Abstract
Kisspeptin, encoded by Kiss1, is essential for reproduction in mammals. Kiss1 expression is regulated by estrogen via histone acetylation in the Kiss1 promotor region. Thus, elucidation of histone modification factor(s) involved in the regulation of Kiss1 expression is required to gain further understanding of the mechanisms of its control. The RNA-seq analysis of isolated kisspeptin neurons, obtained from the arcuate nucleus (ARC) of female rats, revealed that Rbbp7, encoding retinoblastoma binding protein 7 (RBBP7), a member of histone modification and chromatin remodeling complexes, is highly expressed in the ARC kisspeptin neurons. Thus, the present study aimed to investigate whether RBBP7 is involved in Kiss1 expression. Histological analysis using in situ hybridization (ISH) revealed that Rbbp7 expression was located in several hypothalamic nuclei, including the ARC and the anteroventral periventricular nucleus (AVPV), where kisspeptin neurons are located. Double ISH for Rbbp7 and Kiss1 showed that a majority of kisspeptin neurons (more than 85%) expressed Rbbp7 mRNA in both the ARC and the AVPV of female rats. Further, Rbbp7 mRNA knockdown significantly decreased in vitro expression of Kiss1 in a mouse immortalized kisspeptin neuronal cell line (mHypoA-55). Estrogen treatment significantly decreased and increased Kiss1 mRNA levels in the ARC and AVPV of ovariectomized female rats, respectively, but failed to affect Rbbp7 mRNA levels in both the nuclei. Taken together, these findings suggest that RBBP7 is involved in the upregulation of Kiss1 expression in kisspeptin neurons of rodents in an estrogen-independent manner.
Keywords: Kisspeptin neuron, Kiss1, Retinoblastoma binding protein 7
Accumulating evidence suggests that kisspeptin-GPR54 (a kisspeptin receptor) signaling plays an essential role in regulation of reproductive functions via direct stimulation of gonadotropin-releasing hormone (GnRH) and consequent gonadotropin release in mammals, including rodents [1, 2], ruminants [3], primates [4], pigs [5], and musk shrews [6]. Mutations of Kiss1 (encoding kisspeptin) or Gpr54 cause hypogonadotropic hypogonadism in female rodents and humans [1, 2, 7,8,9]. Kisspeptin neurons are mainly located in two regions in the hypothalamus: one is located in the arcuate nucleus (ARC) in the mediobasal hypothalamus and the other is located in rostral hypothalamic regions, such as preoptic area (POA) in primates [10], ruminants [11, 12], and musk shrew [6], periventricular nucleus in pigs [5] or anteroventral periventricular nucleus (AVPV) in rodents [13, 14]. Kisspeptin neurons located in the ARC are suggested to be involved in follicular development and steroidogenesis via generation of pulsatile GnRH/gonadotropin secretion [15,16,17,18,19], while AVPV/POA kisspeptin neurons are suggested to be responsible for the ovulation via induction of GnRH/luteinizing hormone (LH) surge [14, 20,21,22,23,24].
During the few past years, emerging evidence has suggested that epigenetic mechanisms play a role in regulating Kiss1 gene expression in the both ARC and AVPV [25,26,27]. We previously suggested that histone H3 acetylation of the Kiss1 promoter region is involved in the upregulation of Kiss1 mRNA expressions in both the nuclei in mice [27]: ARC Kiss1 mRNA expression increases along with histone H3 acetylation of the Kiss1 promoter region in the absence of estrogen, while estrogen increases AVPV Kiss1 mRNA expression along with histone H3 acetylation of the Kiss1 promoter region. Further, in vitro treatment with trichostatin A, an inhibitor of histone deacetylation, upregulated Kiss1 mRNA expression in the mouse hypothalamic non-Kiss1-expressing cell line. Previous studies have also implied the epigenetic mechanisms underlying a pubertal increase in Kiss1 mRNA expression in rats [28, 29]. Thus, polycomb repressive complex 2 (PRC2), a well-known transcriptional repressor complex, and sirtuin 1 (SIRT1), a histone deacetylase, are suggested to be involved in the prepubertal suppression of ARC Kiss1 mRNA expression in rats [28, 29]. It is also reported that SIRT1 interacts with the PRC2 to decrease Kiss1 promoter activity during the prepubertal period [29]. PRC2 reportedly catalyzes histone H3 trimethylation (also known as H3K27me3), a repressive histone modification [30]. Chromatin immunoprecipitation assay revealed a pubertal decrease in binding of embryonic ectoderm development (EED), a component of PRC2 [31], to the Kiss1 promoter region [28]. The overexpression of EED causes suppression of Kiss1 expression and subsequent GnRH secretion in rats [28]. For a further understanding of epigenetic mechanism underlying the regulation of Kiss1 expression, roles of other histone modification-related proteins in Kiss1 regulation should be investigated.
Retinoblastoma binding protein 7 (RBBP7), also named as retinoblastoma-associated protein 46 (RBAP46), has been reported to function as a histone chaperone in chromatin assembly and disassembly [32, 33]. RBBP7 is also known as a component of several histone modifications and chromatin remodeling complexes. It is well known that RBBP7 coupled with histone acetyltransferase 1 (HAT1) forms type B histone acetylation complex (HAT-B) that plays a key role in histone H4 acetylation in newly-synthesized histones in the cytoplasm [34]. Several reports indicate that RBBP7 forms two histone deacetylation complexes, NuRD and SIN3, that serve as the major transcription repressors in mammals [35]. Besides, RBBP7 also functions as a component of above-mentioned PRC2 [30]. Thus, we hypothesized that RBBP7 could be involved in the regulation of Kiss1 expression in the hypothalamus.
The present study aimed to investigate the epigenetic mechanisms of Kiss1 expression, focusing on the histone modification pathway. For this purpose, we first analyzed the expression of genes, products of which are related to the histone modification pathway using RNA-seq data of the isolated visualized kisspeptin neurons, obtained from the ARC of adult female Kiss1-tdTomato heterozygous rats, as previously described [36]. The analysis of the RNA-seq data revealed that substantial levels of Rbbp7 mRNA were expressed in the ARC kisspeptin neurons. Thus, we performed histological analyses of the localization of Rbbp7 mRNA in the hypothalamus and analyzed co-expression of Rbbp7 and Kiss1 transcripts in the ARC, as well as in the AVPV of female rats. Since we found that a majority of kisspeptin neurons co-expressed Rbbp7 mRNA in both the ARC and the AVPV, we further determined the influence of RBBP7 on the regulation of Kiss1 mRNA expression by analyzing the effect of siRNA-mediated knockdown of Rbbp7 on Kiss1 expression in vitro in a mouse immortalized ARC kisspeptin neuronal model (mHypoA-55 cells).
Materials and Methods
Animals and treatments
Female Wistar-Imamichi rats, 9–12 weeks of age (224–295 g body weight), were maintained under a controlled environment (14 h light and 10 h darkness; lights on at 0500 h; 22 ± 3°C) with free access to food (CE2; CLEA Japan, Tokyo, Japan) and water. Vaginal smears were checked daily to determine the estrous cyclicity and animals with at least two consecutive estrous cycles were selected for the experiment.
Five female rats were bilaterally ovariectomized (OVX) for 2 weeks to serve as OVX rats. Eleven female rats were OVX and immediately received subcutaneous Silastic tubing (1.57 mm inner diameter; 3.18 mm outer diameter; 25 mm in length; Dow Corning, Midland, MI, USA) filled with 17β-estradiol (E2; Sigma-Aldrich, St. Louis, MO, USA), dissolved in peanut oil (Sigma-Aldrich) at 20 μg/ml for 1 week to serve as OVX + low E2 rats (diestrous model). The low E2 treatment was previously confirmed to produce a negative feedback effect on LH pulses, but not to induce LH surges [37]. Five female rats were OVX and immediately received subcutaneous Silastic tubing filled with low E2 for 5 days, followed by Silastic tubing (1.57 mm inner diameter; 3.18 mm outer diameter; 28 mm in length) filled with E2, dissolved in peanut oil at 1000 μg/ml for 2 days to serve as the OVX + high E2 rats (proestrous model). The high E2 treatment was confirmed to induce daily LH surges (unpublished data). The OVX + low E2 rats were chosen so as to analyze the co-expression of the Rbbp7 and Kiss1 mRNA in both the ARC and the AVPV, because OVX + low E2 rats were confirmed to show Kiss1 expression in both the nuclei [14]. All the surgical operations were performed under anesthesia of ketamine (27 mg/kg; Fujita, Tokyo, Japan) and xylazine (5.3 mg/kg; Bayer AG, Leverkusen, Germany) mixtures and isoflurane (1–3%, Pfizer, New York, NY, USA). All rats were injected with antibiotics (Meiji Seika Pharma, Tokyo, Japan) after surgery.
Care of the animals and experimental procedures were approved by the Committee on Animal Experiments of the Graduate School of Bioagricultural Sciences, Nagoya University, Japan.
Preparation of female rat brain sections for in situ hybridization
The OVX + low E2 rats were deeply anesthetized with sodium pentobarbital (40 mg/kg; Kyoritsu Seiyaku, Tokyo, Japan) and perfused with 0.05 M phosphate-buffered saline, followed by 4% paraformaldehyde in 0.05 M phosphate buffer (PB). The brain was removed from the skull, postfixed with the same fixative, overnight at 4°C, and kept in 30% sucrose in 0.05 M PB for 3–4 days at 4°C under ribonuclease (RNase)-free conditions. Frozen coronal sections (50 μm in thickness) of the hypothalamus (from 0.60 mm anterior to 4.56 mm posterior to the bregma) according to a rat brain atlas [38] were obtained by using a cryostat, and subjected to single or double in situ hybridization (ISH).
Analysis of histone modification-related protein gene expression in ARC kisspeptin neurons obtained from Kiss1-tdTomato rats
Expression profiles of genes encoding histone modification-related proteins in ARC kisspeptin neurons were analyzed by using the RNA-seq data of isolated visualized kisspeptin neurons taken from the ARC of female Kiss1-tdTomato heterozygous rats, as described previously [36]. Gene symbols of histone modification-related proteins were obtained from the pathway portal of Rat Genome Database (histone modification pathway, http://rgd.mcw.edu/rgdweb/pathway/pathwayRecord.html?acc_id=PW:0001338). Since Rbbp7 was highly expressed in ARC kisspeptin neurons, we further focused on the role of Rbbp7 in the regulation of Kiss1 expression.
Analysis of localization of Rbbp7 expression in the hypothalamus of female rats by single ISH for Rbbp7
Free-floating ISH for Rbbp7 was performed by using the rat hypothalamic sections collected from OVX + low E2 rats (n = 3), as previously described [5, 39]. Briefly, Rbbp7-specific digoxigenin (DIG)-labeled cRNA probe (nucleotide 310-1424; GenBank accession no. NM_031816.1) was synthesized by using a DIG-labeling kit (Roche Diagnostics, Rotkreuz, Switzerland). The hypothalamic sections were hybridized overnight at 60°C with 1 μg/ml Rbbp7-specific DIG-labeled anti-sense cRNA probes. The specificity of the cRNA probe was confirmed by performing ISH by using anti-sense and sense probes. No signal was detected in the brain sections treated with a sense probe. After hybridization, the DIG-labeled probe was detected with alkaline phosphatase-conjugated anti-DIG antibody (1:500; Roche Diagnostics), and a chromogen solution (0.338 mg/ml 4-nitroblue tetrazolium chloride (NBT; Roche Diagnostics) and 0.175 mg/ml 5-bromo-4-chloro-3-indolyl-phosphate, 4-toluidine salt (BCIP; Roche Diagnostics). The sections were mounted on gelatin-coated slides and examined under a light microscope (BX53; Olympus, Tokyo, Japan).
Analysis of co-localization of Rbbp7 and Kiss1 expression in the ARC and the AVPV of female rats by double ISH for Rbbp7 and Kiss1
Free-floating double ISH for Rbbp7 and Kiss1 was performed by using the hypothalamic sections including the AVPV (from the 0.48 mm anterior to 0.96 mm posterior to the bregma, every second section) and the ARC (from the 1.56 to 4.44 mm posterior to the bregma, every fourth section) gained from OVX + low E2 rats (n = 3), as previously described [36, 39]. Briefly, the Rbbp7-specific DIG-labeled cRNA probe and the Kiss1-specific fluorescein isothiocyanate (FITC)-labeled cRNA probe (position 33–348; GenBank accession no. NM_181692.1) were synthesized. The sections were hybridized overnight at 60°C with 1 μg/ml DIG-labeled Rbbp7 anti-sense cRNA probes and 1 μg/ml FITC-labeled Kiss1 anti-sense cRNA probes. After the hybridization, the FITC-labeled Kiss1 probe on the sections was visualized by peroxidase (POD)-conjugated anti-FITC antibody (Roche Diagnostics) and TSA Plus Fluorescein System (1:100; PerkinElmer, Shelton, CT, USA). After inactivation of peroxidase in the sections with 0.1 N hydrochloric acid for 30 min, the DIG-labeled Rbbp7 probe on the sections was visualized by POD-conjugated anti-DIG antibody (Roche Diagnostics), TSA Plus Biotin Kit (1:100; PerkinElmer) and Dylight 594-conjugated streptavidin (Thermo Fisher Scientific, Waltham, MA, USA). The sections were mounted to slides and the fluorescent images were examined under a fluorescence microscope with the ApoTome optical sectioning (Carl Zeiss, Oberkochen, Germany). The Rbbp7- and Kiss1-positive cells were counted bilaterally under the microscope, three times, by two persons and the average was calculated. No positive signal for Rbbp7 or Kiss1 mRNA was detected in the brain sections hybridized with the corresponding sense probes.
In vitro analysis of the role of Rbbp7 in regulating Kiss1 expression by Rbbp7 knockdown in mHypoA-55 cell line, a model for ARC kisspeptin neurons of mice
Immortalized mouse neuronal cells, mHypoA-55 cells (CVCL_D416; CELLutions Biosystems, Burlington, ON, Canada) were used as a model for ARC kisspeptin neurons [40] for in vitro analysis to investigate the role of Rbbp7 in the regulation of Kiss1 expression. The cells were cultured in phenol red-free Dulbecco's Modified Eagle Medium (Thermo Fisher Scientific) supplemented with 200 mM L-glutamine, 1% penicillin/streptomycin and 5% charcoal-stripped FBS (Biowest, Nuaillé, France) with or without E2 for 2 or 4 h. The E2 concentrations (10 or 100 nM) and incubation period (2 or 4 h) were chosen according to a previous study [40], in which Treen et al. showed that the 4 h treatment of 100 nM E2 decreased Kiss1 expression in mHypoA-55 cells, as well as to our preliminary investigations that revealed decreasing tendency of Kiss1 expression upon E2 treatment using the same cell lines.
The effect of Rbbp7 knockdown on in vitroKiss1 expression in the mHypoA-55 cells was determined using Silencer Select Pre-Designed siRNA (10 nM), corresponding to mouse Rbbp7 (siRNA ID: s110672 (#1); s110673 (#2), Thermo Fisher Scientific). The 2 sets of Rbbp7-siRNAs or negative control siRNA (NC siRNA) were transfected into the cells by Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific). Following this, 48 h after the transfection, the cells were treated with 100 nM E2 for 2 h, and the cells were collected for analyses for Kiss1 and Rbbp7 mRNA or RBBP7 protein levels. The E2 treatment was chosen due to the observed tendency of a decrease in Kiss1 expression following 100 nM E2 treatment for 2 h.
Quantitative RT-PCR analyses for Rbbp7 and Kiss1 mRNA expression levels in the ARC and the AVPV regions of the rats and mHypoA-55 cells
To analyze Rbbp7 and Kiss1 mRNA levels in the ARC or AVPV regions of OVX, OVX + low E2, and OVX + high E2 rats (n = 5 in each group), animals were decapitated between 1300 h and 1500 h. The ARC-median eminence (ME) region was dissected from the brain with a microblade and the AVPV region was punched out with 18-gauge stainless-steel tubing as previously described [41]. Rbbp7 and Kiss1 mRNA levels were also analyzed in the mHypoA-55 cells described above.
DNA-free total RNA was purified from the ARC and AVPV tissues, as well as mHypoA-55 cells by using ISOGEN (Nippon Gene, Tokyo, Japan). For ARC or AVPV tissues, DNase I (Thermo Fisher Scientific) and a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) were used to synthesize full-length cDNAs. For mHypoA-55 cells, ReverTra Ace (TOYOBO, Osaka, Japan) was used to synthesize full-length cDNAs. The obtained cDNA was used as a template in PCR reactions using primers for Rbbp7, Kiss1 and Actb. Forward and reverse primers for rat Kiss1 were 5’-AGCTGCTGCTTCTCCTCTGT-3’ and 5’-AGGCTTGCTCTCTGCATACC-3’, respectively (GenBank accession no. NM_181692.1); for mouse Kiss1, 5’-TGCTGCTTCTCCTCTGT-3’ and 5’-ACCGCGATTCCTTTTCC-3’ (GenBank accession no. NM_178260.3); for rat and mouse Rbbp7, 5’-TCCCAATGATGATGCACAGT-3’ and 5’-CTGGTTTTGCAGGGTGTTTT-3’ (GenBank accession no. NM_031816.1 and NM_009031.3); for rat Actb, 5’-TGTCACCAACTGGGACGATA-3’ and 5’-GGGGTGTTGAAGGTCTCAAA-3’ (GenBank accession no. NM_031144.3); for mouse Actb, 5’-GGTGGGAATGGGTCAGAAGG-3’ and 5’-GTACATGGCTGGGGTGTTGA-3’ (GenBank accession no. NM_007393.5). Real-time PCR analysis was performed by using an ABI 7500 real-time system (Thermo Fisher Scientific) with THUNDERBIRD qPCR Mix (TOYOBO). The cycling protocol was as follows: pre-denature for 1 min at 95°C, 40 cycles amplification for 15 sec at 95°C and 1 min at 60°C. The specificity of the amplification products was confirmed by dissociation curve analysis (60 to 95°C) after 40-cycle amplification. A distinct single peak was considered as proof that only a single DNA sequence was amplified. The expression levels of Rbbp7 and Kiss1 genes were obtained by normalization to the expression levels of Actb.
Verification of Rbbp7 knockdown experiment in mHypoA-55 cells by western blotting for RBBP7
The expression levels of RBBP7 and β-actin proteins in the mHypoA-55 cells after 48 h of siRNA transfection were confirmed by western blotting. The cells cultured with Rbbp7-siRNAs or NC siRNA were treated with 10% (v/v) trichloroacetic acid (Nakarai tesque, Kyoto, Japan) at 4°C for 30 min, and scraped off. These cells were then suspended in UTD buffer [9 M Urea (Sigma-Aldrich), 2% (v/v) Triton X-100 (Sigma-Aldrich), 1% (w/v) DL-Dithiothreitol (DTT; Sigma-Aldrich)] and 2% (w/v) lithium dodecyl sulfate (Sigma-Aldrich). The whole cell lysate was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 4–20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad Laboratories, Hercules, CA, USA), and transferred onto polyvinylidene difluoride membranes (GE Healthcare, Chicago, IL, USA). The membranes were incubated with rabbit anti-human RBBP7 polyclonal antibody (1:4,000 Abcam, Cambridge, UK) at room temperature for 2 h. The specificity of the antibody was confirmed by He et al. [42]. The membranes were then incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (1:4,000 Santa Cruz Biotechnology, Dallas, TX, USA) at room temperature for 1 h. Chemiluminescence was visualized using an ECL Prime Western Blotting Detection Reagent (GE Healthcare), and recorded with a Light-Capture II (AE-6982; Atto, Tokyo, Japan). The membranes were stripped with Restore PLUS Western Blot Stripping Buffer (Thermo Fisher Scientific) for re-probing with rabbit anti-human β-actin polyclonal antibody (1:10,000 Abcam). The expression of β-actin protein was detected by using the same method as stated above and its levels were used as an internal control. Band intensities were estimated by densitometry using Image J software v. 1.52k (NIH, Bethesda, MD, USA).
Statistical analysis
Statistical differences in Rbbp7 and Kiss1 mRNA and RBBP7 protein levels in the mHypoA-55 cells, transfected with siRNAs and treated with E2 were determined by two-way ANOVA (siRNA transfection and E2 treatment as main effects) with Bonferroni post-hoc test. Statistical differences in Rbbp7 and Kiss1 mRNA levels in the ARC and the AVPV of rats or in the mHypoA-55 cells treated with three different doses of E2 were determined by one-way ANOVA with Bonferroni post-hoc test. P < 0.05 was considered to indicate a significant difference. All analyses were performed by using R Statistical Computing, release 3.5.1 (http://www.R-project.org).
Results
Examination of histone modification factor gene expression in the ARC kisspeptin neurons by analyzing the RNA-seq data obtained from isolated rat ARC kisspeptin neurons
The RNA-seq analysis showed that some major histone modification factor genes (Rat Genome Database, http://rgd.mcw.edu/rgdweb/pathway/pathwayRecord.html?acc_id=PW:0001338), such as Rbbp7 (210 RPKM), Hdac1 (encoding histone deacetylase 1, 63 RPKM), Hdac2 (encoding histone deacetylase 2, 35 RPKM), Ep300 (encoding E1A binding protein P300, 1.42 RPKM), and Sirt1 (Sirtuin 1, 0 RPKM) were detected in ARC Kiss1-tdTomato cells. Among these genes, Rbbp7 was expressed at high levels in the cells. Table 1 shows genes whose transcripts form histone modification complexes together with RBBP7, such as NuRD, SIN3, PRC2, and HAT-B complexes. ARC Kiss1-tdTomato cells expressed the component genes of NuRD and HAT-B complexes reported previously [35, 43]. On the other hand, few component genes of SIN3 and PRC2 complexes, such as Sap30 and Eed [35, 44], were detected in the ARC Kiss1-tdTomato cells.
Table 1. Genes whose transcripts form histone modification complexes together with retinoblastoma binding protein 7 (RBBP7).
| Gene symbols | Relative expression levels * | Histone modification complexes |
|---|---|---|
| Rbbp7 | 209.55 | NuRD, SIN3, PRC2, HAT-B |
| Rbbp4 | 127.84 | NuRD, SIN3, PRC2, HAT-B |
| Hdac1 | 62.70 | NuRD, SIN3 |
| Hdac2 | 34.67 | NuRD, SIN3 |
| Gatad2b (p66β) | 23.26 | NuRD |
| Chd4 | 17.15 | NuRD |
| Mbd3 | 12.14 | NuRD |
| Gatad2a (p66α) | 9.94 | NuRD |
| Chd3 | 6.73 | NuRD |
| Mta1 | 0.56 | NuRD |
| Mta2 | 0.06 | NuRD |
| Sap18 | 150.35 | SIN3 |
| Sin3b | 69.22 | SIN3 |
| Sin3a | 4.18 | SIN3 |
| Sap30 | 0.00 | SIN3 |
| Suz12 | 3.10 | PRC2 |
| Ezh1 | 0.00 | PRC2 |
| Ezh2 | 0.00 | PRC2 |
| Eed | 0.00 | PRC2 |
| Hat1 | 8.83 | HAT-B |
* Data are expressed as reads per kilobase per million mapped reads (RPKM). Symbols and names of gene or histone modification complexes are as follows: Chd3, Chromodomain helicase DNA binding protein 3; Chd4, Chromodomain helicase DNA binding protein 4; Eed, Embryonic ectoderm development; Ezh1, Enhancer of zeste 1 polycomb repressive complex 2 subunit; Ezh2, Enhancer of zeste 2 polycomb repressive complex 2 subunit; HAT-B, Type B histone acetyltransferases; Hat1, Histone acetyltransferase 1; Hdac1, Histone deacetylase 1; Hdac2, Histone deacetylase 2; Mbd3, Methyl-CpG binding domain protein 3; Mta1, Metastasis-associated proteins 1; Mta2, Metastasis-associated proteins 2; NuRD, Nucleosome remodeling and deacetylation; Gatad2a (p66α), GATA zinc finger domain containing 2A (Transcriptional repressor p66-alpha); Gatad2b (p66β), GATA zinc finger domain containing 2B (Transcriptional repressor p66-beta); PRC2, Polycomb repressive complex 2; Rbbp4, Retinoblastoma binding protein 4; Rbbp7, Retinoblastoma binding protein 7; Sap18, Sin3A associated protein 18; Sap30, Sin3A associated protein 30; SIN3, Sin3 histone deacetylase complex; Sin3a, SIN3 transcription regulator family member A; Sin3b, SIN3 transcription regulator family member B; Suz12, SUZ12 polycomb repressive complex 2 subunit.
Distribution of the Rbbp7 mRNA-expressing cells in the hypothalamus of female rats
Figure 1 shows the Rbbp7 mRNA expression in the hypothalamus of a representative OVX + low E2 rat. A number of Rbbp7-positive cells were found in the AVPV (Fig. 1A), suprachiasmatic nucleus (SCN) (Fig. 1B), supraoptic nucleus (SON) (Fig. 1C), paraventricular nucleus (PVN) (Fig. 1D), anterior part of the ventromedial hypothalamic nucleus (VMH) and the ARC (Fig. 1E), and the posterior part of the ARC (Fig. 1F).
Fig. 1.
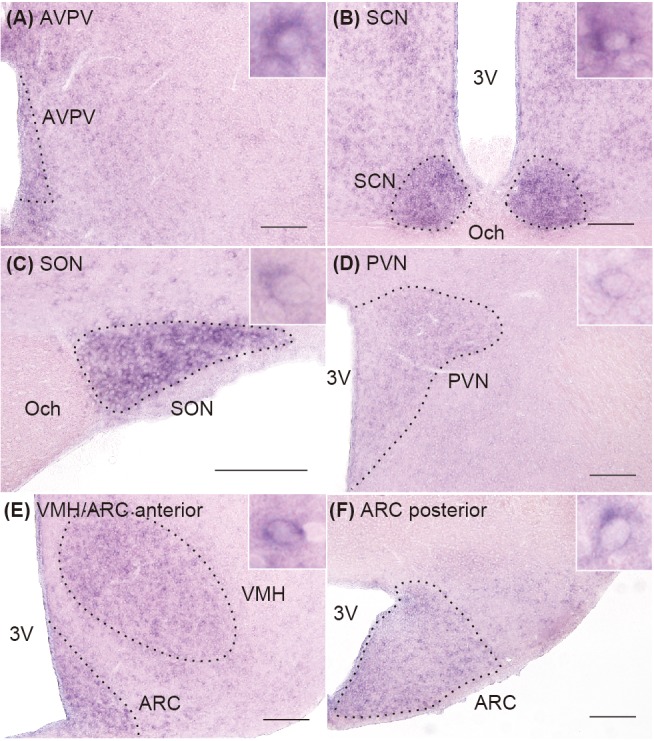
Distribution of Rbbp7-positive cells in the hypothalamus of ovariectomized + low 17β-estradiol (OVX + low E2) rats. Representative photomicrographs showing cells expressing Rbbp7 in the anteroventral periventricular nucleus (AVPV) (A), suprachiasmatic nucleus (SCN) (B), supraoptic nucleus (SON) (C), paraventricular nucleus (PVN) (D), anterior part of the ventromedial hypothalamic nucleus (VMH) and arcuate nucleus (ARC) (E), and posterior part of the ARC (F). The insets indicate Rbbp7-positive cells at a higher magnification. Scale bars: 200 μm. 3V, third cerebroventricle; Och, optic chiasm.
Co-expression of Rbbp7 in Kiss1-positive cells in the ARC and AVPV of female rats
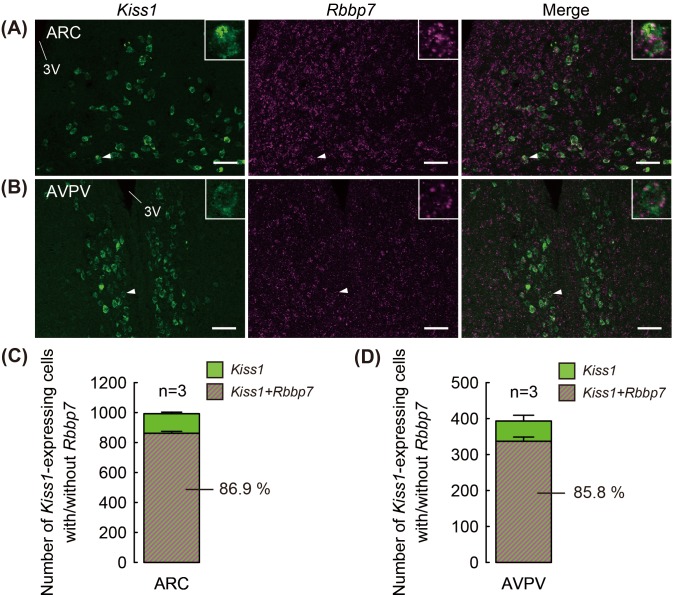
Figures 2A and 2B show Rbbp7- and Kiss1-positive cells, visualized by a double ISH in the ARC and AVPV of representative OVX + low E2 rats. Cell-shaped Kiss1-positive signals were found in the both ARC and AVPV and dot-like Rbbp7-positive signals were widely found in the ARC and the AVPV and outside of the nuclei. Merged images exhibit that most of the Kiss1-positive cells showed co-expression of Rbbp7-positive signals in both, the ARC (Fig. 2A) and the AVPV (Fig. 2B). Quantitative analysis revealed that the majority of Kiss1-positive cells co-expressed Rbbp7 in the ARC (86.9%, Fig. 2C) and the AVPV (85.8%, Fig. 2D) of the OVX + low E2 rats.
Fig. 2.
Co-expression of Kiss1 and Rbbp7 in the arcuate nucleus (ARC) and anteroventral periventricular nucleus (AVPV) of ovariectomized + low 17β-estradiol (OVX + low E2) rats. Representative photomicrographs showing Kiss1-positive cells (green) and Rbbp7-positive signals (magenta) in the ARC (A) and AVPV (B). The insets indicate expression of Rbbp7-positive signals in the Kiss1-positive cells at a higher magnification (arrowheads). The numbers of Kiss1- and Rbbp7-positive cells out of the Kiss1-positive cells were quantified in the ARC (C) and AVPV (D). Data are the mean ± SEM (n = 3). Scale bars: 100 μm. 3V, third cerebroventricle.
Suppression of Kiss1 expression by Rbbp7 silencing in the mHypoA-55 cell line
Both Rbbp7-targeting siRNAs #1 and #2 significantly reduced Rbbp7 mRNA (Fig. 3A) and protein (Fig. 3B) expression, regardless of E2 treatment in the mHypoA-55 cells (P < 0.05, two-way ANOVA with Bonferroni post-hoc test). The knockdown of Rbbp7 mRNA by the siRNA #1 or #2 significantly decreased the Kiss1 mRNA levels in the cells, regardless of E2 treatment (Fig. 3C; P < 0.05, two-way ANOVA with Bonferroni post-hoc test).
Fig. 3.
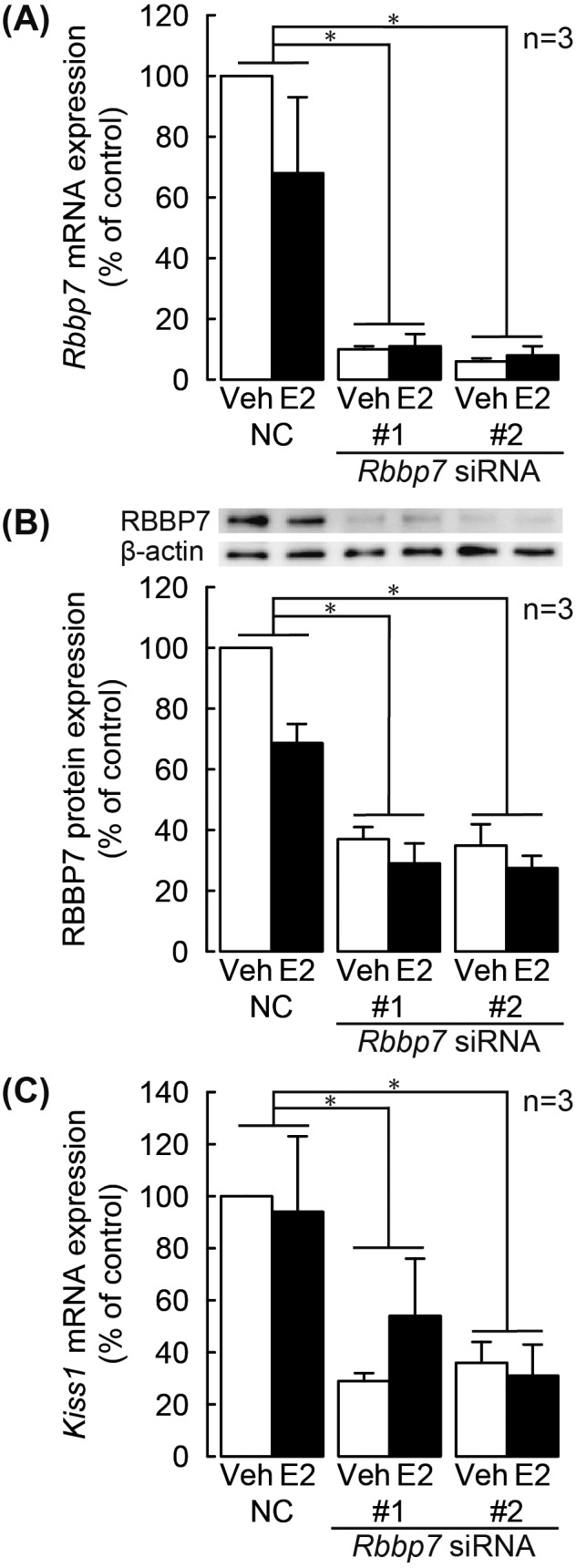
Knockdown of retinoblastoma binding protein 7 (RBBP7) expression repressed Kiss1 mRNA expression in the mHypo-A55 cells. Rbbp7 mRNA expression in mHypoA-55 cells treated with E2 following Rbbp7 siRNA transfection (A). NC, negative control siRNA; #1 and #2, two sets of siRNA against Rbbp7. The mRNA levels were measured by quantitative RT-PCR. Representative western blot bands and results of densitometric analysis of RBBP7 protein expression (B). The band intensities were estimated by densitometry using Image J. Kiss1 mRNA expression in mHypoA-55 cells treated with E2 following Rbbp7 siRNA transfection (C). Expression levels were normalized to Actb mRNA or β-actin protein. Data are the mean ± SEM (n = 3) and are expressed as % of controls treated with vehicle (Veh) and NC siRNA. * P < 0.05, two-way ANOVA with Bonferroni post-hoc test.
Quantitative analysis of Rbbp7 and Kiss1 expression in the mHypoA-55 cells treated with/without E2 and in the ARC and the AVPV of OVX rats treated with/without diestrous (low) and proestrous (high) levels of E2
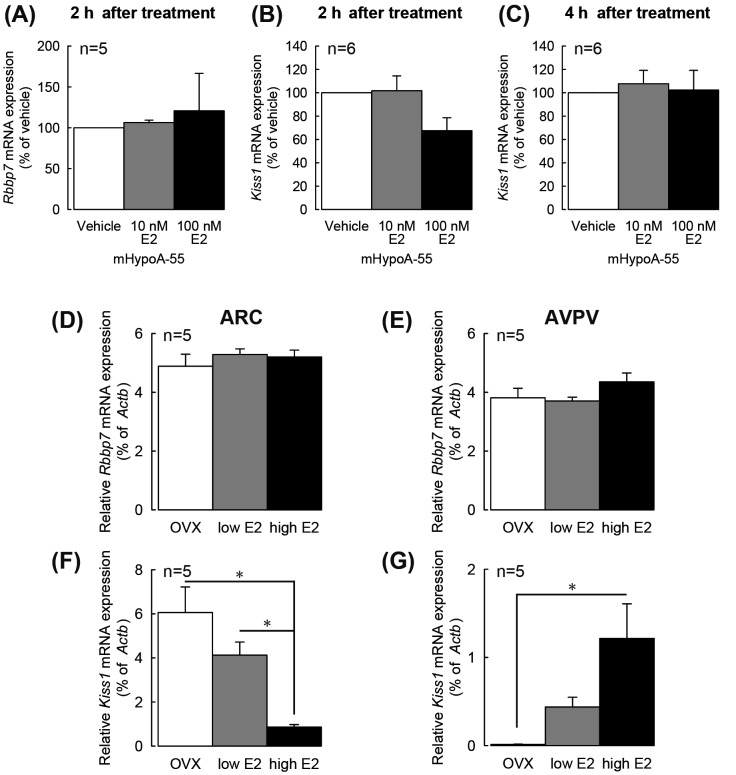
Figures 4 A–C show Rbbp7 and Kiss1 expression levels determined by quantitative RT-PCR in vehicle- or E2-treated mHypoA-55 cells for 2 h or 4 h. Expression of Rbbp7 was detected in the mHypoA-55 cells treated with vehicle, 10 nM or 100 nM E2 for 2 h, and no significant difference in Rbbp7 expression between the groups was detected (Fig. 4A). On the other hand, 100 nM E2 treatment for 2 h tended to decrease Kiss1 expression in the mHypoA-55 cells, compared with vehicle-treated cells (Fig. 4B). There was no significant difference in Kiss1 mRNA levels at 4 h after treatment with vehicle, 10 nM or 100 nM E2 in the mHypoA-55 cells (Fig. 4C).
Fig. 4.
Rbbp7 and Kiss1 expression in mHypoA-55 cells and in the arcuate nucleus (ARC) and anteroventral periventricular nucleus (AVPV) regions of ovariectomized (OVX) rats treated with E2. Rbbp7 expression in mHypoA-55 cells treated with E2 for 2 h (A). Kiss1 expression levels in mHypoA-55 cells treated with E2 for 2 h (B) and 4 h (C), respectively. Expression levels were normalized to Actb mRNA level. Data are the mean ± SEM (n = 5–6) and are expressed as % of vehicle-treated controls. Relative Rbbp7 expression levels in the ARC (D) and AVPV (E) of OVX rats treated with diestrous levels (low) or proestrous levels (high) of E2. Relative Kiss1 expression levels in the ARC (F) and AVPV (G) of OVX rats treated with low or high E2. The mRNA levels were measured by quantitative RT-PCR. Data are the mean ± SEM (n = 5–6) and are expressed as the relative ratio of Rbbp7 or Kiss1 mRNA to Actb mRNA. * P < 0.05, one-way ANOVA with Bonferroni post-hoc test.
Figures 4 D–G show Rbbp7 and Kiss1 expression levels as determined by quantitative RT-PCR in the ARC and the AVPV regions obtained from OVX rats and low- or high-E2-treated OVX rats. Expression of Rbbp7 was detected in both the ARC and the AVPV of the animals in all groups, and there was no significant difference in Rbbp7 expression between the groups (Figs. 4D and 4E). On the other hand, Kiss1 levels in the ARC of OVX rats were decreased by E2 treatment, in a dose-dependent manner, resulting in significant differences in Kiss1 levels between OVX group and OVX + low E2 or OVX + high E2 groups (Fig. 4F; P < 0.05, one-way ANOVA with Bonferroni post-hoc test). E2 treatment increased Kiss1 expression in the AVPV in a dose-dependent manner, resulting in significant difference in Kiss1 level between OVX and OVX + high E2 groups (Fig. 4G; P < 0.05, one-way ANOVA with Bonferroni post-hoc test).
Discussion
The present study demonstrates that RBBP7 would be largely involved in the positive regulation of Kiss1 mRNA expression in the hypothalamic kisspeptin neurons in rodents, because majority (more than 85%) of kisspeptin neurons in both the ARC and the AVPV regions expressed Rbbp7 in female rats, and siRNA-mediated Rbbp7 knockdown significantly decreased Kiss1 expression in the mouse immortalized kisspeptin neuronal cell line. It should be noted that the Rbbp7 knockdown reported in this study, resulted in a profound decrease in the levels of RBBP7 protein. We had previously showed that histone acetylation of the Kiss1 promoter region is positively associated with Kiss1 expression in the ARC and the AVPV of female mice [27]. It was reported that Rbbp7 silencing decreased histone H4K5, H4K8, H4K12 acetylation at the whole-genome level in the mouse stromal cells [42]. Thus, the Rbbp7 mRNA silencing may attenuate histone acetylation of the Kiss1 promoter and, subsequently, decrease Kiss1 expression in kisspeptin neurons. Taken together, the present results suggest that RBBP7 could be involved in upregulation of Kiss1 mRNA by affecting histone acetylation of Kiss1 promoter region.
The present study showed that in vivo estrogen treatment at diestrous and proestrous levels failed to affect Rbbp7 expression in the rat ARC and AVPV regions, although the estrogen treatments downregulated and upregulated Kiss1 expressions in the ARC and the AVPV, respectively, in a dose-dependent manner. Further, in vitro estrogen treatment also failed to affect Rbbp7 mRNA levels in the mHypoA-55 cells, a mouse ARC kisspeptin neuronal model. The present study showed that the Rbbp7 knockdown largely suppressed Kiss1 expression, compared to that in the NC siRNA-transfected cells, regardless of estrogen treatment. Taken together, these results suggest that RBBP7 mediates positive regulation of Kiss1 expression in an estrogen-independent manner. Thus, RBBP7 may have a permissive role for the control of Kiss1 levels, but not be involved in estrogen negative/positive feedback for Kiss1 expression. In other words, RBBP7 could be constitutively involved in the upregulation of Kiss1 expression, but not mediate the estrogen positive and negative feedback for Kiss1 expression. Interestingly, previous studies reported that estrogen affects Rbbp7 expression in a bidirectional manner: estrogen increased Rbbp7 mRNA and protein expression in MCF7 cells [45], whereas it inhibited Rbbp7 expression in progression of transformed breast epithelial cells, MCF10AT3B cells [46]. Since our study implies that estrogen is unlikely to be involved in RBBP7 expression in the rodent hypothalamus, further studies are required to determine factor(s), mediating the estrogen positive/negative feedback for Kiss1 expression, along with the factor(s), regulating Rbbp7 and the protein expressions in hypothalamic kisspeptin neurons.
The current histological analysis with ISH for Rbbp7 mRNA in the rat hypothalamus revealed that Rbbp7 expression was located in several nuclei, such as the SCN, SON, PVN, and VMH in addition to the ARC and the AVPV. Considering the upregulating role of RBBP7 in Kiss1 expression in our study, the widely-distributed RBBP7 in the hypothalamic nuclei implies that RBBP7 could also be involved in the gene expression of some neurotransmitters, such as arginine vasopressin (AVP) and vasoactive intestinal peptide in the SCN, oxytocin and AVP in the SON and PVN, corticotropin-releasing hormone in the PVN, and pituitary adenylate cyclase-activating polypeptide in the VMH [47, 48]. Further studies are required to clarify the role of RBBP7 in the gene expressions of these neurotransmitters in the nuclei.
The present study suggests that RBBP7 may act as a component of NuRD or HAT-B complexes in kisspeptin neurons, because the current RNA-seq data showed that major components of NuRD and HAT-B were detected in rat ARC kisspeptin neurons [35, 43]. It is unlikely that RBBP7 could act as a component of SIN3 and PRC2 complexes in kisspeptin neurons, because the RNA-seq data failed to detect major components of SIN3 and PRC2, such as Sap30 and Eed, respectively [35, 44]. Previous reports [35, 43] suggest that NuRD serves as a major histone deacetylase complex in the nucleus, and HAT-B plays a role in the acetylation of histones H3 and H4 in newly-synthesized histones in cytoplasm. Thus, RBBP7, which may form the NuRD complex in the kisspeptin neurons, may repress the expression of some inhibitory gene(s) of Kiss1 transcription, indirectly leading to an elevation of Kiss1 expression. It would be interesting to examine whether RBBP7 and/or NuRD are involved in the pubertal increase in ARC Kiss1 expression during peripubertal period. Further studies are needed to elucidate the mechanisms of RBBP7-mediated regulation of Kiss1 expression in kisspeptin neurons.
In conclusion, the present findings suggest that RBBP7 is involved in the upregulation of Kiss1 mRNA expression in kisspeptin neurons of rodents by increasing the histone acetylation level of Kiss1 promoter region or repressing the expression of some inhibitory gene(s) for Kiss1 transcription.
Acknowledgments
We recognize and respect the great contributions of the late Kei-ichiro Maeda with his expert advice and supervision of the study. This work was supported in part by grants-in-aid for a research fellow of the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grants 17J05577 to K H; 18H03973 to HT). We thank Shima Motooka for her technical support. We thank Dr Satoshi Ohkura and his laboratory members of Laboratory of Animal Production Science (Nagoya University, Aichi, Japan) for providing the western blotting tools.
References
- 1.Uenoyama Y, Nakamura S, Hayakawa Y, Ikegami K, Watanabe Y, Deura C, Minabe S, Tomikawa J, Goto T, Ieda N, Inoue N, Sanbo M, Tamura C, Hirabayashi M, Maeda KI, Tsukamura H. Lack of pulse and surge modes and glutamatergic stimulation of luteinising hormone release in Kiss1 knockout rats. J Neuroendocrinol 2015; 27: 187–197. [DOI] [PubMed] [Google Scholar]
- 2.d’Anglemont de Tassigny X, Fagg LA, Dixon JP, Day K, Leitch HG, Hendrick AG, Zahn D, Franceschini I, Caraty A, Carlton MB, Aparicio SA, Colledge WH. Hypogonadotropic hypogonadism in mice lacking a functional Kiss1 gene. Proc Natl Acad Sci USA 2007; 104: 10714–10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messager S, Chatzidaki EE, Ma D, Hendrick AG, Zahn D, Dixon J, Thresher RR, Malinge I, Lomet D, Carlton MB, Colledge WH, Caraty A, Aparicio SA. Kisspeptin directly stimulates gonadotropin-releasing hormone release via G protein-coupled receptor 54. Proc Natl Acad Sci USA 2005; 102: 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topaloglu AK, Tello JA, Kotan LD, Ozbek MN, Yilmaz MB, Erdogan S, Gurbuz F, Temiz F, Millar RP, Yuksel B. Inactivating KISS1 mutation and hypogonadotropic hypogonadism. N Engl J Med 2012; 366: 629–635. [DOI] [PubMed] [Google Scholar]
- 5.Tomikawa J, Homma T, Tajima S, Shibata T, Inamoto Y, Takase K, Inoue N, Ohkura S, Uenoyama Y, Maeda K, Tsukamura H. Molecular characterization and estrogen regulation of hypothalamic KISS1 gene in the pig. Biol Reprod 2010; 82: 313–319. [DOI] [PubMed] [Google Scholar]
- 6.Inoue N, Sasagawa K, Ikai K, Sasaki Y, Tomikawa J, Oishi S, Fujii N, Uenoyama Y, Ohmori Y, Yamamoto N, Hondo E, Maeda K, Tsukamura H. Kisspeptin neurons mediate reflex ovulation in the musk shrew (Suncus murinus). Proc Natl Acad Sci USA 2011; 108: 17527–17532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O’Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med 2003; 349: 1614–1627. [DOI] [PubMed] [Google Scholar]
- 8.de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA 2003; 100: 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lapatto R, Pallais JC, Zhang D, Chan YM, Mahan A, Cerrato F, Le WW, Hoffman GE, Seminara SB. Kiss1-/- mice exhibit more variable hypogonadism than Gpr54-/- mice. Endocrinology 2007; 148: 4927–4936. [DOI] [PubMed] [Google Scholar]
- 10.Smith JT, Shahab M, Pereira A, Pau KY, Clarke IJ. Hypothalamic expression of KISS1 and gonadotropin inhibitory hormone genes during the menstrual cycle of a non-human primate. Biol Reprod 2010; 83: 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franceschini I, Lomet D, Cateau M, Delsol G, Tillet Y, Caraty A. Kisspeptin immunoreactive cells of the ovine preoptic area and arcuate nucleus co-express estrogen receptor alpha. Neurosci Lett 2006; 401: 225–230. [DOI] [PubMed] [Google Scholar]
- 12.Matsuda F, Nakatsukasa K, Suetomi Y, Naniwa Y, Ito D, Inoue N, Wakabayashi Y, Okamura H, Maeda KI, Uenoyama Y, Tsukamura H, Ohkura S. The luteinising hormone surge-generating system is functional in male goats as in females: involvement of kisspeptin neurones in the medial preoptic area. J Neuroendocrinol 2015; 27: 57–65. [DOI] [PubMed] [Google Scholar]
- 13.Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, Seminara S, Clifton DK, Steiner RA. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology 2004; 145: 4073–4077. [DOI] [PubMed] [Google Scholar]
- 14.Adachi S, Yamada S, Takatsu Y, Matsui H, Kinoshita M, Takase K, Sugiura H, Ohtaki T, Matsumoto H, Uenoyama Y, Tsukamura H, Inoue K, Maeda K. Involvement of anteroventral periventricular metastin/kisspeptin neurons in estrogen positive feedback action on luteinizing hormone release in female rats. J Reprod Dev 2007; 53: 367–378. [DOI] [PubMed] [Google Scholar]
- 15.Navarro VM, Gottsch ML, Chavkin C, Okamura H, Clifton DK, Steiner RA. Regulation of gonadotropin-releasing hormone secretion by kisspeptin/dynorphin/neurokinin B neurons in the arcuate nucleus of the mouse. J Neurosci 2009; 29: 11859–11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinsey-Jones JS, Grachev P, Li XF, Lin YS, Milligan SR, Lightman SL, O’Byrne KT. The inhibitory effects of neurokinin B on GnRH pulse generator frequency in the female rat. Endocrinology 2012; 153: 307–315. [DOI] [PubMed] [Google Scholar]
- 17.Lehman MN, Coolen LM, Goodman RL. Minireview: kisspeptin/neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: a central node in the control of gonadotropin-releasing hormone secretion. Endocrinology 2010; 151: 3479–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohkura S, Takase K, Matsuyama S, Mogi K, Ichimaru T, Wakabayashi Y, Uenoyama Y, Mori Y, Steiner RA, Tsukamura H, Maeda KI, Okamura H. Gonadotrophin-releasing hormone pulse generator activity in the hypothalamus of the goat. J Neuroendocrinol 2009; 21: 813–821. [DOI] [PubMed] [Google Scholar]
- 19.Clarkson J, Han SY, Piet R, McLennan T, Kane GM, Ng J, Porteous RW, Kim JS, Colledge WH, Iremonger KJ, Herbison AE. Definition of the hypothalamic GnRH pulse generator in mice. Proc Natl Acad Sci USA 2017; 114: E10216–E10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinoshita M, Tsukamura H, Adachi S, Matsui H, Uenoyama Y, Iwata K, Yamada S, Inoue K, Ohtaki T, Matsumoto H, Maeda K. Involvement of central metastin in the regulation of preovulatory luteinizing hormone surge and estrous cyclicity in female rats. Endocrinology 2005; 146: 4431–4436. [DOI] [PubMed] [Google Scholar]
- 21.Kauffman AS, Gottsch ML, Roa J, Byquist AC, Crown A, Clifton DK, Hoffman GE, Steiner RA, Tena-Sempere M. Sexual differentiation of Kiss1 gene expression in the brain of the rat. Endocrinology 2007; 148: 1774–1783. [DOI] [PubMed] [Google Scholar]
- 22.Clarkson J, d’Anglemont de Tassigny X, Moreno AS, Colledge WH, Herbison AE. Kisspeptin-GPR54 signaling is essential for preovulatory gonadotropin-releasing hormone neuron activation and the luteinizing hormone surge. J Neurosci 2008; 28: 8691–8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homma T, Sakakibara M, Yamada S, Kinoshita M, Iwata K, Tomikawa J, Kanazawa T, Matsui H, Takatsu Y, Ohtaki T, Matsumoto H, Uenoyama Y, Maeda K, Tsukamura H. Significance of neonatal testicular sex steroids to defeminize anteroventral periventricular kisspeptin neurons and the GnRH/LH surge system in male rats. Biol Reprod 2009; 81: 1216–1225. [DOI] [PubMed] [Google Scholar]
- 24.Tsukamura H, Homma T, Tomikawa J, Uenoyama Y, Maeda K. Sexual differentiation of kisspeptin neurons responsible for sex difference in gonadotropin release in rats. Ann N Y Acad Sci 2010; 1200: 95–103. [DOI] [PubMed] [Google Scholar]
- 25.Uenoyama Y, Tomikawa J, Inoue N, Goto T, Minabe S, Ieda N, Nakamura S, Watanabe Y, Ikegami K, Matsuda F, Ohkura S, Maeda K, Tsukamura H. Molecular and epigenetic mechanism regulating hypothalamic Kiss1 gene expression in mammals. Neuroendocrinology 2016; 103: 640–649. [DOI] [PubMed] [Google Scholar]
- 26.Semaan SJ, Kauffman AS. Emerging concepts on the epigenetic and transcriptional regulation of the Kiss1 gene. Int J Dev Neurosci 2013; 31: 452–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomikawa J, Uenoyama Y, Ozawa M, Fukanuma T, Takase K, Goto T, Abe H, Ieda N, Minabe S, Deura C, Inoue N, Sanbo M, Tomita K, Hirabayashi M, Tanaka S, Imamura T, Okamura H, Maeda K, Tsukamura H. Epigenetic regulation of Kiss1 gene expression mediating estrogen-positive feedback action in the mouse brain. Proc Natl Acad Sci USA 2012; 109: E1294–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomniczi A, Loche A, Castellano JM, Ronnekleiv OK, Bosch M, Kaidar G, Knoll JG, Wright H, Pfeifer GP, Ojeda SR. Epigenetic control of female puberty. Nat Neurosci 2013; 16: 281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vazquez MJ, Toro CA, Castellano JM, Ruiz-Pino F, Roa J, Beiroa D, Heras V, Velasco I, Dieguez C, Pinilla L, Gaytan F, Nogueiras R, Bosch MA, Rønnekleiv OK, Lomniczi A, Ojeda SR, Tena-Sempere M. SIRT1 mediates obesity- and nutrient-dependent perturbation of pubertal timing by epigenetically controlling Kiss1 expression. Nat Commun 2018; 9: 4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298: 1039–1043. [DOI] [PubMed] [Google Scholar]
- 31.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 2009; 10: 697–708. [DOI] [PubMed] [Google Scholar]
- 32.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 1996; 87: 95–104. [DOI] [PubMed] [Google Scholar]
- 33.Philpott A, Krude T, Laskey RA. Nuclear chaperones. Semin Cell Dev Biol 2000; 11: 7–14. [DOI] [PubMed] [Google Scholar]
- 34.Parthun MR. Histone acetyltransferase 1: More than just an enzyme? Biochim Biophys Acta 2012; 1819: 256–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 2000; 16: 351–356. [DOI] [PubMed] [Google Scholar]
- 36.Assadullah IN, Ieda N, Kawai N, Ishii H, Ihara K, Inoue N, Uenoyama Y, Tsukamura H. Co-expression of the calcitonin receptor gene in the hypothalamic kisspeptin neurons in female rats. Reprod Med Biol 2018; 17: 164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cagampang FR, Maeda KI, Tsukamura H, Ohkura S, Ota K. Involvement of ovarian steroids and endogenous opioids in the fasting-induced suppression of pulsatile LH release in ovariectomized rats. J Endocrinol 1991; 129: 321–328. [DOI] [PubMed] [Google Scholar]
- 38.Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates, 5th Edition. San Diego: Elsevier Academic Press; 2005. [Google Scholar]
- 39.Sugimoto A, Tsuchida H, Ieda N, Ikegami K, Inoue N, Uenoyama Y, Tsukamura H. Somatostatin-somatostatin receptor 2 signaling mediates LH pulse suppression in lactating rats. Endocrinology 2019; 160: 473–483. [DOI] [PubMed] [Google Scholar]
- 40.Treen AK, Luo V, Chalmers JA, Dalvi PS, Tran D, Ye W, Kim GL, Friedman Z, Belsham DD. Divergent regulation of ER and Kiss genes by 17β-estradiol in hypothalamic ARC versus AVPV models. Mol Endocrinol 2016; 30: 217–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takase K, Uenoyama Y, Inoue N, Matsui H, Yamada S, Shimizu M, Homma T, Tomikawa J, Kanda S, Matsumoto H, Oka Y, Tsukamura H, Maeda KI. Possible role of oestrogen in pubertal increase of Kiss1/kisspeptin expression in discrete hypothalamic areas of female rats. J Neuroendocrinol 2009; 21: 527–537. [DOI] [PubMed] [Google Scholar]
- 42.He H, Kong S, Liu F, Zhang S, Jiang Y, Liao Y, Jiang Y, Li Q, Wang B, Zhou Z, Wang H, Huo R. Rbbp7 is required for uterine stromal decidualization in mice. Biol Reprod 2015; 93: 13. [DOI] [PubMed] [Google Scholar]
- 43.Parthun MR. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene 2007; 26: 5319–5328. [DOI] [PubMed] [Google Scholar]
- 44.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011; 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Creekmore AL, Walt KA, Schultz-Norton JR, Ziegler YS, McLeod IX, Yates JR, Nardulli AM. The role of retinoblastoma-associated proteins 46 and 48 in estrogen receptor α mediated gene expression. Mol Cell Endocrinol 2008; 291: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang TF, Yu SQ, Wang ZY. RbAp46 inhibits estrogen-stimulated progression of neoplastigenic breast epithelial cells. Anticancer Res 2007; 27(5A): 3205–3209. [PubMed] [Google Scholar]
- 47.Tohyama M, Takatsuji K. Atlas of Neuroactive Substances and Their Receptors in the Rat. Oxford: Oxford University Press; 1998: 226–317. [Google Scholar]
- 48.Dürr K, Norsted E, Gömüç B, Suarez E, Hannibal J, Meister B. Presence of pituitary adenylate cyclase-activating polypeptide (PACAP) defines a subpopulation of hypothalamic POMC neurons. Brain Res 2007; 1186: 203–211. [DOI] [PubMed] [Google Scholar]