Abstract
From a root bark of Lespedeza bicolor Turch we isolated two new (7 and 8) and six previously known compounds (1–6) belonging to the group of prenylated polyphenols. Their structures were elucidated using mass spectrometry, nuclear magnetic resonance and circular dichroism spectroscopy. These natural compounds selectively inhibited human drug-resistant prostate cancer in vitro. Prenylated pterocarpans 1–3 prevented the cell cycle progression of human cancer cells in S-phase. This was accompanied by a reduced expression of mRNA corresponding to several human cyclin-dependent kinases (CDKs). In contrast, compounds 4–8 induced a G1-phase cell cycle arrest without any pronounced effect on CDKs mRNA expression. Interestingly, a non-substituted hydroxy group at C-8 of ring D of the pterocarpan skeleton of compounds 1–3 seems to be important for the CDKs inhibitory activity.
Keywords: Polyphenolic compounds, Lespedeza bicolor, pterocarpans, prostate cancer, apoptosis, cell cycle
1. Introduction
Lespedeza bicolor Turcz. is a shrub plant belonging to the Leguminosae family. It is widely distributed in East Asia, including the Primorye region of the Russian Far East [1]. In traditional folk medicine, this plant is used for the treatment of nephritis, azotemia, inflammation, hyperpigmentation, energy depletion, diabetes and diuresis [2,3,4,5,6]. In addition, L. bicolor extracts possess antioxidant, anti-tyrosinase, anti-inflammatory, estrogenic, antimicrobial and antifungal activities [6,7,8].
Recently, a group from Korea has shown that L. bicolor extracts exert a potent memory-enhancing effect when treating cognitive dysfunction induced by amyloid β peptide (25–35) in mice models [9]. Additionally, this extract has been described as a promising therapeutic tool to prevent diabetic nephropathy in methylglyoxal (MGO)-induced models both in vitro and in vivo [10]. Remarkably, the L. bicolor extract reduced hyperglycemia-induced hepatic damage, hepatic oxidative stress, and inflammation, as well as liver fibrosis [11]. Finally, this extract inhibited the growth of lung carcinoma LU-1 and prostate cancer LNCaP cells [12]. However, only a little information is available on the compounds responsible for the biological activity of the extract and even less is known about mechanisms of action of these substances.
We have previously isolated several polyphenolic compounds from L. bicolor stem bark, which were able to inhibit the growth of human cancer HTB-19, Kyse-30, and HEPG-2 cells [13]. First insights into the mechanism of action were reported for pterocarpans, coumestans, and arylbenzofurans recently isolated from L. bicolor. These natural compounds were found to promote cell death via induction of a G1 cell cycle arrest, reduction of Bcl-2 levels, and induction of PARP cleavage in Jurkat blood cancer cells [14]. However, to date no significant reports on the mechanisms of action of the purified L. bicolor metabolites are available.
In the current study, we further investigated the metabolites of L. bicolor root bark collected in the Primorye Region (Russian Federation). Therefore, we isolated several new as well as previously known prenylated polyphenolic compounds, investigated their cytotoxic properties and the mechanism of action in human drug-resistant prostate cancer cells.
2. Materials and Methods
2.1. General Experimental Procedures
Optical rotations were measured on a PerkinElmer 343 polarimeter (Perkin Elmer, Waltham, MA, USA). The UV spectra were obtained using a UV-1601 PC spectrophotometer (Shimadzu, Kyoto, Japan). The CD spectra were obtained on a Chirascan-plus Quick Start CD Spectrometer (Applied Photophysics Limited, Leatherhead, UK) (acetonitrile, 20 °C). The IR spectrum was recorded on a Vector 22 fourier-transform infrared spectrometer spectrophotometer (Bruker, Rheinstetten, Germany). The 1H, 13C, and two-dimensional (2D) NMR spectra were recorded in CDCl3 at 30 °C using NMR Bruker AVANCE III DRX-700 and DRX-500 instruments (Bruker, Karlsruhe, Germany). The chemical shift values (δ) and the coupling constants (J) are given in parts per million and Hz, respectively.
2.2. Plant Material
L. bicolor was collected in Khasansky District (Andreevka village) of the Primorye Region (The Russian Federation) from a grassy dry meadow in August 2016 by academician P.G. Grovoy. Voucher specimen No. 103608 is preserved in the herbarium of the Laboratory of Chemotaxonomy (G.B. Elyakov Pacific Institute of Bioorganic Chemistry, FEB RAS).
2.3. Analytical and Preparative HPLC
The analytical HPLC was carried out using an Agilent Technologies 1100 series HPLC system (Agilent Technologies, Waldbronn, Germany) equipped with a VWD detector (λ = 280 nm). The extracts were analyzed using a Supelco Analytical HS-C18 (Supelco Analytical, Bellefonte, PA, USA) column (3 μm, 4.6, 75 mm) thermostated at 30 °C. The mobile phase consisted of 1% aqueous acetic acid (A) and acetonitrile containing 1% acetic acid (B). For the analysis, the following gradient steps were programmed: 0–2 min—5% B, 2–4 min—5–20% B, 5–17 min—20–50% B, 18–23 min—50–90% B, 24–25 min—90–100% B, 16–27 min—100% B, 28–33 min—100–5% B. The flow rate was 0.8 mL/min. The data were analyzed with the ChemStation software (v. 09, Agilent Technologies, Waldbronn, Germany).
The preparative HPLC was carried out using a Shimadzu HPLC system equipped with an LC-20AT pump and SPD-20A detector (λ = 280 nm) (Shimadzu, Kyoto, Japan). The polyphenolic compounds were purified using a Supelco Analytical HS-C18 (Supelco Analytical, Bellefonte, PA, USA) column (3 μm, 4.6, 75 mm). The mobile phase consisted of 1% aqueous acetic acid (A) and acetonitrile containing 1% acetic acid (B). For the analysis, the following gradient steps were programmed: 0–2 min—5–10% B, 2–4 min—10–20% B, 4–7 min—20–30% B, 7–10 min—30–40% B, 10–12 min—40–50% B, 12–17 min—50–100% B, 17–25 min—100–5% B. The flow rate was 1 mL/min.
The data were analyzed with the LabSolutions software (v. 1.0, Shimadzu, Kyoto, Japan).
2.4. HR-ESI-MS
HR-ESI-MS experiments were carried out using a Shimadzu hybrid ion trap–time of flight mass spectrometer (Shimadzu, Kyoto, Japan). The operating settings of the instrument were as follows: electrospray ionization (ESI) source potential, −3.8 and 4.5 kV for negative and positive polarity ionization, respectively, drying gas (N2) pressure—200 kPa, nebulizer gas (N2) flow—1.5 L/min, temperature for the curved desolvation line (CDL) and heat block—200 °C, detector voltage—1.5 kV and the range of detection—100–900 m/z. The mass accuracy was below 4 ppm. The data were acquired and processed using Shimadzu LCMS Solution software (v.3.60.361, Shimadzu, Kyoto, Japan).
2.5. Extraction and Isolation
Air-dried root bark of L. bicolor (150 g) was extracted twice under reflux with a CHCl3–EtOH mixture at a volume ratio of 3:1 for 3 h (60 °C). The obtained extract of L. bicolor root bark (1.7 g) was chromatographed on a polyamide column (80 g, 50–160 µm, Sigma-Aldrich, St. Louis, MI, USA). The column was eluted with a hexane–CHCl3 solution system with gradually increasing CHCl3 amounts (hexane/CHCl3, 1:0, 100:1, 50:1, 40:1) to give fractions 1–13 and then with a CHCl3–EtOH solution system with gradually increasing EtOH amounts (CHCl3/EtOH, 1:0, 100:1, 50:1, 40:1, v/v) to give fractions 14–20. The fractions containing polyphenolic compounds according to TLC HPLC data were subjected to further purification. TLC plates were treated with a 3% solution of FeCl3 in ethanol to reveal the presence of polyphenolic compounds. Fraction 17 (287 mg) eluted with CHCl3–EtOH (50:1) was chromatographed twice over a silica gel column (40–63 µm, Sigma-Aldrich, St. Louis, MI, USA). The column was eluted with a benzene–ethylacetate solution system with gradually increasing ethylacetate amounts (benzene/EtOAc, 1:0, 200:1, 100:1, 50:1, 40:1, v/v) to obtain compound 1 (10.7 mg). Fraction 18 (176 mg), eluted with CHCl3–EtOH (40:1), was also chromatographed on a silica gel column using the same solution system to obtain compounds 1 (5.5 mg), 2 (9.2 mg). Fraction 20 (281 mg), eluted with CHCl3 – EtOH (20:1), was also chromatographed on a silica gel column using the same solution system to obtain compounds 6 (15.0 mg) and 8 (2.5 mg).
Fraction 14 (82 mg), eluted with CHCl3, was subsequently chromatographed over a silica gel column (40–63 µm) eluted twice with a benzene–ethylacetate solution system with gradually increasing ethylacetate amounts (benzene/EtOAc, 1:0, 200:1, 100:1, 50:1, 40:1, v/v) resulting in compound 3 (6.6 mg). Fraction 10 (67 mg), eluted with CHCl3, was subsequently chromatographed over a silica gel (40–63 µm) column twice using the same solution system to afford compounds 4 (6.1 mg), 5 (8.5 mg), and 7 (3.5 mg).
2.5.1. (6aR,11aR)-8-O-Methyl-6a,11a-Dihydrolespedezol A2 (7)
White, amorphous powder; [α]D24 − 201°; (1 mg/mL, MeOH); UV (MeOH), λmax 206, 288, 333 nm; CD (1.89 × 10−4 M, CH3CN) λmax (Δε) 192 (+80899), 206 (–12011), 216 (–109803), 234 (–59308), 292 (+19668); 1H and 13C NMR data, see Table 1; HR-ESI-MS m/z 421.2007 [M-H]− (calcd for [C26H29O5]− 421.2020,), m/z 423.2157 [M+H]+ (calcd for [C26H31O5]+ 423.2166).
Table 1.
1H (700 MHz), 13C (175 MHz), and HMBC NMR data for compound 7 (δ in ppm, J in Hz).
| N | δH (J in Hz) | δC | HMBC | ROESY |
|---|---|---|---|---|
| 1 | 7.40 d (8.4), 1H | 132.4 | C-3, 4, 4a | 2 |
| 2 | 6.55dd (2.6, 8.4), 1H | 109.5 | C-3, 4, 11b | 1 |
| 3 | 156.7 | |||
| 4 | 6.40 dd (2.6), 1H | 103.5 | C-2, 3, 4a, 11b | |
| 4a | 156.6 | |||
| 6 | 4.22 dd (5.0, 11,0), 1H | 66.6 | C-4a, 6a, 6b, 11a | 6, 6a, 7 |
| 3.66 t (11.0), 1H | 66.6 | C-4a, 6a, 6b, 11a | 6 | |
| 6a | 3.48 m, 1H | 40.7 | C-6, 6b, 10a | 6, 11a |
| 6b | 115.6 | |||
| 7 | 6.66 s, 1H | 105.1 | C-6, 6a, 6b, 8, 9, 10, 10a | 6, OCH3-8 |
| 8 | 141.0 | |||
| 9 | 144.5 | |||
| 10 | 111.8 | |||
| 10a | 152.4 | |||
| 11a | 5.40 d (7.0), 1H | 77.4 | C-1, 4a, 6, 6a, 6b, 11b | 6a |
| 11b | 113.3 | |||
| 1’ | 3.32 m, 2H | 23.0 | C-8, 9, 10, 2’, 3’ | 2’, 9’ |
| 2’ | 5.30 t (7.2), 1H | 121.5 | C-10, 1’, 4’, 9’ | 1’, 4’ |
| 3’ | 135.6 | |||
| 4’ | 1,96 m, 2H | 39.8 | C-2’, 3’, 5’, 6’, 9’ | 2’, 6’ |
| 5’ | 2.04 m (8.3, 7.4, 7.2), 2H | 26.7 | C-3’, 4’, 6’, 7’ | 6’ |
| 6’ | 5.07 t (6.8), 1H | 124.4 | C-5’, 8’, 10’ | 4’, 5’ |
| 7’ | 131.1 | |||
| 8’ | 1.64 s, 3H | 25.6 | C-6’, 7’, 10’ | |
| 9’ | 1.76 s, 3H | 16.1 | C-2’, 3’, 4’ | 1’ |
| 10’ | 1.56 s, 3H | 17.6 | C-6’, 7’, 8 | |
| OH-3 | 4.78 bs, 1H | |||
| OCH3-8 | 3.85, 3H | 56.9 | C-8 | 7 |
| OH-9 | 5.69 s, 1H | C-8, 9, 10, 10ª |
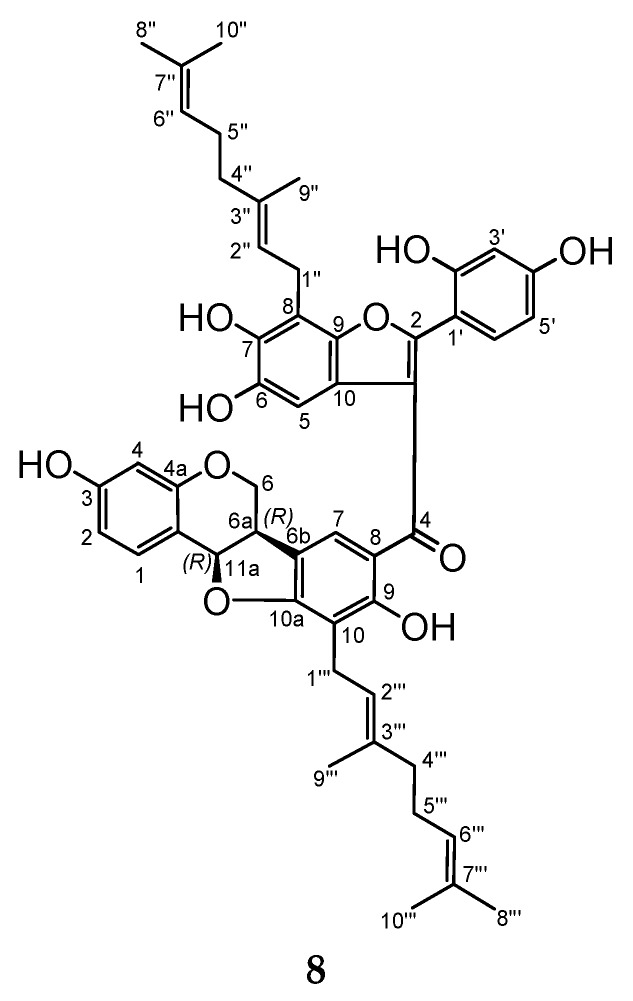
2.5.2. Lespebicolin A (8)
Yellow, amorphous powder; [α]D24 − 95°; (1 mg/mL, MeOH); UV (MeOH), λmax 204, 269, 311 nm; CD (2.46 × 10−4 M, CH3CN) λmax (Δε) 198 (+40706), 211 (–37771), 235 (–33085), 257(+1494), 271 (–7833), 294 (–7058), 329 (–10749), 392 (+3822); 1H and 13C NMR data, see Table 2; HR-ESI-MS m/z 811.3488 [M-H]− (calcd for [C50H51O10]− 811.3488,), m/z 813.3634 [M+H]+ (calcd for [C50H53O10]+ 813.3633).
Table 2.
1H (700 MHz), 13C (175 MHz) and HMBC NMR data for compound 8 (δ in ppm, J in Hz).
| N | δH (J in Hz) | δC | HMBC |
|---|---|---|---|
| Upper | |||
| 2 | 153.7 | ||
| 3 | 119.5 | ||
| 4 | 194.6 | ||
| 5 | 6.82 s, 1H | 103.4 | C-6, 7, 8, 9, 10 |
| 6 | 141.5 | ||
| 7 | 147.3 | ||
| 8 | 111.0 | ||
| 9 | 141.9 | ||
| 10 | 116.9 | ||
| 1’ | 110.7 | ||
| 2’ | 156.6 | ||
| 3’ | 6.50 d (2.3), 1H | 105.0 | C-1’, 2’, 4’, 5’ |
| 4’ | 158.6 | ||
| 5’ | 6.41 dd (2.3, 8.5), 1H | 108.6 | C-1’, 3’ |
| 6’ | 7.26 d (8.5), 1H | 132.2 | C-2, 2’, 4’ |
| 1″ | 3.69 d (7.3), 2H | 23.4 | C-7, 8, 9, 2″, 3″ |
| 2″ | 5.42 t (7.3), 1H | 120.4 | C-4″, 9″ |
| 3″ | 139.3 | ||
| 4″ | 2.11 m, 2H | 39.7 | C-2″, 3″, 5″, 6″, 9″ |
| 5″ | 2.13 m, 2H | 26.4 | C-4″, 6″, 7″ |
| 6″ | 5.07 m, 1H | 123.8 | |
| 7″ | 132.1 | ||
| 8″ | 1.68 s, 3H | 25.7 | C-6″, 7″, 10″ |
| 9″ | 1.86 s, 3H | 16.3 | C-2″, 3″, 4″ |
| 10″ | 1.60 s, 3H | 17.7 | C-6″, 7″, 8″ |
| Lower | |||
| 1 | 7.38 d (8.5), 1H | 132.4 | C-3, 4a,11a |
| 2 | 6.56 dd (2.4, 8.5), 1H | 110.1 | C-4, 11b |
| 3 | 157.2 | ||
| 4 | 6.38 d (2.4), 1H | 103.6 | C-2, 3, 4a, 11b |
| 4a | 156.4 | ||
| 6 | 4.01 m, 1H | 66.3 | |
| 3.56 m, 1H | 66.3 | ||
| 6a | 3.46 m, 1H | 39.6 | |
| 6b | 118.4 | ||
| 7 | 7.34 s, 1H | 127.1 | C-4 (upper), 6a, 9, 10a |
| 8 | 113.4 | ||
| 9 | 164.8 | ||
| 10 | 112.3 | ||
| 10a | 164.8 | ||
| 11a | 5.58 d (7.3), 1H | 79.2 | C-1, 4a, 6, 11b |
| 11b | 112.5 | ||
| 1‴ | 3.32 d (7.2), 2H | 22.2 | C-9, 10, 10a, 2‴, 3‴ |
| 2‴ | 5.26 t (7.2), 1H | 121.0 | C-10, 1‴, 4‴, 9‴ |
| 3‴ | 136.0 | ||
| 4‴ | 1.98 m, 2H | 39.8 | C-2‴, 3‴, 5‴, 6‴, 9‴ |
| 5‴ | 2.04 m, 2H | 29.7 | C-3‴, 4‴, 6‴, 7‴ |
| 6‴ | 5.07 m, 1H | 124.4 | C-8‴, 10‴ |
| 7‴ | 131.3 | ||
| 8‴ | 1.64 s, 3H | 25.6 | C-6‴, 7‴, 10‴ |
| 9‴ | 1.77 s, 3H | 16.2 | C-2‴, 3‴, 4‴ |
| 10‴ | 1.56 s, 3H | 17.6 | C-6‴, 7‴, 8‴ |
| OH-9 | 12.83, s, 1H | ||
2.6. Cell Lines and Conditions
The human prostate cancer cell lines PC-3 [docetaxel-resistant, androgen-independent, AR-FL(-), AR-V7(-)], 22Rv1 [docetaxel-sensitive, androgen-independent, AR-FL(+), AR-V7(+)], and the human non-cancer fibroblast cell line MRC-9 were used. PC-3 and 22Rv1 were purchased from ATCC (Manassas, VA, USA). MRC-9 cells were kindly donated by Prof. Dr. med. Sonja Loges (University Medical Center Hamburg-Eppendorf, Hamburg, Germany). Cells were cultured according to the manufacturers’ protocol in 10% FBS/RPMI (PC-3 and 22Rv1) or 10% FBS/DMEM (MRC-9) medium.
2.7. MTT Assay
In vitro drug sensitivity MTT assay was performed as previously described [15]. In total, 6000 cells/well were seeded in 96-well plates, incubated overnight, and treated with the investigated drugs for 48 h.
2.8. Flow Cytometry Analysis
The effect of the compounds on apoptosis induction and cell cycle progression was analyzed by flow cytometry technique using PI staining as reported before [15]. Cells (0.2 × 106 cells/well) were seeded in six-well plates, incubated overnight, and treated with the investigated drugs for 48 h. The cells were further harvested, fixed with 70% EtOH/H2O, stained with propidium iodide (PI), and analyzed using a FACS Calibur machine (BD Bioscience, San Jose, CA, USA). The results were quantified using BD Bioscience Cell Quest Pro software (v.5.2.1., BD Bioscience).
2.9. Quantitative Real-Time PCR (qPCR)
The CDKs gene expression was measured using qPCR technique. PC-3 cells were seeded in Petri dishes (1 × 106 cells per ø 6 cm dish in 5 mL) in standard culture media, incubated overnight and treated with the investigated drugs in the fresh culture media for 24 h. Cells were harvested using the cell scraper, washed with PBS and homogenized using QIAshredder (QIAGEN, Hilden, Germany). The total RNA was isolated using PureLink® RNA Mini Kit (Invitrogen, Carlsbad, CA, USA) and the on-column DNA digestion using PureLink™ DNase (Invitrogen). Correspondent RNA (2 µg in 30 µL) and was transcribed into cDNA using Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Scientific, Vilnius, Lithuania). The following qPCR was performed using 2X KAPA SYBR FAST qPCR Master Mix Optimized for Roche LightCycler 480 (KAPA biosystems, Worburn, MA, USA) according to the manufacturer’s protocol. In total, 20 ng of the template cDNA and 2 pmol of primers were used for each reaction. The PCR conditions were 30 s at 95 °C, followed by 40 cycles of 15 s at 95 °C, 5 s at melting temperature (Tm), and 26 s at 72 °C (fluorescence measurement). Melting curve analysis was performed under the following conditions directly after the PCR run: 10 s at 95 °C, 60 s at 65 °C and 1 s at 97 °C. Relative gene expression was calculated using the 2-∆∆CT method. The expression of CDKs was normalized to GAPDH gene expression. The analysis was performed using primers, purchased from Eurofins MWG-Biotech AG (Ebersberg, Germany). Primers sequences and melting temperatures (Tm) is presented in Table 3.
Table 3.
The sequences and melting temperatures (Tm) of the primers used for qPCR.
| Primer | Sequence | Tm [°C] |
|---|---|---|
| CDK1_for | ACAGGTCAAGTGGTAGCCATGA | 60 |
| CDK1_rev | ACCTGGAATCCTGCATAAGCA | |
| CDK2_for | TTCTCATCGGGTCCTCCACC | 61 |
| CDK2_rev | TCGGTACCACAGGGTCACCA | |
| CDK4_for | CTGTGCCACATCCCGAACTG | 61 |
| CDK4_rev | GCCTCTTAGAAACTGGCGCA | |
| CDK5_for | CCACAACATCCCTGGTGAACGT | 62 |
| CDK5_rev | CCTCTTCTGCTGAGATACGCTG | |
| CDK6_for | CCGAAGTCTTGCTCCAGTCC | 61 |
| CDK6_rev | GGGAGTCCAATCACGTCCAA | |
| CDK7_for | TCACATCTTCAGTGCAGCAGG | 59 |
| CDK7_rev | TGGCAGCTGACATCCAGGT | |
| CDK8_for | AGCGGGTCGAGGACCTGTTT | 62 |
| CDK8_rev | CATGCCGACATAGAGATCCCAG | |
| CDK3_for | TCGCTGCTCAAGGAACTGAAGC | 62 |
| CDK3_rev | GTCCTGGCTGAGGAACTCAAAC | |
| CDK9_for | CCATTACAGCCTTGCGGGAGAT | 62 |
| CDK9_rev | CAGCAAGGTCATGCTCGCAGAA |
3. Results and Discussion
3.1. Isolation and Structure Elucidation of Compounds 1–8
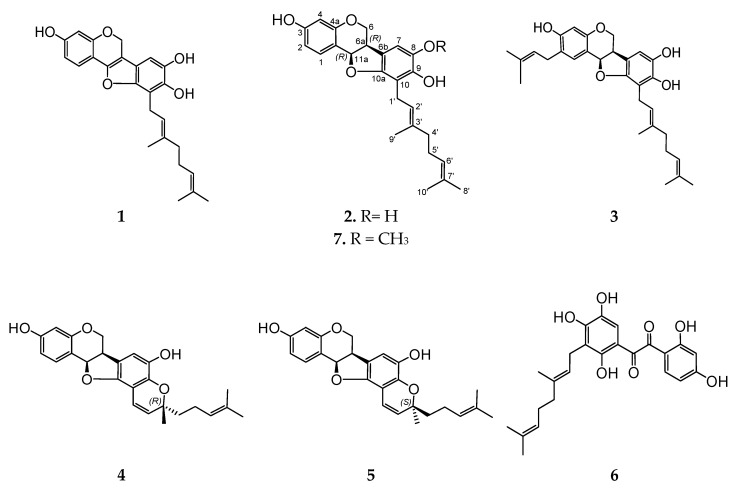
We used a column chromatography on polyamide sorbent to isolate the fractions of the bioactive prenylated polyphenolic compounds. The fractions were tested for the presence of polyphenolics using TLC plates treated with FeCl3 and an HPLC–PDA–MS technique. The fractions evaluated positively in a FeCl3 test were subsequently separated using silica gel column. Individual compounds were further purified using preparative HPLC. Compounds 1–6 were identified by comparison of their HPLC-PDA-MS and NMR spectra with previously published data [13]. Apart from prenylated polyphenolic compounds 1–6, we isolated and identified two new prenylated pterocarpan derivatives 7 and 8 (Figure 1).
Figure 1.
The structures of polyphenolic compounds 1–8 isolated from L. bicolor root bark and stem bark.
The structures of the isolated compounds were elucidated using mass spectrometry, NMR and CD spectroscopy (for the detailed experimental spectral data of the new compounds please see Supplementary data). Optically active prenylated polyphenolic compound 7 was isolated from L. bicolor root bark as a white amorphous powder. Its molecular formula was determined as C26H30O5 based on HRESI-MS data. We observed the [M-H]− ion at m/z 421.2007 (calc. for [C26H29O5]− 421.2020) in the negative ion mode, whereas the spectrum in the positive ion mode revealed the presence of the [M+H]+ ion at m/z 423.2157 (calc. 423.2166 for [C26H31O5]−). The 13C NMR spectrum contained 26 carbon atoms: 15 carbon atoms of the pterocarpan skeleton, 10 carbon atoms of the geranyl side chain and one carbon atom of the methoxy group (Table 1). 1H NMR spectrum also revealed characteristic signals of protons of the pterocarpan skeleton at δH 3.48 (1H, m), 3.66 (1H, t, J = 11.0), 4.22 (1H, dd, J = 5.0, 11.0) and 5.40 (1H, d, J = 7.0) assigned to H-6a, two H-6, and H-11a protons, respectively. The signals at δH 7.40 (1H, d, J = 8.4), 6.55 (1H, dd, J = 2.6, 8.4), and 6.40 (1H, d, J = 2.6) belonged to H-1, H-2, and H-4 protons of ring A, respectively, and formed a spin system. The singlet signal at δH 6.66 (1H, s) was assigned to the aromatic proton at C-7 of ring D. The location of the geranyl side chain at C-10 of the pterocarpan skeleton was determined based on the correlation between the proton signal of 2H-1’ at δH 3.32 and carbon signals of C-9, C-10 и C-10a at δC 144.5, 111.8, and 152.4, respectively, in the HMBC spectrum of 7. The position of the methoxy group was determined as C-8 based on the observed correlation fbetween the signal of its protons at δH 3.85 and the signal of C-8 at δC 141.0 in the HMBC spectrum of 7 (Table 1). The signals in the 1H and 13C spectra were completely assigned using HSQC, ROESY, and HMBC data (Table 1). Negative optical rotation value ([α]D24 − 201º) [16], as well as characteristic Cotton effects in the CD spectrum of 7 (positive bands at λ 192 nm and 292 nm, and negative bands at 216 and 234 nm), confirmed the 6aR,11aR configuration of the asymmetric centers in 7 [2,17]. Thus, the structure of 7 was determined as (6aR,11aR)-8-O-methyl-6a,11a-dihydrolespedezol A2.
Compound 8 was obtained as a yellow amorphous powder. Its HR-ESI-MS spectrum contained the [M-H]− ion at m/z 811.3469 in the negative ion mode (calc. 811.3488 for [C50H51O10]−) and the [M+H]+ ion at m/z 813.3634 (calc. 813.3633 for [C50H53O10]+) in the positive ion mode. Thus, the molecular formula of 8 was determined as C50H52O10. Both positive and negative MS/MS spectra revealed the intensive signals of the ions [M+H-H2O]+ at m/z 795.3521 (calc. 795.3528) and 793.3360 [M-H-H2O]− (calc. 793.3382) due to water molecule loss (Figures S11 and S13, Supplementary data). The presence of the pterocarpan and 2-arylbenzofuran fragments, linked with the carbonyl group, was confirmed using the tandem mass spectrometry technique. The negative MS/MS product ions at m/z 419.1476 with the composition [C25H23O6]− (сalc. 419.1500) and at m/z 393.1663 with the composition [C24H25O5]− (calc. 393.1707) resulted from the carbonyl bridge degradation and indicated a neutral loss of a pterocarpan fragment (Figures S11 and S12, Supplementary data). The signals at m/z 419.1870 (calc. 419.1853) and m/z 377.1711 (calc. 377.1747) corresponded to the ions [C26H27O5]+ and [C24H25O4]−, respectively, and indicated the loss of a neutral pterocarpan fragment. The 13C NMR spectrum revealed the presence of 50 carbon atoms: 15 atoms constituted the pterocarpan skeleton, 14 carbon atoms formed the 2-arylbezofuran fragment, 20 atoms belonged to the two geranyl side chains, and one carbon atom belonged to the carbonyl group. The presence of the pterocarpan fragment in 8 was confirmed by the proton signals at δH 3.46 (1H, m), 3.56 (1H, m), 4.01 (1H, m), and 5.58 (1H, d, J = 7.3) assigned to H-6a, two H-6, and H-11a protons, respectively. The chemical shift values of these signals were very similar to the signals of H-6a, two H-6, and H-11a protons in the 1H spectrum of compounds 1–5 and 7. The geranyl side chain was attached to C-10 of the pterocarpan moiety, which was confirmed by the presence of the HMBC-correlation between the signal of 2H-1‴ at δH 3.32 and the carbon signals of C-9, C-10 и C-10a at δC 164.8, 112.3, and 164.8, respectively.
In the 1H spectrum of 8, we have observed a singlet signal at δH 6.82 assigned to the aromatic proton H-5 and the signals of the ABX proton system in the 2-arylbezofuran fragment at δH 6.50 (1H, d, J = 2.3), 6.41 (1H, dd, J = 2.3, 8.5), and 7.26 (1H, d, J = 8.5) of H-3’, H-5’, and H-6’ protons, respectively. The second geranyl side chain was located at C-8 of the 2-arylbezofuran fragment, which was demonstrated by the presence of the HMBC-correlation between the signals of 2H-1″ at δH 3.69 (2H, d, J = 7.3) and the signals of C-7, C-8 and C-9 at δc 147.3, 111.0, and 141.9, respectively. The chemical shift values of the other atoms of 2-arylbezofuran fragment in 1H and 13C NMR spectra were similar to those for lespecyrtin H2 [18].
We observed a correlation between the proton signal of H-7 at δH 7.34 (1H, s) of the pterocarpan fragments and the carbon signal of C-4 of the carbonyl group at δC 194.6 in the HMBC spectrum of 8. Thus, we concluded that the carbonyl group was attached to C-8 of a pterocarpan fragment. The signal of the hydroxy group proton at C-9 of the pterocarpan fragment had a δH of 12.83, which also confirmed that the carbonyl group was located at C-8 and formed a hydrogen bond with this hydroxy group. The signals in 1H and 13C NMR spectra of 8 were assigned using HSQC, HMBC, and ROESY experiments. Thus, compound 8, named lespebicolin A, was assumed to be a dimeric flavonoid consisting of the 2-arylbenzofuran and the pterocarpan fragments linked via carbonyl group.
The absolute configurations of the asymmetric centers at C-6a and C-11a in the pterocarpan fragment of 8 were determined based on the negative optical rotation value ([α]D24 − 95º) and CD spectral data similar to those of lespecyrtins H1–H4 [18].
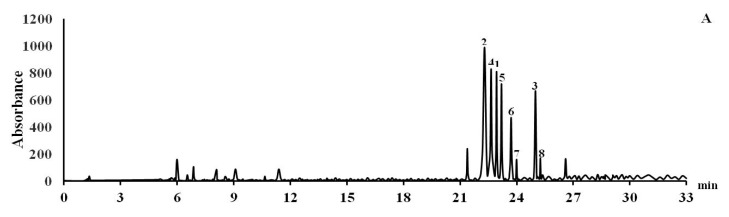
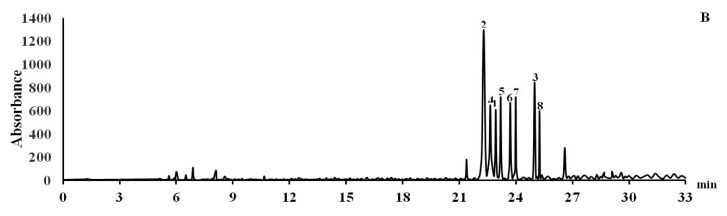
Thus, in addition to the six polyphenolic compounds 1–6 previously isolated from L. bicolor stem bark [13], we isolated two new pterocarpans 7 and 8 from L. bicolor root bark. A comparison of the HPLC profiles of L. bicolor stem bark and root bark showed that compounds 7 and 8 are also present in L. bicolor stem bark but in smaller amounts than in root bark (Figure 2).
Figure 2.
HPLC profiles of L. bicolor stem bark (A) and root bark (B) ethanolic extracts.
Recently, Korean colleagues isolated five pterocarpans, two new coumestans, and two new arylbenzofurans with prenyl and geranyl substituents in their structures [14]. These isolated compounds contain a methoxy group at C-1 and exhibited antiproliferative effects on human leukemia cells. Of note, we could not find these compounds in the stem bark and root bark of L. bicolor harvested in the South of the Primorskiy region of the Russian Far East. The polyphenolic compounds isolated by us from the stem bark and root bark of L. bicolor also contained the pterocarpan skeleton and geranyl side chains in their structures. However, they did not have a methoxy group at C-1. Moreover, for the first time, we have isolated a new dimeric flavonoid lespebicolin A (8) harboring two geranyl side chains. Previously, the related dimeric flavonoids had been found in L. homoloba, L. floribunda and L. cyrtobotria [18,19]. Thus, the chemical compositions of polyphenolic compounds of L. bicolor growing in the Primorskiy region and South Korea differed significantly. This may be because L. bicolor samples collected in the Primorye Region (the South of the Russian Far East) and South Korea belong to different variants of this species [20].
3.2. Investigation of Anticancer In Vitro Activity of the Compounds 1–8
Next, we examined the cytotoxic activity of the isolated compounds in prostate cancer cell lines as well as in non-cancer cells. To get a first impression on the impact of these novel compounds for the treatment of advanced prostate cancer, human drug-resistant cell lines PC-3 and 22Rv1 were used. Both cell lines are known to be castration-resistant (androgen-independent). In addition, the cell lines are known to exhibit resistance to novel second-generation androgen receptor (AR) targeting drugs, e.g., abiraterone and enzalutamide, due to the loss or alternative splicing of the AR, respectively [21,22]. Moreover, PC-3 cells have been reported to be rather resistant to docetaxel, and therefore, are known as one of the most aggressive prostate cancer cell lines used as in vitro and in vivo models. Notably, all eight isolated compounds exhibited a cytotoxic activity in both prostate cancer cell lines in the micromolar range, while non-malignant MRC-9 cells were less affected under the treatment (Table 4). The selectivity index evaluation (SI; MRC-9 vs. PC-3 cells) has revealed compounds 1, 2, 3 as well as 6 and 8 to be the most promising and selective in human drug-resistant prostate cancer cells (SI = 2.5 ~ 8) (Table 4). Remarkably, a well-established cytotoxic chemotherapeutic drug cisplatin applied for the treatment of different cancer types and used in the current research as a positive control was less selective (SI = 0.72) (Table 4). For further investigations on the mechanisms of action, we have chosen the aggressive and drug-resistant prostate cancer PC-3 cell line.
Table 4.
Inhibition concentrations 50% (IC50s) of the investigated compounds in different cell lines. The cells were treated for 48 h. Cisplatin was used as a positive control. Selectivity index (SI) was calculated as [IC50 in MRC-9 cells] / [IC50 in PC-3 cells].
| IC50, µM | ||||||||||
| Compound | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Cisplatin | |
| Cell line | PC-3 | 6.73 | 5.24 | 2.1 | 16.37 | 15.23 | 19.26 | 14.86 | 9.24 | 21.0 |
| 22Rv1 | 20.14 | 13.85 | 6.38 | 29.53 | 35.81 | 46.9 | 30.51 | 17.51 | 4.53 | |
| MRC-9 | 48.51 | 37.57 | 16.82 | 27.18 | 24.61 | >50 | 24.4 | 23.95 | 15.12 | |
| Selectivity index (MRC-9 vs. PC-3) |
7.21 | 7.17 | 7.99 | 1.66 | 1.62 | >2.6 | 1.64 | 2.59 | 0.72 | |
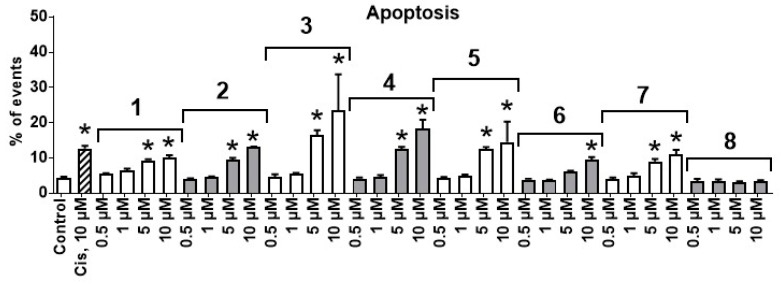
To investigate the mechanism of action contributing to the cytotoxicity in PC-3 cells, we investigated the effects on DNA fragmentation, a well-established marker of cellular apoptosis. Therefore, we evaluated the effects of the isolated compounds at four different concentrations of 0.5 µM, 1 µM, 5 µM, and 10 µM by flow cytometry (Figure 3). All the substances apart from structurally different lespebicolin A (8) revealed a significant apoptosis induction at the tested concentrations (Figure 3). Therefore, prenylated dimeric flavonoid 8 may either reveal a non-apoptotic character of cell death or a predominantly antiproliferative rather than cytotoxic effect. Note, compound 3 exhibited the most pronounced apoptosis-inducing effect.
Figure 3.
Effect of natural compounds on DNA fragmentation of PC-3 cells. Cells were treated with the investigated compounds for 24 h. The effects were analyzed by flow cytometry. Cells detected as sub-G1 population were considered to be apoptotic. Cisplatin (Cis) was used as a positive control. * p ≤ 0.05 (Student’s t-test).
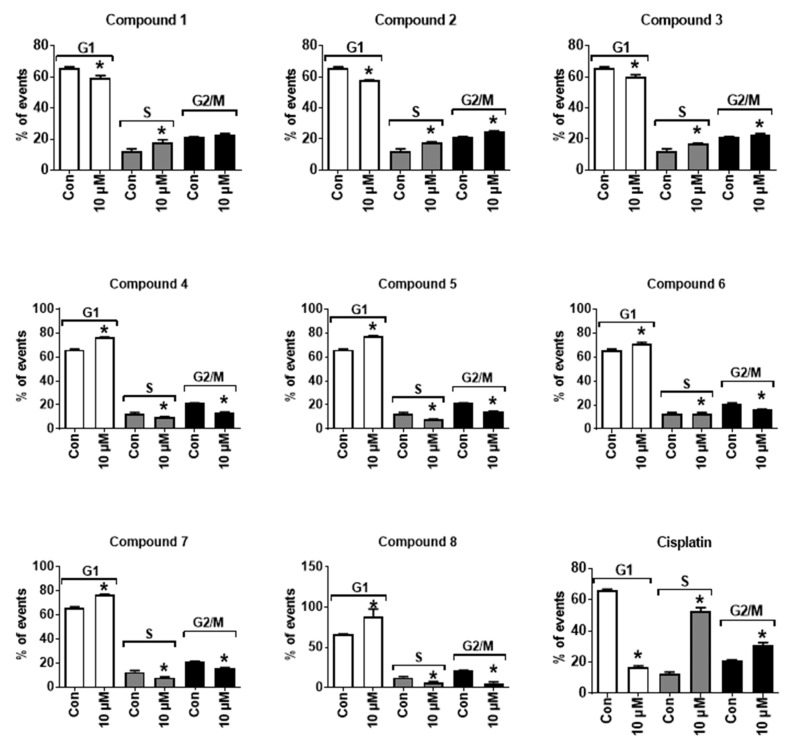
Next, we examined the effects of the isolated compounds on the cell cycle progression of PC-3 cells. For compounds 1–3, a significant accumulation of the cells in S-phase as well as slight accumulation in G2/M-phase was observed (Figure 4). Thus, these compounds mainly induced a S-phase arrest, whereas the compounds 4–8 led to a pronounced G1-phase cell cycle arrest (Figure 4).
Figure 4.
Treatment effects on the progression of the cell cycle of PC-3 cells. Cells were treated with the investigated compounds for 24 h. The effects were analyzed by flow cytometry. Cisplatin was used as a positive control. * p ≤ 0.05 (Student’s t-test).
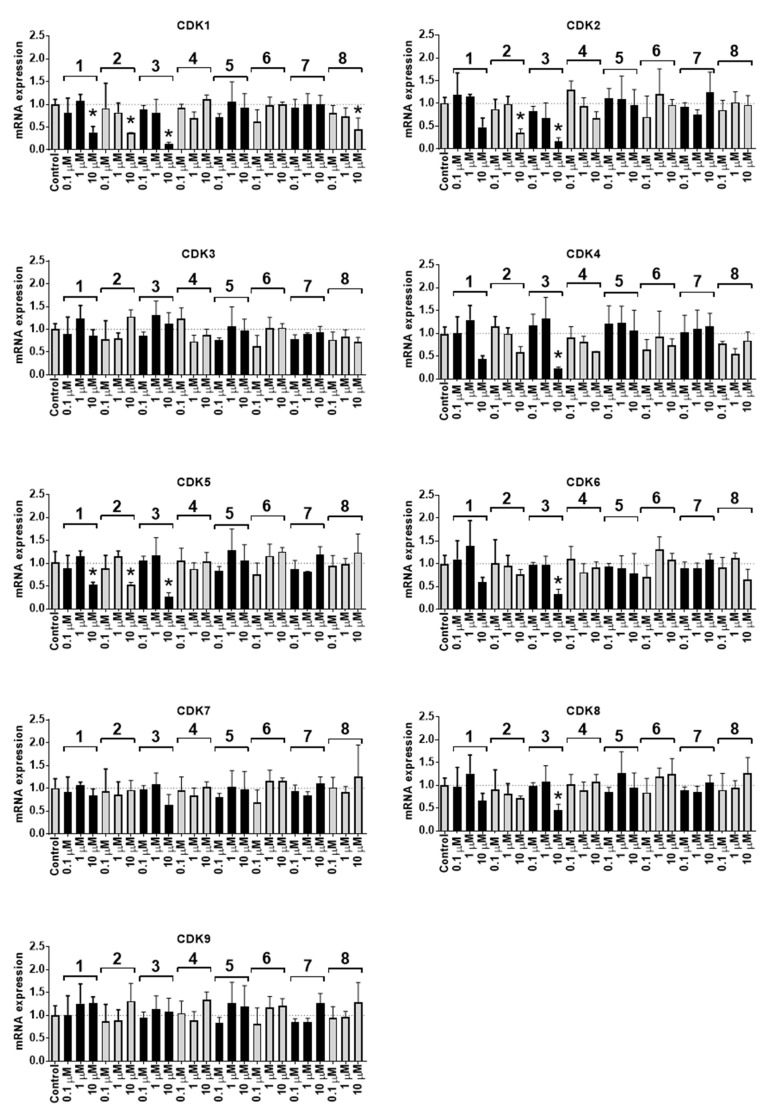
Due to the significant alterations of the cell cycle, we further investigated the effects of the isolated compounds on the cyclin-dependent kinases (CDKs). These proteins are known to control proliferation as well as some apoptotic processes. We have examined the expression of mRNAs corresponding to the nine most studied CDKs known to be involved in the cell cycle progression of cancer cells, namely, CDK1–CDK9. Indeed, we detected the inhibitory effects of compounds 1, 2, and 3 on the expression of mRNA corresponding to several essential CDKs (Figure 5). The most pronounced effects were observed for CDK1, CDK2, CDK4, and CDK5. Remarkably, these effects correlated well with the cytotoxicity of the isolated compounds. Thus, the strongest inhibitory effect on the corresponding mRNA expression was observed with the most cytotoxic compound 3 (IC50 = 2.1 µM in PC-3 cells, Table 3).
Figure 5.
Expression of mRNA correspondent to CDK1-9 in PC-3 cells treated with the isolated compounds 1–8 for 24 h. The target gene expression was normalized to GAPDH gene expression. * p ≤ 0.05 (Student’s t-test).
CDKs are known to play an important role in the development and progression of several cancer types, including human prostate cancer. These kinases are involved in the cell cycle control and are often overexpressed or mutated in cancer cells [23]. Therefore, CDKs are an attractive target in anticancer therapy and are currently actively explored [23]. In particular, CDK1 and CDK6 are known to phosphorylate AR and activate its transcriptional activity at different phosphorylation sites finally leading to a poorer clinical prognosis [24,25,26]. Expression and activity of CDK2 are associated with prostate cancer relapse in patients and with cancer cell invasion [27]. In addition, the particular importance and clinical relevance of CDK5 for prostate cancer growth have been highlighted in different publications. Thus, it was reported to phosphorylate and stabilize AR leading to the promotion of its transcriptional activity [28,29]. Moreover, CDK5 stimulates the growth of AR-negative prostate cancer cells via Akt activation [30]. Of note, the CDK4/6 inhibitor is currently undergoing clinical trials in metastatic castration-resistant prostate cancer. They can promote cell senescence as well as disruption of cancer cells in vivo by the induction of cytotoxic T cell-mediated cell death. Moreover, inhibition of CDK4/6 may cause S-phase cell cycle arrest, which has been observed for compounds 1–3 (Figure 4) [31,32].
Of note, the other compounds 4–8 did not exhibit pronounced effects on CDKs expression (Figure 5) but still inhibited cell cycle progression, however, in the G1 phase (Figure 4). This effect may result from the other processes related either to apoptosis induction or inhibition of different CDKs at the post-transcriptional (inhibition of CDKs protein expression) or even post-translational levels (inhibition of CDKs activity). Indeed, a free hydroxyl group at C-8 (e.g., pterocarpans 1–3) seems to be important for the inhibition of CDKs mRNA expression by these natural compounds. Hence, compound 2, containing 8-OH was fairly active in this experiment, however, very structurally similar new compound 7 containing a methylated hydroxyl group (8-OCH3) did not exhibit this activity; moreover, it was less cytotoxic and had a different effect on cell cycle progression (i.e., G1-phase arrest instead of S-phase arrest). In addition, new compound 8 has a pterocarpan fragment related to compounds 1, 2 and 3. However, this compound is substituted at C-8 and therefore similar to 7 did not exhibit any pronounced effect on CDKs mRNA expression and could not induce an S-phase cell cycle arrest.
4. Conclusions
In conclusion, we isolated two new (7 and 8) and six recently reported (1–6) polyphenolic compounds from the root bark of L. bicolor. Structures were established using mass spectrometry and NMR spectroscopy. The natural compounds were active and selective in human drug-resistant prostate cancer cells in vitro. Prenylated pterocarpans 1–3 effectively inhibited cell cycle progression of human cancer cells in S-phase. This was associated with the inhibition of mRNA expression corresponding to several human CDKs. Compounds 4–8 induced G1-phase cell cycle arrest without any pronounced effect on CDK mRNA expression. The non-substituted hydroxy group at C-8 in ring D of the pterocarpan skeleton seems to be important for the CDKs inhibitory activity.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/10/3/451/s1, Figures S1–S20.
Author Contributions
Conceptualization, S.A.D., D.T., and S.F.; methodology, S.A.D., D.T., S.F., J.H., and G.v.A.; validation, S.A.D., D.T., S.F., T.B., M.K., M.V., A.K., J.H., V.G., and N.K.; formal analysis, all authors; investigation, S.A.D., D.T., S.F., T.B., M.K., M.V., A.K., J.H., V.G., and N.K., V.G.; resources, P.G., C.B., M.G., and G.v.A.; data curation, S.A.D., D.V., S.F., T.B., M.K., M.V., A.K., J.H., V.G., N.K., and V.G.; writing—original draft preparation, S.A.D., D.T., S.F., and G.v.A.; writing—review and editing, all authors; visualization, S.A.D., D.T., and S.F.; supervision, S.F. and G.v.A.; project administration, S.F., and G.v.A.; funding acquisition, D.T., C.B., M.G., and G.v.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Russian Foundation for Basic Research (grant 18-34-00502 mol_a).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Sun H., Lei Y.N., Deng W., Wang H.M., Teng Y.O., Zhao H.Y., Yao Q.W., Yu P. First Total Synthesis and Cytotoxicity of Naturally Occurring Lespedezol E1. Chem. Nat. Compd. 2016;52:896–898. doi: 10.1007/s10600-016-1807-0. [DOI] [Google Scholar]
- 2.Woo H.S., Kim D.W., Curtis-Long M.J., Lee B.W., Lee J.H., Kim J.Y., Kang J.E., Park K.H. Potent inhibition of bacterial neuraminidase activity by pterocarpans isolated from the roots of Lespedeza bicolor. Bioorg. Med. Chem. Lett. 2011;21:6100–6103. doi: 10.1016/j.bmcl.2011.08.046. [DOI] [PubMed] [Google Scholar]
- 3.Maximov O.B., Kulesh N.I., Stepanenko L.S., Dmitrenok P.S. New prenylated isoflavanones and other constituents of Lespedeza bicolor. Fitoterapia. 2004;75:96–98. doi: 10.1016/j.fitote.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Glyzin V.I., Ban’kovskii A.I., Zhurba O.V., Sheichenko V.I. Flavonoids of Lespedeza bicolor. Chem. Nat. Compd. 1970;6:487–488. doi: 10.1007/BF00564266. [DOI] [Google Scholar]
- 5.Jae-Hak T.A.L., Jin-Woo T.A.J. Antioxidant activity of different parts of Lespedeza bicolor and isolation of antioxidant compound. Korean J. Food Sci. Technol. 2012;44:763–771. [Google Scholar]
- 6.Do M.H., Lee J.H., Wahedi H.M., Pak C., Lee C.H., Yeo E.J., Lim Y., Ha S.K., Choi I., Kim S.Y. Lespedeza bicolor ameliorates endothelial dysfunction induced by methylglyoxal glucotoxicity. Phytomedicine. 2017;36:26–36. doi: 10.1016/j.phymed.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y.-S., Chang Z., Park S.-C., Rim N.-R., Kim N.-W. Antioxidative activity and irritation response of Lespedeza bicolor. Toxicol. Res. 2005;21:115–119. [Google Scholar]
- 8.Lee S.J., Hossaine M.D., Park S.C. A potential anti-inflammation activity and depigmentation effect of Lespedeza bicolor extract and its fractions. Saudi J. Biol. Sci. 2016;23:9–14. doi: 10.1016/j.sjbs.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ko Y.H., Shim K.Y., Kim S.K., Seo J.Y., Lee B.R., Hur K.H., Kim Y.J., Kim S.E., Do M.H., Parveen A., et al. Lespedeza bicolor Extract Improves Amyloid Beta25–35-Induced Memory Impairments by Upregulating BDNF and Activating Akt, ERK, and CREB Signaling in Mice. Planta Med. 2019;85:1363–1373. doi: 10.1055/a-1018-5402. [DOI] [PubMed] [Google Scholar]
- 10.Do M.H., Lee J.H., Cho K., Kang M.C., Subedi L., Parveen A., Kim S.Y. Therapeutic Potential of Lespedeza bicolor to Prevent Methylglyoxal-Induced Glucotoxicity in Familiar Diabetic Nephropathy. J. Clin. Med. 2019;8:1138. doi: 10.3390/jcm8081138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Y., Lee H., Kim S.Y., Lim Y. Effects of Lespedeza Bicolor Extract on Regulation of AMPK Associated Hepatic Lipid Metabolism in Type 2 Diabetic Mice. Antioxidants. 2019;8:599. doi: 10.3390/antiox8120599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullah S. Methanolic extract from Lespedeza bicolor: Potential candidates for natural antioxidant and anticancer agent. J. Tradit. Chin. Med. 2017;37:444–451. doi: 10.1016/S0254-6272(17)30150-4. [DOI] [PubMed] [Google Scholar]
- 13.Tarbeeva D.V., Fedoreyev S.A., Veselova M.V., Blagodatski A.S., Klimenko A.M., Kalinovskiy A.I., Grigorchuk V.P., Berdyshev D.V., Gorovoy P.G. Cytotoxic polyphenolic compounds from Lespedeza bicolor stem bark. Fitoterapia. 2019;135:64–72. doi: 10.1016/j.fitote.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Thuy N.T.T., Lee J.E., Yoo H.M., Cho N. Antiproliferative Pterocarpans and Coumestans from Lespedeza bicolor. J. Nat. Prod. 2019;82:3025–3032. doi: 10.1021/acs.jnatprod.9b00567. [DOI] [PubMed] [Google Scholar]
- 15.Dyshlovoy S.A., Tabakmakher K.M., Hauschild J., Shchekaleva R.K., Otte K., Guzii A.G., Makarieva T.N., Kudryashova E.K., Fedorov S.N., Shubina L.K., et al. Guanidine alkaloids from the marine sponge Monanchora pulchra show cytotoxic properties and prevent EGF-induced neoplastic transformation in vitro. Mar. Drugs. 2016;14:133. doi: 10.3390/md14070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goel A., Kumar A., Raghuvanshi A. Synthesis, stereochemistry, structural classification, and chemical reactivity of natural pterocarpans. Chem. Rev. 2013;113:1614–1640. doi: 10.1021/cr300219y. [DOI] [PubMed] [Google Scholar]
- 17.Veitch N.C. Isoflavonoids of the leguminosae. Nat. Prod. Rep. 2013;30:988–1027. doi: 10.1039/c3np70024k. [DOI] [PubMed] [Google Scholar]
- 18.Mori-Hongo M., Yamaguchi H., Warashina T., Miyase T. Melanin synthesis inhibitors from Lespedeza cyrtobotrya. J. Nat. Prod. 2009;72:63–71. doi: 10.1021/np800535g. [DOI] [PubMed] [Google Scholar]
- 19.Miyase T., Sano M., Yoshino K., Nonaka K. Antioxidants from Lespedeza homoloba (II) Phytochemistry. 1999;52:311–319. doi: 10.1016/S0031-9422(99)00194-6. [DOI] [PubMed] [Google Scholar]
- 20.Lee T.B. Illustrated Flora of Korea Seoul. Hyangmunsa; Seoul, Korea: 1993. [Google Scholar]
- 21.Dyshlovoy S.A., Otte K., Tabakmakher K.M., Hauschild J., Makarieva T.N., Shubina L.K., Fedorov S.N., Bokemeyer C., Stonik V.A., von Amsberg G. Synthesis and anticancer activity of the derivatives of marine compound rhizochalin in castration resistant prostate cancer. Oncotarget. 2018;9:16962–16973. doi: 10.18632/oncotarget.24764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miao L., Yang L., Li R., Rodrigues D.N., Crespo M., Hsieh J.T., Tilley W.D., de Bono J., Selth L.A., Raj G.V. Disrupting Androgen Receptor Signaling Induces Snail-Mediated Epithelial-Mesenchymal Plasticity in Prostate Cancer. Cancer Res. 2017;77:3101–3112. doi: 10.1158/0008-5472.CAN-16-2169. [DOI] [PubMed] [Google Scholar]
- 23.Vijayaraghavan S., Moulder S., Keyomarsi K., Layman R.M. Inhibiting CDK in Cancer Therapy: Current Evidence and Future Directions. Target. Oncol. 2018;13:21–38. doi: 10.1007/s11523-017-0541-2. [DOI] [PubMed] [Google Scholar]
- 24.Willder J.M., Heng S.J., McCall P., Adams C.E., Tannahill C., Fyffe G., Seywright M., Horgan P.G., Leung H.Y., Underwood M.A., et al. Androgen receptor phosphorylation at serine 515 by Cdk1 predicts biochemical relapse in prostate cancer patients. Br. J. Cancer. 2013;108:139–148. doi: 10.1038/bjc.2012.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S., Xu Y., Yuan X., Bubley G.J., Balk S.P. Androgen receptor phosphorylation and stabilization in prostate cancer by cyclin-dependent kinase 1. Proc. Natl. Acad. Sci. USA. 2006;103:15969–15974. doi: 10.1073/pnas.0604193103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim J.T., Mansukhani M., Weinstein I.B. Cyclin-dependent kinase 6 associates with the androgen receptor and enhances its transcriptional activity in prostate cancer cells. Proc. Natl. Acad. Sci. USA. 2005;102:5156–5161. doi: 10.1073/pnas.0501203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin X., Yu J., Zhou Y., Wang C., Jiao Z., Qian Z., Sun H., Chen B. Identification of CDK2 as a novel target in treatment of prostate cancer. Future Oncol. 2018;14:709–718. doi: 10.2217/fon-2017-0561. [DOI] [PubMed] [Google Scholar]
- 28.Hsu F.N., Chen M.C., Chiang M.C., Lin E., Lee Y.T., Huang P.H., Lee G.S., Lin H. Regulation of androgen receptor and prostate cancer growth by cyclin-dependent kinase 5. J. Biol. Chem. 2011;286:33141–33149. doi: 10.1074/jbc.M111.252080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oner M., Lin E., Chen M.-C., Hsu F.-N., Shazzad Hossain Prince G.M., Chiu K.-Y., Teng C.-L.J., Yang T.-Y., Wang H.-Y., Yue C.-H., et al. Future Aspects of CDK5 in Prostate Cancer: From Pathogenesis to Therapeutic Implications. Int. J. Mol. Sci. 2019;20:3881. doi: 10.3390/ijms20163881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindqvist J., Imanishi S.Y., Torvaldson E., Malinen M., Remes M., Orn F., Palvimo J.J., Eriksson J.E. Cyclin-dependent kinase 5 acts as a critical determinant of AKT-dependent proliferation and regulates differential gene expression by the androgen receptor in prostate cancer cells. Mol. Biol. Cell. 2015;26:1971–1984. doi: 10.1091/mbc.E14-12-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schettini F., De Santo I., Rea C.G., De Placido P., Formisano L., Giuliano M., Arpino G., De Laurentiis M., Puglisi F., De Placido S., et al. CDK 4/6 Inhibitors as Single Agent in Advanced Solid Tumors. Front. Oncol. 2018;8:608. doi: 10.3389/fonc.2018.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batra A., Winquist E. Emerging cell cycle inhibitors for treating metastatic castration-resistant prostate cancer. Expert Opin. Emerg. Drugs. 2018;23:271–282. doi: 10.1080/14728214.2018.1547707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.