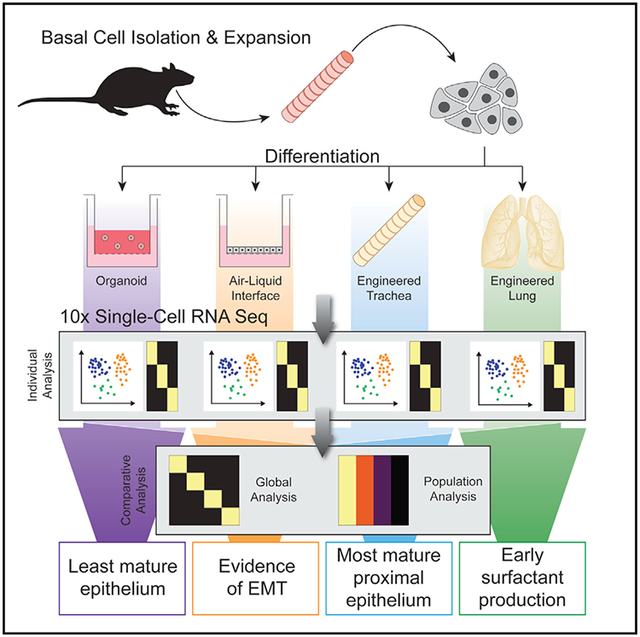
SUMMARY
Cell-based therapies have shown promise for treating myriad chronic pulmonary diseases through direct application of epithelial progenitors or by way of engineered tissue grafts or whole organs. To elucidate environmental effects on epithelial regenerative outcomes in vitro, here, we isolate and culture a population of pharmacologically expanded basal cells (peBCs) from rat tracheas. At peak basal marker expression, we simultaneously split peBCs into four in vitro platforms: organoid, air-liquid interface (ALI), engineered trachea, and engineered lung. Following differentiation, these samples are evaluated using single-cell RNA sequencing (scRNA-seq) and computational pipelines are developed to compare samples both globally and at the population level. A sample of native rat tracheal epithelium is also evaluated by scRNA-seq as a control for engineered epithelium. Overall, this work identifies platform-specific effects that support the use of engineered models to achieve the most physiologic differential outcomes in pulmonary epithelial regenerative applications.
Graphical Abstract
In Brief
Greaney et al. compare pulmonary epithelial regeneration across multiple modalities in vitro, finding that decellularized scaffolds achieved the most physiologic differentiation over more artificial platforms. scRNA-seq enables high-resolution comparison between engineered and native cell populations, thereby better gauging progress toward the generation of a tissue that may function on implantation.
INTRODUCTION
Pulmonary epithelium is a heterogeneous and dynamic cell lining that imparts critical functionality from large conducting airways to terminal alveoli. To regenerate critically damaged tissues with cell-based therapies, it is necessary to draw from an expandable population of epithelial cells that are capable of a range of functional differentiation. Basal cells (BCs) are considered the progenitor cells of proximal airway epithelium, giving rise to secretory, ciliated, and tuft cells under homeostatic conditions in vivo (Rock et al., 2009). Recently, studies of severe lung injury have revealed a basal-like, Tp63-lineage, bipotent progenitor cell that may migrate to alveoli and participate in the regeneration of distal-like epithelium unique from proximal phenotypes (Vaughan et al., 2015; Xi et al., 2017; Kumar et al., 2011; Ray et al., 2016), as well as other epithelial progenitors with similar regenerative potential (Zacharias et al., 2018; Kim et al., 2005; Liu et al., 2019). This observed phenomenon suggests that environmental cues throughout the airway tree, such as localized matrix bioactivity, air exposure, or mechanical cues, may influence the fate of such basal-like cells. For that reason, a better understanding of these extracellular cues could be exploited to drive the differentiation of pulmonary epithelial progenitors in regenerative medicine.
The potential of primary BCs for such regenerative applications has been enhanced by the development of pharmacological expansion techniques for epithelial cells. While pluripotent stem cell-derived epithelial progenitors find great utility in developmental modeling (Huang et al., 2015; McCauley et al., 2018), pharmacologic expansion facilitates the use of primary adult BCs for regenerative medicine by enabling growth to greater cell numbers while maintaining plasticity, without the use of exogenous genetic reprogramming factors or irradiated feeder cells. By this method, the addition of small molecule inhibitors of PAK1-ROCK-myosin II and transforming growth factor β (TGF-β) signaling results in a >1 trillion-fold expansion of human epithelial stem and progenitor cells from skin, airway, mammary, and prostate glands (Zhang et al., 2018).
The regenerative capacity of various pulmonary epithelial cell populations is often evaluated in vitro in any number of culture platforms. One common method is growing single cells suspended in Matrigel, thereby allowing cells to clonally expand and organize into spheroids or organoids (Rock et al., 2009; Barkauskas et al., 2013; Lee et al., 2014; Nichane et al., 2017). Proximal airway epithelial cells grown at an air-liquid interface (ALI) on a filter insert tend to display robust ciliation and self-organize into a pseudostratified epithelial layer (Nichane et al., 2017; Whitcutt et al., 1988; Schoch et al., 2004; Randell et al., 2011). Culturing rat tracheal epithelial cells in the lumens of denuded rat tracheas has demonstrated the inherent regional regenerative capacity of proximal epithelium (Liu et al., 1994). This regional specificity of proximal epithelial populations has been supported by two recent evaluations in engineered lung constructs, which produced little evidence of functional alveolar restoration (LaRanger et al., 2018; Gilpin et al., 2016). While it is well known that the physical and bioactive environments of differentiation have a huge influence on stem cell fate (McBeath et al., 2004; Bissell et al., 1982), this concept has been understudied in the pulmonary field.
In this report, we demonstrate robust expansion and differentiation of rat pharmacologically expanded basal cells (peBCs) and the evaluation of platform-specific effects on regenerative outcomes. Using a single line of peBCs, we conduct a focused evaluation of differentiation across four in vitro culture platforms, including organoids, ALIs, and tracheal and lung regeneration models. Furthermore, we use 10× Drop-Seq single-cell RNA sequencing (scRNA-seq) to evaluate peBC heterogeneity and differential outcomes as a function of culture platform, including global platform-based variability, as well as population-level comparisons. For this analysis, we developed computational pipelines for transcriptomic comparison between engineered samples, relating these results to a native rat tracheal epithelium control. We conclude that peBC differentiation in engineered tissue constructs results in more mature epithelial populations in vitro, as compared to standard organoid and ALI culture platforms. These natural matrix effects are region specific, with engineered trachea producing the most mature proximal epithelium and engineered lung encouraging distalization in the form of early surfactant production. peBC differentiation in artificial culture platforms, organoid and ALI, showed evidence of non-physiologic cellular response related to cellular assimilation in these non-native environments. Overall, this work advances our understanding of platform effects on pulmonary epithelial progenitor cells, including scRNA-seq evaluation of engineered tissues generated by decellularization and recellularization, while also demonstrating the utility of peBC for regenerative applications.
RESULTS
Single-Cell Evaluation of Native Rat Tracheal Epithelium
To contextualize peBC phenotype and differentiation, we sequenced a sample of native rat tracheal epithelium (1,049 cells) obtained by an adapted epithelial scraping technique (Fulcher et al., 2005). Clustering using Seurat and single-cell analysis in Python (SCANPY) methodologies (Satija et al., 2015; Wolf et al., 2018) yielded expected canonical cell types, as identified by top differentially expressed gene (DEG) lists, including basal, secretory, submucosal gland (SMG) secretory, and ciliated and tuft cell populations (immune, stromal, and endothelial populations were removed) (Figures 1A–1C). There were a small number of Foxi1+ and Ascl1+ cells potentially representing ionocytes and pulmonary neuroendocrine cells, respectively; however, they were too few to resolve in unsupervised clustering and therefore not considered in further analysis (data not shown).
Figure 1. Single-Cell Evaluation of Native Rat Tracheal Epithelium.
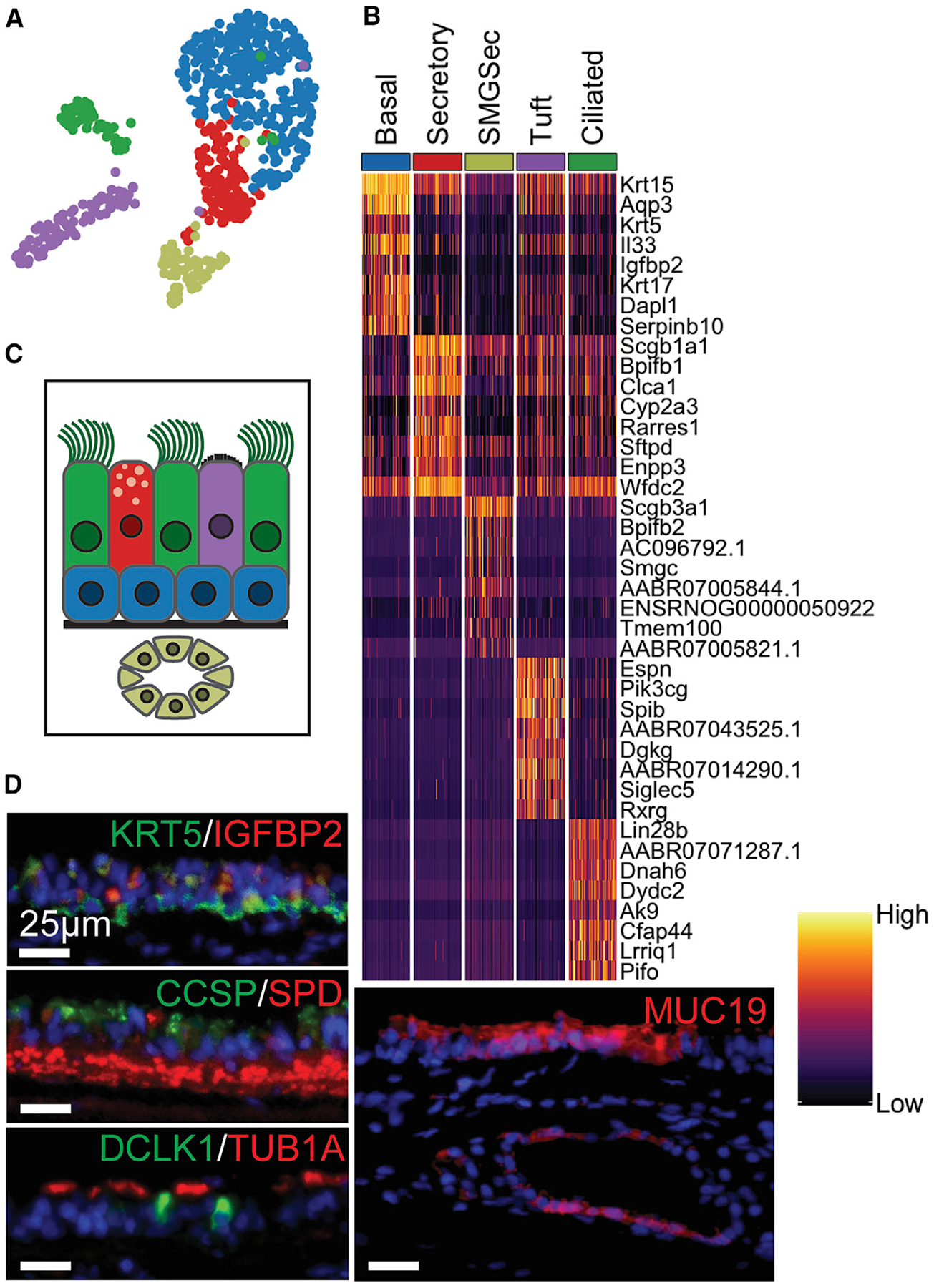
(A) Uniform Manifold Approximation and Projection (UMAP) of epithelial cell clusters.
(B) Heatmap of top DEGs for each cluster (lowest p value, see Method Details); columns grouped by cluster representing 60 randomly selected cells per cluster to show cluster heterogeneity; rows represent unity normalized expression of top 8 DEGs per cluster.
(C) Diagram of native rat tracheal epithelium, color-coded to UMAP clusters.
(D) IF of key markers for each cluster, including KRT5/IGFBP2 (BCs), CCSP/SPD (secretory; red background staining in lamina propria), DCLK1(tuft)/Tub1α(ciliated), and MUC19 (SMG secretory) (scale bars, 25 μm).
See also Figure S1.
The top defining markers for BCs (blue in Figures 1A–1C and S1A) include Krt5 and Igfbp2 (Figure 1D). Native secretory cells (red in Figures 1A–1C and S1B) represent an intermediate differential state between BCs and all other mature populations and consequently share some character with these cell types. This population is most uniquely identified by the expression of Clca1 and Sftpd (Figure 1D), both of which are involved in mucous secretion in proximal airways (Hegab et al., 2004; Madsen et al., 2000). The SMG secretory population (khaki in Figures 1A–1C and S1C) is defined by AC096792.1 (Muc19) and Smgc (Das et al., 2010), both found in submucosal glands, and ENSR-NOG00000050922 (Nupr1), which is expressed in Paneth cells of small intestinal crypts (Mead et al., 2018). Tuft and ciliated cells (purple and green in Figures 1A–1C, S1D, and S1E, respectively) were identified by the expression of distinctive markers such as Dclk1 and luminal Tub1α, respectively (Figure 1D). The tuft cell population in the rat expressed many of the defining markers that were recently reported in mice and humans (Plasschaert et al., 2018; Montoro et al., 2018). This characterization of rat tracheal epithelium served as the native control in subsequent scRNA-seq analyses.
Primary BC Character and Transcriptomic Heterogeneity over the Course of Pharmacologic Expansion
We isolated primary tracheal epithelial cells from adult rats using the same adapted epithelial scraping technique as for the native sample (Fulcher et al., 2005). peBCs selectively expand out of bulk epithelium in the defined expansion medium EpiX (Propagenix) on collagen-coated plates, with doubling times of roughly 4.25 days. Expanded cells were identified by nearly ubiquitous expression of BC markers, although the corresponding native origin of the peBC that expand in culture remains unclear. From the first passage, peBCs were more expressive of canonical BC markers Krt5, Krt14, and Tp63 than the starting bulk native tracheal epithelial sample (Figures 2A and 2B). The expression of these basal-specific genes was significantly greater in cells expanded in EpiX medium than in traditional bronchial epithelial growth medium (BEGM, Lonza) (Figures S2A–S2C). Bulk RNA expression profiles of rat tracheal epithelium expanded in EpiX versus BEGM vary (Figure S2D), implying differing phenotypic outcomes for freshly isolated cells cultured in the two media. BCs maintain Krt5, Krt14, and Tp63 expression at least up to 10 passages (Figure 2B) and 15 population doublings (Figure S2E), thereby allowing for suitably large cell numbers to support regenerative medicine applications.
Figure 2. Primary BC Character and Heterogeneity over Pharmacologic Expansion.
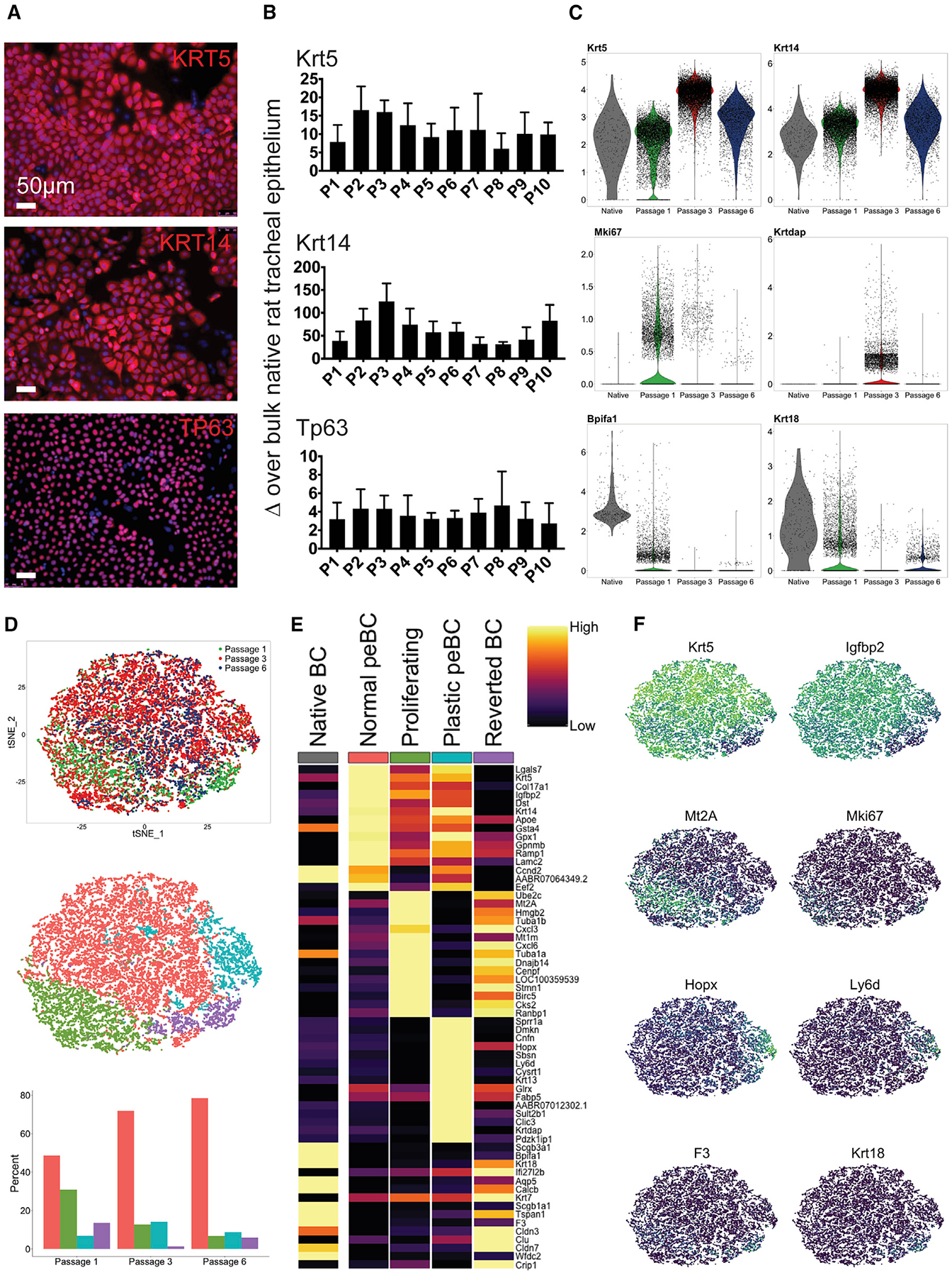
(A) IF of peBCs grown in EpiX at post-natal day 0 (P0) for KRT5, KRT14, and TP63 (scale bars, 50 μm).
(B) qRT-PCR of peBCs (n = 3 lines) over passage for Krt5, Krt14, and Tp63 relative to expression in bulk native rat tracheal epithelium; data represented as means ± SDs.
(C) Violin plots of native BCs (Figure 1) and peBCs at P1, P3, and P6 (all scaled together) for Krt5, Krt14, Mki67, Krtdap, Bpifa1, and Krt18.
(D) t-Distributed stochastic neighbor embedding (t-SNE) showing alignment of peBC at P1, P3, and P6 (top), sub-clustering of aligned sample (center), and proportions of each cluster comprising passage samples (bottom).
(E) Heatmap of top DEGs (greatest average log fold-change, see Method Details) for each cluster, with comparison to native BCs; columns show the average expression for each cluster; rows represent unity normalized expression of top DEGs per cluster.
(F) Featureplots of key genes per cluster, including Krt5, Igfbp2, Mt2A, Mki67, Hopx, Ly6d, F3 (tissue factor), and Krt18. See also Figure S2.
Samples from a single line of peBC were evaluated over the course of expansion with scRNA-seq (P1: 7,557 cells, P3: 10,955 cells, P6: 2,234 cells). Cells were sequenced and clustered using Seurat and SCANPY methodologies (Satija et al., 2015; Wolf et al., 2018). Analysis of individual passage samples reveals a contaminating Vim+ non-BC population at P1 that is lost over passage, as indicated by Vim negativity at both P3 and P6, further indicating a lack of epithelial-mesenchymal transition (EMT) over time in culture (Figure S2F). For consistency, this Vim+ population was removed from the P1 sample for subsequent analysis of peBC heterogeneity. Analysis over passage also reveals a peak in Krt5, Krt14, and Krtdap expression at P3; decreasing Mki67 (proliferation marker) expression over passage; and a minimum of Bpifa1 and Krt18 expression at P3 (Figure 2C).
scRNA-seq samples of peBCs at P1, P3, and P6 were aligned using standard canonical correlation analysis (CCA) methodology in Seurat to correct for batch effects (Butler et al., 2018). The aligned sample was further clustered into four subpopulations, with their cellular proportions varying over passage (Figure 2D). Analysis of the top DEGs of the subpopulations revealed putative functional roles for each, which will require further experimental validation that is beyond the scope of this work (Figures 2E and 2F). The largest subpopulation of the aligned, passaged samples was called “normal peBC” and is defined by the relative greatest expression of BC markers Krt5 and Igfbp2. The relative proportion of this normal peBC subpopulation increased over passage. A population of “proliferating” peBC was identified by proliferation markers Mki67, Tub1α1, and Tub1αb, as well as metallothionein genes Mt2A and Mt1m, and was found to decrease in proportion over passage. A subpopulation uniquely defined by plasticity markers Hopx (Jain et al., 2015; Takeda et al., 2011), Ly6d (Zuo et al., 2015), Krt13 (Montoro et al., 2018; Plasschaert et al., 2018), and Krtdap (Hegab et al., 2011) was called “plastic peBC” and was found to peak in proportion at P3. A final subpopulation was called “reverted BC” for its unique expression of secretory markers such as Scgb3a1, Bpifa1, and Scgb1a1, and shared character with native BC. This shared character suggests that the reverted BCs may have exited the reprogrammed state and reverted to expressing markers typical of homeostatic regenerative pathways, whereby BCs pass through a secretory intermediate before generating other terminally differentiated populations. Reverted BCs trend opposite to plastic BCs in terms of proportion, reaching a minimum at P3.
This scRNA-seq evaluation of peBC heterogeneity suggests that the pharmacologic expansion process broadly induces a unique set of phenotypes, including normal, plastic, and proliferating peBC populations, as well as a small population of reverted BCs that appears to have reverted out of (or possibly never entered) the reprogrammed state. This characterization of our starting peBCs strongly supports their use in regenerative medicine applications by the nature of their expandability and maintenance of BC markers over passage. The peak in basal and plasticity gene expression observed by PCR (and later corroborated with scRNA-seq) informed our decision to split cells out at P3 for differentiation in four separate differentiation platforms.
peBCs Achieve Robust Proximal Epithelial Differentiation in Three Platforms
Three of the four tested culture platforms encouraged proximal differentiation of peBCs when grown in Propagenix airway differentiation medium (PADM), complete PneumaCult ALI medium (Stemcell Technologies), and airway differentiation supplement (Propagenix). Differentiation as organoids, at ALI, and in engineered trachea consistently yielded pseudostratified epithelial layers with full mucociliary differentiation, as demonstrated by positive immunofluorescence (IF) staining for KRT5, MUC5AC (secretory cell marker), and Tub1α in cilia (Figures 3A–3C).
Figure 3. peBCs Achieve Robust Proximal Epithelial Differentiation in 3 Platforms and More Distal Character in Engineered Lung.
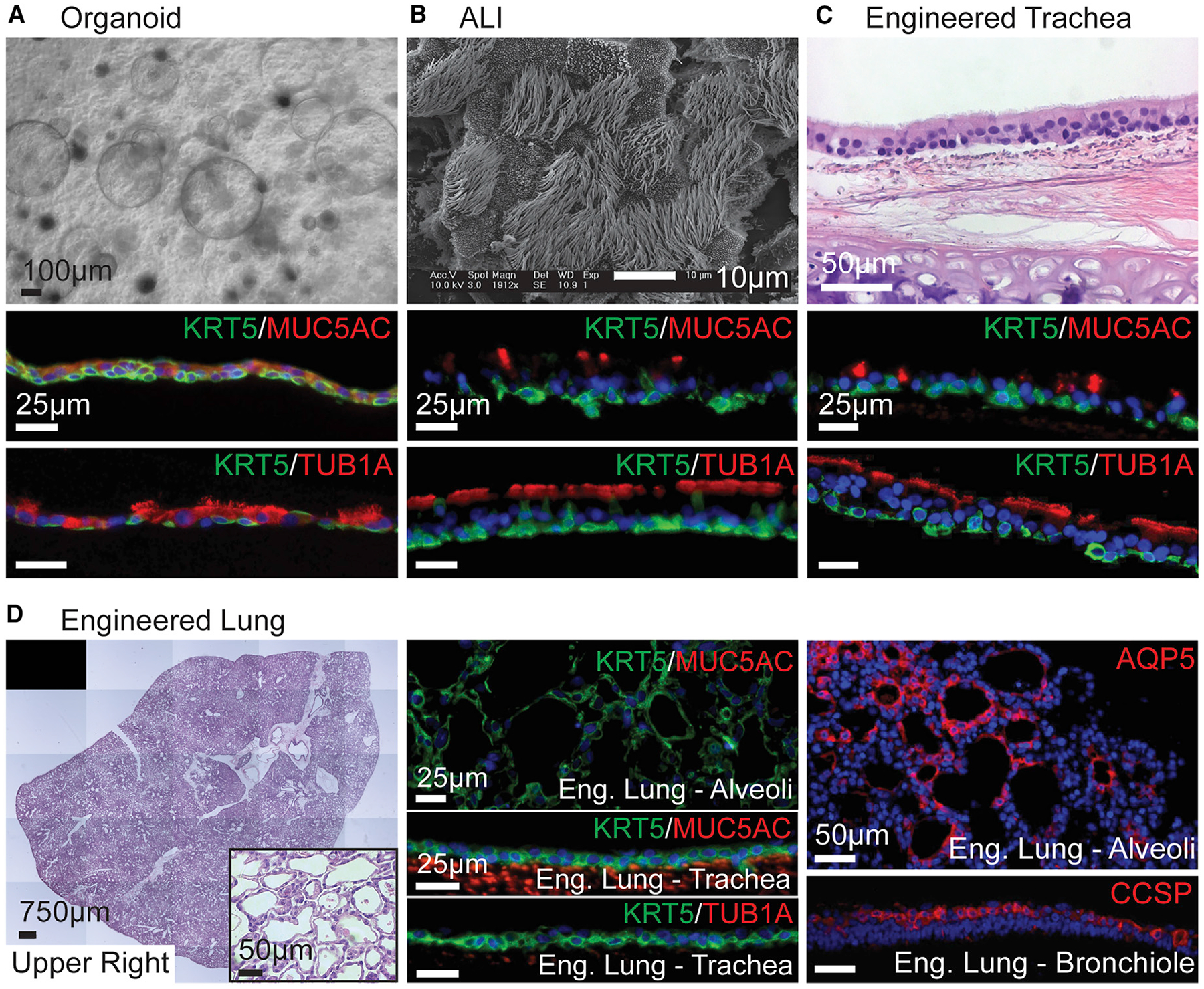
(A–D) peBC differentiated as (A) organoid, (B) ALI, (C) engineered trachea, and (D) engineered lung.
(A) Bright field of organoids at 2 weeks in Matrigel (scale bar, 100 μm).
(B) SEM of ALI at 3 weeks (scale bar, 10 μm).
(C) H&E of engineered trachea at 4 weeks (scale bar, 50 μm).
(D) Stitched H&E image and inset of the upper right lobe of engineered lung at 1 week taken with EVOS system (scale bars 750 μm, 50 μm, resp.); IF of KRT5/ MUC5AC in alveolar region; KRT5/MUC5AC and KRT5/Tub1α in the trachea showing a lack of secretory and ciliated cells (scale bars, 25 μm); IF of AQP5 in alveolar region, CCSP in bronchiole (scale bars, 50 μm).
In (A)–(C), IF of KRT5/MUC5AC and KRT5/Tub1α in organoid, ALI and engineered trachea; organoid lumens and apical cell surfaces oriented up (scale bars, 25 μm).
See also Figures S3–S5.
In organoid culture, peBCs formed spherical, hollow, thinwalled organoids filled with secretions and visible cilia by 2 weeks (Figure 3A). A BC population expressing KRT5 is maintained on the outside of the organoids, with mature cells expressing some MUC5AC and Tub1α differentiating luminally. By 4 weeks in ALI culture, peBCs develop mature cilia, as visualized with scanning electron microscopy (SEM) (Figure 3B) and bright-field video, in which cilia could be seen cycling secreted mucus around the ALI surface (Video S1). Similar to organoids, cells self-organize with a basal BC population and luminal ciliated and secretory cells at the air interface.
To evaluate the effects of native tracheal matrix on differentiation, we developed an in vitro culture protocol for peBCs on a decellularized trachea scaffold. Native rat tracheas were decellularized using a detergent-enzymatic method adapted from a whole-lung decellularization protocol (Balestrini et al., 2015). peBCs were cultured in the lumen of the engineered trachea for 2 days to allow for cell adhesion, followed by luminal perfusion in a custom bioreactor. After 4 weeks, the peBC formed a pseudostratified epithelial layer and achieved full mucociliary differentiation resembling native tracheal epithelium (Figure 3C).
When cultured in these three platforms, peBC consistently formed pseudostratified epithelium and mucociliary differentiation of varying degrees of organization and maturity. The expressions of Foxj1 (ciliated marker) and Muc5ac following the differentiation of multiple peBC lines were both significantly greater in engineered tracheas than in organoid or ALI (Figure S3). This supports the use of decellularized tracheal scaffolds to achieve proximal regeneration over organoid and ALI.
peBCs Gain More Distal Character in Engineered Lung
To evaluate proximal versus distal scaffold effects on differentiation, we also seeded peBCs into an engineered lung construct using established protocols (Balestrini et al., 2015; Ghaedi et al., 2013). Adult native rat lungs were decellularized using a detergent-enzymatic technique to remove all of the cellular components while preserving the extracellular matrix (ECM), a procedure established and rendered highly repeatable in our lab. This method has been shown to preserve key ECM proteins to which epithelial cells adhere (Hill et al., 2015; Calle et al., 2016), as well as retaining key mechanical characteristics and much of the original alveolar and microvascular architecture of native lungs (Petersen et al., 2012). A total of 100 million peBCs were seeded via the trachea into the most distal regions of the air compartment by applying negative pressure to the decellularized lung scaffold mounted inside a closed bioreactor system (as a reference, a set of native rat lungs contains ~300 million epithelial cells; Stone et al., 1992; Crapo et al., 1982). The recellularized lung was then cultured for 1 week, perfusing PADM through the vasculature via the pulmonary artery. Pressures were measured continuously at the pulmonary artery, vein, trachea, and bioreactor interior for the week of culture and extrapolated to flows and resistances throughout the organ, using methods reported previously (Figure S4) (Engler et al., 2019).
After 1 week of growth, peBCs fully repopulated the airways and alveoli of the engineered lung (Figure 3D). Alveolar barrier function of the organ, as extrapolated from measured pressure differentials in the vasculature and airway, greatly improved as compared to the starting baseline barrier of the decellularized organ (Figure S4D). This improved barrier implies that peBCs took up residence in the alveoli and contributed to resistance to transmural fluid flow. peBCs in engineered lung achieved different phenotypic outcomes than in other platforms, as they did not form columnar or ciliated structures of proximal airways. This could have been due to the shorter duration of culture, as negative staining for MUC5AC and Tub1α extended from alveolar regions through the trachea of the engineered lung, which had been repopulated with KRT5+ BC, but without yet achieving the epithelial maturation expected for that region. Flow conditions in the engineered trachea and the trachea of the engineered lung were comparable, with the engineered trachea being perfused at 4 mL/min and the outflow from the trachea of the engineered lung hovering around 4–5 mL/min (Figure S4C). KRT5 expression was noticeably reduced in alveolar regions compared to large airways, suggesting maturation and loss of basal character distally (Figure 3D). Cells in small airways of the distal lung showed evidence of differentiation into secretory cells by positive IF staining for Clara cell secretory protein (CCSP). Cells in alveolar regions of the lung did not appear to co-express canonical markers for alveolar type I (ATI) or type II (ATII) cells by IF. Rings of AQP5 were identified in alveoli but lacked the co-expression of other ATI markers, such as podoplanin, which suggests a different functional role for these cells (Raina et al., 1995) (Figure 3D). Engineered lung with peBCs was repeated to similar results (Figure S5). Lack of proximal epithelial differentiation and unique morphology and expression of distal markers in engineered lung suggests a localized influence of natural matrix on peBC differentiation.
Conserved and Distinct Populations in Differentiated peBCs by scRNA-Seq
A single line of peBCs was expanded to P3 in EpiX and 121 million cells were simultaneously split into organoids, ALI, engineered tracheas, and engineered lung, with samples of these starting cells saved for scRNA-seq and PCR. A total of 21,790 differentiated cells were sequenced, and Seurat and SCANPY methodologies applied to cluster each differentiated sample (Satija et al., 2015; Wolf et al., 2018). Clusters were identified by top DEGs.
There appeared to be consistent populations among organoid, ALI, and engineered trachea, specifically basal, cycling basal, secretory, tuft, and ciliated cell populations (Figures 4A–4C). The shared top DEGs for these populations across the three platforms include Krt5 and Dapl1 (Sun et al., 2006) in BCs; Tuba1b and Tubb5 in cycling BCs; Scgb1a1, Bpifb1, and Sftpd in secretory cells; Dclk1, AABR07062481.1 (Lrmp), and Rgs13 in tuft cells (Plasschaert et al., 2018; Montoro et al., 2018); and Tppp3, Dynlrb2, and Cfap126 in ciliated cells (Mukherjee et al., 2019; Thomas et al., 2010; Gegg et al., 2014).
Figure 4. Conserved and Distinct Populations in Differentiated peBCs by scRNA-Seq.
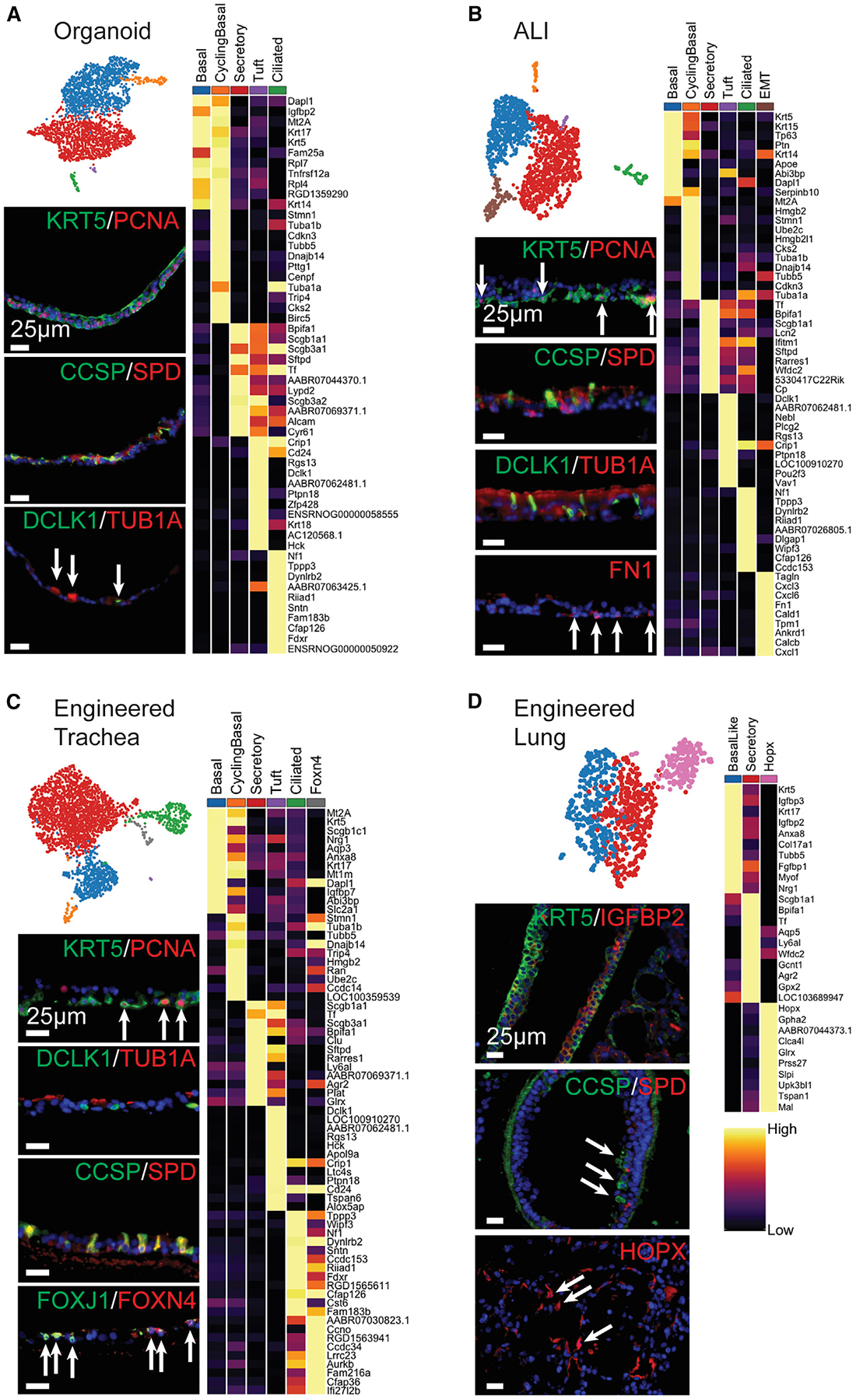
(A–D) UMAPs and heatmaps of top DEGs per cluster (greatest average log fold-change, see Method Details) for each differentiated scRNA-seq sample: (A) organoid, (B) ALI, (C) engineered trachea, and (D) engineered lung. Heatmap columns show the average expression for each cluster; rows represent unity normalized expression of top DEGs per cluster. IF of each sample for (A)–(C) KRT5/PCNA (BC and cycling BC), (A)–(D) CCSP/SPD (secretory), (A)–(C) DCLK1(tuft)/Tub1α(ciliated), (B) FN1 (EMT), (C) FOXJ1/FOXN4 (Foxn4 ciliated), (D) KRT5/IGFBP2 (basal-like), and HOPX (Hopx population) (scale bars, 25 μm). The arrows indicate sparse positive staining, organoid lumens, and apical cell surfaces oriented up.
See also Figure S6.
ALI gained a unique population defined by a mesenchymal character commonly associated with EMT. DEGs in this putative EMT cluster include Tagln and Fn1, among other mesenchymal and matrix-associated genes (Thiery, 2003; Yu et al., 2008) (Figure S6A). These markers were not expressed in the starting peBC population at P3 and hence must have developed during the culture period. It is possible that the significantly stiffer substrate that the peBC experienced on the polyethylene terephthalate (PET) filter insert at ALI, as compared to the other culture platforms that did not develop this response, encouraged this EMT phenotype (Wei et al., 2015). IF shows FN1 positivity in basolateral cells contacting the substrate (Figure 4B).
Engineered trachea uniquely gained a ciliated subpopulation defined by the unique expression of Foxn4 (Figures 4C and S6B). This population has been identified in human epithelial cells differentiated at ALI and was hypothesized to represent an intermediate state of multiciliated cell differentiation (Plasschaert et al., 2018). Our data support this, since the Foxn4 population shares much of the character of the mature ciliated population, except for mature functional genes such as Wipf3 and Sntn (Suetsugu et al., 2007; Tadokoro et al., 2014).
Engineered lung shared some character with other differentiated samples, but with several notable differences. There were basal-like and secretory populations that shared some top genes with those in other culture platforms, such as Krt5 in the basal-like population and Scgb1a1 and Bpifa1 in the secretory population (Figure 4D). The Krt5-expressing population in lung was characterized as “basal-like” for its reduced Pearson correlation with the native BC population (Figure 5A). There were no ciliated or tuft populations identified in engineered lung. Engineered lung uniquely gained a population defined by Hopx expression (Figures 4D and S6C), a marker typically associated with ATI cells (Barkauskas et al., 2013) and involved in the regulation of pulmonary maturation (Yin et al., 2006), although it has also been found to be upregulated in early alveolar injury in mice and humans (Ota et al., 2018). While this Hopx population in engineere lung does not co-express other canonical ATI markers, it also distinctly lacks Igfbp2 (Figure S6C), much like plastic ATI cells (Wang et al., 2018). This prominent Hopx expression in engineered lung may suggest a role in epithelial differentiation. While peBCs did not regenerate mature ATI cells, this Hopx population implies potential distal matrix effects on progenitor gene expression to promote localized niche regeneration of lung epithelium after injury.
Figure 5. Validation of Putative Analogous Engineered Populations Relative to Native.
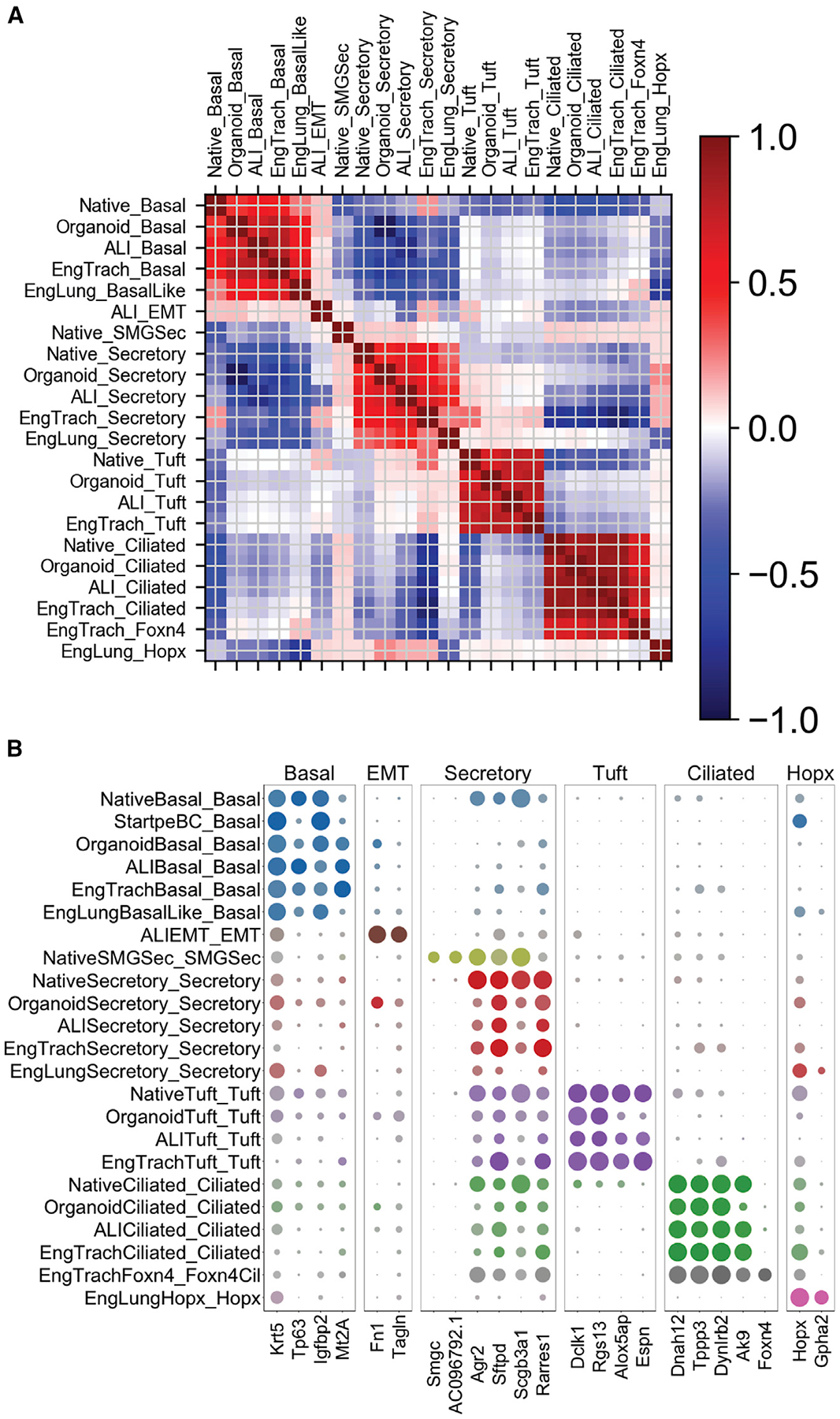
(A) Heatmap of Pearson correlation of all native and engineered clusters on variable genes used to cluster the native sample (2,424 genes).
(B) Dot plot of key genes across all of the native and engineered clusters.
This observed variation in transcriptomic character across four differentiation platforms demonstrates that there exists significant population-level variability in regenerative outcomes reliant on in vitro culture conditions.
Validation of Putative Engineered Populations Relative to Native
Following supervised cluster naming of engineered scRNA-seq data, we validated our labels against native proximal epithelial clusters to determine whether populations were truly analogous, before cross-sample comparisons. To that end, we ran a Pearson correlation between all native and engineered subpopulations using the variable gene list used to cluster the native tracheal epithelium (Figure 5A). This yielded strong correlations among predicted analogous basal, secretory, tuft, and ciliated cell populations. We observed weak correlations among populations expected to be unique to engineered or native samples, including EMT in ALI, native SMG secretory, and Hopx in engineered lung. Once overall correlation was confirmed for a large set of defining genes, we performed a split dot plot on key defining genes to compare expression level and specificity among subpopulations (Figure 5B). We thereby confirmed appropriate labeling of engineered subpopulations relative to native proximal epithelium, allowing appropriate comparison of these populations in subsequent analyses.
Engineered Platforms Encourage Localized Physiologic Differentiation
To capture global functional transcriptomic variation between peBCs differentiated in each platform, we performed differential gene expression combined with Gene Ontology (GO) (Database for Annotation, Visualization, and Integrated Discovery [DAVID]) (Huang et al., 2009a, 2009b). Comparison of the four bulk engineered scRNA-seq samples with GO reveals functional groupings of DEGs for each sample (Figures 6A and 6B). Organoids tended to uniquely express genes that promote cell-to-cell adhesion (Yarden, 2001; Solecki et al., 2002; Hoover et al., 2019), blastocyst development (Li et al., 2015; Kanadia et al., 2003; Zimmerman et al., 1986), and cell polarity (Hable and Kropf, 2005), all of which are necessary for 3-dimensional (3D) clonogenic development (Figure 6C). ALI expressed genes enabling adhesion and development on such a platform, including those involved in hair follicle morphogenesis (Petiot et al., 2003; Vauclair et al., 2005; Sennett and Rendl, 2012), cell-matrix adhesion (Bai et al., 2015), and establishment of epithelial cell polarity (Zen et al., 2009) (Figure 6D).
Figure 6. Engineered Platforms Encourage Localized Physiologic Differentiation.
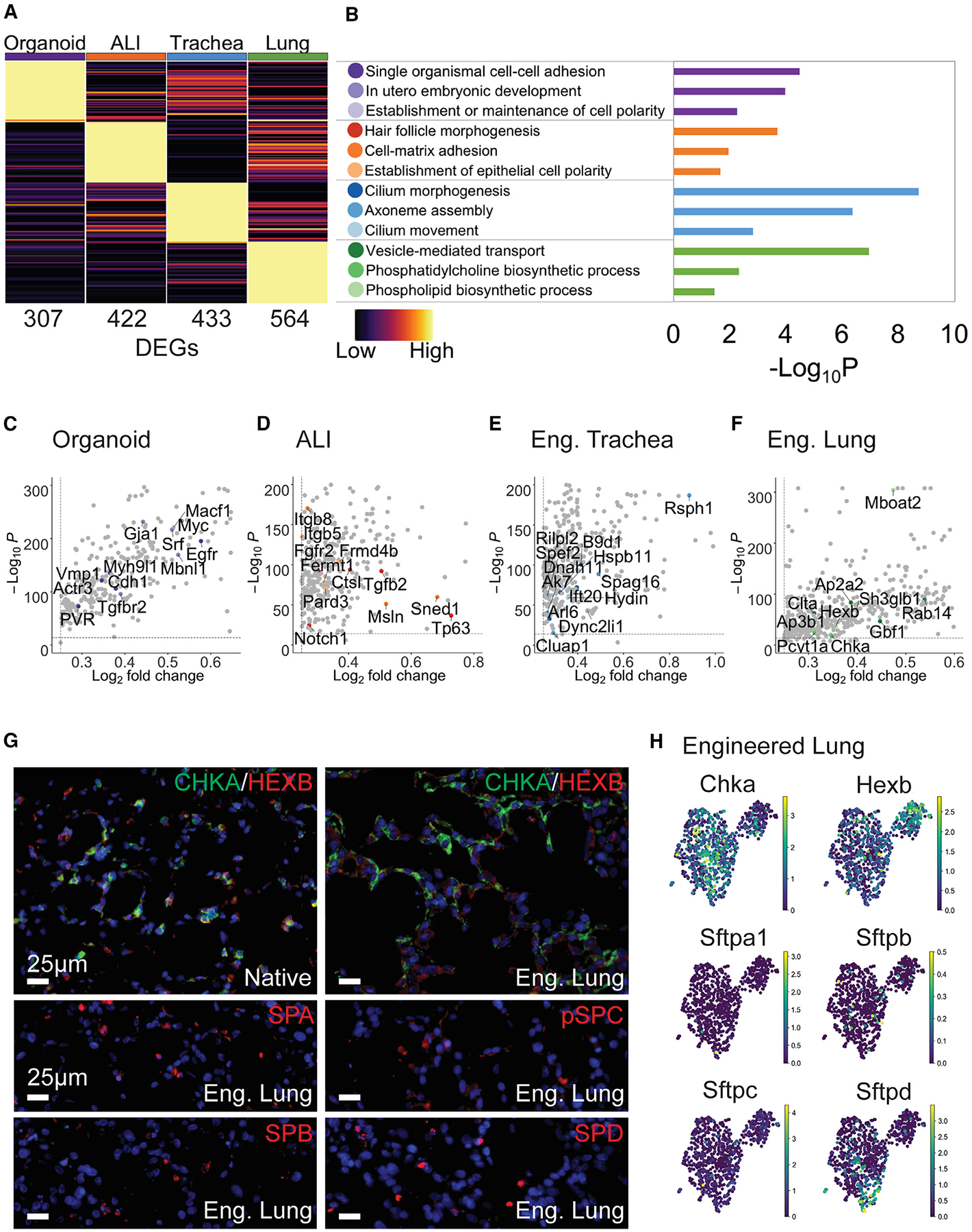
(A) Heatmap of top DEGs between samples; columns represent average expression per sample, rows represent unity normalized expression of top DEGs (lowest p value, see Method Details); number of DEGs per sample listed under each column.
(B) GO (DAVID) analysis of top DEGs yield functional groupings with p values. (C–F) Scatterplots (significance versus expression) of key genes in each GO category per sample (color legend in B); all other DEGs in gray; (C) organoid, (D) ALI, (E) engineered trachea, and (F) engineered lung
(G) IF of native and engineered lung for CHKA/HEXB and sparse staining of engineered lung for SPA, pro-SPC, SPB, and SPD (scale bars, 25 μm).
(H) Featureplots of engineered lung for Chka, Hexb, Sftpa1, Sftpb, Sftpc, and Sftpd.
For engineered trachea, groupings of top DEGs included mediators of cilium morphogenesis (Garcia-Gonzalo et al., 2011; Taylor et al., 2015; Keady et al., 2012), movement (Schwabe et al., 2008; Lechtreck et al., 2008), and axoneme assembly (Botilde et al., 2013; Fernandez-Gonzalez et al., 2009; Kott et al., 2013) (Figure 6E), suggesting that this platform encourages more mature ciliation. Engineered lung uniquely expresses genes associated with phosphatidylcholine (Dobbs et al., 1987; Agassandian and Mallampalli, 2013; Stern et al., 1976) and phospholipid biosynthesis (Buccoliero et al., 2004; Matsuda et al., 2008), as well as vesicle-mediated transport, several of which have been specifically linked to surfactant transport (Rückert et al., 2003; Ridsdale et al., 2011; Gou et al., 2008; Guttentag et al., 2005; Bouvet et al., 2013) (Figure 6F). Pulmonary surfactant consists of ~90% lipid (Weaver and Whitsett, 1991), with phosphatidylcholine comprising 70%–80% of this lipid component (Batenburg, 1992). Phosphatidylcholine is synthesized using key enzymes choline kinase α (Chka) and cholinephosphate cytidylyltransferase (Pcyt1a) (Agassandian and Mallampalli, 2013; Stern et al., 1976), both of which are top DEGs for engineered lung in this analysis. Another top unique gene, Hexb, is involved in the regulation of phospholipid synthesis (Buccoliero et al., 2004). These genes associated with surfactant production were most upregulated in engineered lung over other platforms. This suggests that peBC culture in engineered lung uniquely encourages “distalization” into epithelium that bears some functional resemblance to alveolar epithelium, as evidenced by increased phospholipid synthesis and handling associated with surfactant production (Figures 6G and 6H). We also see some cells expressing surfactant proteins A1, B, C, and D in engineered lung, although notably fewer than in native, while this expression is absent in other platforms.
We have elucidated several platform-specific effects on epithelial regeneration. The “artificial” scaffolds, organoid and ALI, produce proximal epithelial phenotypes that possess transcriptomic artifacts due to the non-physiologic nature of these platforms, including a tendency toward EMT in ALI. The two natural scaffolds, engineered trachea and lung, both show a unique tendency toward region-specific functional regenerative outcomes, including ciliation in the trachea and a tendency toward surfactant production and Hopx expression in distal lung. This enhanced physiologic regeneration supports the use of these platforms as more native-like in vitro model systems to study epithelial differentiation.
Similarities and Differences among Engineered Cell Populations
To directly compare cell types across platforms, analogous populations were pulled from native and engineered scRNA-seq samples, merged, and re-scaled. The top DEGs of native populations were used for heatmaps of relative expression (Figures 7A–7D) and dot plots of overall expression level and specificity (Figure 7E). peBCs at P3 were included in the BC comparison as an initial control for differentiated samples. There were no tuft or ciliated populations in engineered lung, so this condition was excluded from those comparisons.
Figure 7. Similarities and Differences among Engineered Cell Populations.
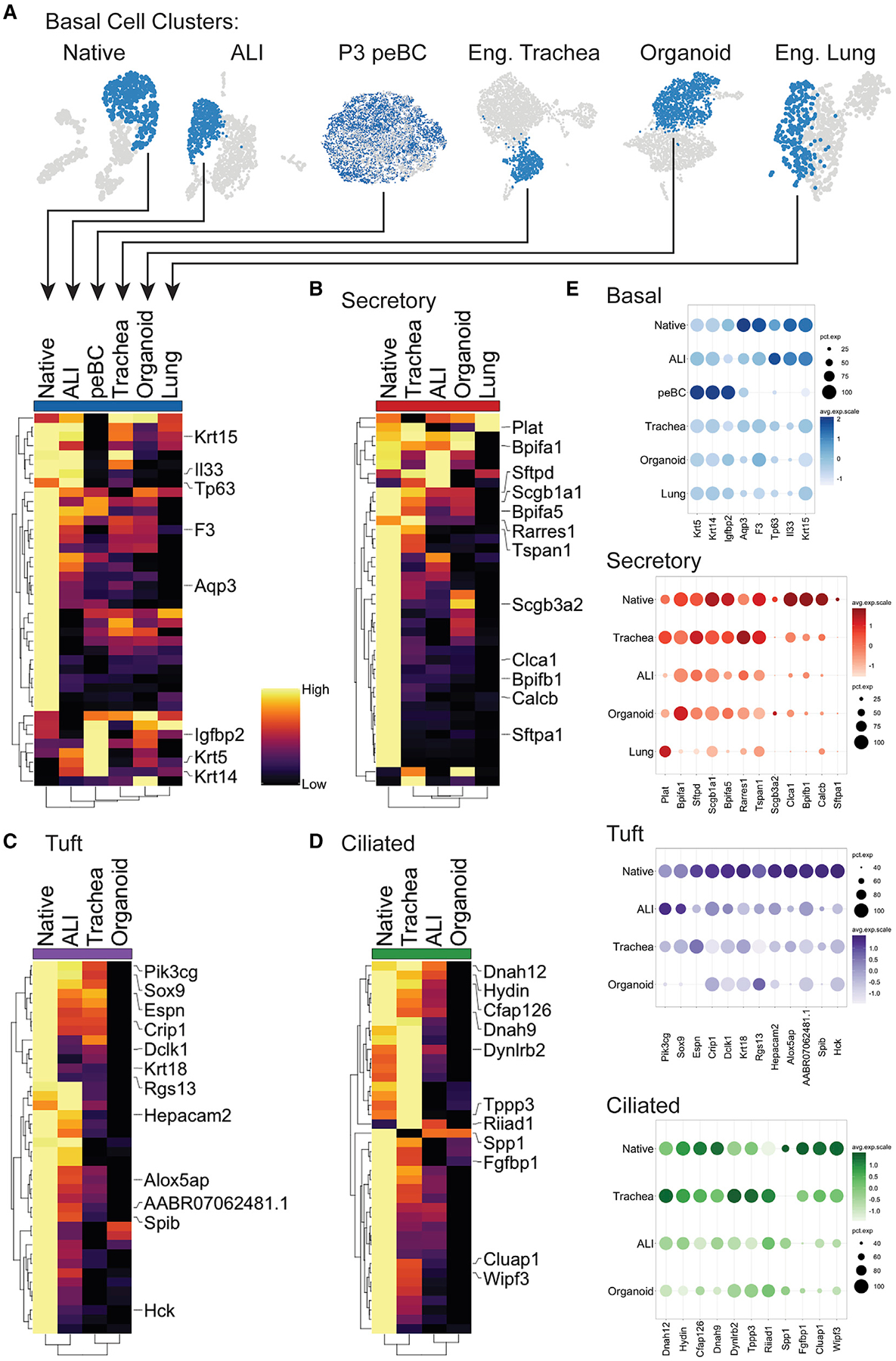
(A–D) Cell-type clusters were merged and re-scaled (A). Heatmaps of comparisons for (A) basal, (B) secretory, (C) tuft, and (D) ciliated cells. Heatmap columns are labeled with cluster origin and represent the average expression per cluster; rows represent the unity normalized expression of the top 40 DEGs in native (greatest average log fold-change, see Method Details) for the given cell type, with key genes labeled; rows and columns are ordered by dendograms.
(E) Dot plots of key genes for basal, secretory, tuft, and ciliated cells.
See also Figure S7.
In the BC comparison, peBCs at P3 appear to overexpress certain genes that define native BC, including Krt5, Igfbp2, and Krt14 (Figure 7A). Krt14 is considered a marker of regenerative activation in BCs (Cole et al., 2010; Hong et al., 2004). In a computational evaluation of mouse and human BC heterogeneity, the expression of IGFBP family members correlated with Krt14 (Plasschaert et al., 2018). Therefore, peBCs may be gaining an activated regenerative phenotype compared to native isolated BC. ALI has the most native-like BC population by dendrogram clustering, which also places the basal-like cells of engineered lung least similar to native. This would suggest that alveolar matrix composition does not support the maintenance of a true proximal BC population. A focused evaluation of heterogeneity in engineered BCs relative to native and initial peBCs reveals culture platform-specific tendencies toward certain BC subpopulations, which could influence downstream differential outcomes (Figure S7).
Secretory populations in organoid, ALI, and engineered trachea gain near-native expression of a few key markers defining native luminal secretory cells, including Scgb1a1, Sftpd, and Bpifa1, but are relatively lacking in the expression of Sftpa1, Calcb, Clca1, and Bpifb1 (Figure 7B). Dendrogram clustering of secretory populations place all engineered samples distinct from native, with the engineered lung secretory population as the least native-like, again possibly due to distal matrix effects.
Tuft cells in ALI are the most similar to native, clustering together by dendrogram, whereas engineered trachea and organoid cluster separately (Figure 7C). Top shared genes between ALI and native tuft cells include Pik3cg, Sox9, Hepacam2, and Alox5ap (Montoro et al., 2018), whereas tuft cells in organoids show the least expression of most native genes.
Ciliated cells in engineered trachea clustered together with native by dendrogram, while ALI and organoid clustered separately (Figure 7D). Engineered trachea gained the most native-like expression of top native markers, such as Dnah12, Hydin, and Cfap126, while somewhat surpassing native cells in the expression of Dynlrb2, Tppp3, and Riiad1. ALI produced intermediate expression of some genes, whereas organoid had the lowest expression. This finding suggests that the effects of natural scaffolding on peBCs in this experimental setup was more important for cilia maturation than air exposure, since the engineered trachea was a submerged culture and yet produced the most similar gene expression profiles to native ciliated cells. This contrasts previous dogma in the field (de Jong et al., 1994; Ostrowski and Nettesheim, 1995).
DISCUSSION
We conducted a controlled evaluation using peBCs to identify the effects of culture platform on epithelial progenitor cell differentiation in vitro. We further used scRNA-seq to compare these differentiated cell populations to native controls.
We first isolated and characterized a highly proliferative and plastic population of peBCs from the rat. In evaluating the heterogeneity of the starting cells, we found that they clustered into four subpopulations, three of which were unique to the culture-expanded peBC and the fourth resembling native BCs. One peBC subpopulation was defined by the unique expression of multiple plasticity markers, including Hopx, Ly6d, Krt13, and Krtdap, possibly suggesting enhanced plasticity in this population, which also appeared to be maintained or enhanced in the engineered lung condition (Figure S7). peBC proved to be an effective cell type for pulmonary regenerative applications for its expandability, while maintaining plasticity in recapitulating all of the proximal epithelial populations in vitro.
We then differentiated the peBCs in four in vitro culture platforms, including two traditional artificial platforms (organoid and ALI) and two engineered tissue models (engineered trachea and lung), holding the cell type and differentiation medium consistent. Following differentiation, we evaluated these samples using scRNA-seq. Globally, we found that artificial culture platforms are uniquely associated with transcriptomic patterns reflecting cellular assimilation to the artificial culture environment, including 3D clonogenic expansion (organoid) and matrix adhesion and polarity (ALI). Differentiation at ALI also uniquely produced a non-physiologic EMT population that did not occur in other platforms, perhaps due to the super-physiologic stiffness of the PET filter substrate.
Global expression patterns of samples on native scaffolds (engineered trachea and lung) showed more physiologic differential outcomes, suggesting the importance of localized matrix effects on peBC differentiation. Engineered trachea encouraged the generation of the most mature cilia, as evidenced by the unique prevalence of genes associated with ciliation in the global analysis, as well as the most native-like expression of the top ciliated cell genes in the population analysis. Engineered lung demonstrated evidence of distalization in the form of unique gene expression that is associated with phospholipid synthesis and handling, which is typically associated with surfactant production in ATII cells and Hopx expression. Engineered lung also lacked proximal hallmarks, such as ciliation and expression of related genes. Via individual population analysis, engineered trachea generated secretory and ciliated cells that most resembled native proximal epithelium, whereas ALI produced the most native-like basal and tuft cell populations, possibly representing competing differentiation cues by native matrix and air exposure, respectively. By the same metric, engineered lung produced cell populations that least resembled the native proximal epithelium, generating the least-physiologic basal and secretory populations, while completely lacking tuft or ciliated populations. Following engineered lung, organoids produced the least physiologic differential outcomes.
Finally, this work represents the evaluation of engineered tissues using scRNA-seq. This technology allows for unprecedented resolution in isolating highly specific cellular responses within constructs. scRNA-seq has already revolutionized our understanding of native, developing, and diseased organ systems. In our hands, scRNA-seq also has proven to be a tremendously powerful tool for understanding tissue regeneration in vitro. By resolving and evaluating cell populations in an engineered organ over the course of differentiation, we gain the most complete view of this process of any technology to date. The ability to conduct quantitative comparisons between analogous engineered and native populations with multidimensional data is the best method to measure progress toward regenerating a tissue that could function upon implantation. scRNA-seq also enables the detection of rare cell types in engineered constructs, such as tuft cells, cells expressing the EMT phenotype, and Foxn4-expressing cells in our engineered datasets. These rare cell types would have been difficult to resolve or characterize previously. More broadly, scRNA-seq opens the door to GO to filter out functional patterns in gene expression, computational lineage tracing (Trapnell et al., 2014), a process which has been previously unavailable in non-murine cell populations, and connectomic analyses that can identify essential and aberrant pathways in regenerating tissues (Raredon et al., 2019).
It is a known concept in tissue engineering that the physical differentiation environment has a critical impact on the fate and function of progenitor cells. This can include the mechanical properties and chemical composition of the substrate, as well as physical and mechanical inputs and bioactive cues. Using a controlled approach, we characterized these platform-specific effects holistically, using a population of differentiating epithelium across four in vitro culture platforms. We found unique benefits of engineered model platforms, as well as drawbacks associated with standard artificial culture platforms. While artificial platforms are highly useful for high-throughput screening studies, it will be important to consider and control for possible environmental artifacts in such studies, particularly in the application of scRNA-seq, which has the potential to parse and amplify misleading signals. It remains to be investigated how specific aspects of these model systems (e.g., bioactive cues, stiffness, shear stress) affect differential outcomes in isolation. Overall, these findings advocate for the utility of engineered lung and tracheal constructs as powerful model systems for the most physiologic evaluation of epithelial development and regeneration available in vitro.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Allison M. Greaney (allison.greaney@yale.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Source Organisms
All animal procedures in this study were approved by the Yale University Institutional Animal Care and Use Committee and complied with the NIH Guidelines for the Care and Use of Laboratory Animals. Male WT Sprague Dawley rats (Rattus norvegicus) were obtained from Charles River (8–12 weeks old, 300–350 g) and used for peBC isolation (n = 15), tracheal decellularization (n = 16) or whole-lung decellularization (n = 1). Animal sex was not expected to have a significant influence on outcomes.
Primary Cultures
All animal procedures in this study were approved by the Yale University Institutional Animal Care and Use Committee and complied with the NIH Guidelines for the Care and Use of Laboratory Animals. Rat peBC were isolated and cultured as described in the Method Details of this paper. Male WT Sprague Dawley rats (Rattus norvegicus) were obtained from Charles River (n = 15, 12 weeks old, 325–350 g) and used for peBC isolation. Animal sex was not expected to have a significant influence on outcomes.
METHOD DETAILS
peBC isolation & culture
All animal procedures in this study were approved by the Yale University Institutional Animal Care and Use Committee and complied with the NIH Guidelines for the Care and Use of Laboratory Animals. BC were isolated from the tracheas of adult rat (12wks, 370 ± 39 g, male WT Sprague Dawley) using a method adapted from an isolation protocol for human airway epithelium (Fulcher et al., 2005). Briefly, tracheas were isolated from larynx to carina into an enzyme solution (1% Protease XIV, 0.01% DNase) diluted 1:8 in wash media (MEM supplemented with 1% L-glutamine, 1% penn/strep, 0.08% gentamicin) and incubated on a rocker at 4°C for 24hrs. Enzyme was then quenched with 10% Fetal Bovine Serum (FBS) and the luminal plane of each trachea was thoroughly scraped with a scalpel blade. Cells were pooled, spun at 233 g for 5min at 4°C and washed in wash media once. Cells were then further dissociated in 2 mM EDTA, 0.5 mg/mL DTT, 0.25 mg/mL collagenase, 10 μg/mL DNase on a rocker for 10min at 37°C. Cell solution was quenched in 10% FBS, spun again, resuspended in EpiX Medium (Propagenix) with antibiotics (1% P/S, 0.1% gentamicin, 0.1% Amphotericin B) and plated on Col-I+III-coated plates (Advanced Biomatrix). Medium was changed every other day and BC passaged approximately every 8 days. BC were passaged with two rounds of 0.025% trypsin (0.05% diluted in PBS) for 5min each at 37°C, followed by a third rinse in PBS.
Differentiation in organoid
peBC were suspended at a concentration of 500,000 cells/ml in growth factor-reduced Matrigel (Corning) diluted in medium 6:1, in cell culture inserts (Millipore). Constructs were grown in Propagenix Airway Differentiation Medium (Propagenix, PADM) in the well beneath the insert, which was changed every other day for the duration of culture. Culture times for organoid, ALI and engineered trachea were selected for the earliest time point that mature ciliation could be identified histologically, indicating that seeded peBC had completed the full trajectory of maturation (from basal to secretory to ciliated).
Differentiation at ALI
peBC were seeded on Col-I+III-coated cell culture inserts (Corning) at 1 million cells/ml and cultured submerged in PADM medium for 48 hours until cells formed sufficient barrier to expose apical surface to air for the duration of culture. Medium was changed every other day for the duration of culture.
Differentiation in engineered trachea
Native tracheas of adult rats (8wks, 279 ± 30 g, male WT Sprague Dawley) were isolated and decellularized using a method adapted from a lung decellularization protocol developed in our lab (Balestrini et al., 2015). Briefly, tracheas were isolated from larynx to just above the carina, cannulated and hooked up to a peristaltic pump to luminally perfuse the following solutions serially at 50ml/min: antibiotics (10% penn/strep, 4% amphotericin B and 2% gentamycin in PBS), sodium deoxycholate (SDC, 0.1%), benzonase, and Triton X-100, with intermittent PBS rinses. Final sterilization of the tissues involve perfusing with antibiotics/antimycotics for 48 hours at 37°C. Notably chondrocytes are not completely removed in this protocol in order to maintain mechanical integrity of the construct. Decellularized tracheas were then coated in fibronectin (Sigma, 50 μg/ml) luminally overnight at 37°C before seeding to encourage cell adhesion. We utilized fibronectin on the decellularized trachea graft because we have seen that while our decellularization protocol largely preserves collagens, there is a greater loss of fibronectin during this procedure (Balestrini et al., 2015). 2 million peBC in 100 μL PADM medium were introduced to the lumen of each trachea and the ends capped. Trachea was rotated to allow cells to adhere to both sides for 30min each at 37°C before the ends were uncapped and incubated in PADM medium.
Tracheas were cultured statically in a tissue culture flask with a stir bar for three days to repopulate the scaffold before moving into a bioreactor system perfused at 4ml/min for the duration of culture. Engineered tracheas were cultured for up to four weeks with half media changes (50ml) every three days.
Differentiation in engineered lung
Native lungs of adult rat (8wks, 300–350 g, male WT Sprague Dawley) were isolated and decellularized using a whole-lung decellularization protocol developed in our lab (Balestrini et al., 2015). Briefly, rat lungs were extracted and perfused with antibiotics/antimycotics and PBS containing heparin and sodium nitroprusside to clear the organ of blood. Lungs were then decellularized with serial washes with SDC (0.1%, 0.5% and 1%), Triton X-100 and benzonase. Following decellularization, the constructs were sterilized by perfusing with antibiotics/antimycotics for 48 hours at 37°C. The scaffolds could then be stored at 4°C for up to one month until recellularization. For reseeding with peBC, decellularized lungs were hooked up in a custom bioreactor system that enables vascular perfusion through the pulmonary artery, oxygenation of medium with a hollow fiber cartridge, sterile media changes and continuous pressure monitoring at the pulmonary artery and vein, trachea and bioreactor interior. Continuous pressure data was then extrapolated to resistances and flows through the system using models developed in our lab (Yuan et al., 2019). peBC were vacuum-seeded into the air compartment of the decellularized construct following procedures established in our lab. Briefly, 100 million peBC were suspended in 10ml PADM medium and added to a 10ml syringe hooked up to the trachea cannulation in the bioreactor. Using a vacuum regulator, a negative pressure of −3mmHg was pulled inside the bioreactor around the lung to “inhale” the cell suspension into the air compartment of the lung. Vacuum was maintained to allow static adhesion for 1 hour before vascular perfusion was gradually ramped up to 20ml/min over an hour. Barrier measurements were taken at least every 12 hours. Lung was cultured in PADM with medium changes daily based on measured glucose consumption and lactate production. On takedown, lower right lobe was dissociated for 10× scRNaseq, middle right and accessory lobes were snap frozen in liquid nitrogen for PCR and protein analysis and upper right and left lobes were perfusion-fixed in formalin for histological analysis. 100MM peBC were seeded as this is approximately 1/3 of the total native epithelial cell numbers in rat (Crapo et al., 1982, Stone et al., 1992), with the knowledge that our expanding peBC have a doubling time of about 4.25 days on tissue culture plastic. Notwithstanding obvious variation in substrate, culture conditions, media, cellular behavior, etc., we used this information to roughly estimate that there would be 313MM cells by 7 days of culture, which would approximately constitute a fully repopulated airway compartment. While we did appear to achieve full repopulation along the length of the airway compartment, as well as evidence of epithelial differentiation, this shorter time point could have curtailed further cellular maturation. We decided not to control culture time between conditions in this experiment due to inherent variation in maturation rates among platforms, choosing instead to compare similar observed differential outcomes, thereby encompassing temporal aspects in our holistic platform comparison.
Quantitative real-time reverse transcription-PCR
Total mRNA was isolated from native tracheal epithelial scrapings, cultured cells, or differentiated samples using an RNA isolation kit (QIAGEN) and cDNA was synthesized from resulting templates (BioRad). mRNA levels of Krt5, Krt14, Tp63, Foxj1, Muc5ac and β-actin (Table S1) in rat were quantified using SYBR green (BioRad) and a Real-Time PCR system (BioRad CFX96). Relative changes in mRNA expression of target genes were determined by calculating ΔΔCt and each sample was internally normalized to housekeeping gene β-actin.
Microarray gene expression
Bulk transcriptomic profiles of rat tracheal epithelial cells isolated into BEGM and EpiX were compared using a microarray. peBC were isolated from three groups of three native rats each (12wks, 370 ± 39 g, male WT Sprague Dawley) to generate three replicate lines of cells. On isolation, cells from each line were split into either BEGM or EpiX. After 8 days of culture, cells were harvested and mRNA samples isolated using the previously described RNA isolation kit (QIAGEN), with added DNase digestion step. Samples were then processed at the Yale Center for Genome Analysis (YCGA) and analyzed using the Transcriptome Analysis Console (TAC) Software (ThermoFisher Scientific).
Immunofluorescent staining of cultured cells
To detect BC marker expression in cells expanded in EpiX, 660,000 cells/well were isolated into collagen-coated 6-well plates. After 8 days in culture, cells were fixed in 4% paraformaldehyde for 10 minutes, then permeabilized and blocked with 10% FBS and 0.2% Triton X-100 in PBS for 45 minutes at RT. Cells were probed with primary antibodies for KRT5, KRT14 and TP63 for 45 minutes at RT. Wells were rinsed three times with PBS and incubated in secondary antibodies for 45 minutes at RT, before rinsing and counter-staining with DAPI. All samples were imaged by fluorescence microscopy (Leica 6000).
Immunofluorescent staining of embedded samples
Samples of native rat trachea, organoids, ALI, engineered trachea and engineered lung were fixed in formalin for 3–4 hours, then paraffin-embedded and sectioned at Yale Pathology Tissue Services (YPTS). Hematoxylin and eosin (H&E) staining of sections was carried out by YPTS. To localize protein expression in paraffin-embedded samples, sections were de-waxed in xylene and ethanol, followed by soaking in antigen retrieval solution (0.05% Tween-20, 1 mM EDTA and 10 mM Tris (pH = 9)) at 75°C for 20 min. Sections were blocked (10% FBS and 0.2% Triton X-100, RT, 30min) and stained with primary antibodies 1 hour at RT for the following: KRT5, IGFBP2, DCLK1, Tub1α, SCGB1A1, SPD, MUC5AC, AQP5, PCNA, FN1, CHKA, HEXB, SPA, SPB, pSPC, FOXN4 and HOPX. Following PBS rinses, sections were incubated in Alexa Fluor conjugated secondary antibodies at 4 mg/ml for 1 hour at RT and DAPI (1 min). All samples were imaged by fluorescence microscopy (Leica 6000).
Scanning Electron Microscopy
All EM sample preparation and imaging was carried out by the Yale School of Medicine Center for Cellular and Molecular Imaging (CCMI) Electron Microscopy (EM) core facility.
Single cell dissociation of samples for scRNaseq
All differentiation methods were developed and tested for line-to-line consistency prior to sampling for scRNaseq evaluation. Native rat trachea epithelium, cultured cells and differentiated samples were rendered into single-cell suspensions for single-cell transcriptomic evaluation using distinct protocols most suited to each platform. All dissociated samples were counted, assessed for viability, filtered to reduce doublets and diluted to the desired concentration in filtered 0.01% BSA in PBS in for single cell library preparation. The native rat trachea epithelial sample started with the tissue extraction and enzyme solution (Protease XIV and DNase) digestion in wash medium (MEM, L-glutamine, antibiotics) as for isolating peBC, however the enzyme incubation was shortened to 1 hour at RT before luminal scraping. Epithelial clumps were then further dissociated in 50 μg/ml liberase (Roche) in PBS on a rocker for 10min at 37°C before passing through a 40 μm filter, counting and resuspension. Cultured peBC at passages 1, 3 and 6 were passaged with trypsin as normal and passed through a 40 μm filter before counting and resuspension. Organoids were cultured for three weeks before dissociation following an established protocol (Barkauskas et al., 2013). Briefly, culture inserts (n = 3 inserts) were incubated with 5U/mL dispase on a rocker for 30min at 37°C. Following a rinse in PBS, inserts were incubated in 0.05% trypsin on a rocker for 30min at 37°C. Samples were then rinsed in PBS, passed through a 40 μm filter, counted and resuspended. peBC were differentiated at ALI for four weeks before dissociation by passaging. Briefly, culture inserts (n = 3 inserts) were incubated with 0.025% trypsin (0.05% diluted in PBS) for 5min at 37°C twice, followed by a third 5min incubation in 0.05% trypsin. Samples were then rinsed in PBS, passed through a 40 μm filter, counted and resuspended. peBC differentiated in engineered tracheas (n = 3 constructs) were isolated after three weeks of culture by incubation in 0.05% trypsin on a rocker for 40min at 37°C, followed by luminal scraping as in the cell isolation procedure. Cells from the constructs were pooled and passed through a 40 μm filter before counting and resuspension. The peBC engineered lung construct was cultured for one week before dissociation. The lower right lobe of the recellularized lung was dissociated in 10ml digestion solution (DMEM High Glucose (GIBCO) + 1mg/ml Collagenase/Dispase (Roche) + 3U/ml Elastase (Worthington) + 20U/ml DNase). The lobe was injected with digestion solution, minced into a fine homogenate and the enzyme and tissue incubated on a rocker for 20min at 37°C. The tissue was strained and pooled in cold medium (DMEM High Glucose + 10% FBS), then rinsed three times in 0.01% BSA in PBS, spinning at 300 g for 5min at 4°C.
10× scRNaseq library preparation, sequencing and alignment
Single cell suspensions were converted to scRNaseq libararies with the Chromium Single Cell 3′ v2 assay (10× Genomics). All samples were diluted to a stock concentration of 1000 cells/μl for a targeted cell recovery of 5000–10000 cells per lane. Libraries generated with this method were then sequenced on the Hiseq4000 platform (Illumina) to a depth of 160–220 million reads per library. Read 1 sequencing was 26 bp long, read 2 sequencing was 98 bp long. FASTQ read 2 sequences were trimmed of the reverse complement sequence of the template switch oligo, as well as for poly(A) contaminants. Paired reads with a trimmed read 2 less than 30 bp long were discarded. Downstream processing was conducted with zUMIs v 0.4 (44) with the default parameters; the top 10,000 cell barcodes ranked by reads were output for each sample. Ensembl Rnor6.0 release 92 was used to align the data to the rat transcriptome.
scRNaseq data filtration, normalization and scaling
Aligned data was then processed using Seurat v2.3 (Satija et al., 2015). Default parameters were used for each function unless otherwise specified. After creating Seurat objects for each sample, cells were filtered based on number of unique mapped genes to remove potential partial cells and multiplets. All samples were also filtered on percent mitochondrial genes in order to exclude damaged and dying cells, with dissociated cells (native tracheal scraping and engineered samples) receiving further filtering on nUMI due to the more aggressive dissociations. Passaged peBC cells were accepted as expressing 1,000–4,000 genes (as they displayed a relatively narrow range) with less than 5% mitochondrial mapping. Cells of the native tracheal scraping were accepted as expressing 500–10,000 genes, less than 10% mitochondrial mapping and a minimum of 4,000 nUMI. Cells of engineered samples were accepted as expressing 500–7,500 cells, less than 10% mitochondrial mapping and minimum of 1,000 nUMI. Expression values were then log normalized to 10,000 transcripts per cell. Data were scaled using Seurat’s ScaleData function with regression of nUMI and percent mitochondrial transcripts to ignore variation based on the quality of each dissociation. Following all filtering steps, cell numbers for each sample are as follows – peBC Passage 1: 7,557, Passage 3: 10,955, Passage 6: 2,234, Native tracheal scraping: 1,049, Organoid: 2,387, ALI: 1,566, Engineered trachea: 2,613, Engineered lung: 778.
scRNaseq clustering
Initial clustering was performed on each sample independently. Variable genes were determined for each sample using Seurat’s FindVariableGenes function with input cutoffs for expression and dispersion based on sample-specific distributions. Each sample was then clustered using the FindClusters function based on an input number of principal components (PC) and resolution, with greater number of PC and higher resolution for samples expected to be more heterogeneous. Non-epithelial clusters (immune, stromal and endothelial cells) in the native tracheal epithelial scraping were removed and the resulting cells (640 cells) reclustered for subsequent analyses. For improved visualization, Seurat objects were converted to AnnData for use in SCANPY (Wolf et al., 2018). Objects were re-clustered in SCANPY using the same parameters calculated in Seurat (nUMI, percent mitochondrial, variable genes, PC) and visualized by plotting UMAPs. Canonical Correlation Analysis (CCA) was performed to align passaged peBC samples (Figure 2) and engineered BC populations (Figure S7) using the RunMultiCCA function and roughly following the published pipeline (Butler et al., 2018). 5 CC’s were applied to align passaged peBC’s and 3 CC’s were used for engineered BC’s.
Cluster identification by DEGs
The FindAllMarkers function in Seurat was applied to each sample to identify DEGs for each cluster relative to all the other clusters in the object. These marker lists were used to apply cluster labels based on top defining genes of canonical cell types in the literature, as well as observed functional patterns in gene expression. Heatmaps of relative gene expression among clusters or samples (in subsequent analyses) were generated with ComplexHeatmap (Gu et al., 2016). Plotted genes were selected from these markers lists based on expression level (avg_logFC) or specificity (p_val), depending on the application and specified in figure captions. Expression per cluster or sample is expressed by a number of randomly-selected individual cells, or as an average across all cells in a sample. All samples were unity normalized by row, using a custom function, to visualize relative gene expression among samples rather than overall expression levels. This process sets the greatest expressing sample as the maximum (“High”) and the lowest expressing sample as the minimum (“Low”), while preserving intermediate variation. This representation controls for inherent variation in overall expression magnitude of selected genes. Putative cluster identifications of differentiated samples were compared to those of the native tracheal epithelial control to evaluate relatedness to native clusters by Pearson correlation (Figure 5A, Bioinfokit). The same correlation plotting method was applied to compare native BC, starting peBC and engineered BC in Figure S7E. Dot plots and violin plots were generated using the DotPlot or SplitDotPlotGG and VlnPlot functions, respectively. Most plot aesthetics were modified for publication using ggplot2.
Global sample comparisons
Entire scRNaseq objects of engineered samples (organoid, ALI, engineered trachea, engineered lung) were merged and re-clustered in Seurat to observe functional global shifts in gene expression based on differentiation platform. Differential gene expression analysis was performed between samples and the resulting gene list was analyzed by gene ontology (DAVID) (Huang et al., 2009b, 2009a). Generated gene categories were investigated for relevance in the literature and genes with the strongest precedent were plotted (EnhancedVolcano).
Population comparisons
For comparison of clusters across samples, individual clusters were subset, merged together and re-clustered in Seurat to observe differences in expression across analogous cell types in different platforms. Heatmaps were generated using ComplexHeatmap with top DEGs of the native tracheal epithelial clusters. Row and column clustering was generated in that function using standard parameters and visualized with dendograms.
QUANTIFICATION AND STATISTICAL ANALYSIS
All indicated sample sizes (n) represent biological replicates for in vitro experiments and number of individuals for animal use. All bar graphs of qRT-PCR data represent mean expression of biological triplicates, as evaluated in technical triplicate and internally normalized to the housekeeping gene (β-actin), with error bars representing SD (Figures 2B, S2A–S2C, S3A, and S3B). Significant differences in PCR expression between experimental groups was analyzed in Prism (v7.0a) by Student’s t test and one-way ANOVA, with degree of significance indicated in figure captions. Pearson correlation of native variable gene expression among native and engineered sample clusters was performed using Bioinfokit in python (Figures 5A and S7E).
DATA AND CODE AVAILABILITY
Datasets generated in this study are available at NCBI Gene Expression Omnibus (GEO). scRNaseq data: GEO: GSE145517. Microarray data: GEO: GSE145516. Other requests for data or software may be directed to and will be fulfilled by the Lead Contact, Allison Greaney (allison.greaney@yale.edu).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-Cytokeratin 14 (clone RCK107) | Abcam | Cat #ab9220; RRID: AB_307087 |
| Rabbit Anti-Cytokeratin 5 | Abcam | Cat #ab53121; RRID: AB_869889 |
| Rabbit Anti-p63 | Fitzgerald | Cat #70R-50620 |
| Mouse Anti-IGFBP2 (C-10) | Santa Cruz | Cat #sc-25285; RRID: AB_2123220 |
| Rabbit Anti-DCAMKL1 | Abcam | Cat #ab31704; RRID: AB_873537 |
| Mouse Anti-alpha Tubulin (acetyl K40) [6–11B-1] | Abcam | Cat #ab24610; RRID: AB_448182 |
| Rabbit Anti-CCSP | Dr. Barry Stripp | n/a |
| Goat Anti-Muc19 | Abcam | Cat #ab121014; RRID: AB_10971842 |
| Mouse Anti-Surfactant protein D [VIF11] (Biotin) | Abcam | Cat #ab15696; RRID: AB_302044 |
| Mouse Anti-Muc5AC (45M1) | Thermo Fisher Scientific | Cat #MA5–12178; RRID: AB_10978001 |
| Rabbit Anti-Aquaporin 5 | Millipore | Cat #AB3559-50UL; RRID: AB_2141915 |
| Mouse Anti-PCNA [PC10] | Abcam | Cat #ab29; RRID: AB_303394 |
| Rabbit Anti-Fibronectin | Abcam | Cat #ab23751; RRID: AB_447656 |
| Mouse Anti-Hop (E-1) | Santa Cruz | Cat #sc-398703; RRID: AB_2687966 |
| Rabbit Anti-Choline kinase alpha | Abcam | Cat #ab88053; RRID: AB_10675356 |
| Mouse Anti-HEXB B chain (D-9) | Santa Cruz | Cat #sc-376781 |
| Rabbit Anti-SPA (H-148) | Santa Cruz | Cat #sc-13977; RRID: AB_661294 |
| Rabbit Anti-SPB (H-300) | Santa Cruz | Cat #sc-13978; RRID: AB_2185370 |
| Rabbit Anti-Prosurfactant Protein C | Millipore | Cat #AB3786; RRID: AB_91588 |
| Mouse Anti-FOXN4 (G-1) | Santa Cruz | Cat #sc-377166 |
| Rabbit Anti-FOXJ1 | Sigma | Cat #AV38038; RRID: AB_1849069 |
| Goat Alexa Fluor 488 Anti-Rabbit IgG | Invitrogen | Cat #A11034; RRID: AB_2576217 |
| Goat Alexa Fluor 555 Anti-Mouse IgG | Invitrogen | Cat #A21424; RRID: AB_141780 |
| Goat Alexa Fluor 555 Anti-Rabbit IgG | Invitrogen | Cat #A21429; RRID: AB_141761 |
| Donkey Alexa Fluor 555 Anti-Goat IgG | Invitrogen | Cat #A21432; RRID: AB_141788 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Protease XIV from Streptomyces griseus | Sigma | Cat #P5147 |
| Deoxyribonuclease I from bovine pancreas | Sigma | Cat #DN25 |
| Minimum Essential Medium Eagle | Sigma | Cat #M8028 |
| Penn/Strep | GIBCO | Cat #15140–122 |
| Fetal Bovine Serum | HyClone | Cat #SH30071.03 |
| EDTA, 0.5M solution, pH 8.0 | American Bio | Cat #ab00502–01000 |
| Collagenase | Worthington | Cat #LS004196 |
| Amphotericin B | HyClone | Cat #SV30078.01 |
| PureCol Bovine Type I Atelo-Collagen Solution | Advanced Biomatrix | Cat #5005-100ML |
| Bronchial epithelial cell growth medium | Lonza | Cat #CC-3170 |
| EpiX Medium | Propagenix | Cat #276–00A-D |
| Airway Differentiation Supplement | Propagenix | https://www.propagenix.com/index.php/research-products |
| Pneumacult ALI Medium | StemCell Technologies, Inc. | Cat #05001 |
| Heparin | StemCell Technologies, Inc. | Cat #07980 |
| Hydrocortisone | StemCell Technologies, Inc. | Cat #07925 |
| 0.05% Trypsin-EDTA | GIBCO | Cat #25300–054 |
| Growth factor-reduced Matrigel | Corning | Cat #356231 |
| Fibronectin bovine plasma | Sigma | Cat #F4759 |
| DMEM High Glucose | GIBCO | Cat #11965–092 |
| Collagenase/Dispase | Roche | Cat #11097113001 |
| Elastase | Worthington | Cat #LS002292 |
| Liberase TM | Sigma | Cat #5401119001 |
| Bovine Serum Albumin | Gemini Bio-Products | Cat #700–100P |
| RNase-Free DNase | QIAGEN | Cat #79254 |
| DAPI | Thermo Fisher Scientific | Cat #62247 |
| Dispase II | GIBCO | Cat #17105041 |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | QIAGEN | Cat #74106 |
| iScript cDNA Synthesis Kit | BioRad | Cat #1708891 |
| iQ SYBR Green | BioRad | Cat #1708882 |
| Chromium Single Cell 3′ Library & Gel Bead Kit v2 | 10x Genomics | Cat #PN-120237 |
| Chromium Single Cell A Chip Kit | 10x Genomics | Cat #PN-120236 |
| Chromium i7 Multiplex Kit | 10x Genomics | Cat #PN-120262 |
| Clariom S Assay, rat | Thermo Fisher Scientific | Cat #902934 |
| Deposited Data | ||
| scRNaseq data | This paper | GEO: GSE145517 |
| Microarray data | This paper | GEO: GSE145516 |
| Experimental Models: Organisms/Strains | ||
| SAS Sprague-Dawley Rattus norvegicus | Charles River | Strain code: 400 |
| Oligonucleotides | ||
| Primers for β-actin, Foxj1, Krt14, Krt5, Muc5ac, Tp63, see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| Seurat (v2.3) | Satija et al., 2015 | https://satijalab.org/seurat/ |
| SCANPY (v1.4) | Wolf et al., 2018 | https://scanpy.readthedocs.io/en/stable/ |
| Bioinfokit | Renesh Bedre, Ph.D. | https://github.com/reneshbedre/bioinfokit |
| ComplexHeatmap (v2.1.0) | Gu et al., 2016 | http://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html |
| DAVID Gene Ontology | Huang et al., 2009a, 2009b | https://david.ncifcrf.gov/ |
| EnhancedVolcano | https://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html | |
| MATLAB custom code | Engler et al., 2019 | N/A |
| Transcriptome Analysis Console (TAC) Software | Thermo Fisher Scientific | https://www.thermofisher.com/us/en/home/life-science/microarray-analysis/microarray-analysis-instruments-software-services/microarray-analysis-software/affymetrix-transcriptome-analysis-console-software.html |
| Other | ||
| Millicell cell culture insert (12mm, 12 μm) | Millipore | Cat #PIXP01250 |
| Falcon permeable support for 24-well plate (0.4 μm) | Corning | Cat #353095 |
Highlights.
peBCs are an expandable plastic cell type to study pulmonary epithelial regeneration
scRNA-seq enables high-res comparison of engineered tissues to native benchmarks
peBCs in organoid and ALI show evidence of non-physiologic assimilation by scRNA-seq
peBCs in engineered lung and trachea yield more physiologic epithelial regeneration
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (1R01HL13854001 and 1R21EB024889 to L.E.N., U01HL145567 and R01HL127349 to N.K., and T32GM007205 and 1F30HL14390601A1 to M.S.B.R.). We thank the Yale Center for Genome Analysis (YCGA), the Yale Pathology Tissue Services (YPTS), the Yale Stem Cell Center, and the Yale School of Medicine Center for Cellular and Molecular Imaging (CCMI). We would like to acknowledge Barry Stripp and Gianni Carraro for their help in developing single-cell analysis methodology and the kind gift of CCSP primary antibody and Katherine Leiby for helpful discussions.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.03.004.
DECLARATION OF INTERESTS
L.E.N. is a founder of and shareholder in Humacyte, which is a regenerative medicine company. Humacyte produces engineered blood vessels from allogeneic smooth muscle cells for vascular surgery. L.E.N.’s spouse has equity in Humacyte, and L.E.N. serves on Humacyte’s board of directors. L.E.N. is listed as an inventor on patents that are licensed to Humacyte and that produce royalties for L.E.N. L.E.N. has received an unrestricted research gift to support research in her laboratory at Yale University. Humacyte did not fund these studies and Humacyte did not influence the conduct, description, or interpretation of the findings in this article. N.K. served as a consultant to Boehringer Ingelheim, Third Rock, Pliant, Samumed, NuMedii, Indaloo, Theravance, Life-Max, Three Lake Partners, Optikira, and CohBar over the last 3 years and received non-financial support from MiRagen. N.K. is listed as an inventor on several patents and patent applications, none of which are relevant to this manuscript.
REFERENCES
- Agassandian M, and Mallampalli RK (2013). Surfactant phospholipid metabolism. Biochim. Biophys. Acta 1831, 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Yang M, Fan Z, Li S, Gao T, and Fang Z (2015). Associations of chemo- and radio-resistant phenotypes with the gap junction, adhesion and extracellular matrix in a three-dimensional culture model of soft sarcoma. J. Exp. Clin. Cancer Res 34, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestrini JL, Gard AL, Liu A, Leiby KL, Schwan J, Kunkemoeller B, Calle EA, Sivarapatna A, Lin T, Dimitrievska S, et al. (2015). Production of decellularized porcine lung scaffolds for use in tissue engineering. Integr. Biol 7, 1598–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BLM (2013). Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest 123, 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batenburg JJ (1992). Surfactant phospholipids: synthesis and storage. Am. J. Physiol 262, L367–L385. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Hall HG, and Parry G (1982). How does the extracellular matrix direct gene expression? J. Theor. Biol 99, 31–68. [DOI] [PubMed] [Google Scholar]
- Botilde Y, Yoshiba S, Shinohara K, Hasegawa T, Nishimura H, Shiratori H, and Hamada H (2013). Cluap1 localizes preferentially to the base and tip of cilia and is required for ciliogenesis in the mouse embryo. Dev. Biol 381, 203–212. [DOI] [PubMed] [Google Scholar]
- Bouvet S, Golinelli-Cohen MP, Contremoulins V, and Jackson CL (2013). Targeting of the Arf-GEF GBF1 to lipid droplets and Golgi membranes. J. Cell Sci 126, 4794–4805. [DOI] [PubMed] [Google Scholar]
- Buccoliero R, Ginzburg L, and Futerman AH (2004). Elevation of lung surfactant phosphatidylcholine in mouse models of Sandhoff and of Niemann-Pick A disease. J. Inherit. Metab. Dis 27, 641–648. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle EA, Hill RC, Leiby KL, Le AV, Gard AL, Madri JA, Hansen KC, and Niklason LE (2016). Targeted proteomics effectively quantifies differences between native lung and detergent-decellularized lung extracellular matrices. Acta Biomater 46, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole BB, Smith RW, Jenkins KM, Graham BB, Reynolds PR, and Reynolds SD (2010). Tracheal basal cells: a facultative progenitor cell pool. Am. J. Pathol 177, 362–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crapo JD, Barry BE, Gehr P, Bachofen M, and Weibel ER (1982). Cell number and cell characteristics of the normal human lung. Am. Rev. Respir. Dis 126, 332–337. [DOI] [PubMed] [Google Scholar]
- Das B, Cash MN, Hand AR, Shivazad A, Grieshaber SS, Robinson B, and Culp DJ (2010). Tissue distibution of murine Muc19/smgc gene products. J. Histochem. Cytochem 58, 141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong PM, van Sterkenburg MAJA, Hesseling SC, Kempenaar JA, Mulder AA, Mommaas AM, Dijkman JH, and Ponec M (1994). Ciliogenesis in human bronchial epithelial cells cultured at the air-liquid interface. Am. J. Respir. Cell Mol. Biol 10, 271–277. [DOI] [PubMed] [Google Scholar]
- Dobbs LG, Wright JR, Hawgood S, Gonzalez R, Venstrom K, and Nellenbogen J (1987). Pulmonary surfactant and its components inhibit secretion of phosphatidylcholine from cultured rat alveolar type II cells. Proc. Natl. Acad. Sci. USA 84, 1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Raredon MSB, Le AV, Yuan Y, Oczkowicz YA, Kan EL, Baevova P, and Niklason LE (2019). Non-invasive and real-time measurement of microvascular barrier in intact lungs. Biomaterials 217, 119313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez A, Kourembanas S, Wyatt TA, and Mitsialis SA (2009). Mutation of murine adenylate kinase 7 underlies a primary ciliary dyskinesia phenotype. Am. J. Respir. Cell Mol. Biol 40, 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, and Randell SH (2005). Well-differentiated human airway epithelial cell cultures. Methods Mol. Med 107, 183–206. [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Corbit KC, Sirerol-Piquer MS, Ramaswami G, Otto EA, Noriega TR, Seol AD, Robinson JF, Bennett CL, Josifova DJ, et al. (2011). A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet 43, 776–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg M, Böttcher A, Burtscher I, Hasenoeder S, Van Campenhout C, Aichler M, Walch A, Grant SGN, and Lickert H (2014). Flattop regulates basal body docking and positioning in monoand multiciliated cells. eLife 3, e03842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaedi M, Calle EA, Mendez JJ, Gard AL, Balestrini J, Booth A, Bove PF, Gui L, White ES, and Niklason LE (2013). Human iPS cell-derived alveolar epithelium repopulates lung extracellular matrix. J. Clin. Invest 123, 4950–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin SE, Charest JM, Ren X, Tapias LF, Wu T, Evangelista-Leite D, Mathisen DJ, and Ott HC (2016). Regenerative potential of human airway stem cells in lung epithelial engineering. Biomaterials 108, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou D, Mishra A, Weng T, Su L, Chintagari NR, Wang Z, Zhang H, Gao L, Wang P, Stricker HM, and Liu L (2008). Annexin A2 interactions with Rab14 in alveolar type II cells. J. Biol. Chem 283, 13156–13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, and Schlesner M (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. [DOI] [PubMed] [Google Scholar]
- Guttentag SH, Akhtar A, Tao JQ, Atochina E, Rusiniak ME, Swank RT, and Bates SR (2005). Defective surfactant secretion in a mouse model of Hermansky-Pudlak syndrome. Am. J. Respir. Cell Mol. Biol 33, 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hable WE, and Kropf DL (2005). The Arp2/3 complex nucleates actin arrays during zygote polarity establishment and growth. Cell Motil. Cytoskeleton 61, 9–20. [DOI] [PubMed] [Google Scholar]
- Hegab AE, Sakamoto T, Uchida Y, Nomura A, Ishii Y, Morishima Y, Mochizuki M, Kimura T, Saitoh W, Massoud HH, et al. (2004). CLCA1 gene polymorphisms in chronic obstructive pulmonary disease. J. Med. Genet 41, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegab AE, Ha VL, Gilbert JL, Zhang KX, Malkoski SP, Chon AT, Darmawan DO, Bisht B, Ooi AT, Pellegrini M, et al. (2011). Novel stem/progenitor cell population from murine tracheal submucosal gland ducts with multipotent regenerative potential. Stem Cells 29, 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RC, Calle EA, Dzieciatkowska M, Niklason LE, and Hansen KC (2015). Quantification of extracellular matrix proteins from a rat lung scaffold to provide a molecular readout for tissue engineering. Mol. Cell. Proteomics 14, 961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Watkins S, Fuchs E, and Stripp BR (2004). In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am. J. Physiol. Lung Cell. Mol. Physiol 286, L643–L649. [DOI] [PubMed] [Google Scholar]
- Hoover M, Runa F, Booker E, Diedrich JK, Duell E, Williams B, Arellano-Garcia C, Uhlendorf T, La Kim S, Fischer W, et al. (2019). Identification of myosin II as a cripto binding protein and regulator of cripto function in stem cells and tissue regeneration. Biochem. Biophys. Res. Commun 509, 69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, and Lempicki RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, and Lempicki RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Huang SXL, Green MD, de Carvalho AT, Mumau M, Chen YW, D’Souza SL, and Snoeck HW (2015). The in vitro generation of lung and airway progenitor cells from human pluripotent stem cells. Nat. Protoc 10, 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Barkauskas CE, Takeda N, Bowie EJ, Aghajanian H, Wang Q, Padmanabhan A, Manderfield LJ, Gupta M, Li D, et al. (2015). Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat. Commun 6, 6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Urbinati CR, Crusselle VJ, Luo D, Lee YJ, Harrison JK, Oh SP, and Swanson MS (2003). Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Gene Expr. Patterns 3, 459–462. [DOI] [PubMed] [Google Scholar]
- Keady BT, Samtani R, Tobita K, Tsuchya M, San Agustin JT, Follit JA, Jonassen JA, Subramanian R, Lo CW, and Pazour GJ (2012). IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev. Cell 22, 940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CFB, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, and Jacks T (2005). Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835. [DOI] [PubMed] [Google Scholar]
- Kott E, Legendre M, Copin B, Papon JF, Dastot-Le Moal F, Montantin G, Duquesnoy P, Piterboth W, Amram D, Bassinet L, et al. (2013). Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am. J. Hum. Genet 93, 561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PA, Hu Y, Yamamoto Y, Hoe NB, Wei TS, Mu D, Sun Y, Joo LS, Dagher R, Zielonka EM, et al. (2011). Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 147, 525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRanger R, Peters-Hall JR, Coquelin M, Alabi BR, Chen C, Wright WE, and Shay J (2018). Reconstituting Mouse Lungs with Conditionally Reprogrammed Human Bronchial Epithelial Cells. Tissue Eng. Part A 24, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Delmotte P, Robinson ML, Sanderson MJ, and Witman GB (2008). Mutations in Hydin impair ciliary motility in mice. J. Cell Biol 180, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Bhang DH, Beede A, Huang TL, Stripp BR, Bloch KD, Wagers AJ, Tseng YH, Ryeom S, and Kim CF (2014). Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell 156, 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Lin MH, Hwu YM, Lu CH, Yeh LY, Chen YJ, and Lee RKK (2015). Correlation of cumulus gene expression of GJA1, PRSS35, PTX3, and SERPINE2 with oocyte maturation, fertilization, and embryo development. Reprod. Biol. Endocrinol 13, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JY, Nettesheim P, and Randell SH (1994). Growth and differentiation of tracheal epithelial progenitor cells. Am. J. Physiol 266, L296–L307. [DOI] [PubMed] [Google Scholar]
- Liu Q, Liu K, Cui G, Huang X, Yao S, Guo W, Qin Z, Li Y, Yang R, Pu W, et al. (2019). Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat. Genet 51, 728–738. [DOI] [PubMed] [Google Scholar]
- Madsen J, Kliem A, Tornoe I, Skjodt K, Koch C, and Holmskov U (2000). Localization of lung surfactant protein D on mucosal surfaces in human tissues. J. Immunol 164, 5866–5870. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Inoue T, Lee HC, Kono N, Tanaka F, Gengyo-Ando K, Mitani S, and Arai H (2008). Member of the membrane-bound O-acyltransferase (MBOAT) family encodes a lysophospholipid acyltransferase with broad substrate specificity. Genes Cells 13, 879–888. [DOI] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, and Chen CS (2004). Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6, 483–495. [DOI] [PubMed] [Google Scholar]
- McCauley KB, Alysandratos KD, Jacob A, Hawkins F, Caballero IS, Vedaie M, Yang W, Slovik KJ, Morley M, Carraro G, et al. (2018). Single-Cell Transcriptomic Profiling of Pluripotent Stem Cell-Derived SCGB3A2+ Airway Epithelium. Stem Cell Reports 10, 1579–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead BE, Ordovas-Montanes J, Braun AP, Levy LE, Bhargava P, Szucs MJ, Ammendolia DA, MacMullan MA, Yin X, Hughes TK, et al. (2018). Harnessing single-cell genomics to improve the physiological fidelity of organoid-derived cell types. BMC Biol 16, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, et al. (2018). A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee I, Roy S, and Chakrabarti S (2019). Identification of Important Effector Proteins in the FOXJ1 Transcriptional Network Associated With Ciliogenesis and Ciliary Function. Front. Genet 10, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichane M, Javed A, Sivakamasundari V, Ganesan M, Ang LT, Kraus P, Lufkin T, Loh KM, and Lim B (2017). Isolation and 3D expansion of multipotent Sox9+ mouse lung progenitors. Nat. Methods 14, 1205–1212. [DOI] [PubMed] [Google Scholar]
- Ostrowski LE, and Nettesheim P (1995). Inhibition of ciliated cell differentiation by fluid submersion. Exp. Lung Res 21, 957–970. [DOI] [PubMed] [Google Scholar]
- Ota C, Ng-Blichfeldt JP, Korfei M, Alsafadi HN, Lehmann M, Skronska-Wasek W, M De Santis M, Guenther A, Wagner DE, and Königshoff M (2018). Dynamic expression of HOPX in alveolar epithelial cells reflects injury and repair during the progression of pulmonary fibrosis. Sci. Rep 8, 12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TH, Calle EA, Colehour MB, and Niklason LE (2012). Matrix composition and mechanics of decellularized lung scaffolds. Cells Tissues Organs (Print) 195, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiot A, Conti FJA, Grose R, Revest JM, Hodivala-Dilke KM, and Dickson C (2003). A crucial role for Fgfr2-IIIb signalling in epidermal development and hair follicle patterning. Development 130, 5493–5501. [DOI] [PubMed] [Google Scholar]
- Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, and Jaffe AB (2018). A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina S, Preston GM, Guggino WB, and Agre P (1995). Molecular cloning and characterization of an aquaporin cDNA from salivary, lacrimal, and respiratory tissues. J. Biol. Chem 270, 1908–1912. [DOI] [PubMed] [Google Scholar]
- Randell SH, Fulcher ML, O’Neal W, and Olsen JC (2011). Primary Epithelial Cell Models for Cystic Fibrosis Research. Methods Mol. Biol 742, 285–310. [DOI] [PubMed] [Google Scholar]
- Raredon MSB, Adams TS, Suhail Y, Schupp JC, Poli S, Neumark N, Leiby KL, Greaney AM, Yuan Y, Horien C, et al. (2019). Single-cell connectomic analysis of adult mammalian lungs. Sci. Adv 5, eaaw3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Chiba N, Yao C, Guan X, Mcconnell AM, Brockway B, Que L, Mcqualter JL, and Stripp BR (2016). Rare SOX2+ Airway Progenitor Cells Generate KRT5+ Cells that Repopulate Damaged Alveolar Parenchyma following Influenza Virus Infection. Stem Cell Rep 7, 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridsdale R, Na CL, Xu Y, Greis KD, and Weaver T (2011). Comparative proteomic analysis of lung lamellar bodies and lysosome-related organelles. PLoS One 6, e16482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, and Hogan BLM (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA 106, 12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rückert P, Bates SR, and Fisher AB (2003). Role of clathrin- and actindependent endocytotic pathways in lung phospholipid uptake. Am. J. Physiol. Lung Cell. Mol. Physiol 284, L981–L989. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch KG, Lori A, Burns KA, Eldred T, Olsen JC, and Randell SH (2004). A subset of mouse tracheal epithelial basal cells generates large colonies in vitro. Am. J. Physiol. Lung Cell. Mol. Physiol 286, L631–L642. [DOI] [PubMed] [Google Scholar]
- Schwabe GC, Hoffmann K, Loges NT, Birker D, Rossier C, de Santi MM, Olbrich H, Fliegauf M, Failly M, Liebers U, et al. (2008). Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations. Hum. Mutat 29, 289–298. [DOI] [PubMed] [Google Scholar]
- Sennett R, and Rendl M (2012). Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Semin. Cell Dev. Biol 23, 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki DJ, Gromeier M, Mueller S, Bernhardt G, and Wimmer E (2002). Expression of the human poliovirus receptor/CD155 gene is activated by sonic hedgehog. J. Biol. Chem 277, 25697–25702. [DOI] [PubMed] [Google Scholar]
- Stern W, Kovac C, and Weinhold PA (1976). Activity and properties of CTP: cholinephosphate cytidylyltransferase in adult and fetal rat lung. Biochim. Biophys. Acta 441, 280–293. [DOI] [PubMed] [Google Scholar]
- Stone KC, Mercer RR, Gehr P, Stockstill B, and Crapo JD (1992). Allometric relationships of cell numbers and size in the mammalian lung. Am. J. Respir. Cell Mol. Biol 6, 235–243. [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Banzai Y, Kato M, Fukami K, Kataoka Y, Takai Y, Yoshida N, and Takenawa T (2007). Male-specific sterility caused by the loss of CR16. Genes Cells 12, 721–733. [DOI] [PubMed] [Google Scholar]
- Sun L, Ryan DG, Zhou M, Sun TT, and Lavker RM (2006). EEDA: a protein associated with an early stage of stratified epithelial differentiation. J. Cell. Physiol 206, 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T, Wang Y, Barak LS, Bai Y, Randell SH, and Hogan BLM (2014). IL-6/STAT3 promotes regeneration of airway ciliated cells from basal stem cells. Proc. Natl. Acad. Sci. USA 111, E3641–E3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, and Epstein JA (2011). Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SP, Dantas TJ, Duran I, Wu S, Lachman RS, University of Washington Center for Mendelian Genomics Consortium; Nelson SF, Cohn DH, Vallee RB, and Krakow D (2015). Mutations in DYNC2LI1 disrupt cilia function and cause short rib polydactyly syndrome. Nat. Commun 6, 7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP (2003). Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol 15, 740–746. [DOI] [PubMed] [Google Scholar]
- Thomas J, Morlé L, Soulavie F, Laurençon A, Sagnol S, and Durand B (2010). Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol. Cell 102, 499–513. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauclair S, Nicolas M, Barrandon Y, and Radtke F (2005). Notch1 is essential for postnatal hair follicle development and homeostasis. Dev. Biol 284, 184–193. [DOI] [PubMed] [Google Scholar]
- Vaughan AE, Brumwell AN, Xi Y, Gotts JE, Brownfield DG, Treutlein B, Tan K, Tan V, Liu FC, Looney MR, et al. (2015). Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature 517, 621–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang Z, Huang H, Li J, Wang Z, Yu Y, Zhang C, Li J, Dai H, Wang F, et al. (2018). Pulmonary alveolar type I cell population consists of two distinct subtypes that differ in cell fate. Proc. Natl. Acad. Sci. USA 115, 2407–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver TE, and Whitsett JA (1991). Function and regulation of expression of pulmonary surfactant-associated proteins. Biochem. J 273, 249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SC, Fattet L, Tsai JH, Guo Y, Pai VH, Majeski HE, Chen AC, Sah RL, Taylor SS, Engler AJ, and Yang J (2015). Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat. Cell Biol 17, 678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcutt MJ, Adler KB, and Wu R (1988). A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell. Dev. Biol 24, 420–428. [DOI] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Kim T, Brumwell AN, Driver IH, Wei Y, Tan V, Jackson JR, Xu J, Lee DK, Gotts JE, et al. (2017). Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat. Cell Biol 19, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y (2001). The EGFR family and its ligands in human cancer. Signalling mechanisms and therapeutic opportunities. Eur. J. Cancer 37 (Suppl 4), S3–S8. [DOI] [PubMed] [Google Scholar]
- Yin Z, Gonzales L, Kolla V, Rath N, Zhang Y, Lu MM, Kimura S, Ballard PL, Beers MF, Epstein JA, and Morrisey EE (2006). Hop functions downstream of Nkx2.1 and GATA6 to mediate HDAC-dependent negative regulation of pulmonary gene expression. Am. J. Physiol. Lung Cell. Mol. Physiol 291, L191–L199. [DOI] [PubMed] [Google Scholar]
- Yu H, Königshoff M, Jayachandran A, Handley D, Seeger W, Kaminski N, and Eickelberg O (2008). Transgelin is a direct target of TGF-beta/Smad3-dependent epithelial cell migration in lung fibrosis. FASEB J 22, 1778–1789. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Engler AJ, Raredon MS, Le A, Baevova P, Yoder MC, and Niklason LE (2019). Epac agonist improves barrier function in iPSC-derived endothelial colony forming cells for whole organ tissue engineering. Biomaterials 200, 25–34. [DOI] [PubMed] [Google Scholar]
- Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, and Morrisey EE (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zen K, Yasui K, Gen Y, Dohi O, Wakabayashi N, Mitsufuji S, Itoh Y, Zen Y, Nakanuma Y, Taniwaki M, et al. (2009). Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene 28, 2910–2918. [DOI] [PubMed] [Google Scholar]
- Zhang C, Lee HJ, Shrivastava A, Wang R, McQuiston TJ, Challberg SS, Pollok BA, and Wang T (2018). Long-Term In Vitro Expansion of Epithelial Stem Cells Enabled by Pharmacological Inhibition of PAK1-ROCK-Myosin II and TGF-β Signaling. Cell Rep 25, 598–610.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman KA, Yancopoulos GD, Collum RG, Smith RK, Kohl NE, Denis KA, Nau MM, Witte ON, Toran-Allerand D, Gee CE, et al. (1986). Differential expression of myc family genes during murine development. Nature 319, 780–783. [DOI] [PubMed] [Google Scholar]
- Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, et al. (2015). p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature 517, 616–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets generated in this study are available at NCBI Gene Expression Omnibus (GEO). scRNaseq data: GEO: GSE145517. Microarray data: GEO: GSE145516. Other requests for data or software may be directed to and will be fulfilled by the Lead Contact, Allison Greaney (allison.greaney@yale.edu).


