cryostat (CM3050S, Leica) for in situ hybridization and stored at −20 °C until use.
Novel Environment
Animals were housed under control cage conditions before being placed into a novel environment. The novel environment consisted of a new standard mouse cage enriched with several objects. Mice were allowed to explore this novel environment without interference for 5 min before they were taken back into their old cage. They were kept in their home cage for 0.5, 2, 4, 6, 8, 12, 24, or 48 h before anesthesia and transcardial perfusion (Fig. 1).
Figure 1.
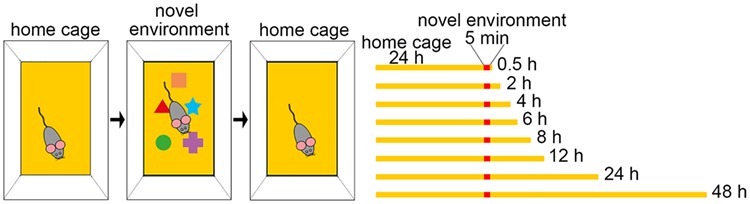
Schematic of behavioral experiments. Mice were housed under control cage conditions for 24 h, before being placed into a cage with a novel environment for 5 min. They were put back into their control cage and killed after 0.5, 2, 4, 6, 8, 12, 24, and 48 h, respectively. N = 5 animals per time point.
Entorhinal Denervation and Control of Lesion Quality
Unilateral transection of the left perforant path was performed under deep anesthesia using a wire knife (David Kopf Instruments) as described previously (Del Turco et al. 2003, 2014). After perfusion, the brains were removed and cut into an anterior block containing most of the dorsal hippocampus and a posterior block containing the ventral hippocampus and the entorhinal cortex. The latter block was cut into horizontal sections containing the lesion site to verify the correct placement of the wire-knife cut. The anterior block was cut into frontal sections and used for analysis. Some sections were used to verify the entorhinal denervation extend based on the appearance of degeneration products or the activation of astrocytes and microglial cells in the outer molecular layer (Del Turco et al. 2003, 2014). Animals without complete lesions or with damage to the hippocampus itself were excluded from the study.
Immunohistochemistry
Free-floating sections were washed several times in 50 mM Tris-buffered saline (TBS) containing 0.1% Triton X-100, incubated in a blocking buffer (0.5% Triton X-100, 5% bovine serum albumin (BSA) in 50 mM TBS) for 30 min at room temperature (RT) and subsequently incubated with the appropriate primary antibody (diluted in 0.1% Triton X-100, 1% BSA in 0.05 M TBS) for 2 days at RT. Guinea pig anti-Arc (1 mg/mL, 1:1000, Synaptic Systems), guinea pig anti-SP (1 mg/mL, 1:2000, Synaptic Systems), rabbit anti-SP (1 mg/ml, 1:2000, Synaptic Systems), rabbit anti-Calbindin D 28k (1 mg/mL, 1:500, Swant), guinea pig anti-AnkG (100 μL lyophilized, 1:500, Synaptic Systems), rabbit anti-α-Actinin2 (0.142 mg/mL, 1:500, Abcam, clone EP2529Y), and rabbit anti-GRP78 (1 mg/mL, 1:500, Biotrend) were used as primary antibodies. After several washing steps, sections were incubated with the appropriate secondary Alexa-conjugated antibodies (2 mg/mL, 1:2000, goat anti guinea pig or goat anti rabbit Alexa Fluor 488 or 568, Invitrogen) for 4 h at RT, counterstained with Draq5 (1:10 000, Thermo Fisher Scientific) to visualize nuclei and finally mounted in Fluorescence Mounting Medium (Dako, Agilent Technologies).
Electron Microscopy
Serial 50 μm frontal brain sections were cut with a vibratome (Leica VT1000S), washed in TBS, incubated in 0.1% NaBH4 (Sigma-Aldrich), and blocked with 5% BSA for 1 h at RT to reduce nonspecific staining. For primary antibodies, rabbit anti-Arc (1 μg/μL, 1:1000; Synaptic Systems), guinea pig anti-Arc (1 mg/mL, 1:1000, Synaptic Systems), rabbit anti-SP (1 mg/mL, 1:2000, Synaptic Systems), and rabbit anti-GRP78 (1 mg/mL, 1:500, Biotrend) were used. Biotinylated specific anti-IgG (1:200; Vector Laboratories; biotinylated goat anti guinea pig or biotinylated goat anti rabbit) was used as secondary antibody. After washing in TBS, sections were incubated in avidin–biotin–peroxidase complex (ABC-Elite; Vector Laboratories) for 90 min at RT and were reacted with diaminobenzidine (DAB) solution (Vector Laboratories) at RT. Sections were silver-intensified by incubation in 3% hexamethylenetetramine (Sigma-Aldrich), 5% silver nitrate (AppliChem), and 2.5% disodium tetraborate (Sigma-Aldrich) for 10 min at 60 °C, in 1% tetrachlorogold (AppliChem) solution for 3 min, and in 2.5% sodium thiosulfate (Sigma-Aldrich) for 3 min. After staining, sections were washed in 0.1 M cacodylate buffer, osmicated (0.5% OsO4 in cacodylate buffer), dehydrated (1% uranyl acetate [Serva] in 70% ethanol), and embedded in Durcupan (Sigma-Aldrich). Sections were collected on single-slot Formvar-coated copper grids that were contrast enhanced with lead citrate for 4 min and examined using a Zeiss electron microscope (Zeiss EM 900).
SP Probe Synthesis
The cDNA of murine SP (Mundel et al. 1997) was subcloned into the pBluescript KS–Vector (Agilent Technologies) using HindIII and ApaI restriction sites. Recombinant plasmids containing SP cDNA were linearized and transcribed with T3 (anti-sense probe) or T7 (sense probe) RNA polymerase using a digoxigenin (DIG) RNA labeling kit (Roche/Sigma-Aldrich). No staining was observed in wild-type brain sections for sense probe or on SP-deficient sections for antisense probe.
Fluorescence in situ Hybridization
For fluorescence in situ hybridization (FISH), sections were incubated in 10 mM citrate buffer (pH 6) for 20 min at 85 °C, washed several times in 2× SSC (LONZA AccuGEN), and prehybridized for 2 h at 60 °C with hybridization buffer (50% formamide, 5× SSC, 5% dextran sulfate, 500 μg/mL DNA MB grade from sperm [Roche], 250 μg/mL t-RNA [Sigma Aldrich] and 1× Denhardt’s [Sigma Aldrich]). After heat treatment of FISH probes for 5 min at 85 °C, sections were incubated with probe in hybridization buffer (1:1000) overnight at 60 °C. Several washing steps (2× SSC for 10 min at RT, 2× SSC/50% formamide [AppliChem] for 15 min at 60 °C, 0.1× SSC/50% formamide for 15 min at 60 °C, 0.1× SSC for 15 min at 60 °C, and TN buffer [0.1 M Tris-HCl, 0.15 M NaCl; pH 7.4] for 5 min at RT) were performed before blocking solution (1% blocking reagent [Roche] in TN buffer) was added for 30 min at RT. After blocking, anti-Digoxigenin-POD (1:2000 in blocking solution; Roche) was added to sections for 2 h at RT. Sections were washed several times with TNT (0.1 M Tris-HCl, 0.15 M NaCl, 0.3% Triton X-100) and incubated in TSA-Plus Cyanine 3 System (Perkin Elmer) diluted in amplification solution (1:50) for 10 min in the dark at RT. After several washing steps with TNT, sections were further processed for immunohistochemistry.
Cell Counting
Quantitative confocal microscopy analysis in combination with Fiji/ImageJ (Schindelin et al. 2012) were used for cell counting. The suprapyramidal blade of the dentate gyrus (dorsal hippocampus; 4 dentate gyri; 40 μm sections) was analyzed per animal (N), except where indicated in the figure caption. Draq5 was used to visualize nuclei in the granule cell layer. In brief, 5–10 single plane confocal images were captured of the suprapyramidal blade of the granule cell layer using a 20× objective and 4× digital zoom (Eclipse C1 Plus, Nikon). Single- and double-labeled Arc and somatic SP-positive cells as well as the total number of nuclei stained with Draq5 were counted using the cell counter plug-in of Fiji/ImageJ. The percentage of single- and double-labeled Arc and somatic SP-positive cells were averaged per animal. To benchmark the semiautomatic analysis technique, counting was performed by two independent investigators (M.P. and M.C.), which yielded comparable results.
Statistics
Statistical analysis was done using GraphPad Prism 6 Software. One-way ANOVA and post hoc multiple comparison (Bonferroni correction) were used. Significance level was set at *P < 0.05. Individual tests and test parameters are indicated in figure captions.
Results
A Subpopulation of Mature Dentate Granule Cells Exhibits SP-Positive Structures in Their Somata
The plasticity-related protein SP is tightly associated with specialized endoplasmic reticulum (ER) organelles, that is, the spine apparatus found in dendritic spines (Deller et al. 2000) and the cisternal organelle found in the AIS (Bas Orth et al. 2007). Using sensitive fluorescence labeling of SP in combination with confocal imaging, we revisited the distribution of SP in the dentate gyrus and confirmed punctate SP labeling in the dentate molecular layer corresponding to spine apparatus organelles (Deller et al. 2000) and string-like SP dots and rods in the dentate granule cell layer corresponding to cisternal organelles (Fig. 2A1,B1; Deller et al. 2000; Bas Orth et al. 2007). In addition, we observed a small number of cells in the granule cell layer exhibiting SP-positive dots and SP-positive rods (Fig. 2B1). These SP-positive structures were arranged in a circular pattern, suggesting a somatic and perinuclear localization of SP. The vast majority of granule cells were devoid of such SP labeling (Fig. 2B1).
Figure 2.
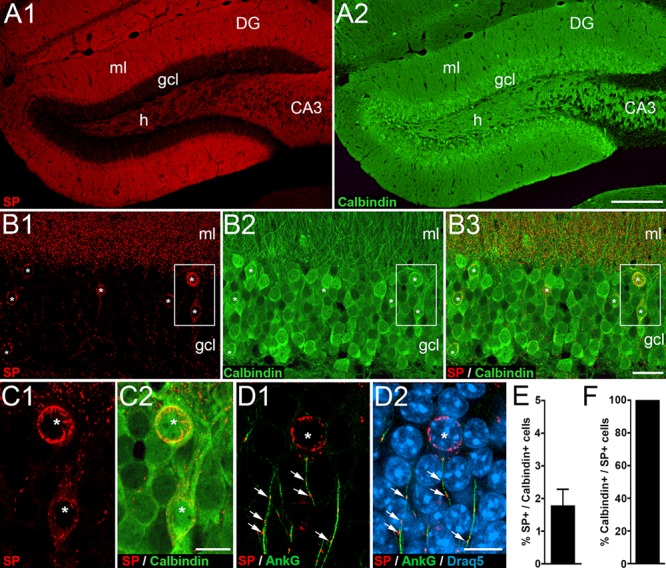
A subpopulation of mature dentate granule cells exhibits SP in their somata. (A) Immunofluorescence staining for SP (A1) and for Calbindin (A2), a marker for mature granule cells in the dentate gyrus (DG). (B) Higher magnification of the suprapyramidal granule cell layer (gcl) stained for SP (B1) and Calbindin (B2). (B3) Overlay shows colocalization of SP and Calbindin in mature granule cells (asterisks). (C) Higher magnifications of boxed area in B. Two granule cells (asterisks) with somatic SP expression (C1) also express Calbindin (C2). (D) Higher magnification of a section stained for SP (red; D1, D2), AnkyrinG, (AnkG, green; marker for the AIS; D1, D2), and Draq5 (nuclear marker; D2). Somatic SP formed circular structures (asterisk) that could be readily distinguished from punctate SP (arrows) in the AIS. CA3, hippocampal subfield CA3; h, hilus; ml, molecular layer. (E) Somatic SP was detected in 1.8 ± 0.5% (mean with SD; N = 4 animals; two dentate gyri sections analyzed per animal) of Calbindin-positive granule cells in the suprapyramidal blade of the DG. (F) All cells exhibiting somatic SP were Calbindin-positive. Scale bars: 200 μm (A2), 25 μm (B3), and 10 μm (C2, D2).
Since the granule cell layer contains several other cell types (e.g., Houser 2007) as well as immature granule cells (Kempermann et al. 2015), we verified that the SP-positive cells exhibiting strong SP labeling in their somata are mature granule cells. Double fluorescence labeling for SP and Calbindin, a marker for mature granule cells (Radic et al. 2017), was employed (Fig. 2A–C). This revealed that 1.8 ± 0.5% of all Calbindin-positive cells are also strongly SP-positive (Fig. 2E). Conversely, all cells exhibiting SP-positive structures in their soma were Calbindin-positive (Fig. 2F). We conclude from these observations that the somatic SP-labeled cells are mature granule cells.
Next, we verified that the circular SP-positive structures are not associated with the AIS of granule cells and used double fluorescence labeling against SP and AnkyrinG (Fig. 2D), a marker for the AIS (Sobotzik et al. 2009). Although we observed the expected double-labeling of SP and AnkyrinG for the vertically oriented string-like SP puncta (Sobotzik et al. 2009), the circular SP-positive structures surrounding nuclei were not double-labeled (Fig. 2D). We conclude from these observations that the circular SP-positive structures are distinct from the SP-positive cisternal organelle in the AIS.
SP-Positive Structures in Mature Granule Cells Are Tightly Associated with the Somatic Endoplasmic Reticulum
Since SP is tightly associated with the ER of spines and the AIS (Deller et al. 2000; Bas Orth et al. 2007), we wondered whether this is also the case for the somatic SP-positive structures. Therefore, we used double immunofluorescence for SP and 78 kDa glucose-regulated protein (GRP78), an ER chaperone that protects cells against ER stress (Thon et al. 2014) to further elucidate the localization of SP in granule cells. Confocal analysis revealed a close apposition of somatic SP-positive structures and GRP78-positive ER within the granule cell body, especially in the perinuclear region (Fig. 3A).
Figure 3.
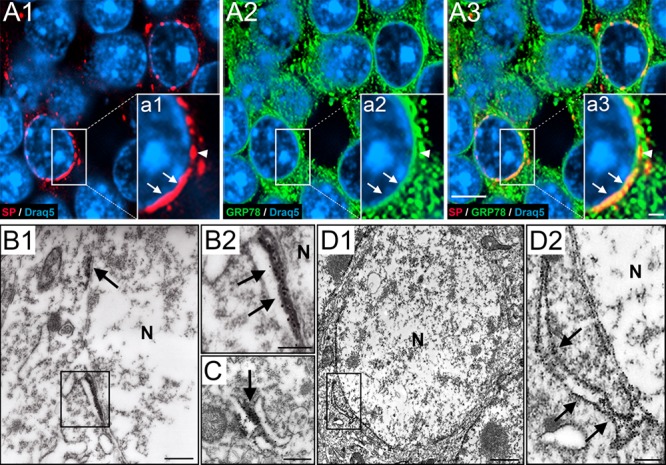
SP associates with perinuclear endoplasmic reticulum of dentate granule cells. (A1) Higher magnification of a portion of the granule cell layer (gcl). Section immunolabeled for SP (red) and Draq5 (nuclei, blue). Somatic SP is typically found close to the nucleus. (A2) The same section labeled for 78-kDa glucose-regulated protein (GRP78, green), a marker for endoplasmic reticulum (ER), and Draq5 (blue). (A3) Triple labeling of SP, GRP78, and Draq5 demonstrates colocalization of SP and GRP78. Double-labeled structures are abundant close to the nucleus. Note that SP is highly colocalized with GRP78, whereas GRP78 is only colocalized with SP in the perinuclear zone. (a1–a3) Insets show higher magnifications of boxed areas. (B–D) Electron microscopy reveals SP- or GRP78-positive ER cisterns. (B1) Part of a granule cell located in the gcl. Next to the nucleus (N), two SP-positive structures (rectangle; arrow) can be seen. (B2) Higher magnification of rectangle in B1. The DAB precipitate has been intensified with silver particles (see methods). Note association of SP with dense material in between the perinuclear ER. (C) SP was also found in dense material between cytoplasmic ER cisterns, forming triad structures of ER-SP-ER similar to those seen in the spine apparatus or the cisternal organelle. (D1) Granule cell located in the gcl stained for GRP78. Immunopositive ER cisterns were found in the soma and perinuclear. (D2) Boxed area in D1 shown at higher magnification. Arrows point to a cistern connected to the perinuclear ER. Scale bars: 5 μm (A3), 1 μm (inset a3) 500 nm (B1, C), 250 nm (B2, D2), and 1 μm (D1).
Using ultrastructural analysis, we verified our light microscopic observations and found SP-positive structures in between somatic ER cisterns (Fig 3B,C). Of note, SP immunoprecipitate did not label ER membranes, as seen following immunolabeling against GRP78 (Fig. 3D). Rather, SP immunoprecipitate was associated with dense material in between the ER membranes (Fig. 3B,C). This localization is similar to the one found in spines and in the AIS, where SP is required for the formation of the dense plates, which stack the ER and which form the spine apparatus and the cisternal organelles (Deller et al. 2003; Bas Orth et al. 2007). In line with these findings, we did not observe stacked perinuclear ER elements in neighboring granule cells, which did not contain somatic SP. We conclude from these observations that somatic SP organizes the perinuclear and somatic ER of a subpopulation of granule cells in the dentate gyrus, similar to its ER-organizing role in the other two compartments.
Colocalization of SP and α-Actinin2, an Actin-Bundling Protein, in Granule Cell Somata
Previous studies identified α-Actinin2 as a binding partner for SP (Asanuma et al. 2005; Kremerskothen et al. 2005). Thus, we searched for a colocalization of α-Actinin2 and SP in the somatic SP-positive granule cells. Indeed, double immunofluorescence labeling for SP and α-Actinin2 revealed that somatic SP-positive granule cells also accumulate α-Actinin2 in their somata (Fig. 4A–C). Furthermore, we confirmed colocalization of SP and α-Actinin2 in dendritic spines (Fig. 4D) and the AIS (Fig. 4E) of granule cells. We conclude from these observations that SP is closely associated with α-Actinin2 in all three SP-containing granule cell subcompartments, that is, spines, AIS, and the soma.
Figure 4.
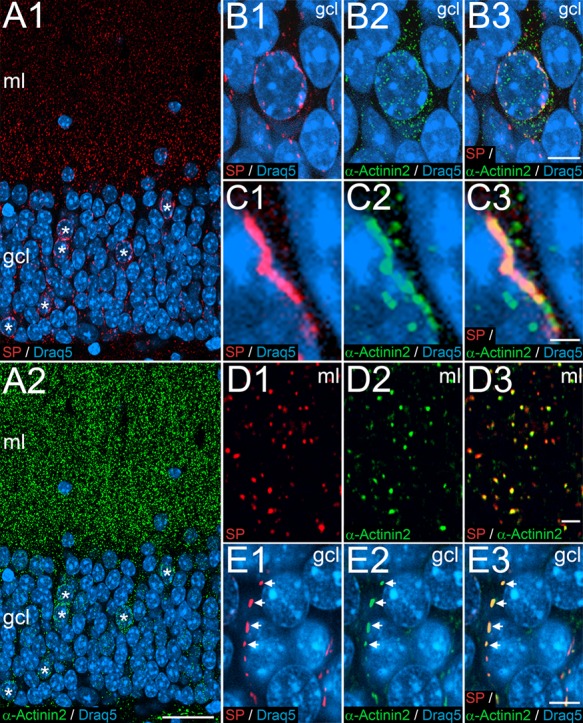
SP and the Actin-bundling protein α-Actinin2 colocalize in dentate granule cells. (A1, A2) Immunofluorescence labeling for SP (red), α-Actinin2 (green), and Draq5 (blue). Double-labeled SP- and alpha-Actinin2-positive cells are indicated (asterisks). (B) Higher magnification of a somatic SP-positive granule cell in the granule cell layer (gcl) of the dentate gyrus. Somatic SP is primarily found near the nucleus (B1), whereas α-Actinin2 is more widely distributed in the granule cell cytoplasm (B2). The merged picture illustrates a high degree of colocalization between SP and perinuclear α-Actinin2 (B3). (C1-C3) Higher magnification of a portion of the somatic SP-positive granule cell illustrated in (B) demonstrating a perinuclear SP-positive structure, which is also α-Actinin2-positive. (D) Immunofluorescence double-labeling for SP (red, D1) and α-Actinin2 (green, D2) in the molecular layer (ml). The merged image reveals a high degree of colocalization of SP and α-Actinin2 puncta (D3). These puncta represent light microscopic correlates of the spine apparatus organelle. (E) Immunofluorescence labeling for SP (red, E1) and α-Actinin2 (green, E2) in the gcl. The merged image reveals a high degree of colocalization of SP and α-Actinin2 puncta and rods arranged in a string-like fashion (E3). These structures correspond to cisternal organelles in the AIS. Scale bars: 25 μm (A2), 5 μm (B3, E3), 1 μm (C3), and 2 μm (D3).
Somatic SP-Positive Granule Cells Express Activity-Regulated Cytoskeleton-Associated Protein
We now wondered what the role of the somatic SP-positive granule cells could be. Since the distribution and the number of somatic SP-positive granule cells resemble the distribution and number of granule cells expressing activity-regulated cytoskeleton-associated protein (Arc), we speculated that there could be an overlap between these two cell populations. Accordingly, we performed double immunofluorescence for SP and Arc (Fig. 5) and found a high degree of overlap between Arc- and SP-positive granule cells: 75.1 ± 6.4% of somatic SP-positive cells were Arc-positive (Fig. 5C) and 60.2 ± 5.3% of Arc-positive cells contained somatic SP (Fig. 5D).
Figure 5.
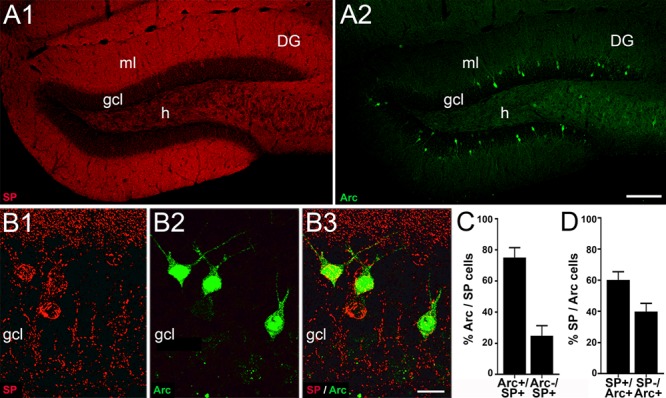
The subpopulation of dentate granule cells exhibiting somatic SP overlaps with the subpopulation of granule cells exhibiting activity-regulated cytoskeleton-associated protein (Arc). (A) Overview of immunofluorescence labeling for SP (red, A1) and Arc (green, A2) in the dentate gyrus (DG). (B1–B3) Higher magnification of a portion of the suprapyramidal blade. Most granule cells exhibiting somatic SP are also Arc-positive. (C) Arc was detected in 75.1 ± 6.4% of somatic SP-positive cells in the suprapyramidal blade of the gcl. Conversely, 24.9 ± 6.4% of somatic SP-positive cells were devoid of Arc (mean with SD, N = 5). (D) 60.2 ± 5.3% of Arc-positive cells in the suprapyramidal blade also contained somatic SP. Conversely, 39.8 ± 5.3% of Arc-positive cells did not contain somatic SP (mean with SD, N = 5). Scale bars: 100 μm (A2) and 15 μm (B3).
Somatic Expression of SP Can Be Induced in Dentate Granule Cells by Placing Mice into a Novel Environment
Arc labels an ensemble of dentate granule cells encoding for the environment of the animal (Temple et al. 2003, Schmidt et al. 2012). Placing an animal into a novel environment activates a “novel” second ensemble of granule cells. These newly activated cells now become Arc-positive, while the granule cells of the original ensemble lose Arc within roughly a day (Schmidt et al. 2012). Since the populations of Arc- and somatic SP-positive granule cells overlap, we speculated that somatic SP-positive granule cells might also be part of environment encoding granule cell ensembles. If this were the case, we would expect an increase in the number of somatic SP-positive cells after placing mice into a novel environment. Indeed, after exposing mice to an enriched environment for 5 min, we saw a rapid increase in the number of Arc and somatic SP-positive granule cells by 2 h (Fig. 6). A detailed time course analysis revealed an almost parallel increase in the number of Arc and somatic SP-positive granule cell populations (Fig. 6C–F). The maximum cell number, which was close to double the original number of cells, was reached between 2 and 6 h after placing the mice into the novel environment. By 12–24 h, Arc- and somatic SP-positive cell numbers had returned to baseline. Induction of additional Arc-positive cells occurred slightly earlier and elevated numbers of Arc-positive cells persisted slightly longer in the dentate gyrus than the numbers of somatic SP-positive cells.
Figure 6.
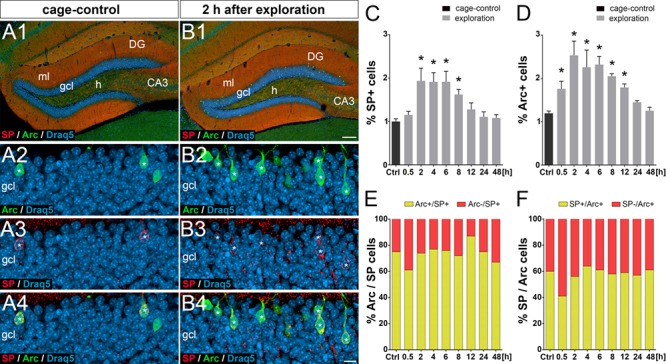
Granule cell ensembles activated by placing mice into a novel environment coexpress Arc and somatic SP. (A, B) Immunofluorescence labeling of the dentate gyrus (DG) for SP (red) and activity-regulated cytoskeleton-associated protein (Arc, green) under control conditions (A) and 2 h after placing the mice into a novel environment (B). Draq5 (blue) was used to label nuclei. (A2–A4) Higher magnification of a portion of the suprapyramidal blade in a cage control animal. Granule cells coexpressing somatic SP and Arc are indicated (asterisks). (B2–B4) Higher magnification of a portion of the suprapyramidal blade 2 h after placing the mouse into a novel environment. Note the increase in the fraction of granule cells expressing Arc and SP. Granule cells coexpressing somatic SP and Arc are indicated (asterisks). ml, molecular layer; h, hilus. Percentage of somatic SP-positive cells (C) and Arc-positive cells (D) in the suprapyramidal blade of the gcl. Cage control (Ctrl) animals and animals at different time points (0.5, 2, 4, 6, 8, 12, 24, and 48 h) after exposure to a novel environment are shown. One-way ANOVA with Bonferroni correction for multiple comparisons: F(8, 36) = 28.31 for (C), F(8, 36) = 29.07 for (D); *P < 0.05; N = 5 for each time point. (E) The percentages of somatic SP-positive cells also containing Arc are illustrated (mean values of N = 5 animals per time point). (F) The percentages of Arc-positive cells also containing SP in their somata are shown (mean values of N = 5 animals per time point). Scale bars: 100 μm (B1), 10 μm (B4).
Somatic SP and Arc Induction in Dentate Granule Cells Require Entorhinal Input
The activation of Arc expressing granule cell ensembles requires entorhinal input in rats (Temple et al. 2003). We wondered whether this is also the case for Arc- and somatic SP-positive granule cell ensembles in mice and removed the entorhinal input to the dentate gyrus by transecting the perforant path (Del Turco et al. 2014). Lesion quality was controlled on horizontal sections (Fig. 7A) and with the help of Fluoro-Jade C as a marker for degenerating terminals (Fig. 7B). Fluorescent labeling for Arc and SP revealed a complete loss of Arc and somatic SP-positive granule cells in the dentate gyrus on the side of the transection (Fig. 7C). In comparison, the contralateral dentate gyrus revealed a normal pattern (Fig. 7D). We conclude that Arc-positive granule cell ensembles in mice depend on input from the entorhinal cortex, as has been previously described in rats (Temple et al. 2003). Furthermore, we conclude that somatic SP expression of granule cells also depends on specific entorhinal input and that granule cells do not constitutively express somatic SP.
Figure 7.
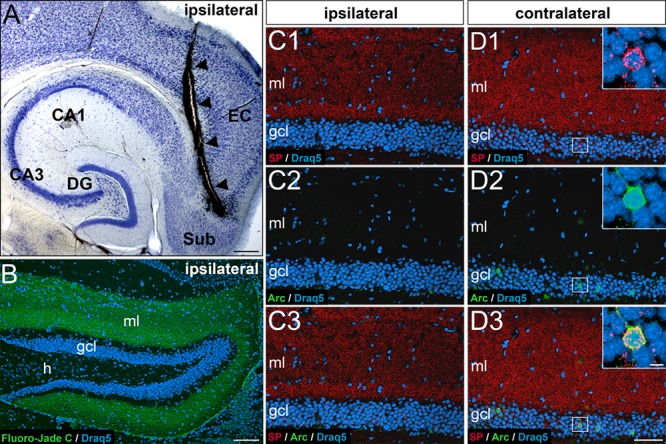
Entorhinal input is necessary for dentate granule cells to express somatic SP or Arc. (A) Horizontal section of a mouse brain showing the wire-knife cut (arrowheads). CA1, CA3, hippocampal subfields CA1, CA3; DG, dentate gyrus; EC, entorhinal cortex; Sub, subiculum. (B) Fluoro-Jade C labeling (green) of degeneration products in the DG (frontal section) ipsilateral to the lesion. Degenerating entorhinal terminals fill the molecular layer (ml) 7 days post lesion. Draq5 (blue) was used as a counterstain to visualize neuronal nuclei. gcl, granule cell layer; h, hilus. (C, D) Portions of the suprapyramidal layer of the DG ipsilateral (C) and contralateral (D) to the lesion site immunolabeled for SP (red) and activity-regulated cytoskeleton-associated protein (Arc, green) and counterstained for Draq5 (blue). Note the absence of SP and Arc-labeling on the side ipsilateral to the lesion (C1-C3; 7 days post lesion) compared to the non-denervated contralateral side (D). The nondenervated contralateral dentate gyrus displays a normal pattern of somatic-SP (D1), Arc (D2), and SP-Arc double-labeled cells (D3). Scale bars: 250 μm (A), 100 μm (B), 80 μm (D3), and 5 μm (D3, inset).
Taken together, behavioral and lesion experiments show that the expression of somatic SP in dentate granule cells is driven by activity, which is relayed via the entorhinal cortex to the dentate gyrus. The number of somatic SP-positive cells can be increased by specific input, for example, following exploration of a novel environment, and the number of somatic SP-positive cells can be reduced by input deprivation, for example, by transecting the perforant path.
High Levels of SP mRNA Are Expressed by Arc-Positive Ensembles of Granule Cells under Control Conditions and after Placement of Mice into a Novel Environment
We then wondered whether we could find evidence for an increased SP synthesis in the activated granule cell ensembles. Since SP mRNA is primarily found in the somata of granule cells and SP protein is abundant in spines and the AIS (Deller et al. 2000; Bas Orth et al. 2007), SP protein appears to be synthesized in the soma and subsequently transported to its target compartments (Deller et al. 2000). Thus, alterations in SP transport and/or a redistribution of SP protein to the soma would be mechanistically sufficient to explain a somatic accumulation of SP without requiring changes in SP synthesis. Alternatively, if an increased de novo synthesis of SP contributes to somatic SP, a stronger SP mRNA signal would be expected in activated granule cell ensembles.
To address this question, we used FISH for SP mRNA and labeled the activated granule cell ensembles with an antibody against Arc protein. We chose three time points for our analysis: baseline expression before stimulation, 2 h after placement into a novel environment (maximum Arc and SP expression), and 24 h after placement into a novel environment (return to baseline). Already under baseline conditions, we observed a small percentage of granule cells expressing high levels of SP mRNA (Fig. 8A). The percentage of these strongly SP mRNA-positive cells (Fig. 8E) corresponds nicely to the number of somatic SP-positive cells (cf. Fig. 2E). Furthermore, coimmunolabeling for Arc protein revealed that 84.5 ± 4.6% of the strongly SP mRNA-labeled cells are also Arc-positive (Fig. 8D) and 81.8 ± 0.8% of Arc-positive cells express SP mRNA (Fig. 8G). Following placement of the mice into a novel environment, the number of cells expressing high levels of SP mRNA (Fig. 8B,E) or Arc (Fig. 8B,H) almost doubled after 2 h. By 24 h after exploration, the numbers of strongly SP mRNA-and of Arc-positive granule cells were back to baseline (Fig 8C,E,H). The fraction of double-labeled granule cells expressing both SP mRNA and Arc protein (Fig. 8F,I) stayed high during the observation period. We conclude from these observations that activated granule cell ensembles express high levels of SP mRNA and that a strong SP mRNA expression can be rapidly induced in newly activated ensembles of dentate granule cells following placement of mice into a novel environment. Thus, an increased SP synthesis is highly likely to contribute to the somatic accumulation of SP in activated granule cell ensembles.
Figure 8.
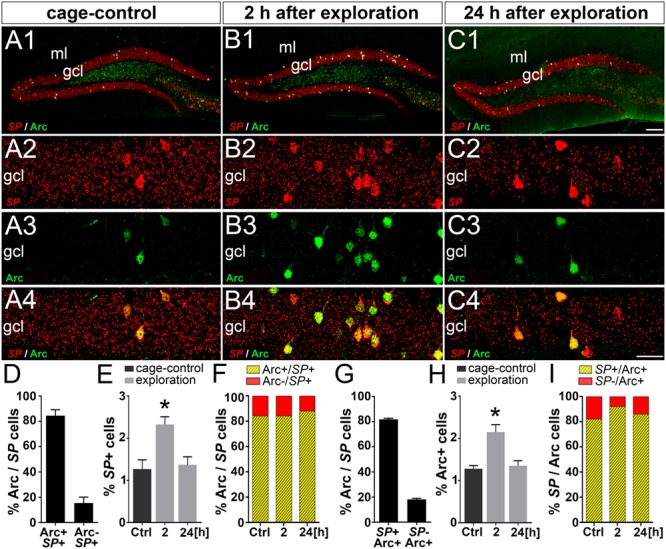
SP mRNA and Arc are coexpressed in ensembles of dentate granule cells activated in a novel environment. (A–C) Fluorescent in situ hybridization for SP mRNA and immunofluorescence (IF) labeling for activity-regulated cytoskeleton-associated protein (Arc) in the dentate gyrus of control mice (A1-A4) and mice 2 h (B1-B4) and 24 h (C1-C4) after placement into a novel environment. (D) In controls, 84.5 ± 4.6% of strongly SP mRNA expressing granule cells were also Arc-positive. Conversely, 15.5 ± 4.6% of strongly SP mRNA-positive granule cells were Arc-negative. (E) In control mice (Ctrl), 1.3 ± 0.2% of all granule cells (suprapyramidal blade) showed a strong SP mRNA signal. Two hours after placement into a novel environment, the number of granule cells expressing high levels of SP mRNA significantly increased to 2.3 ± 0.2%. By 24 h, the number of strongly SP mRNA expressing cells had returned back to control level (1.4 ± 0.2%). One-way ANOVA with Bonferroni correction for multiple comparisons; F(2, 6) = 26.02; *P < 0.05, N = 3 for each time point. (F) Fraction of strongly SP mRNA-positive granule cells coexpressing Arc. (G) In controls, 81.8 ± 0.9% of Arc-positive cells were also strongly SP mRNA-positive. Conversely, 18.2 ± 0.9% of Arc-positive cells were not strongly SP mRNA-positive. (H) Arc–IF followed a similar time course (cage-control: 1.3 ± 0.1%, 2 h: 2.1 ± 0.2%, and 24 h: 1.4 ± 0.1%). One-way ANOVA with Bonferroni correction for multiple comparisons; F(2, 6) = 39.86; *P < 0.05, N = 3 for each time point. (I) Fraction of Arc-positive granule cells strongly coexpressing SP mRNA. Scale bars: 100 μm (C1) and 25 μm (C4). gcl, granule cell layer; ml, molecular layer.
SP Is an Essential Component of Perinuclear ER Stacks
Finally, we wondered whether SP is not only a marker for the perinuclear ER stacks seen in the activated granule cells but, in fact, necessary for their formation. To address this question, we used SP-deficient mice, which do not have somatic SP-positive granule cells (Fig. 9A). In these mice, Arc-positive granule cells are still present, making it possible to identify activated granule cells in the mutants (Fig. 9B). Since ~ 75% of Arc-positive granule cells contain somatic SP structures in the wild-type, we reasoned that we should see perinuclear ER stacks in these cells, should they form in the absence of SP. Conversely, should the perinuclear ER stacks depend on SP, we would expect them to be absent. Accordingly, we examined Arc-positive and SP-deficient granule cells in the electron microscope, and none of these cells exhibited the characteristic perinuclear ER stacks that we had found in the wild-type (Fig. 9C,D). ER cisterns were abundantly present in these cells (Fig. 9D), suggesting that SP is needed to form the dense material between the ER cisterns. This is similar to what we have previously found for the spine apparatus (Deller et al. 2003) and the cisternal organelle (Bas Orth et al. 2007).
Figure 9.
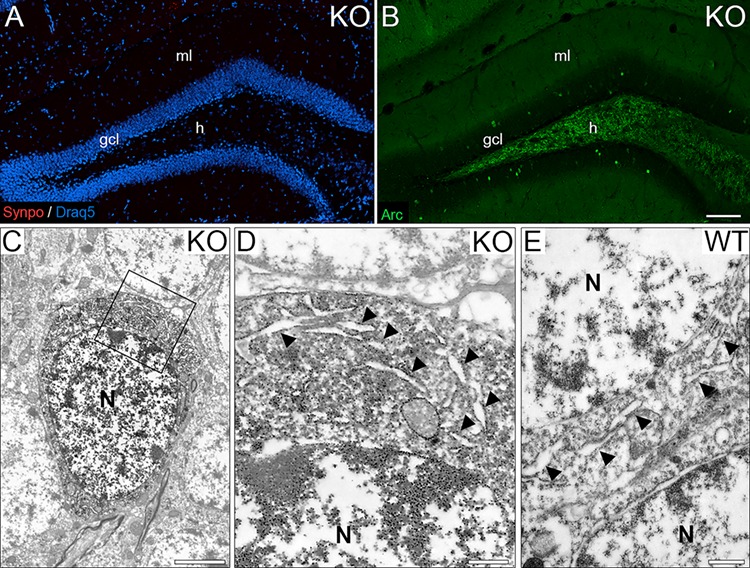
SP is an essential component of perinuclear ER stacks. (A) Dentate gyrus of a SP-deficient mouse stained for SP. Note the absence of staining for SP in all cellular compartments, that is, somatic SP, dendritic SP, and axonal SP. Draq5 was used as a counterstain to visualize cell nuclei. (B) Dentate gyrus of a SP-deficient mouse stained for Arc. Arc-positive (green) granule cell ensembles were still present in the granule cell layer (gcl) of SP-deficient mice. (C) Electron microscopy of an Arc-positive SP-deficient granule cell. DAB immunoprecipitate is seen in the soma. (D) Higher magnification of boxed area in (C). Cisterns of endoplasmic reticulum (ER) are abundant in the granule cell soma (arrow heads). Neither perinuclear ER stacks nor dense plates were observed (N = 3 animals, 6–7 cells per animal). (E) Somatic SP-negative wild-type (WT) granule cell. ER cisterns (arrow heads) were found throughout the soma but neither perinuclear ER stacks nor dense plates were observed (N = 3 animals, 100 SP-negative cells per animal). gcl, granule cell layer; ml, molecular layer; h, hilus; N, nuclei. Scale bars: 100 μm (A, B), 2.5 μm (C), and 500 nm (D, E).
To corroborate these findings in the mutant, we also investigated granule cells without somatic SP in the granule cell layer of wild-type mice stained for SP. We reasoned that perinuclear ER stacks should not form in the soma of granule cells, if SP is absent or scarce in this compartment. In line with this reasoning, we did not find perinuclear ER stacks in somatic SP-negative cells in the granule cell layer of wild-type animals (Fig. 9E). Similar to the situation in the SP-deficient mouse, ER cisterns were found to be abundant (Fig. 9E).
Taken together, our data from SP-deficient granule cells and from somatic SP-negative wild-type granule cells suggest that perinuclear ER stacks require SP for their formation. SP appears to be a necessary component of the dense material linking the ER cisterns but not of the ER cisterns themselves.
Discussion
In this study, we show that the plasticity-related protein SP and its mRNA are rapidly upregulated in Arc-positive granule cell ensembles in the dentate gyrus. Our main findings can be summarized as follows: (1) A subpopulation of ~ 1–2% Calbindin-positive granule cells shows perinuclear accumulations of the plasticity-related protein SP in their somata. (2) The majority (~75%) of these somatic SP-positive cells are also Arc-positive. (3) Upon placement of mice into a novel environment, SP mRNA, somatic SP, and Arc are rapidly upregulated in additional granule cells, transiently almost doubling the number of somatic SP-positive and Arc-positive cells. (4) Lesion experiments revealed that behavioral activation of somatic SP-positive granule cell ensembles requires input from the entorhinal cortex. (5) α-Actinin2, a known binding partner of SP, is coexpressed with SP in the somata of the behaviorally activated cells. (6) Ultrastructural analysis revealed that somatic SP is found on dense plates between perinuclear and cytoplasmic ER cisterns. (7) Perinuclear ER stacks are absent from Arc-positive granule cells in SP-deficient mice, suggesting that SP is required for their formation. In summary, we conclude that the plasticity-related protein SP is rapidly induced in environmentally activated granule cell ensembles. We propose that it plays a role in the plasticity of these ensembles, possibly by reorganizing the cytoplasmic ER and changing transmission of information from the synapse to the nucleus.
Somatic SP Is Found in Arc-Positive Granule Cell Ensembles
While revisiting the distribution of SP in the dentate gyrus using sensitive immunofluorescence confocal imaging, we were struck by the fact that some granule cells stand out with a strong circular SP signal in their somata. These cells may have escaped detection previously, because granule cells exhibit a baseline expression of SP mRNA (Deller et al. 2000) and because some punctate SP immunolabeling is present in the granule cell layer, that is, within the AIS and spines of granule cells (Deller et al. 2000; Bas Orth et al. 2005, 2007). Double-labeling of hippocampal sections with SP and Calbindin, which label mature granule cells and their dendrites (Radic et al. 2017), and SP and AnkyrinG, which label the AIS (Sobotzik et al. 2009), corroborated our findings and demonstrated that the somatic SP signal is clearly distinct from other SP puncta in the granule cell layer. Thus, it is possible to identify somatic SP-positive cells in between their somatic SP-negative peers based on the strength of the somatic SP signal as well as the conspicuous circular arrangement of SP.
The scattered pattern of somatic SP-positive cells and their small number, that is, 1–2% of all granule cells, are strikingly similar to the pattern and number of granule cell ensembles encoding for specific environments or contexts (Guzowski et al. 2001; Chawla et al. 2005; Schmidt et al. 2012). Since Arc is arguably a robust marker of such ensembles (Guzowski et al. 2001; Chawla et al. 2005; Bramham 2007), we double-labeled our sections for SP and Arc and found somatic SP in ~ 75% of Arc-positive cells. Using SP-FISH in combination with Arc immunolabeling, ~ 85% double-labeled cells were found. Such a high degree of colocalization suggests that somatic SP-positive cells are part of this functionally relevant cell population and not a random collection of granule cells expressing higher SP levels coincidentally.
SP mRNA and Somatic SP Are Rapidly Upregulated in Granule Cells Following a Behavioral Experience
After observing a strong somatic SP signal in some granule cells, we wondered whether this somatic protein accumulation goes together with an increase in SP mRNA. Alternatively, a resorting of SP protein within granule cells, for example, a transport of SP from the periphery back to the soma, could explain somatic SP aggregations. Using in situ hybridization for SP mRNA, we found that activated (Arc-positive) granule cells also express higher levels of SP mRNA compared to nonactivated neighboring granule cells, suggesting that a de novo synthesis of SP contributes to its accumulation in the soma.
Next, we wondered whether SP mRNA can also be rapidly induced in a novel ensemble of granule cells, as has been demonstrated for Arc (Chawla et al. 2005). To test this hypothesis, mice were placed in a novel environment and changes in SP mRNA, somatic SP, and Arc were analyzed. Indeed, under these conditions, the number of somatic SP-positive granule cells almost doubled. In line with others (Guzowski et al. 2001; Chawla et al. 2005; Bramham et al. 2008; Schmidt et al. 2012), we interpret this finding as evidence for the activation of a second ensemble of somatic SP-positive and Arc-positive granule cells encoding for the new, that is, second, environment. For a short time period, both sets of granule cells are somatic SP-positive, that is, the first one encoding for the original environment and the second one encoding for the new environment. Within 24 h, the first ensemble of granule cells downregulates SP and its set of immediate early genes, whereas the second ensemble of granule cells continues its gene expression.
Similar to Arc, the somatic expression of SP is inducible and rapid. Using a classification for immediate early genes, which distinguishes between regulatory transcription factors controlling downstream genes and effector immediate early genes, which directly influence cellular functions (Lanahan and Worley, 1998; Kubik et al., 2007), both genes appear to belong into the latter group mediating long-term structural and plasticity-related changes (Guzowski 2002). At present, however, very little is known about the transcriptional regulation of SP, except for the fact that it can be upregulated under conditions of plasticity (Yamazaki et al., 2001; Roth et al., 2001; see Jedlicka and Deller, 2017, for review), leaving room for a re-classification. Similarly, mechanisms regulating the removal of SP from the somata of activated granule cells are unknown. Both degradation and resorting of SP protein could play a role. Future experiments using cell culture models are needed to shed light on these questions.
SP mRNA Expression in Activated Granule Cells Depends on an Intact Entorhino-Hippocampal Projection
The ability of the dentate gyrus to form Arc-positive granule cell ensembles depends on afferent input from the entorhinal cortex in rats (Temple et al. 2003). Similarly, transections of the entorhino-hippocampal projection in mice resulted in the loss of Arc- and somatic SP-positive granule cells (this study). Together with the fact that placement of mice into a novel environment alters entorhinal input to the dentate gyrus (e.g., Fyhn et al. 2007; Leutgeb and Moser 2007; Schmidt et al. 2012), these lesioning experiments suggest that entorhinal input encoding environmental information is necessary to activate both Arc and somatic SP in granule cells.
Of note, such a cell- and compartment-specific upregulation of SP has not yet been described. Using classical experimental paradigms, for example, high-frequency stimulation of the perforant path, we and others observed region- or layer-specific changes in SP mRNA and SP protein (Yamazaki et al. 2001; Deller et al. 2003; Fukazawa et al. 2003; Jedlicka et al. 2009; Vlachos et al. 2009, 2013; Zhang et al. 2013; Jedlicka and Deller 2017). Since SP is part of the downstream machinery involved in the regulation of synaptic strength at excitatory postsynapses (Jedlicka and Deller 2017), an increased expression of SP may also enhance the plasticity of the activated cells. This could, in turn, bind the activated cells into a new and environment-specific granule cell ensemble that can then reform if the specific environmental conditions recur.
SP Is Targeted to the Somata of Granule Cells Where It Colocalizes with α-Actinin2
SP mRNA is abundant in the somata of hippocampal principal cells, whereas SP protein is found in spines and the AIS. This mismatch indicates that SP protein is synthesized in the soma and subsequently transported to its destinations (Deller et al. 2000). Although a mechanistic explanation for this targeting has not yet been provided, local binding partners could trap SP in spines or the AIS. A candidate SP-binding molecule is α-Actinin2 (Asanuma et al. 2005; Kremerskothen et al. 2005), which binds to NMDA-R (Merrill et al. 2007) and SK channels (Zhang et al. 2017) and influences the shape of dendritic spines (Hodges et al. 2014). Of note, α-Actinin also serves as a PSD-95 anchor tethering the AMPAR-PSD-95 complex to the postsynapse (Matt et al. 2018). In the hippocampus, α-Actinin2 is abundant and found in the spine compartment (Hodges et al. 2014) and the AIS (Sanchez-Ponce et al. 2012). After finding the somatic SP-positive cells, we speculated that these cells could also accumulate α-Actinin2 in their somata, and, indeed, α-Actinin2 was found enriched in the same locations as somatic SP in these cells. In line with the observation that α-Actinin2 is downregulated in SP-deficient mice (Asanuma et al. 2005), this concomitant upregulation and colocalization of SP and α-Actinin2 suggest that α-Actinin2 could play a role in the sorting of SP to the somata of granule cells. Alternatively, it could link SP-positive structures to the neuronal cytoskeleton or the postsynaptic density, for example, by binding to SP as well as PSD-95 (Matt et al. 2018). Which signals control the sorting of α-Actinin2 to the soma or the sorting of SP to the perinuclear ER is presently unclear.
SP Organizes ER in Three Neuronal Subcompartments
In our previous work, we have described SP in dendritic spines (Deller et al. 2000) and inside the AIS (Bas Orth et al. 2007). Both compartments are sites of neuronal plasticity and both contain a specialized SP-positive organelle consisting of stacked ER, that is, the spine apparatus and the cisternal organelle. SP is an essential component of the dense material stacking the ER cisterns since mice lacking SP still exhibit ER cisterns but do not form the stacked ER organelles (Deller et al. 2003; Bas Orth et al. 2007). In the present study, we report an inducible accumulation of SP in a third neuronal compartment, that is, the soma. Very similar to the situation in spines and the AIS, SP in the soma is again associated with electron-dense material connecting cisterns of ER and SP-deficient mice lack the dense material stacking the perinuclear ER cisterns. Both localization and the formation of an ER structure, that is, by and large homologous to the two organelles found in spines and the AIS, suggest a specific function for SP in the soma.
What could be the functional role of SP in the soma? The ER of neurons has been shown to change its organization dynamically and is able to form junctional membrane complexes locally (Takeshima et al. 2015). Such reorganizations may result in channel crosstalk or may alter Ca2+ trafficking in particular between extracellular and intracellular Ca2+ stores (Moccia et al. 2015). Of note, SP-positive dense material was often found between the outer nuclear envelope (perinuclear ER) and neighboring somatic ER cisterns. This may be of functional relevance, since the neuronal ER labyrinth is contiguous (Ramirez and Couve 2011; Chen et al. 2013) and Ca2+ signals from the synapse may reach the nucleus via ER of spines, dendrites, and the soma (Ch’ng and Martin 2011; Bading 2013). Thus, we speculate that SP may cause changes in somatic ER geometry, which subsequently alters Ca2+ trafficking in the somatic compartment, similar to what has been described for SP and Ca2+ trafficking in spines (Vlachos et al. 2009; Korkotian and Segal 2011; Korkotian et al. 2014) and the AIS (Segal 2018).
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG DE 2741/1–1; DFG DE 551/13–1; CRC 1080) and the Fachbereich Medizin (Heinrich und Fritz Riese-Stiftung to M.H.P.; scholarship to M.C.).
Notes
We thank Heike Korff, Ute Fertig, and Anke Biczysko for expert technical assistance. Conflict of Interest: None declared.
References
- Asanuma K, Kim K, Oh J, Giardino L, Chabanis S, Faul C, Reiser J, Mundel P. 2005. Synaptopodin regulates the actin-bundling activity of alpha-actinin in an isoform-specific manner. J Clin Invest. 115:1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bading H. 2013. Nuclear calcium signalling in the regulation of brain function. Nat Rev Neurosci. 14:593–608. [DOI] [PubMed] [Google Scholar]
- Bas Orth C, Schultz C, Muller CM, Frotscher M, Deller T. 2007. Loss of the cisternal organelle in the axon initial segment of cortical neurons in synaptopodin-deficient mice. J Comp Neurol. 504:441–449. [DOI] [PubMed] [Google Scholar]
- Bas Orth C, Vlachos A, Del Turco D, Burbach GJ, Haas CA, Mundel P, Feng G, Frotscher M, Deller T. 2005. Lamina-specific distribution of Synaptopodin, an actin-associated molecule essential for the spine apparatus, in identified principal cell dendrites of the mouse hippocampus. J Comp Neurol. 487:227–239. [DOI] [PubMed] [Google Scholar]
- Bramham CR. 2007. Control of synaptic consolidation in the dentate gyrus: mechanisms, functions, and therapeutic implications. Prog Brain Res. 163:453–471. [DOI] [PubMed] [Google Scholar]
- Bramham CR, Worley PF, Moore MJ, Guzowski JF. 2008. The immediate early gene arc/arg3.1: regulation, mechanisms, and function. J Neurosci. 28:11760–11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch'ng TH, Martin KC. 2011. Synapse-to-nucleus signaling. Curr Opin Neurobiol. 21:345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla MK, Guzowski JF, Ramirez-Amaya V, Lipa P, Hoffman KL, Marriott LK, Worley PF, McNaughton BL, Barnes CA. 2005. Sparse, environmentally selective expression of Arc RNA in the upper blade of the rodent fascia dentata by brief spatial experience. Hippocampus. 15:579–586. [DOI] [PubMed] [Google Scholar]
- Chen S, Novick P, Ferro-Novick S. 2013. ER structure and function. Curr Opin Cell Biol. 25:428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Turco D, Schlaudraff J, Bonin M, Deller T. 2014. Upregulation of APP, ADAM10 and ADAM17 in the denervated mouse dentate gyrus. PLoS One. 9:e84962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Turco D, Woods AG, Gebhardt C, Phinney AL, Jucker M, Frotscher M, Deller T. 2003. Comparison of commissural sprouting in the mouse and rat fascia dentata after entorhinal cortex lesion. Hippocampus. 13:685–699. [DOI] [PubMed] [Google Scholar]
- Deller T, Korte M, Chabanis S, Drakew A, Schwegler H, Stefani GG, Zuniga A, Schwarz K, Bonhoeffer T, Zeller R et al. 2003. Synaptopodin-deficient mice lack a spine apparatus and show deficits in synaptic plasticity. Proc Natl Acad Sci U S A. 100:10494–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deller T, Merten T, Roth SU, Mundel P, Frotscher M. 2000. Actin-associated protein synaptopodin in the rat hippocampal formation: localization in the spine neck and close association with the spine apparatus of principal neurons. J Comp Neurol. 418:164–181. [DOI] [PubMed] [Google Scholar]
- Diamantaki M, Frey M, Berens P, Preston-Ferrer P, Burgalossi A. 2016. Sparse activity of identified dentate granule cells during spatial exploration. Elife. 5:e20252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y, Saitoh Y, Ozawa F, Ohta Y, Mizuno K, Inokuchi K. 2003. Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron. 38:447–460. [DOI] [PubMed] [Google Scholar]
- Fyhn M, Hafting T, Treves A, Moser MB, Moser EI. 2007. Hippocampal remapping and grid realignment in entorhinal cortex. Nature. 446:190–194. [DOI] [PubMed] [Google Scholar]
- Guzowski JF. 2002. Insights into immediate-early gene function in hippocampal memory consolidation using antisense oligonucleotide and fluorescent imaging approaches. Hippocampus. 12:86–104. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Setlow B, Wagner EK, McGaugh JL. 2001. Experience-dependent gene expression in the rat hippocampus after spatial learning: a comparison of the immediate-early genes Arc, c-fos, and zif268. J Neurosci. 21:5089–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges JL, Vilchez SM, Asmussen H, Whitmore LA, Horwitz AR. 2014. Alpha-Actinin-2 mediates spine morphology and assembly of the post-synaptic density in hippocampal neurons. PLoS One. 9:e101770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser CR. 2007. Interneurons of the dentate gyrus: an overview of cell types, terminal fields and neurochemical identity. Prog Brain Res. 163:217–232. [DOI] [PubMed] [Google Scholar]
- Jedlicka P, Deller T. 2017. Understanding the role of synaptopodin and the spine apparatus in Hebbian synaptic plasticity - new perspectives and the need for computational modeling. Neurobiol Learn Mem. 138:21–30. [DOI] [PubMed] [Google Scholar]
- Jedlicka P, Schwarzacher SW, Winkels R, Kienzler F, Frotscher M, Bramham CR, Schultz C, Bas Orth C, Deller T. 2009. Impairment of in vivo theta-burst long-term potentiation and network excitability in the dentate gyrus of synaptopodin-deficient mice lacking the spine apparatus and the cisternal organelle. Hippocampus. 19:130–140. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Song H, Gage FH. 2015. Neurogenesis in the adult hippocampus. Cold Spring Harb Perspect Biol. 7:a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkotian E, Frotscher M, Segal M. 2014. Synaptopodin regulates spine plasticity: mediation by calcium stores. J Neurosci. 34:11641–11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkotian E, Segal M. 2011. Synaptopodin regulates release of calcium from stores in dendritic spines of cultured hippocampal neurons. J Physiol. 589:5987–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremerskothen J, Plaas C, Kindler S, Frotscher M, Barnekow A. 2005. Synaptopodin, a molecule involved in the formation of the dendritic spine apparatus, is a dual actin/alpha-actinin binding protein. J Neurochem. 92:597–606. [DOI] [PubMed] [Google Scholar]
- Kubik S, Miyashita T, Guzowski JF. 2007. Using immediate-early genes to map hippocampal subregional functions. Learn Mem. 14:758–770. [DOI] [PubMed] [Google Scholar]
- Lanahan A, Worley P. 1998. Immediate-early genes and synaptic function. Neurobiol Learn Mem. 70:37–43. [DOI] [PubMed] [Google Scholar]
- Leutgeb JK, Moser EI. 2007. Enigmas of the dentate gyrus. Neuron. 55:176–178. [DOI] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, Tonegawa S. 2012. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 484:381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Tonegawa S. 2014. Inception of a false memory by optogenetic manipulation of a hippocampal memory engram. Philos Trans R Soc Lond B Biol Sci. 369: 20130142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt L, Kim K, Hergarden AC, Patriarchi T, Malik ZA, Park DK, Chowdhury D, Buonarati OR, Henderson PB, Gokcek Sarac C et al. 2018. Alpha-Actinin anchors PSD-95 at postsynaptic sites. Neuron. 97:1094–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill MA, Malik Z, Akyol Z, Bartos JA, Leonard AS, Hudmon A, Shea MA, Hell JW. 2007. Displacement of alpha-actinin from the NMDA receptor NR1 C0 domain by Ca2+/calmodulin promotes CaMKII binding. Biochemistry. 46:8485–8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moccia F, Zuccolo E, Soda T, Tanzi F, Guerra G, Mapelli L, Lodola F, D'Angelo E. 2015. Stim and Orai proteins in neuronal Ca(2+) signaling and excitability. Front Cell Neurosci. 9:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundel P, Heid HW, Mundel TM, Kruger M, Reiser J, Kriz W. 1997. Synaptopodin: an actin-associated protein in telencephalic dendrites and renal podocytes. J Cell Biol. 139:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Cushman JD, Pelkey KA, Renaudineau S, Buhl DL, McHugh TJ, Rodriguez Barrera V, Chittajallu R, Iwamoto KS, McBain CJ et al. 2012. Young dentate granule cells mediate pattern separation, whereas old granule cells facilitate pattern completion. Cell. 149:188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo-Suzuki R, Okada D, Sekiguchi M, Inokuchi K. 2008. Synaptopodin maintains the neural activity-dependent enlargement of dendritic spines in hippocampal neurons. Mol Cell Neurosci. 38:266–276. [DOI] [PubMed] [Google Scholar]
- Pinaud R, Penner MR, Robertson HA, Currie RW. 2001. Upregulation of the immediate early gene arc in the brains of rats exposed to environmental enrichment: implications for molecular plasticity. Brain Res Mol Brain Res. 91:50–56. [DOI] [PubMed] [Google Scholar]
- Radic T, Jungenitz T, Singer M, Beining M, Cuntz H, Vlachos A, Deller T, Schwarzacher SW. 2017. Time-lapse imaging reveals highly dynamic structural maturation of postnatally born dentate granule cells in organotypic entorhino-hippocampal slice cultures. Sci Rep. 7: 43724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Amaya V, Angulo-Perkins A, Chawla MK, Barnes CA, Rosi S. 2013. Sustained transcription of the immediate early gene Arc in the dentate gyrus after spatial exploration. J Neurosci. 33:1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, Barnes CA. 2005. Spatial exploration-induced arc mRNA and protein expression: evidence for selective, network-specific reactivation. J Neurosci. 25:1761–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez OA, Couve A. 2011. The endoplasmic reticulum and protein trafficking in dendrites and axons. Trends Cell Biol. 21:219–227. [DOI] [PubMed] [Google Scholar]
- Ramirez S, Liu X, Lin PA, Suh J, Pignatelli M, Redondo RL, Ryan TJ, Tonegawa S. 2013. Creating a false memory in the hippocampus. Science. 341:387–391. [DOI] [PubMed] [Google Scholar]
- Redondo RL, Kim J, Arons AL, Ramirez S, Liu X, Tonegawa S. 2014. Bidirectional switch of the valence associated with a hippocampal contextual memory engram. Nature. 513:426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SU, Sommer C, Mundel P, Kiessling M. 2001. Expression of synaptopodin, an actin-associated protein, in the rat hippocampus after limbic epilepsy. Brain Pathol. 11:169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Ponce D, Blazquez-Llorca L, DeFelipe J, Garrido JJ, Munoz A. 2012. Colocalization of alpha-actinin and synaptopodin in the pyramidal cell axon initial segment. Cereb Cortex. 22:1648–1661. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B, Marrone DF, Markus EJ. 2012. Disambiguating the similar: the dentate gyrus and pattern separation. Behav Brain Res. 226:56–65. [DOI] [PubMed] [Google Scholar]
- Segal M. 2018. Calcium stores regulate excitability in cultured rat hippocampal neurons. J Neurophysiol. 120:2694–2705. [DOI] [PubMed] [Google Scholar]
- Severa W, Parekh O, James CD, Aimone JB. 2017. A combinatorial model for dentate Gyrus sparse coding. Neural Comput. 29:94–117. [DOI] [PubMed] [Google Scholar]
- Sobotzik JM, Sie JM, Politi C, Del Turco D, Bennett V, Deller T, Schultz C. 2009. AnkyrinG is required to maintain axo-dendritic polarity in vivo. Proc Natl Acad Sci U S A. 106:17564–17569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Hoshijima M, Song LS. 2015. Ca(2)(+) microdomains organized by junctophilins. Cell Calcium. 58:349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple MD, Worley PF, Steward O. 2003. Visualizing changes in circuit activity resulting from denervation and reinnervation using immediate early gene expression. Journal of Neuroscience. 23:2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon M, Hosoi T, Yoshii M, Ozawa K. 2014. Leptin induced GRP78 expression through the PI3K-mTOR pathway in neuronal cells. Sci Rep. 4:7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Strien NM, Cappaert NL, Witter MP. 2009. The anatomy of memory: an interactive overview of the parahippocampal-hippocampal network. Nat Rev Neurosci. 10:272–282. [DOI] [PubMed] [Google Scholar]
- Vlachos A, Ikenberg B, Lenz M, Becker D, Reifenberg K, Bas-Orth C, Deller T. 2013. Synaptopodin regulates denervation-induced homeostatic synaptic plasticity. Proc Natl Acad Sci U S A. 110:8242–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos A, Korkotian E, Schonfeld E, Copanaki E, Deller T, Segal M. 2009. Synaptopodin regulates plasticity of dendritic spines in hippocampal neurons. J Neurosci. 29:1017–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Matsuo R, Fukazawa Y, Ozawa F, Inokuchi K. 2001. Regulated expression of an actin-associated protein, synaptopodin, during long-term potentiation. J Neurochem. 79:192–199. [DOI] [PubMed] [Google Scholar]
- Zhang XL, Poschel B, Faul C, Upreti C, Stanton PK, Mundel P. 2013. Essential role for synaptopodin in dendritic spine plasticity of the developing hippocampus. J Neurosci. 33:12510–12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ledford HA, Park S, Wang W, Rafizadeh S, Kim HJ, Xu W, Lu L, Lau VC, Knowlton AA et al. 2017. Distinct subcellular mechanisms for the enhancement of the surface membrane expression of SK2 channel by its interacting proteins, alpha-actinin2 and filamin a. J Physiol. 595:2271–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]


