Summary
The amyloid-like aggregation propensity present in most globular proteins is generally considered to be a secondary side effect resulting from the requirements of protein stability. Here, we demonstrate, however, that mutations in the globular and amyloid state are thermodynamically correlated rather than simply associated. In addition, we show that the standard genetic code couples this structural correlation into a tight evolutionary relationship. We illustrate the extent of this evolutionary entanglement of amyloid propensity and globular protein stability. Suppressing a 600-Ma-conserved amyloidogenic segment in the p53 core domain fold is structurally feasible but requires 7-bp substitutions to concomitantly introduce two aggregation-suppressing and three stabilizing amino acid mutations. We speculate that, rather than being a corollary of protein evolution, it is equally plausible that positive selection for amyloid structure could have been a driver for the emergence of globular protein structure.
Keywords: protein folding, protein stability, amyloid, evolution
Graphical Abstract
Highlights
-
•
Mutations in the globular and amyloid state are thermodynamically correlated
-
•
The genetic code tightens this relationship between the amyloid and native state
-
•
Strongly amyloidogenic sequences in globular proteins are deeply conserved
-
•
Positive selection of amyloid propensity will favor globular protein stability
Langenberg et al. show that amyloid propensity favors protein stability. This results from the energetic correlation of mutation in the native and amyloid state. The genetic code tightens this relationship so that stable amyloidogenic sequences are deeply conserved. Positive selection of amyloidogenic sequences could therefore have favored the evolution of globular structure.
Introduction
The Anfinsen postulate states that protein folding is a thermodynamically determined process and, thus, that all of the information required to adopt the native conformation is encoded in the amino acid sequence of a protein (Anfinsen, 1973, Anfinsen and Haber, 1961). For most proteins, however, this principle is embedded in a more complex reality dictated by physico-chemical constraints on protein folding kinetics and thermodynamics. Local structural propensities can conflict with the native state conformation, resulting in structural frustration (Dill et al., 2008). These local versus global structural contradictions are a source of protein misfolding and lead to less efficient protein folding. For some proteins, this results in folding kinetics that are too complex to be resolved without the help of molecular chaperones (Horwich et al., 1990, Jayaraj et al., 2020). A major form of structural frustration is the tendency of local sequence fragments to form intermolecular interactions with the identical sequence fragment of another protein chain by β strand association, resulting in formation of amyloid-like assemblies (Iadanza et al., 2018).
Amyloid assembly is therefore a competitive side reaction of protein folding that significantly affects folding efficiency and requires a significant amount of metabolic energy to avoid aggregation (Landreh et al., 2016). It is not only detrimental to protein function, but the amyloid conformation is also associated with toxic gain of function in a range of degenerative pathologies (Dobson et al., 2020). It is therefore considered that the high aggregation propensity of proteomes across all kingdoms of life is a negative side effect of globular protein structure and their requirement of a hydrophobic core (Ganesan et al., 2016, Rousseau et al., 2006). As discussed above, this strained relationship between protein stability and aggregation led to a redefinition of the Anfinsen postulate, where the native folding landscape is in competition with a second aggregation-determining energetic landscape (Jahn and Radford, 2008). These competing landscapes are characterized by very different structural topologies that are dominated by different non-covalent interactions (Iadanza et al., 2018). The amyloid conformation is a multimolecular assembly that is dominated by backbone-backbone interactions, whereas the globular structure is generally a monomolecular conformation that is predominantly stabilized by side-chain interactions (Fitzpatrick et al., 2011, Eisenberg and Sawaya, 2017).
As a result, aggregation is concentration dependent, whereas folding is concentration independent. Thus, the lower the aggregation propensity of a protein, the higher concentration at which it can be safely expressed. Many proteins, we now know, have expression levels reaching or exceeding their critical concentration so that, in effect, the native state is a meta-stable conformation (Ciryam et al., 2015, Tartaglia et al., 2007). It is still unclear why proteins live at the limit of or above their intrinsic solubility and why protein folding is, in fact, controlled kinetically rather than thermodynamically. To approach these questions, we need to have a better understanding of the structure-activity relationship between the amyloid and the globular state of proteins. Here we set out to explore the biophysical, structural, and evolutionary degrees of freedom that are available to thermodynamically favor globular structure with respect to the amyloid state.
We find that, rather than being simply associated by overlap of average biophysical properties, mutations in the globular and amyloid state are thermodynamically correlated. In addition, we show that the genetic code couples this correlation into a tight evolutionary relationship. We discuss the structural and evolutionary implications of this underestimated thermodynamic relationship. Finally, we speculate how this could have contributed to an amyloid origin of the globular protein universe.
Results
Proteins with High Thermal Stability Are Enriched in Amyloid Sequences
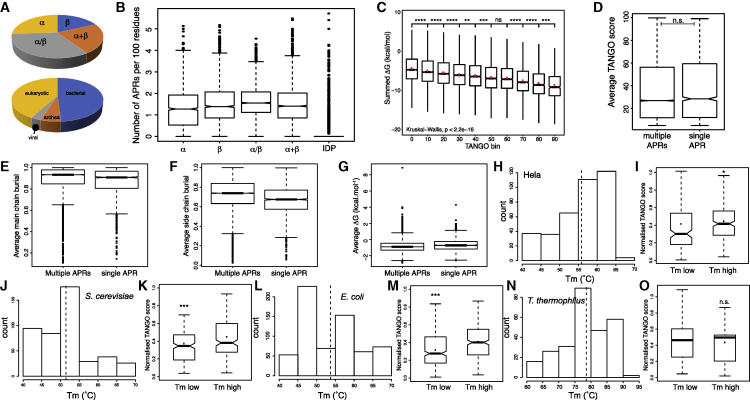
It is well established that amyloidogenic segments (aggregation-prone regions [APRs]) are mostly located in buried positions in the native structure, but they can, to a lesser extent, also be found at exposed sites of functional importance, such as those required for binding or catalysis (Ganesan et al., 2016, Castillo and Ventura, 2009, Buck et al., 2013). Here we investigated the relationship between the aggregation propensity of amyloidogenic sequences and the free energy contribution of the same amino acid residues to the stability of the native state. To have a set of protein domains that is representative of the diversity of protein folds, we employed the SCOPe (structural classification of proteins–extended) database (release 2.06; Chandonia et al., 2017) and filtered for single-chain globular domains and 40% sequence identity using the CD-hit (cluster database at high identity with tolerance) algorithm (Fu et al., 2012). This yielded a dataset of 9,017 PDB structures of single-protein domains, amounting to 16,4791 residues in 23,329 amyloidogenic segments detected by the TANGO algorithm (Fernandez-Escamilla et al., 2004). The four main SCOP (structural classification of proteins) classes (α, β, α/β, and α+β) are well sampled (Figure 1A, top panel) and contain similar amounts of sequences of pro- and eukaryotic origin (Figure 1A, bottom panel), showing that the set reflects a broad sample of protein sequence space. We first compared the APR frequency in the different SCOPe classes of globular protein folds, expressed as the number of APRs per 100 amino acids, and compared it with a set of intrinsically disordered proteins obtained from DisProt (Hatos et al., 2020; release 8.0.1, excluding ambiguous and obsolete regions, length larger than 25, resulting in 1,392 intrinsically disordered protein (IDP) regions from 1,039 proteins, amounting to 145,770 amino acid residues in total; Figure 1B). This showed that the frequency of APRs is very similar across SCOPe fold classes, although there is a slight increase in the topologically most complex α/β class, and it is only really reduced in intrinsically disordered proteins, confirming earlier observations (Linding et al., 2004). At the same time, the analysis revealed many outliers in the IDP category (i.e., IDPs containing APRs; Figure 1B), consistent with well-known aggregation-prone IDPs, including yeast prions and amyloid disease-associated human proteins (Chiti and Dobson, 2017). This basic observation already suggests that globular structure and aggregation propensity are associated properties. To probe this further, we focused on the globular SCOPe classes and calculated the thermodynamic contribution of APR residues to the stability of the native state, for which we employed the empirical force field FoldX, which computes local protein stability contributions per residue as part of its overall stability calculation (Schymkowitz et al., 2005), whereas the aggregation strength was determined using TANGO. We observed a clear association (Pearson’s correlation coefficient = −0.27 ± 0.01, p < 2.2e−16) between the average contribution of an APR to the stability of its native structure and the aggregation strength of that same region (Figure 1C), again suggesting an association between globular protein structure and aggregation. To exclude that the presence of multiple APRs per domain would influence our analysis, we used TANGO to compare the intrinsic aggregation propensity of APRs in domains with a single APR with those containing more APRs (Figure 1D) and found no significant differences (Wilcoxon-Mann-Whitney test). Also, the observation that APRs tend to reside in buried positions (Figures 1E and 1F), where they contribute to the thermodynamic stability of the protein (Figure 1G), is even more pronounced in proteins that contain more than one APR. To address this further, we turned to a recent dataset on the proteome-wide determination of thermal protein stability in four species: E. coli, T. thermophiles, S. cerevisiae, and H. sapiens (HeLa cells) (Leuenberger et al., 2017). We filtered the raw data by LocTree3 subcellular localization prediction (Goldberg et al., 2014) to obtain melting temperature (Tm) values of 1,726 proteins with cytoplasmic or nuclear (“chromosomal” for bacteria) localization. For each species, we divided the proteins into two groups (Figures 1H–1O): one with proteins that have Tm values above the average for that species and the other with Tm values below the average (Table S1). We then calculated the sequence length normalized total TANGO score for each protein and compared the distribution of aggregation propensities in the high- and low-Tm groups (Figures 1H–1O). For the mesophilic HeLa cells (Figures 1H and 1I), S. cerevisiae (Figures 1J and 1K), and E. coli (Figures 1L and 1M), the amyloid-like aggregation propensity of proteins from the high-Tm group was significantly higher than of proteins from the low-Tm group. Interestingly, in the extremophile T. thermophilus (Figures 1N and 1O), which has an optimal growth temperature of about 65°C (Henne et al., 2004), the average length-normalized TANGO score of all proteins is equivalent to the high-Tm group in mesophilic organisms. No further increase in TANGO score was obtained by splitting the proteins of this extremophile into low-and high-Tm groups (Figure 1O), indicating that the hydrophobicity and the associated aggregation propensity are maximized in the entire proteome.
Figure 1.
Stability and Aggregation Propensity Are Related
(A) Class and kingdom composition of the SCOPe dataset.
(B) Boxplot representation of the distribution of APRs in the SCOPe and IDP datasets.
(C) Boxplot showing the contribution of APRs to the stability of the native state calculated by FoldX in the SCOPe dataset in function of the predicted aggregation propensity by TANGO.
(D–G) Boxplots comparing APRs occurring in domains with one APR to those occurring in domains with more than one APR: the distribution TANGO score of APRs (D), the average main-chain burial (E), the average side-chain burial (F), and the average contribution of an APR to native-state stability (G, ΔG calculated by FoldX, in kilocalories per mole).
(H, J, L, and N) Histograms of the melting temperature (Tm) observed in whole-proteome protein stability measurements (Leuenberger et al., 2017) for HeLa cells (H), S. cerevisiae (J), E. coli (L), and T. thermophilus (N). The dotted line indicates the mean Tm of the proteome in question.
(I, K, M, and O) Boxplots comparing the normalized TANGO scores of proteins with a high or low Tm value in HeLa cells (I), S. cerevisiae (K), E. coli (M), and T. thermophilus (O).
The bottoms and tops of the boxes are the first and third quartiles, and the band inside the box represents the median; the dot indicates the mean. The whiskers encompass the minimum and maximum of the data. Significant differences were computed using a Wilcox rank test. Asterisks denote level of significance: n.s., not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
The source files (Data S1) and R-scripts (Data S2) used to generate this figure are available.
This analysis shows that within mesophilic species and between mesophiles and extremophiles, high thermal stability is associated with a high amyloid-like aggregation propensity, suggesting that protein stability and amyloid propensity are entangled properties.
Correlated Thermodynamic Response to Mutation between Tertiary Structure and the Amyloid State
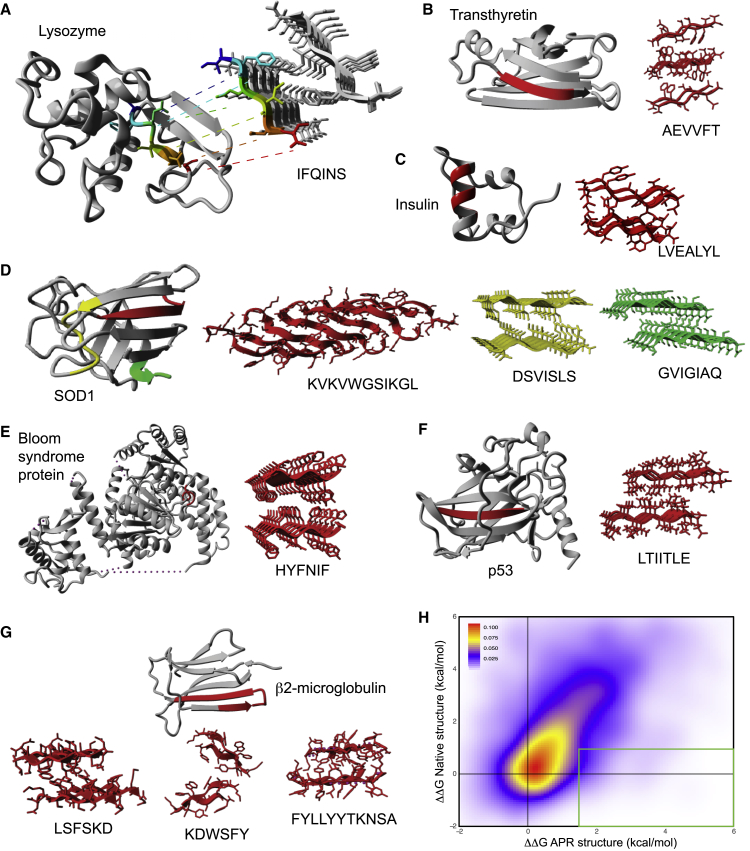
To investigate the interdependence of protein stability and aggregation propensity in more detail, we compared the thermodynamic effect of point mutations in the native and amyloid conformation. Searching for proteins for which high-resolution crystallographic information was available for the globular native fold as well as for its amyloid state yielded a set of 11 amyloidogenic fragments derived from seven proteins (Figures 2A–2G): Bloom syndrome protein, transthyretin, insxulin, super oxide dismutase (SOD1), p53, β2-microglobulin, and lysozyme. We employed FoldX (Schymkowitz et al., 2005) to perform saturation mutagenesis (i.e., mutating each amino acid of the APRs to every other amino acid in the native and the amyloid structure) and calculated the associated change in free energy (ΔΔG values in kilocalories per mole) of both structures. The heatmap summarizing these 1.368 mutations (Figure 2H) clearly shows that the effects of mutation on the stability of native and amyloid structures are correlated (correlation coefficient = 0.46, p < 2.2e–16, Pearson’s product moment correlation test), in line with amyloid and globular structure being associated properties. It is also clear from Figure 2H that there are outliers; i.e., the regions of the plot corresponding to mutations that destabilize the amyloid state without overly perturbing the native state. However, even when considering liberal thresholds for tolerated native-state destabilization (<1.0 kcal/mol), only 53 of 1.368 mutations or 3.87% significantly destabilize the amyloid (>1.5 kcal/mol; i.e., the equivalent of loss of one backbone-backbone hydrogen bond; indicated by the green box in Figure 2H). Thus, the number of mutations that destabilize the amyloid state without simultaneously disrupting the structure of its conjugate native tertiary protein is inherently restricted by the context of globular protein structure. Strikingly, when we considered the DNA sequences of the corresponding genes of the proteins under study, we found that, of these 53 mutations, only 15 or 1.10% of all mutations are accessible by a single-base-pair substitution. This suggests that codon usage further enforces the coupling of these thermodynamically correlated structural conformations.
Figure 2.
Mutational Energies of APRs and Native Structures Are Coupled
(A–G) Schematic representations of the crystal structures of APRs and their cognate native states for lysozyme (A), transthyretin (B), insulin (C), SOD1 (D), Bloom syndrome protein (E), p53 (F), and β2-microglobulin (G). The APR segment is highlighted in color in an otherwise gray native-state structure. The PDB identifiers used are listed in STAR Methods.
(H) Density plot of the free energy change in thermodynamic stability (ΔΔG, FoldX, in kilocalories per mole) associated with mutation in the native state versus the APR state. The green box encompasses the APR-destabilizing mutations (ΔΔG at least 1.5 kcal/mol) that may be tolerated at the native structure level at a cutoff of ΔΔG no larger than 1 kcal/mol. The source files (Data S1) and R-script (Data S2) used to generate this panel are available.
Thermodynamic Coupling between the Native and Amyloid State by the Genetic Code
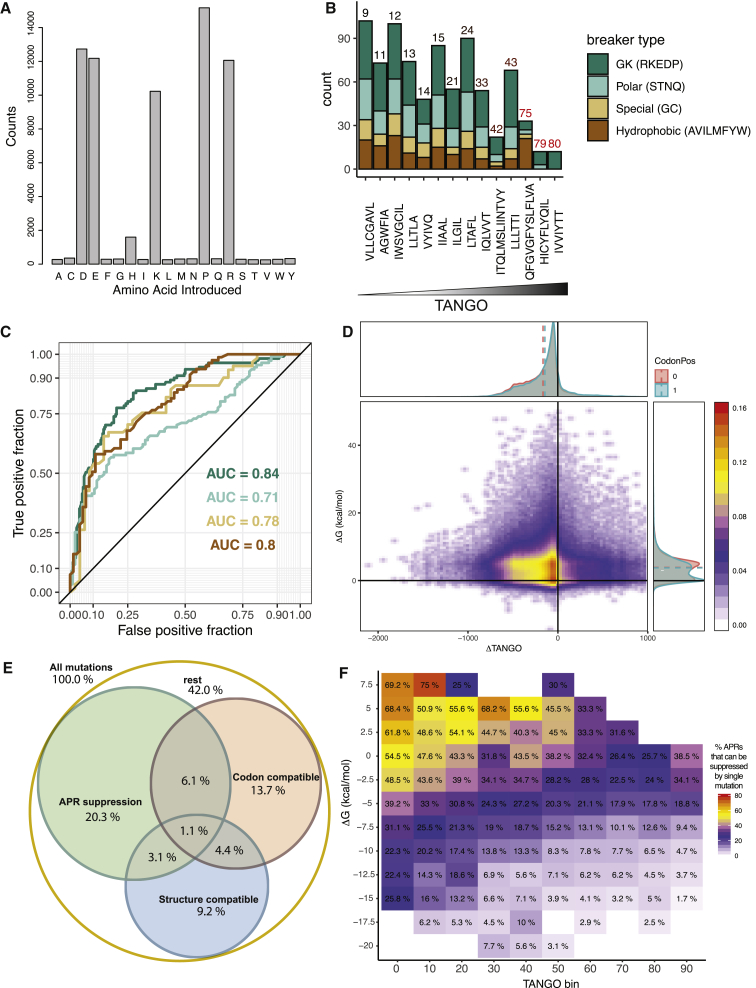
To understand the mechanisms and degree of coupling between the native and amyloid state by codon usage, we analyzed the conservation of both properties by computationally comparing the ability to suppress amyloid propensity by single amino acid mutations in a large variety of protein folds. To do this, we set out to mutate, in silico, each amino acid in each APR in the SCOPe set described above to glutamate, aspartate, lysine, proline, and arginine. These amino acids have been shown previously to have the strongest aggregation-reducing effects of amino acid substitutions and are collectively called gatekeepers (GKs) (Ganesan et al., 2016, Rousseau et al., 2006, van der Kant et al., 2017, Van Durme et al., 2016). To further illustrate this aspect here, we performed an exhaustive mutation scan of the SCOP dataset, selected those that fully suppress (reduce to zero) an APR with an average TANGO score above 50 (to restrict the analysis to the strongest regions) and analyzed the type of substitutions this involved. These data confirm that mutation to P, R, K, D, and E is required to suppress the stronger APRs, allowing us to restrict our in silico mutation screening to these residues (Figure 3A). Then we reanalyzed a published dataset of deep mutational scanning of 10 different proteins, where each mutation was annotated with a normalized fitness score in a recent meta-analysis (Gray et al., 2017). First, we calculated which mutations would fully suppress the intrinsic aggregation propensity of each of the APRs in this dataset, taking only the TANGO score into account and ignoring the structural or codon context (Figure 3B). This showed again that, for stronger APRs, mutations to GK residues are the only effective option, whereas for weaker APRs, other options are possible. Next, we used a receiver operating characteristic (ROC) curve analysis to assess how well FoldX ΔΔG values could predict the fitness outcome (tolerated or disruptive) of each suppressing mutation (Figure 3C). In this analysis, a mutation was classified as tolerated when their normalized fitness effect was reported to be above 0.8. In a ROC curve, the fraction of correct binary classifications (tolerated or disruptive) at each threshold of the predictor (FoldX) is plotted against the fraction of false classifications. The area under the curve gives an indication of the performance of the predictor, in this case FoldX, at the classification problem, in this case classifying mutations into tolerated or disruptive. This showed (Figure 3C) that FoldX performance is overall good at this task but is best in the case of GK residues. Given these results and the fact that, for the current analysis, we were mostly interested in high-scoring, conserved APRs, we restricted subsequent in silico screens for APR-suppressing mutations to P, R, K, D, and E. For each mutation, we evaluated its effect on native-state stability using FoldX and its effect on aggregation propensity using TANGO. In addition, we mapped the residues back to their corresponding codons in the gene sequence to evaluate effects from codon usage bias. We restricted the dataset to APRs that had a length of at least five amino acid residues and an average TANGO score equal to or larger than 5.0% and classified each amino acid change by accessibility with a single DNA mutation (Figure 3D). Ultimately, this yielded a dataset of 821,420 mutations to the GK residues D, E, K, R, or P in 23,263 APRs (totaling 164,284 amino acid positions) derived from 7,876 domains. Of all mutations, 30.7% significantly suppressed aggregation strength (TANGO < 5%), 25.4% were codon accessible, and 17.8% did not disrupt the native structure (ΔΔG < 0.5 kcal/mol) (Figure 3D). Cross-sectioning these three requirements of the restrictions imposed by codon usage and the structural context leaves only 1.1% of mutations that can suppress aggregation using codon-allowed single-base mutations without compromising the native structure compared to 4.2% when also considering artificial mutations to any of the GKs (Figure 3E). Of the five GK residues, arginine is easiest to place, and the four others appear with approximately half of its frequency in the group of the possible mutations (Figure S1). Thus, on a mutation-per-residue basis, it is apparent that codon usage further strengthens the thermodynamic correlation between the amyloid and native state.
Figure 3.
Codon Usage and Native-State Stability Reduce the Number of Possible Aggregation-Suppressing Mutations in a Systematic Mutation Scan of the SCOPe Set of Protein Structures
(A) Mutational screen for mutations that completely suppress an APR (TANGO score of mutant = 0) for all APRs with a TANGO score of more than 50 in the SCOPe set. The height of the bars indicates the number of times each amino acid is found in the suppressed mutant.
(B) Types of suppressing mutations in the deep mutational scanning dataset. Bars depict the number of mutations predicted to be suppressing to the APRs identified in the dataset. Colors indicate amino acid type, and APRs are ordered along the x axis according to their TANGO score.
(C) Receiver operating characteristic (ROC) curve for the prediction of the fitness outcome of a mutation (with a fitness score of more than 0.8 corresponding to a tolerated mutation) from the ΔΔG calculated through FoldX. ROC curves were produced for each amino acid type (color-coded as in B), and area under the curve (AUC) is reported for each prediction.
(D) Density plot of the change of free energy of the native state (ΔΔG, FoldX, in kilocalories per mole) upon mutation versus the associated change in predicted aggregation propensity (TANGO). The top and right insets show histogram projections of the same data, split by whether (CodonPos = 1) or not (CodonPos = 0) the mutations are accessible through single-base changes in the cognate genes. The segmented lines indicate the means of the respected groups.
(E) Venn diagram grouping the mutations from (A) into three groups: APR suppression (TANGO reduced to 0 by the mutation), codon compatible (mutation accessible through a single-base change of the cognate gene), and structure compatible (mutations with a ΔΔG value [FoldX] of less than 0.5 kcal/mol).
(F) Heatmap of the percentage of APRs for which suppressing mutations can be identified in function of the contribution to the native-state stability of the APRs (ΔΔG, FoldX, in kilocalories per mole) and its aggregation propensity (TANGO).
The source files (Data S1) and R-scripts (Data S2) used to generate this figure are available.
Because each amyloidogenic segment can potentially be altered by many single mutations, we can also estimate the quantity of amyloidogenic segments that can be fully suppressed by codon-allowed and structurally conservative GK mutations. Plotting the percentage of suppressible segments per native stability bin, we can see that, although globally approximately one-fourth (24.5%) of all APRs can, in principle, be suppressed by single-nucleotide mutations, the majority of these consist of APRs with low aggregation propensity in regions that also have a low contribution to the stability of the native structure (Figure 3F). It is, however, much more difficult to uncouple both properties when the thermodynamic stability of both states is high (Figure 3F). It is therefore to be expected that sequences that are strongly coupled by high amyloid propensity and high native-state stability are also strongly conserved.
To illustrate this further, we created a network of the transitions that can occur between amino acids, based on single-base-pair substitutions and codon usage, the basic evolutionary operator. The substitution frequency between amino acids is indicated by colored lines (Figure S2; red and yellow are frequent substitutions, and blue is rare.). When functional protein evolution is considered, the amino acids are grouped into hydrophobic (HP), polar (PO), and charged (+/−). Single-base-pair changes favor a conservation of class, which has been noted before and is generally rationalized to favor mutations that retain biological function by replacing like with like but also allows a low frequency of changes between classes, allowing evolutionary changes. When this scheme is considered from the point of view of amyloid-like aggregation, the amino acids can be grouped very similarly into APR, neutral (NT), or aggregation GK. This side of the plot clearly shows that codon bias also tends to conserve amyloid-like propensity along with biological function, including globular structure.
Amyloid Addiction of the p53 Core Domain Fold
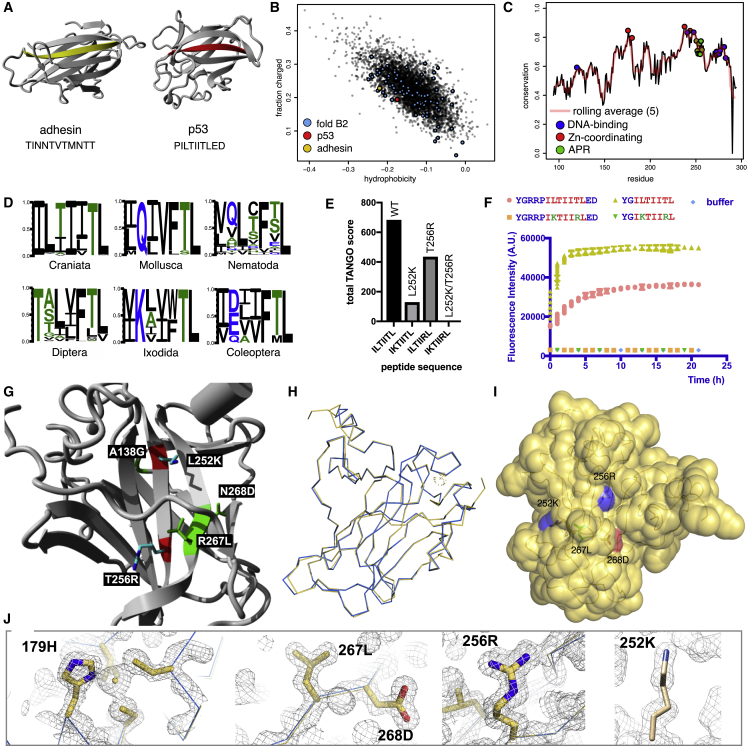
To understand the consequences of codon-enforced thermodynamic entanglement of native and amyloid-like interactions during protein evolution, we took a closer look at fold b.2 (“common fold of diphtheria toxin/transcription factors/cytochrome f”) of the SCOPe database, which contains the superfamily b.2.5 (“p53-like transcription factors”). For p53 itself, we previously identified a very-high-scoring amyloidogenic segment in its HP core (Xu et al., 2011) that was later confirmed by others (Soragni et al., 2016, Wang and Fersht, 2017, Das and Makarov, 2016; Figure 4A, red), although it has been also shown that the protein, in addition, can undergo phase separation (Boija et al., 2018) and amorphous aggregation (Wang and Fersht, 2017). The b.2 fold has 75 unique PDB entries (corresponding to 78 unique sequences) in the SCOPe set, two of which do not have an amyloid-prone sequence according to TANGO (at a threshold of 1.0%): Invasin AfaD of Escherichia coli and adhesin SdrG of Staphylococcus epidermidis, both in superfamily b.2.3. Figure 4A shows the structures of adhesin SdrG (PDB: 1R17; amino acids 425–582) and human p53 (PDB: 2AC0; a monomer with the DNA-binding α helix removed; amino acids 106–275), with the β strand containing the prototypical APR sequence PILTIITLED of p53 colored red and the APR-free corresponding strand in the adhesin colored yellow (the residues constituting the core of the APR are in bold font, the flanking GK residues are in normal font). Structural alignment (utilizing MUSTANG; Konagurthu et al., 2006) of the two superfamilies has a root-mean-square deviation (RMSD) of 1.815 Å over 45 aligned residues with just 2.22% sequence identity. Both folds are close together at an edge position in a plot of the fraction of charged amino acids over total hydrophobicity (Figure 4B), showing that their overall amino acid composition is very similar. None of the 20 proteins in the SCOPe set of the p53-like superfamily b.2.5 is APR free, according to the TANGO algorithm, suggesting that the aggregation propensity of the p53 APR ILTIITL (or equivalent) is an exquisitely conserved feature in this superfamily.
Figure 4.
Evolutionary and Protein Engineering Analysis of an APR in the p53DBD
(A) Structures of the APR-free adhesin fold (PDB: 1R17) and of the p53-family fold (PDB: 2AC0) with the APR highlighted red and the equivalent strand in the adhesin highlighted yellow.
(B) Fraction of charged amino acids and hydrophobicity for the entire fold class B in dataset 1, with fold class B2 members highlighted blue, human p53 red, and bacterial adhesin yellow.
(C) Degree of sequence conservation of the p53 DNA-binding domain (DBD) in all craniates per residue (black line) and as a rolling average per five residues (red). Functional residues are marked in blue (DNA-binding) and red (Zn ion coordination), and APR residues are marked in green.
(D) APR sequence logos for several phylogenetic groups in dataset 5.
(E) Total TANGO score of the ILTIITL sequence and selected mutants.
(F) Aggregation kinetics of the indicated peptides monitored by Thioflavin-T fluorescence.
(G) Location of selected mutations in the native structure of p53DBD.
(H) Backbone alignment of the crystal structures of p53cc (gold) and the WT (blue).
(I) Surface rendering of the crystal structure of p53cc with the introduced side chains highlighted. A blue surface marks a positive charge and a red surface a negative charge.
(J) Electron density maps (contoured to 1σ) around the zinc binding site (H179) and mutated residues in p53cc.
To analyze this in more detail, we wanted to find out whether p53 homologs without the ILTIITL amyloid can be identified altogether. To this end, we performed a HMMER (Prakash et al., 2017) search of UniProtKB with the p53DBD (DNA-binding domain). At an E-value of <0.03 for hits, this yielded 1,278 sequences. We filtered this set at 95% sequence identity, and to ensure that we only compared bona fide homologous structures, we removed sequences missing the entire amyloid-containing segment as well as those that lacked more than one equivalent of the zinc-ion-coordinating residues C176, H179, C238, and C242 of the (human) p53DBD. Of the final 337 proteins (dataset 5), we find that only six have a TANGO score of less than 1.0% in the equivalent of the ILTIITL stretch, whereas another 10 score between 1.0% and 5.0% (Table S2). Of the six, the closest divergence time from humans is about 800 million years ago (Ma), and of the 10, about 700 Ma (www.timetree.org; Kumar et al., 2017). The identity with the human p53DBD ranges between 27% and 40%. We find that the ILTIITL sequence is highly conserved throughout nearly the entire subphylum Craniata (average TANGO score, 76 ± 13 in 136 members; Table S3), which, in dataset 5, covers an evolutionary distance to humans of 615 Ma at an average sequence identity of 66%. Of all amino acids in the DBD, the amyloidogenic residues are among the most highly conserved (Figure 4C). At increasing evolutionary distance to craniates, the sequence diversity of that segment generally increases, and along with it, the TANGO score standard deviation for members of a clade (Table S3; Figure 4D).
The ILTIITL segment thus appears to be an integral part of the p53 family DBD structure. p53-like folds with weaker variants of it, or even entirely lacking this sequence stretch, can only be found at very long evolutionary distances. On the other hand, the evolutionarily unrelated instances of the p53-like fold that lack this segment altogether show that it is not required to generate a functional instance of this fold. Thus it appears that the amyloidogenicity of this segment is in an evolutionary cul-de-sac; by its strong contribution to the stability of the native state, its amyloidogenic propensity was also conserved over long evolutionary distances.
Suppressing a 600-Ma-Old APR Requires Multiple Concomitant Aggregation-Inhibiting and Compensatory Stabilizing Mutations
To validate that the coupling between the stability of the amyloid and native states is evolutionarily enforced, we investigated whether it is possible to uncouple amyloid aggregation and native-state stability using protein engineering. Given that the p53 fold is possible without the ILTIITL sequence, we wondered how many mutational steps it would take to eliminate the aggregation propensity of the ILTIITL segment from the p53 sequence itself: is this feasible by small steps, or does it require evolutionary improbable alterations? We calculated the predicted energy changes based on the DNA-bound tetrameric structure (PDB: 2AC0; Kitayner et al., 2006) using FoldX. Given its high average TANGO score (89%), strong contribution to native-state stability (i.e., low summed free energy contribution according to FoldX, ΔGsum = −12.4 kcal/mol), and very strong evolutionary conservation, the p53DBD APR represents a case of high amyloid-native state coupling that appears difficult to suppress by codon- and structure-compatible mutations. Indeed, there is not a single mutation to a GK residue that is predicted to be compatible with the thermodynamic stability of the native state (ΔΔG of < 0.5 kcal/mol). Of the two least destabilizing aggregation-reducing mutations (T256R and L252K; Figure 4E; Table S4), only the less effective one (T256R) is accessible by a single base change, whereas L252K would require changing all three bases. Moreover, each mutation is predicted by FoldX to still significantly destabilize the folded p53DBD domain (2AC0) by an extent that has been shown previously to lead to misfolding and aggregation of the protein (De Smet et al., 2017, Xu et al., 2011) (ΔΔG T256R = 1.88 kcal/mol and ΔΔG L252K = 1.24 kcal/mol per monomeric unit). Hence, neither mutation is likely to be selected during the course of evolution, and moreover, TANGO suggests that the combination of both is required to completely suppress aggregation of this APR (Figure 4E). We corroborated experimentally that the aggregation kinetics of the double mutant version of the ILTIITL peptide were indeed suppressed (Figure 4F), both with and without the natural sequence context of the peptide.
To introduce both mutations while maintaining the native state of the protein, the ΔΔG analysis above showed that, for each aggregation-reducing mutation, at least one concomitant compensatory mutation would need to be found to rescue the thermodynamic stability of the native p53DBD. A FoldX scan for compensatory mutations identified R267L and N268D on the adjacent β strand and A138G in a distal loop. So to completely suppress aggregation while maintaining protein structure and DNA binding, we had to introduce two aggregation-suppressing mutations (L252K/T256R) and three compensating stabilizing mutations (A138G, R267L, and N268D) (Figure 4G). We dubbed the final quintuple mutant (A138G/L252K/T256R/R267L/N268D) “p53 charged core” or p53cc. The need for combining two aggregation-breaking residues with three compensatory stabilizing mutations again illustrates the high degree of coupling between both properties and makes it highly unlikely for natural variation to eliminate the amyloid propensity of this segment; this would require four concomitant single-base substitutions to suppress the amyloid propensity together with three additional single-base substitutions to maintain a stable native fold.
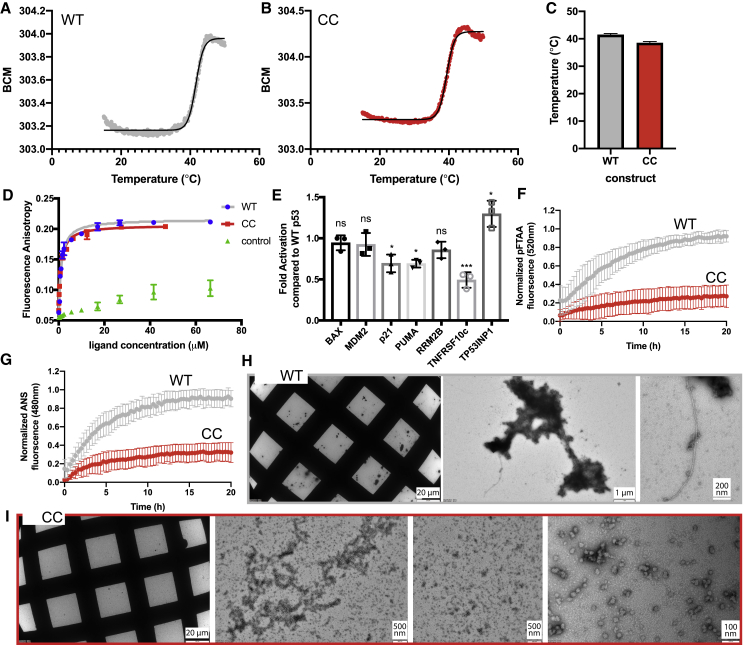
To verify the structural integrity of p53cc, we expressed the p53DBD (residues 89–311) containing the five-amino-acid-residue mutations in bacteria and purified and crystallized the protein. We collected data from a single crystal that diffracted to 1.63 Å. The structure was solved by molecular replacement using the p53 wild-type (WT) DBD structure already deposited in the PDB as a search model (accession code of the search model, PDB: 2XWR; chain A, resulting structure, PDB: 6SL6; Table S5). The backbone alignment of p53WT and p53cc has an RMSD of 0.49 Å over 199 aligned residues (Figure 4H). We find all mutations to be well resolved and placed without energetic clashes (Figures 4I and 4J). The structures of other central features of the p53DBD, such as the zinc ion and its coordinating residues, are virtually identical to the WT. Importantly, as predicted by FoldX, the charges at the end of the long side chains of arginine and lysine that we placed in the APR in the core of the DBD reach the surface of the structure (Figure 4I). When we measured the thermal stability of the WT and mutant p53, we observed a lower Tm value for the mutant (Figure 5A; Tm = 37.7°C ± 0.55°C) than the WT (Figure 5B; Tm = 42.4°C ± 0.50°C) (Figure 5C). To assess the functional integrity of the mutant p53, we first measured the DNA-binding activity of the purified DBD by two orthogonal methods using fluorescently labeled DNA oligonucleotides: microscale thermophoresis (MST; Figure S3) and fluorescence anisotropy (FA; Figure 5D). Both methods are in general agreement and show that the affinity of the mutant for an oligo containing the consensus p53 DNA-binding motif is the same as the WT (mutant KD = 1.28 μM ± 0.28 μM; WT KD = 1.23 μM ± 0.20 μM in FA). To test the ability of p53 to engage with its target promoters in a natural context, which requires more complex steps than just DNA binding and, hence, tests the mutant in a more complex biological function, we transfected full-length WT or p53cc into the p53-negative human cell line Saos-2 (Sarcoma Osteogenic-2) and measured promoter activation for seven different genes by qPCR (Figure 5E). Across the seven promoters, p53cc had an average activity of 0.95 compared with the WT protein, a difference that was not statistically significant, although individual genes showed some differences. In summary, these results demonstrate that we did not disrupt protein structure and function by reducing amyloid propensity.
Figure 5.
Biophysical Analysis of p53cc
(A) Heat denaturation of WT p53DBD, monitored by intrinsic fluorescence plotted as the barycentric mean (BCM) of the emission spectrum. The black line indicates the fit performed to determine the midpoint of the transition.
(B) Same as (A) but for the CC mutant.
(C) Average Tm values obtained from five biological replicate measurements.
(D) DNA-binding affinity of p53cc and the WT, measured by FA. Error bars indicate standard deviation of five biological replicates, each measured in three technical replicates.
(E) Degree of promoter activation of p53cc relative to WT p53 in Saos-2 cells, measured by qPCR. Error bars indicate standard deviation of three biological replicates, each measured in three technical replicates.
(F) Aggregation kinetics of p53cc and the WT, measured by p-FTAA fluorescence.
(G) Aggregation kinetics of p53cc and the WT, measured by ANS fluorescence.
(F and G) Error bars indicate standard deviation of five biological replicates, each measured in three technical replicates.
(H) Transmission electron microscopy (TEM) analysis of aggregates of WT p53.
(I) TEM analysis of p53cc.
(H and I) Representative images from three biological replicates.
We determined the aggregation kinetics of WT and mutant DBD using the amyloid-specific dye pentameric formyl thiophene acetic acid (p-FTAA; Figure 5C) and 8-anilinonaphthalene-1-sulfonic acid (ANS; Figure 5F), a dye that shows increased fluorescence upon binding to exposed HP patches. The dye binding kinetics for p-FTAA and ANS show a similar decrease in the aggregation kinetics for the mutant compared with the WT protein (Figures 5F and 5G). Moreover, according to transmission electron microscopy performed at the end of the kinetics studies, the aggregates formed by WT DBD are larger, and fibrillar structures can be observed (Figure 5H), whereas for mutant DBD, only smaller, spherical structures were seen (Figure 5I), suggesting non-amyloid aggregation or phase separation. Taken together, these data show that a small number of mutations is sufficient to eliminate an important amyloidogenic segment from the p53 structure without disrupting DNA binding and transcriptional activation. Moreover, the case shows that such mutations can be detected with reasonable accuracy using our computational approach. However, our data also illustrate why a buried APR is an evolutionary cul-de-sac; to reach this combination of mutations by natural variation would involve intermediate sequences that would be severely functionally impaired. In addition, one of the crucial mutations (L252K) requires changing all three bases of the codon at once, making an evolutionary path toward this solution extremely improbable.
Generalization: Topological Invariance of Amyloid Addiction
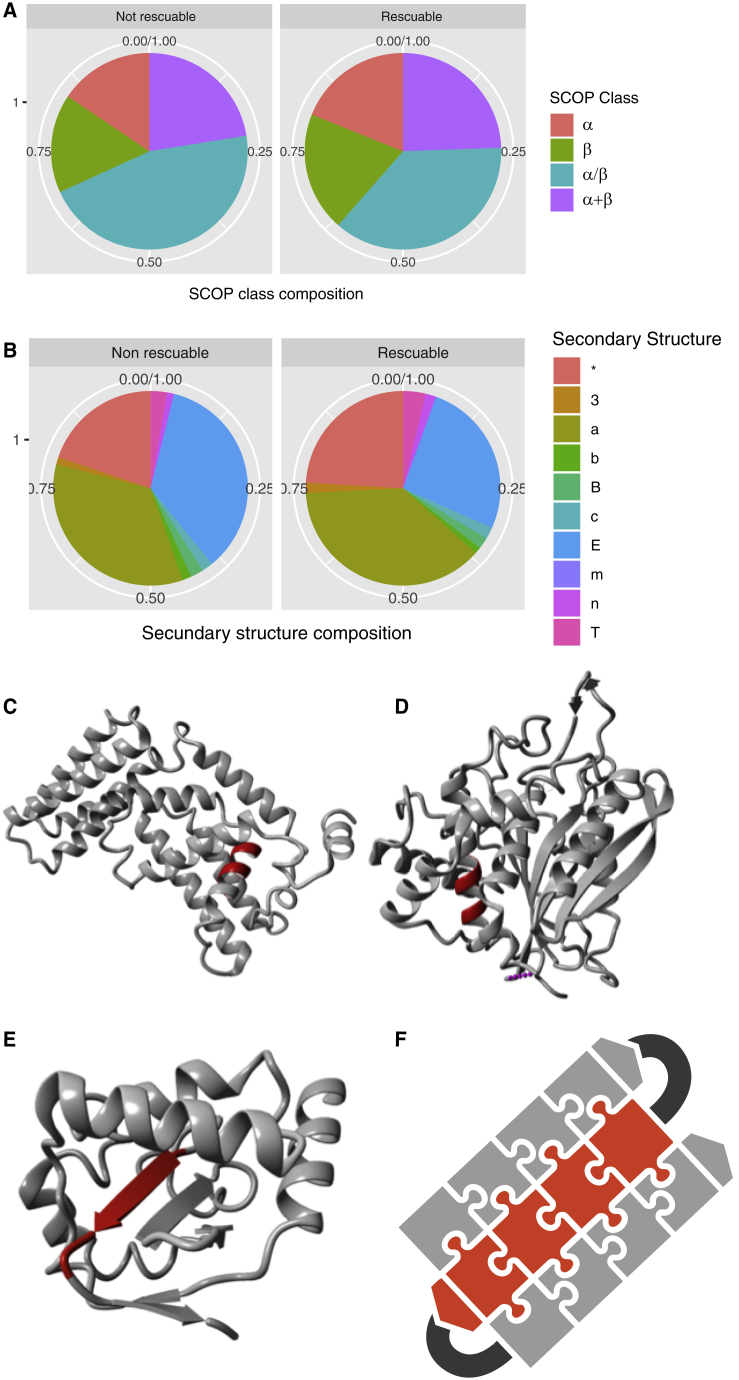
To investigate whether the results obtained with p53 are representative of many globular folds, we investigated, in the SCOPe set, the difference between APRs that can be fully suppressed (TANGO reduced to zero) by a single, codon-compatible, and structure-compatible (FoldX ΔΔG < 0.5 kcal/mol) mutation (called rescuable) versus those that cannot (called non-rescuable). First of all, we found a similar frequency of non-rescuable APRs in each of the four main SCOP classes (Figure 6A) and also found no apparent bias toward any specific secondary structure element of the segment in the native state (Figure 6B). This is in agreement with earlier observations that APRs occur in all elements of secondary structure despite the fact that the segment will adopt a β sheet conformation in the aggregated state (Rousseau et al., 2006, Linding et al., 2004). Because we already showed that intrinsic aggregation propensity, codon usage, and the native state structural context are strong determinants of how easily an APR can be suppressed (Figure 3C), we used these parameters to take a closer look at three cases from the main SCOP classes other than the one to which p53DBD belongs (b). To this end we searched the SCOP set for human domains with an APR with a high TANGO score (>80%), a high average side-chain burial (>0.8), and a minimum length of 7 amino acids that have no mutation that reduces the TANGO score to zero with a predicted ΔΔG below 1.5 kcal/mol. In the all-α-helical folds (class a), we identified the VPS9 domain of the Rab5 guanosine diphosphate (GDP)/guanosine triphosphate (GTP) exchange factor (Figure 6C). This contains an α-helical stretch with the sequence TLIYIVL that has a TANGO score of 86.5 per residue and an average side-chain burial of 0.91 in the native state and contributes – 13.1 kcal/mol to native state stability. A HMMER search for homologs with an E-value cutoff, followed by redundancy filtering at 95% (CD-hit), left 2,302 unique sequences. Of these, only 8 had a TANGO score below 1, with a closest divergence time of 600 Ma to humans. In the α/β class (c), which is generally regarded as the topologically most complex class that is enriched in obligate chaperone clients (Kerner et al., 2005), we further looked at glutaminyl-peptide cyclotransferase (Figure 6D), which contains an APR with the sequence ILQVFVL, a native helical conformation and an average TANGO score of 95.2% per residue, an average side-chain burial of 0.93 in the native state, and a contribution to its thermodynamic stability of −14.4 kcal/mol. HMMER searching with redundancy filtering yielded 1,285 homologous sequences, 117 of which have a TANGO score below 1, of which the closest evolutionary distance to human was 800 Ma. Finally, in the α+β fold class, we investigated the E3 ubiquitin protein ligase NRDP1 (Figure 6E). This contains an APR with a native β strand conformation and the sequence LVMIFA, with a TANGO score of 85.5% per residue, a side-chain burial in the native state of 0.84, and a contribution to its stability of −9.91 kcal/mol. HMMER searching followed by redundancy filtering identified 109 homologous sequences, of which only one had a TANGO score below 1, corresponding to an evolutionary distance of 800 Ma.
Figure 6.
Topological Invariance of Amyloid Addiction
(A and B) Comparison of APRs that can be fully suppressed by a single codon-compatible mutation (rescuable) with those that cannot, in terms of SCOP class (A) and secondary structure element (B) in which they occur, following the FoldX code for secondary structure annotation (∗, unclassified; 3, 3-10 helix; a, α helix; b, parallel beta conformation; B, antiparallel beta conformation; E, β strand; c, random coil; m, helix C-cap; n, helix N-cap; T, turn).
(C) Example of a conserved APR in an all-α fold (SCOP class a): the VPS9 domain of Rab5 (Homo sapiens; PDB: 1TXU).
(D) Example of a conserved APR in the α/β fold (SCOP class c): glutaminyl-peptide cyclotransferase (Homo sapiens; PDB: 2AFW).
(E) Example of a conserved APR in an α+β fold (SCOP class d): E3 ubiquitin-protein ligase NRDP1 (Homo sapiens; PDB: 2FZP).
(F) Schematic representation illustrating the APR addiction principle.
The source files (Data S1) and R-scripts (Data S2) used to generate this figure are available.
Taken together, these cases and the wider studies shown in Figures 6A and 6B clearly show that the evolutionary and thermodynamic profile of the APR studied in the p53 cases is not an exception, and a general picture emerges that regions with a high aggregation propensity and a high contribution to native state stability show high evolutionary conservation (Figure 6F).
Discussion
Here, we find that cognate globular and amyloid structures possess correlated thermodynamic responses toward mutation, which is in line with earlier results obtained by Lee et al. (2010) that translationally optimal codons associate with APRs. In addition, we find that the genetic code couples this thermodynamic correlation into an evolutionary relationship. These findings entail several structural and evolutionary implications. First, they suggest that the observed association between protein stability and protein aggregation is not a mere side effect of similar average biophysical properties (hydrophobicity, β strand propensity, charge) but is determined by fundamental and highly sequence-dependent structural similarities between both conformations. This explains why only a minority of mutations in a protein can lower amyloid-like propensity without affecting stability (Ganesan et al., 2016). Second, the restrictions on amino acid substitutions by the genetic code further tighten this relationship by favoring mutations that conserve both structural properties. Accordingly, we find that the amyloidogenicity of segments that are strong contributors to protein stability are also strongly conserved and cannot be uncoupled by single-base substitutions. Third, this means that evolutionary pressure toward increased protein stability will also increase the amyloid-like propensity of proteins, and we find here that thermostable proteins in mesophiles and the entire proteome of thermophiles are indeed more amyloidogenic. Fourth, more stable proteins will therefore be less efficient at folding because of kinetic competition with aggregation. As a result, allowing selection of stable protein variants will require the support of mechanisms for kinetic compartmentalization between folding and aggregation, including aggregation GKs (De Baets et al., 2014, Reumers et al., 2009, Rousseau et al., 2006, Monsellier and Chiti, 2007) and chaperones (Ramakrishnan et al., 2020, Scior et al., 2016). In agreement, it has been shown recently that inhibition of Hsp90 function results in selection of polio virus variants with reduced aggregation but at the expense of protein stability (Geller et al., 2018). This suggests Hsp90 indeed supports the selection of stabilizing protein mutations by managing their correlated aggregation propensity. Finally, from all of the above, we speculate that amyloid propensity could have been a driver for the evolution of globular protein structure by reciprocal selective enrichment.
Indeed, the hypothesis for an amyloid-driven origin of life has been posited recently by several groups (Dale, 2006, Maury, 2009, Greenwald and Riek, 2012). Amyloid peptides possess many of the properties that would make them stronger candidates than RNA for being the first self-replicating and catalytic molecules of life. Small amyloidogenic peptides can be spontaneously generated by organic chemical reactions under conditions prevailing on prebiotic earth (Miller, 1953, Parker et al., 2014, Bada, 2013). Amyloids can condensate under such harsh conditions (Greenwald et al., 2016), conformationally perpetuating their sequence information in a selective manner (Maury, 2015), but also allow variation and divergent evolution (Wickner et al., 2009). Additionally, it has been demonstrated recently that amyloid fibrils can catalyze peptide bond condensation in a templated manner (Rout et al., 2018). From a functional perspective, amyloid assembly can provide catalytic surfaces that further promote their proliferation (Cohen et al., 2013) as well as diverse enzymatic activities (Tena-Solsona et al., 2016, Omosun et al., 2017, Al-Garawi et al., 2017, Ivnitski et al., 2014). Their mode of assembly can also provide binding interfaces stabilizing nucleic acid interactions and vice versa (Braun et al., 2011, Macedo and Cordeiro, 2017). Amyloid assembly can mediate compartmentalization and membrane formation (Boke et al., 2016, Mahalka et al., 2011, Domanov and Kinnunen, 2008). Finally, nature still exploits the amyloid conformation for functional purposes (Fowler et al., 2007, Otzen and Riek, 2019). It has therefore been proposed that the amyloid state is a “common ancestor” fold from which the globular protein universe could have emerged (Greenwald and Riek, 2012, Maury, 2009), but a proximate structural mechanism that could drive the transition from linear amyloid peptides to globular protein-like structures has not yet been identified. Our current findings suggest that the intrinsic structural properties of amyloid and globular structure possess similar thermodynamic proclivities that could easily provide the gradient for their co-evolutionary selection. Amyloid toxic gain of function in human disease is believed to be largely exerted by meta-stable soluble amyloid-like oligomers (Breydo and Uversky, 2015, Chiti and Dobson, 2017, Benilova et al., 2012). The globular-like structure of these oligomers (Laganowsky et al., 2012) suggests that the same mechanism leading to amyloid toxicity in human disease could have served the emergence of globular structure. A large fraction of proteins supersaturated under physiological conditions (Ciryam et al., 2015, Tartaglia et al., 2007) could be a relic of this process, along with the requirement of tight protein quality control and the devastating effect of the loss thereof with aging (Hipp et al., 2019, Labbadia and Morimoto, 2015).
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| PWO DNA polymerase | Sigma | 11644947001 |
| restriction enzymes Nde1 and BamH1 | NEB | R0111 and R3136 |
| HEPES | Merck / Sigma-Aldrich | RDD002 |
| n-octyl-β-D-glucoside | Merck / Sigma-Aldrich | O8001 |
| imidazole | Merck / Sigma-Aldrich | 792527 |
| NaCl | Merck / Sigma-Aldrich | S7653 |
| IPTG | Duchefa | DUC.I1401.0025 |
| zinc chloride | Merck / Sigma-Aldrich | 229997 |
| beta-mercaptoethanol | Merck / Sigma-Aldrich | 6250 |
| DTT | Duchefa | DUC.D1309.0025 |
| Triton X-100 | Merck / Sigma-Aldrich | 1.08603 |
| sodium deoxycholate | Merck / Sigma-Aldrich | D6750 |
| glycerol | VWR | 444485B |
| ammonium bisulfate | Merck / Sigma-Aldrich | 09849 |
| Critical Commercial Assays | ||
| Q5 Site-Directed Mutagenesis Kit | NEB | E0554S |
| GenElute mammalian total RNA extraction kit | Merck / Sigma-Aldrich | RTN70 |
| RevertAid H Minus First Strand cDNA Synthesis Kit | Thermo Fisher Scientific | K1631 |
| GoTaq Probe qPCR Master Mix | Promega | A6101 |
| Deposited Data | ||
| Structure of p53cc | this paper | PDB: 6SL6 |
| Experimental Models: Cell Lines | ||
| Saos-2 | ATCC | ATB-85 |
| Oligonucleotides | ||
| p53 consensus binding site: TGGTGTTTTGCAGGCA TGTCTAGGCATGTCT |
this paper | Integrated DNA Technologies |
| p53 binding site control: TGGTGTTTTGCAGGACGT TCTAGGACGTTCT |
this paper | Integrated DNA Technologies |
| T256R mutagenesis primer forward cccatcctcacca tcatcagactggaagactcc |
this paper | Integrated DNA Technologies |
| T256R mutagenesis primer reverse ggagtcttccagt ctgatgatggtgaggatggg |
this paper | Integrated DNA Technologies |
| L252K (+T256R) mutagenesis primer forward catgaaccggaggcccatcaagaccatcatcagactggaag | this paper | Integrated DNA Technologies |
| L252K (+T256R) mutagenesis primer reverse cttccagtctgatgatggtcttgatgggcctccggttcatg | this paper | Integrated DNA Technologies |
| A138G mutagenesis primer forward gttttgccaactg ggcaagacctgccctg |
this paper | Integrated DNA Technologies |
| A138G mutagenesis primer reverse cagggcaggt cttgcccagttggcaaaac |
this paper | Integrated DNA Technologies |
| R267L mutagenesis primer forward tggtaatctact gggactgaacagctttgaggtgc |
this paper | Integrated DNA Technologies |
| R267L mutagenesis primer reverse gcacctcaaag ctgttcagtcccagtagattacca |
this paper | Integrated DNA Technologies |
| N268D (+R267L) mutagenesis primer forward acgcacctcaaagctgtccagtcccagtagattac | this paper | Integrated DNA Technologies |
| N268D (+R267L) mutagenesis primer reverse gtaatctactgggactggacagctttgaggtgcgt | this paper | Integrated DNA Technologies |
| RRM2B qPCR assay forward primer: ggtcttatgc caggactcac |
this paper | Integrated DNA Technologies |
| RRM2B qPCR assay reverse primer: caatgatctc cctgaccctttc, probe:ctgtgactttgcttgcctgatgttcc |
this paper | Integrated DNA Technologies |
| RRM2B qPCR assay hydrolysis probe: ctgtgactt tgcttgcctgatgttcc |
this paper | Integrated DNA Technologies |
| TNFRSF10B qPCR assay forward primer: accacgac cagaaacacag |
this paper | Integrated DNA Technologies |
| TNFRSF10B qPCR assay reverse primer: cattcgatgt cactccaggg |
this paper | Integrated DNA Technologies |
| TNFRSF10B qPCR assay hydrolysis probe: acaatca ccgaccttgaccatccc |
this paper | Integrated DNA Technologies |
| TP53INP1 qPCR assay forward primer: ctcattgaac atcccagcatg |
this paper | Integrated DNA Technologies |
| TP53INP1 qPCR assay reverse primer: atttcattttgagc ttccactctg |
this paper | Integrated DNA Technologies |
| TP53INP1 qPCR assay hydrolysis probe: ctgtgcataa ctcctgccctggt |
this paper | Integrated DNA Technologies |
| BAX qPCR assay forward primer: gacatgttttctgac ggc aac | this paper | Integrated DNA Technologies |
| BAX qPCR assay reverse primer: aagtccaatgtccagccc | this paper | Integrated DNA Technologies |
| BAX qPCR assay hydrolysis probe: ctggcaaagtagaaa agggcgacaac |
this paper | Integrated DNA Technologies |
| MDM2 qPCR assay forward primer: tgccaagcttctc tgtgaaag |
this paper | Integrated DNA Technologies |
| MDM2 qPCR assay reverse primer: tccttttgatcactc ccacc | this paper | Integrated DNA Technologies |
| MDM2 qPCR assay hydrolysis probe: acctgagtccga tgattcctgctg |
this paper | Integrated DNA Technologies |
| P21 qPCR assay forward primer: tgtcactgtcttgtacccttg | this paper | Integrated DNA Technologies |
| P21 qPCR assay reverse primer: ggcgtttggagtggtagaa | this paper | Integrated DNA Technologies |
| P21 qPCR assay hydrolysis probe: tctgtcatgctggt ctgccgc |
this paper | Integrated DNA Technologies |
| PUMA qPCR assay forward primer: cctaattgggctc catctcg |
this paper | Integrated DNA Technologies |
| PUMA qPCR assay reverse primer: cgacctcaacgca cagtac |
this paper | Integrated DNA Technologies |
| PUMA qPCR assay hydrolysis probe: atcatgggactcc tgcccttac |
this paper | Integrated DNA Technologies |
| Recombinant DNA | ||
| pET15b-TEV with p53 DBD WT | MerckMillipore, this paper | 69661-3 |
| pET15b-TEV with p53 DBD CC | MerckMillipore, this paper | 69661-3 |
| pCDNA3.1 with full-length p53 WT | Thermo Fisher Scientific / Invitrogen, this paper | V79020 |
| pCDNA3.1 with full-length p53 CC | Thermo Fisher Scientific / Invitrogen, this paper | V79020 |
| Software and Algorithms | ||
| cd-hit | 16731699 | http://weizhongli-lab.org/cd-hit/ |
| AliView | 25095880 | https://github.com/AliView/AliView |
| AGADIR | 7664054 | http://agadir.crg.es/ |
| FoldX | 15980494 | http://foldxsuite.crg.eu/ |
| TANGO | 15313629 | http://tango.crg.es/ |
| YASARA | YASARA Biosciences GmbH | http://www.yasara.org/ |
| XDS | Kabsch, 2010 | http://xds.mpimf-heidelberg.mpg.de/ |
| Phaser | 19461840 | https://www.phaser.cimr.cam.ac.uk/index.php/Phaser_Crystallographic_Software |
| Phenix | 31588918 | https://www.phenix-online.org/ |
| Coot | 20383002 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| MolProbity | 17452350 | http://molprobity.biochem.duke.edu/ |
| PyMOL | The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC. | https://pymol.org/2/ |
| REFMAC | 25075342 | http://www.ccp4.ac.uk/html/refmac5/keywords/xray-principal.html |
| CCP4i2 | 29533233 | http://www.ccp4.ac.uk/ccp4i2/ |
| Origin | OriginLab | https://www.originlab.com/ |
| Cytoscape | 14597658 | https://cytoscape.org/ |
| qbase+ | 17291332 | https://www.qbaseplus.com/ |
| R | The R Project for Statistical Computing | https://www.r-project.org/ |
| R Studio | R Studio | https://rstudio.com/ |
| HMMER | 29220076 | https://www.ebi.ac.uk/Tools/hmmer/search/phmmer |
| Other | ||
| Transfection reagent | Transit X2 | Mirus Bio |
| Complete protease inhibitor | Merck / Sigma-Aldrich | 04693116001 |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact: Frederic Rousseau (frederic.rousseau@kuleuven.be) or by Joost Schymkowitz (joost.schymkowitz@kuleuven.be). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental Model and Subject Details
Cell culture
Saos-2 cells (ATCC ATB-85, osteosarcoma from a 11-year old female Caucasian) were cultured in DMEM with 10% fetal bovine serum (GIBCO) with minimum non-essential amino acid and sodium pyruvate supplements (GIBCO) in a cell culture incubator at 5% CO2 and 37°C. The cells were obtained directly from ATCC, expanded and vials were kept in cryostorage. A fresh vial of cryopreserved cells was thawed out after every 20 passages. The identity of the line was monitored by the absence of p53.
Method Details
Transfection
1.5 μg DNA and 4.5 μL transfection reagent (TransIT X2, Mirus Bio) per reaction were mixed in 200 μL OptiMEM medium (GIBCO), incubated for at least 10 min and then added to 2 mL of a cell suspension at 150000 cells/ml in 6well plates. Cells were harvested 20-24 hours later.
Cloning and mutation of p53 DBD
Mutations A138G, L252K, T256R, R267L and N268D were generated by QuickChange PCR in the CDS of full-length p53 in the pCDNA3 backbone. For protein purification the mutated and wt DBD were amplified (residues 89-311) by PCR and cloned (Nde1/BamH1) into a modified version of the vector pET15b containing an N-terminal HIS-tag followed by a TEV-protease cleavage site (pET15b-TEV). For transfection, the full-length p53 versions in the pCDNA3.1 backbone were used.
Protein expression and purification
Production of p53 DBD wt or cc was induced from an overnight starter culture of freshly transformed E. coli strain NiCo21 (DE3) (NEB) at an OD of 0.8 to 1.0 with 1 mM IPTG at 28°C o/n in the presence of 20 μM ZnCl2. The next day, cells were lysed (in 50 mM HEPES, 300 mM NaCl, 3% glycerol, 5 mM beta-mercaptoethanol and protease inhibitors) using a French Press, followed by sonification and centrifugation at 40,000 g for 30 min. The p53 DBD was purified from the supernatant by IMAC (HisTrapFF 5 ml, GE Healthcare), followed by size exclusion on a HiLoad 26/60 Superdex 75 column (Amersham Biosciences) on an Äkta Pure system (GE Healthcare) in 50 mM HEPES, 300 mM NaCl, 5 mM DTT and 10 mM ammonium bisulfate. The insoluble fraction was purified from the pellet after centrifugation by several washes in increasing detergent concentration (Triton X-100 and sodium deoxycholate), and finally by dissolving in buffered 8 M urea, followed by IMAC. Protein identity was verified by mass spectrometry.
Crystallization and structure determination
Purified p53cc was dialyzed against buffer C (50 mM HEPES, 50 mM NaCl, 10 mM DTT, pH 7.5) containing 0.4% w/v n-octyl-β-D-glucoside for 24 hours at 4°C. After dialysis, the protein was concentrated to 5.1 mg/ml using centrifugal concentrators of MWCO 3000 Da (Millipore) operated at 3500 g at 4°C. The concentrated protein stock was filtered through 0.2 um PVDF filters (Millipore) and kept on ice until use. Crystallization screenings were carried out using the sitting-drop vapor diffusion method and several commercially available crystallization screens from Hampton Research (California, U.S.A.) and Molecular Dimensions (Suffolk, UK). To this end 100 nL drops of protein stock were dispensed and mixed 1:1 with crystallization buffer using a Mosquito nanoliter crystallization robot (TTP Labtech, Melbourn, UK) and incubated at either 4°C or 20°C. Rod-shape crystals appeared after one week and grew to > 50 μm in the longest axis direction over four weeks at 4°C in the following condition: 200 mM ammonium citrate tribasic pH 7.0, 20% w/v polyethylene glycol 3350 (H4 condition of Index screen, Hampton Research). Crystals were flash-cooled in liquid nitrogen after a quick passage through cryo-protection solution (75% crystallization solution plus 25% glycerol). X-ray diffraction data were collected to a resolution of 1.67 Å at the PROXIMA-I beam line of the SOLEIL synchrotron (Saint-Aubin, France). Data were indexed, integrated and scaled with XDS (Kabsch, 2010), and merged with Aimless (Evans and Murshudov, 2013). The phase problem was solved by molecular replacement using Phaser (McCoy, 2007) and the p53 wt structure 2XWR (Natan et al., 2011) as a search model. Only the A chain of 2XWR, of which metal ions and water molecules were removed, was used in molecular replacement. The molecular replacement solution was refined by iterative cycles of manual structure building using Coot (Emsley et al., 2010) and REFMAC5 (Murshudov et al., 2011) until R-factors converged to 16.4% and 18.0% (Rwork and Rfree, respectively). Structure validation was done using MOLPROBITY (Chen et al., 2010) and a final round of structure refinement was carried out in PHENIX (Adams et al., 2010). All figures were prepared using PyMOL (Schrödinger). Aimless, Phaser, REFMAC5 and Coot were used as implemented in CCP4i2 (Potterton et al., 2018). The full statistics of structure building and refinement can be seen in Table S1. The PDB accession code for the p53cc structure is 6SL6.
Dynamic light scattering
Purified protein was diluted to 0.25 mg/ml (in 50 mM HEPES, 300 mM NaCl, 5 mM DTT, 10 mM ammonium bisulfate), filtered using 0.1 mM syringe-tip PVDF filters and measured in a flat-bottom 96-well microclear plate (Greiner, Frickenhausen, Germany) on a Wyatt DynaPro plate reader equipped with a 830 nm laser source (Wyatt, Santa Barbara, CA, USA) every 5 min with auto-attenuation of laser power.
ThT and pFTAA binding
Purified protein was filtered through 0.1 mM PVDF filters, concentrated to 1 mg/ml and dye binding was measured on a FluoStar plate reader in a flat-bottom 96-well microclear plate (Greiner, Frickenhausen, Germany) every 5 min with 30 s shaking prior to each measurement. Filter settings: 440-10 nm excitation / 480-10 nm emission for ThT and 480-10 nm excitation / 520-10 nm emission for pFTAA.
Fluorescence anisotropy
Fluorescence anisotropy (FA) was recorded at room temperature in a FlexStation 3 (Molecular Devices, USA) and a PolarStar Optima plate reader (BMG labtech, Germany), with 490 nm excitation and 525 nm emission filters, using 10 nm band-pass. DNA binding affinities were determined as monitored changes in the anisotropy of 5′-Alexa488-labeled oligomers containing the consensus binding sequence (5′-TGGTGTTTTGCAGGCATGTCTAGGCATGTCT-3′), as well as the control sequence (5′-TGGTGTTTTGCAGGACGTTCTAGGACGTTCT-3′). Measurements were obtained in 50 mM HEPES (pH 7.5), 0.2 M NaCl and 5 mM dithiothreitol (DTT), containing 0.2% Tween20 and 0.4% PEG400. Protein concentration was varied up to 70 μM and labeled-DNA concentration was fixed at 25 nM. Dissociation constants were calculated by fitting the anisotropy (A) experimental data to a one-site binding model to the following quadratic equation:
where A0 and AT are the initial and total fluorescence, KD is the equilibrium constant, [R] is the protein concentration and [L] is the DNA oligomer concentration.
Microscale thermophoresis
DNA binding affinities of the WT DBD domain, as well as the mutant were also determined with microscale thermophoresis (MST). DNA oligomer concentration was kept constant at 25 nM, whereas both proteins were titrated down from 70 μM. Buffer composition was similar to FA measurements. Measurements were recorded on a Monolith NT automated instrument (NanoTemper Technologies GmbH, Germany) with a blue-laser channel at 40% LED excitation power and 60% MST power at ambient conditions. Affinity constants and experimental data fitting was performed using the NanoTemper analysis software (v2.2.4). The thermophoretic movement of bound and unbound state superpose linearly, therefore the fraction bound (f) is described as:
where Fnorm is the normalized fluorescence, Fnorm, unbound corresponds to normalized fluorescence of the unbound state and Fnorm, bound is the normalized fluorescence of the bound state.
Quantitative PCR
Cells were lysed for at least 15 min on ice in 300 μL lysis buffer (1% NP40 in PBS with nuclease (Pierce Universal Nuclease for Cell Lysis)). After centrifugation at 4°C for 15 min in a table-top centrifuge, RNA was extracted from the supernatant using the GenElute mammalian total RNA extraction kit with on-column DNase treatment (Sigma). RNA was reverse transcribed using RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Quantitative PCR was carried out with 6FAM or HEX-labeled probe/primer mixes (ZEN/IBFQ quenched, Integrated DNA Technologies) and GoTaq Probe qPCR Master Mix (Promega) in a C1000Touch/CFX96 cycler (BioRad) using the following settings: 20 s 95°C (1 cycle), 5 s 95°C, 25 s 60°C (40 cycles). Primers and probes were designed using the RealTime qPCR Assay tool (Integrated DNA Technologies). Data were analyzed in qbase+ (Biogazelle) with normalization to two reference genes out of eight tested that performed best in the geNorm analysis in qbase+.
Quantification and Statistical Analysis
SCOPe analysis and artificial protein datasets
The entire SCOPe database (release 2.06; Chandonia et al., 2017) was filtered at 40% identity using cd-hit (Fu et al., 2012), sequences with gaps were removed and finally, because the subject of this study are single chain globular proteins, all folds not belonging to SCOPe classes a (all alpha), b (all beta), c (alpha and beta interspersed - α/β) or d (alpha and beta largely separated - α+β) were removed. Average hydrophobicity per fold was determined according to the Eisenberg scale using a custom plugin to the AGADIR algorithm (Eisenberg et al., 1984).
Evolutionary analysis of p53
Residues 89-311 of the human p53 protein were used to run HMMER (Prakash et al., 2017) (EMBL-EBI) against the UniProtKB (Uniprot, 2008) at significance E-values for sequence and hit of 0.01 and 0.03, respectively. The initial 1294 hits were reduced to 337 by running cd-hit at 95% sequence identity and further manual curation using AliView, removing all sequences with large gaps at or near the ILTIITL APR or lacking more than one of the conserved Zn-coordinating residues of p53 family proteins. Phylogenetic information was extracted from the UniProtKB.
PDB entries of APRs and corresponding full-length protein structures
| Protein | APR | Full-length | PMIDs |
|---|---|---|---|
| Bloom syndrome protein | L. Serpell, personal communication | 4O3M | 23252554, 24816114 |
| Transthyretin | 4XFN | 4QXV | 26459562, 26020516 |
| Insulin | 3HYD | 5E7W | 19864624, 29086855 |
| SOD1 | 5DLI, 4NIN, 4NIP | 2C9V | 29453800, 24344300, 16406071 |
| Tp53 | 4RP6 | 2XWR | 26748848, 21457718 |
| β2-microglobulin | 3LOZ, 4E0K, 4E0L | 2YXF | 21131979, 23213214, 17646174 |
| Lysozyme | 4R0P | 2NWD | 25474758, 17360367 |
Deep mutational scanning analysis for suppressing mutations
Deep mutational scanning information was retrieved from earlier work by Gray et al. (2017). This dataset combines several studies in which protein fitness was assessed upon exhaustive mutations in virtually every position in the primary protein sequence. Gray et al. (2017) gathered these data and performed normalizations on the fitness scores therein, yielding a large database of the effects of amino acid substitutions spanning 10 different proteins. In this dataset, we identified APRs using the TANGO algorithm, after which each APR position was mutated to all other amino acids, and reanalysed through TANGO. Mutations were then classified as “suppressing” when they fully disrupted their parent APR. Next, suppressing mutations were cross-referenced with the deep mutational scanning data to find suppressing mutations for which a fitness score was actually reported. Finally, structure files for each protein were obtained from the PDB, and the list of suppressing mutations was further filtered on their occurrence in a pdb-structure. These filtering steps resulted in a list of 14 APRs from 3 different proteins (uniport IDs P02829, P28482 and P62593), for which a total of 664 suppressing mutations were identified. To analyze the structural effects of these mutations, the corresponding pdb-structures (2cg9, 2y9q and 1erm, respectively) were first repaired in FoldX using the RepairPDB command, after which stability effects of mutations were predicted using the Buildmodel command, with default settings. Statistical analyses on and visualization of these data were performed using the R statistical computing software. The source files and R-scripts used are available in the files Data S1 (Source files) and Data S2 (R-scripts).
Codon usage correlation analysis
Individual weights were attributed to codon transitions induced by single-nucleotide mutation events (AAtransition), using:
where freqperAA is the codon frequency usage per residue, Cprop corresponds to the number of possible single-mutation codon transitions per property (referring to residue side chain physicochemical properties or aggregation propensity) and n is the number of codons encoding for a single amino acid. Network construction and analysis was performed utilizing Cytoscape 3 (Smoot et al., 2011).
Statistical testing
Normality was assessed both visually (using R’s hist and qqnorm functions) and by using the Shapiro test for normality (R function shapiro.test). Significance of the difference between the mean of two groups was then either calculated using Student’s unpaired t test for normal or near-normal distributions, or the unpaired Wilcoxon-Mann-Whitney U test for non-normal distributions (R functions t.test and wilcox.test, respectively). Significance is denoted as ‘∗∗∗’, p value between 0 and 0.001; ‘∗∗’ p value between 0.001 and 0.01; ‘∗’, p value between 0.01 and 0.05; ‘n.s.’, p value > 0.05.
Data and Code Availability
The crystallographic dataset generated during this study is available at the Protein Data Bank (https://www.rcsb.org/) under ID 6SL6.
Acknowledgments
The Switch Laboratory was supported by grants from the European Research Council under European Union Horizon 2020 Framework Program ERC grant agreement 647458 (MANGO) (to J.S.); the Flanders Institute for Biotechnology (VIB, grant no. C0401); the Industrial Research Fund of KU Leuven (“Industrieel Onderzoeksfonds”); the Funds for Scientific Research Flanders (FWO; Hercules Foundation grant AKUL/15/34 - G0H1716N); the Flanders Agency for Innovation by Science and Technology (IWT; SBO grant 60839); and the Federal Office for Scientific Affairs of Belgium (Belspo; IAP grant P7/16). C.U. was supported by KU Leuven Financiering C14/17/093. N.L. was funded by Fund for Scientific Research Flanders Post-doctoral Fellowship (FWO 12P0919N to N.L.). We acknowledge SOLEIL for providing synchrotron radiation facilities under proposal number 20160142, and we would like to thank the PROXIMA-I team for assistance with using their beamline. Béla Z. Schmidt (KU Leuven) helped with editing the manuscript.
Author Contributions
Conceptualization, F.R., J.S., T.L., and R.G.; Methodology and Investigation, T.L., R.G., R.v.d.K., N.L., E.M., R.D.-R., B.H., R.C., H.W., T.G., and J.V.D.; Writing – Original Draft, T.L., J.S., and F.R.; Writing – Review & Editing, T.L., J.S., F.R., R.G., and C.U.; Funding Acquisition, J.S. and F.R.; Resources, F.R., J.S., and C.U.; Supervision, F.R. and J.S.
Declaration of Interests
F.R. and J.S. are scientific founders of Aelin Therapeutics and members of its scientific advisory board.
Published: April 14, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.03.076.
Contributor Information
Frederic Rousseau, Email: frederic.rousseau@kuleuven.be.
Joost Schymkowitz, Email: joost.schymkowitz@kuleuven.be.
Supplemental Information
Contains the source files of the R-scripts used for generating Figures 1, 2, 3, and 6.
Contains the R-scripts used for generating Figures 1, 2, 3, and 6
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Garawi Z.S., McIntosh B.A., Neill-Hall D., Hatimy A.A., Sweet S.M., Bagley M.C., Serpell L.C. The amyloid architecture provides a scaffold for enzyme-like catalysts. Nanoscale. 2017;9:10773–10783. doi: 10.1039/c7nr02675g. [DOI] [PubMed] [Google Scholar]
- Anfinsen C.B. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Anfinsen C.B., Haber E. Studies on the reduction and re-formation of protein disulfide bonds. J. Biol. Chem. 1961;236:1361–1363. [PubMed] [Google Scholar]
- Bada J.L. New insights into prebiotic chemistry from Stanley Miller’s spark discharge experiments. Chem. Soc. Rev. 2013;42:2186–2196. doi: 10.1039/c3cs35433d. [DOI] [PubMed] [Google Scholar]
- Benilova I., Karran E., De Strooper B. The toxic Aβ oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat. Neurosci. 2012;15:349–357. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- Boija A., Klein I.A., Sabari B.R., Dall’Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell. 2018;175:1842–1855.e16. doi: 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boke E., Ruer M., Wühr M., Coughlin M., Lemaitre R., Gygi S.P., Alberti S., Drechsel D., Hyman A.A., Mitchison T.J. Amyloid-like Self-Assembly of a Cellular Compartment. Cell. 2016;166:637–650. doi: 10.1016/j.cell.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S., Humphreys C., Fraser E., Brancale A., Bochtler M., Dale T.C. Amyloid-associated nucleic acid hybridisation. PLoS ONE. 2011;6:e19125. doi: 10.1371/journal.pone.0019125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breydo L., Uversky V.N. Structural, morphological, and functional diversity of amyloid oligomers. FEBS Lett. 2015;589(19 Pt A):2640–2648. doi: 10.1016/j.febslet.2015.07.013. [DOI] [PubMed] [Google Scholar]
- Buck P.M., Kumar S., Singh S.K. On the role of aggregation prone regions in protein evolution, stability, and enzymatic catalysis: insights from diverse analyses. PLoS Comput. Biol. 2013;9:e1003291. doi: 10.1371/journal.pcbi.1003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo V., Ventura S. Amyloidogenic regions and interaction surfaces overlap in globular proteins related to conformational diseases. PLoS Comput. Biol. 2009;5:e1000476. doi: 10.1371/journal.pcbi.1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandonia J.M., Fox N.K., Brenner S.E. SCOPe: Manual Curation and Artifact Removal in the Structural Classification of Proteins - extended Database. J. Mol. Biol. 2017;429:348–355. doi: 10.1016/j.jmb.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017;86:27–68. doi: 10.1146/annurev-biochem-061516-045115. [DOI] [PubMed] [Google Scholar]
- Ciryam P., Kundra R., Morimoto R.I., Dobson C.M., Vendruscolo M. Supersaturation is a major driving force for protein aggregation in neurodegenerative diseases. Trends Pharmacol. Sci. 2015;36:72–77. doi: 10.1016/j.tips.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S.I., Linse S., Luheshi L.M., Hellstrand E., White D.A., Rajah L., Otzen D.E., Vendruscolo M., Dobson C.M., Knowles T.P. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. USA. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale T. Protein and nucleic acid together: a mechanism for the emergence of biological selection. J. Theor. Biol. 2006;240:337–342. doi: 10.1016/j.jtbi.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Das A., Makarov D.E. Effect of Mutation on an Aggregation-Prone Segment of p53: From Monomer to Dimer to Multimer. J. Phys. Chem. B. 2016;120:11665–11673. doi: 10.1021/acs.jpcb.6b07457. [DOI] [PubMed] [Google Scholar]
- De Baets G., Van Durme J., Rousseau F., Schymkowitz J. A genome-wide sequence-structure analysis suggests aggregation gatekeepers constitute an evolutionary constrained functional class. J. Mol. Biol. 2014;426:2405–2412. doi: 10.1016/j.jmb.2014.04.007. [DOI] [PubMed] [Google Scholar]
- De Smet F., Saiz Rubio M., Hompes D., Naus E., De Baets G., Langenberg T., Hipp M.S., Houben B., Claes F., Charbonneau S. Nuclear inclusion bodies of mutant and wild-type p53 in cancer: a hallmark of p53 inactivation and proteostasis remodeling by p53 aggregation. J. Pathol. 2017;242:24–38. doi: 10.1002/path.4872. [DOI] [PubMed] [Google Scholar]
- Dill K.A., Ozkan S.B., Shell M.S., Weikl T.R. The protein folding problem. Annu. Rev. Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson C.M., Knowles T.P.J., Vendruscolo M. The Amyloid Phenomenon and Its Significance in Biology and Medicine. Cold Spring Harb. Perspect. Biol. 2020;12:a033878. doi: 10.1101/cshperspect.a033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanov Y.A., Kinnunen P.K. Islet amyloid polypeptide forms rigid lipid-protein amyloid fibrils on supported phospholipid bilayers. J. Mol. Biol. 2008;376:42–54. doi: 10.1016/j.jmb.2007.11.077. [DOI] [PubMed] [Google Scholar]
- Eisenberg D.S., Sawaya M.R. Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 2017;86:69–95. doi: 10.1146/annurev-biochem-061516-045104. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R.M., Terwilliger T.C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. USA. 1984;81:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla A.M., Rousseau F., Schymkowitz J., Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick A.W., Knowles T.P.J., Waudby C.A., Vendruscolo M., Dobson C.M. Inversion of the balance between hydrophobic and hydrogen bonding interactions in protein folding and aggregation. PLoS Comput. Biol. 2011;7:e1002169. doi: 10.1371/journal.pcbi.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler D.M., Koulov A.V., Balch W.E., Kelly J.W. Functional amyloid--from bacteria to humans. Trends Biochem. Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan A., Siekierska A., Beerten J., Brams M., Van Durme J., De Baets G., Van der Kant R., Gallardo R., Ramakers M., Langenberg T. Structural hot spots for the solubility of globular proteins. Nat. Commun. 2016;7:10816. doi: 10.1038/ncomms10816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller R., Pechmann S., Acevedo A., Andino R., Frydman J. Hsp90 shapes protein and RNA evolution to balance trade-offs between protein stability and aggregation. Nat. Commun. 2018;9:1781. doi: 10.1038/s41467-018-04203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg T., Hecht M., Hamp T., Karl T., Yachdav G., Ahmed N., Altermann U., Angerer P., Ansorge S., Balasz K. LocTree3 prediction of localization. Nucleic Acids Res. 2014;42:W350-5. doi: 10.1093/nar/gku396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray V.E., Hause R.J., Fowler D.M. Analysis of Large-Scale Mutagenesis Data To Assess the Impact of Single Amino Acid Substitutions. Genetics. 2017;207:53–61. doi: 10.1534/genetics.117.300064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald J., Riek R. On the possible amyloid origin of protein folds. J. Mol. Biol. 2012;421:417–426. doi: 10.1016/j.jmb.2012.04.015. [DOI] [PubMed] [Google Scholar]
- Greenwald J., Friedmann M.P., Riek R. Amyloid Aggregates Arise from Amino Acid Condensations under Prebiotic Conditions. Angew. Chem. Int. Ed. Engl. 2016;55:11609–11613. doi: 10.1002/anie.201605321. [DOI] [PubMed] [Google Scholar]
- Hatos A., Hajdu-Soltesz B., Monzon A.M., Palopoli N., Álvarez L., Aykac-Fas B., Bassot C., Benítez G.I., Bevilacqua M., Chasapi A. DisProt: intrinsic protein disorder annotation in 2020. Nucleic Acids Res. 2020;48:D269–D276. doi: 10.1093/nar/gkz975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne A., Brüggemann H., Raasch C., Wiezer A., Hartsch T., Liesegang H., Johann A., Lienard T., Gohl O., Martinez-Arias R. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 2004;22:547–553. doi: 10.1038/nbt956. [DOI] [PubMed] [Google Scholar]
- Hipp M.S., Kasturi P., Hartl F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019;20:421–435. doi: 10.1038/s41580-019-0101-y. [DOI] [PubMed] [Google Scholar]
- Horwich A.L., Neupert W., Hartl F.U. Protein-catalysed protein folding. Trends Biotechnol. 1990;8:126–131. doi: 10.1016/0167-7799(90)90153-o. [DOI] [PubMed] [Google Scholar]
- Iadanza M.G., Jackson M.P., Hewitt E.W., Ranson N.A., Radford S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018;19:755–773. doi: 10.1038/s41580-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivnitski D., Amit M., Rubinov B., Cohen-Luria R., Ashkenasy N., Ashkenasy G. Introducing charge transfer functionality into prebiotically relevant β-sheet peptide fibrils. Chem. Commun. (Camb.) 2014;50:6733–6736. doi: 10.1039/c4cc00717d. [DOI] [PubMed] [Google Scholar]
- Jahn T.R., Radford S.E. Folding versus aggregation: polypeptide conformations on competing pathways. Arch. Biochem. Biophys. 2008;469:100–117. doi: 10.1016/j.abb.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraj G.G., Hipp M.S., Hartl F.U. Functional Modules of the Proteostasis Network. Cold Spring Harb. Perspect. Biol. 2020;12:a033951. doi: 10.1101/cshperspect.a033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner M.J., Naylor D.J., Ishihama Y., Maier T., Chang H.C., Stines A.P., Georgopoulos C., Frishman D., Hayer-Hartl M., Mann M., Hartl F.U. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Kitayner M., Rozenberg H., Kessler N., Rabinovich D., Shaulov L., Haran T.E., Shakked Z. Structural basis of DNA recognition by p53 tetramers. Mol. Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Konagurthu A.S., Whisstock J.C., Stuckey P.J., Lesk A.M. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64:559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Suleski M., Hedges S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Labbadia J., Morimoto R.I. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganowsky A., Liu C., Sawaya M.R., Whitelegge J.P., Park J., Zhao M., Pensalfini A., Soriaga A.B., Landau M., Teng P.K. Atomic view of a toxic amyloid small oligomer. Science. 2012;335:1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landreh M., Sawaya M.R., Hipp M.S., Eisenberg D.S., Wüthrich K., Hartl F.U. The formation, function and regulation of amyloids: insights from structural biology. J. Intern. Med. 2016;280:164–176. doi: 10.1111/joim.12500. [DOI] [PubMed] [Google Scholar]
- Lee Y., Zhou T., Tartaglia G.G., Vendruscolo M., Wilke C.O. Translationally optimal codons associate with aggregation-prone sites in proteins. Proteomics. 2010;10:4163–4171. doi: 10.1002/pmic.201000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuenberger P., Ganscha S., Kahraman A., Cappelletti V., Boersema P.J., von Mering C., Claassen M., Picotti P. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science. 2017;355:6327. doi: 10.1126/science.aai7825. [DOI] [PubMed] [Google Scholar]
- Linding R., Schymkowitz J., Rousseau F., Diella F., Serrano L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- Macedo B., Cordeiro Y. Unraveling Prion Protein Interactions with Aptamers and Other PrP-Binding Nucleic Acids. Int. J. Mol. Sci. 2017;18:5. doi: 10.3390/ijms18051023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalka A.K., Maury C.P.J., Kinnunen P.K.J. 1-Palmitoyl-2-(9′-oxononanoyl)-sn-glycero-3-phosphocholine, an oxidized phospholipid, accelerates Finnish type familial gelsolin amyloidosis in vitro. Biochemistry. 2011;50:4877–4889. doi: 10.1021/bi200195s. [DOI] [PubMed] [Google Scholar]
- Maury C.P. Self-propagating beta-sheet polypeptide structures as prebiotic informational molecular entities: the amyloid world. Orig. Life Evol. Biosph. 2009;39:141–150. doi: 10.1007/s11084-009-9165-6. [DOI] [PubMed] [Google Scholar]
- Maury C.P. Origin of life. Primordial genetics: Information transfer in a pre-RNA world based on self-replicating beta-sheet amyloid conformers. J. Theor. Biol. 2015;382:292–297. doi: 10.1016/j.jtbi.2015.07.008. [DOI] [PubMed] [Google Scholar]
- McCoy A.J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.L. A production of amino acids under possible primitive earth conditions. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- Monsellier E., Chiti F. Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 2007;8:737–742. doi: 10.1038/sj.embor.7401034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natan E., Baloglu C., Pagel K., Freund S.M.V., Morgner N., Robinson C.V., Fersht A.R., Joerger A.C. Interaction of the p53 DNA-binding domain with its n-terminal extension modulates the stability of the p53 tetramer. J. Mol. Biol. 2011;409:358–368. doi: 10.1016/j.jmb.2011.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omosun T.O., Hsieh M.C., Childers W.S., Das D., Mehta A.K., Anthony N.R., Pan T., Grover M.A., Berland K.M., Lynn D.G. Catalytic diversity in self-propagating peptide assemblies. Nat. Chem. 2017;9:805–809. doi: 10.1038/nchem.2738. [DOI] [PubMed] [Google Scholar]
- Otzen D., Riek R. Functional Amyloids. Cold Spring Harb. Perspect. Biol. 2019;11:a033860. doi: 10.1101/cshperspect.a033860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker E.T., Zhou M., Burton A.S., Glavin D.P., Dworkin J.P., Krishnamurthy R., Fernández F.M., Bada J.L. A plausible simultaneous synthesis of amino acids and simple peptides on the primordial Earth. Angew. Chem. Int. Ed. Engl. 2014;53:8132–8136. doi: 10.1002/anie.201403683. [DOI] [PubMed] [Google Scholar]
- Potterton L., Agirre J., Ballard C., Cowtan K., Dodson E., Evans P.R., Jenkins H.T., Keegan R., Krissinel E., Stevenson K. CCP4i2: the new graphical user interface to the CCP4 program suite. Acta Crystallogr. D Struct. Biol. 2018;74:68–84. doi: 10.1107/S2059798317016035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A., Jeffryes M., Bateman A., Finn R.D. The HMMER Web Server for Protein Sequence Similarity Search. Curr. Protoc. Bioinformatics. 2017;60:3.15.1–3.15.23. doi: 10.1002/cpbi.40. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan R., Houben B., Kreft L., Botzki A., Schymkowitz J., Rousseau F. Protein Homeostasis Database: protein quality control in E. coli. Bioinformatics. 2020;36:948–949. doi: 10.1093/bioinformatics/btz628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumers J., Maurer-Stroh S., Schymkowitz J., Rousseau F. Protein sequences encode safeguards against aggregation. Hum. Mutat. 2009;30:431–437. doi: 10.1002/humu.20905. [DOI] [PubMed] [Google Scholar]
- Rousseau F., Serrano L., Schymkowitz J.W. How evolutionary pressure against protein aggregation shaped chaperone specificity. J. Mol. Biol. 2006;355:1037–1047. doi: 10.1016/j.jmb.2005.11.035. [DOI] [PubMed] [Google Scholar]
- Rout S.K., Friedmann M.P., Riek R., Greenwald J. A prebiotic template-directed peptide synthesis based on amyloids. Nat. Commun. 2018;9:234. doi: 10.1038/s41467-017-02742-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382-8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scior A., Juenemann K., Kirstein J. Cellular strategies to cope with protein aggregation. Essays Biochem. 2016;60:153–161. doi: 10.1042/EBC20160002. [DOI] [PubMed] [Google Scholar]
- Smoot M.E., Ono K., Ruscheinski J., Wang P.L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soragni A., Janzen D.M., Johnson L.M., Lindgren A.G., Thai-Quynh Nguyen A., Tiourin E., Soriaga A.B., Lu J., Jiang L., Faull K.F. A Designed Inhibitor of p53 Aggregation Rescues p53 Tumor Suppression in Ovarian Carcinomas. Cancer Cell. 2016;29:90–103. doi: 10.1016/j.ccell.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia G.G., Pechmann S., Dobson C.M., Vendruscolo M. Life on the edge: a link between gene expression levels and aggregation rates of human proteins. Trends Biochem. Sci. 2007;32:204–206. doi: 10.1016/j.tibs.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Tena-Solsona M., Nanda J., Díaz-Oltra S., Chotera A., Ashkenasy G., Escuder B. Emergent Catalytic Behavior of Self-Assembled Low Molecular Weight Peptide-Based Aggregates and Hydrogels. Chemistry. 2016;22:6687–6694. doi: 10.1002/chem.201600344. [DOI] [PubMed] [Google Scholar]
- Uniprot C., UniProt Consortium The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kant R., Karow-Zwick A.R., Van Durme J., Blech M., Gallardo R., Seeliger D., Aßfalg K., Baatsen P., Compernolle G., Gils A. Prediction and Reduction of the Aggregation of Monoclonal Antibodies. J. Mol. Biol. 2017;429:1244–1261. doi: 10.1016/j.jmb.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Durme J., De Baets G., Van Der Kant R., Ramakers M., Ganesan A., Wilkinson H., Gallardo R., Rousseau F., Schymkowitz J. Solubis: a webserver to reduce protein aggregation through mutation. Protein Eng. Des. Sel. 2016;29:285–289. doi: 10.1093/protein/gzw019. [DOI] [PubMed] [Google Scholar]
- Wang G., Fersht A.R. Multisite aggregation of p53 and implications for drug rescue. Proc. Natl. Acad. Sci. USA. 2017;114:E2634–E2643. doi: 10.1073/pnas.1700308114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R.B., Edskes H.K., Shewmaker F., Kryndushkin D., Nemecek J. Prion variants, species barriers, generation and propagation. J. Biol. 2009;8:47. doi: 10.1186/jbiol148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Reumers J., Couceiro J.R., De Smet F., Gallardo R., Rudyak S., Cornelis A., Rozenski J., Zwolinska A., Marine J.C. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat. Chem. Biol. 2011;7:285–295. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains the source files of the R-scripts used for generating Figures 1, 2, 3, and 6.
Contains the R-scripts used for generating Figures 1, 2, 3, and 6
Data Availability Statement
The crystallographic dataset generated during this study is available at the Protein Data Bank (https://www.rcsb.org/) under ID 6SL6.