Abstract
Integrin αIIbβ3 is a predominant type of integrin abundantly expressed on the surface of platelets and its activation regulates the process of thrombosis. Talin and kindlin are cytoplasmic proteins that bind to integrin and modulate its affinity for extracellular ligands. Although the molecular details of talin-mediated integrin activation are known, the mechanism of kindlin involvement in this process remains elusive. Here, we demonstrate that the interplay between talin and kindlin promotes integrin activation. Our all-atomic molecular dynamics simulations on complete transmembrane and cytoplasmic domains of integrin αIIbβ3, talin1 F2/F3 subdomains, and the kindlin2 FERM domain in an explicit lipid-water environment over a microsecond timescale unraveled the role of kindlin as an enhancer of the talin interaction with the membrane proximal region of β−integrin. The cooperation of kindlin with talin results in a complete disruption of salt bridges between R995 on αIIb and D723/E726 on β3. Furthermore, kindlin modifies the molecular mechanisms of inside-out activation by decreasing the crossing angle between transmembrane helices of integrin αIIbβ3, which eventually results in parallelization of integrin dimer. In addition, our control simulation featuring integrin in complex with kindlin reveals that kindlin binding is not sufficient for unclasping the inner-membrane and outer-membrane interactions of integrin dimer, thus ruling out the possibility of solitary action of kindlin in integrin activation.
Significance
Using the newly solved crystal structure of kindlin, we investigated, to the best of our knowledge, for the first time, the molecular mechanism of kindlin-mediated integrin activation through simultaneous binding of talin and kindlin. We demonstrate molecular details of how kindlin cooperates with talin to promote the activation of integrin αIIbβ3.
Introduction
Integrin plays a central role in regulating cell-matrix and cell-cell adhesion and is crucial for various signaling pathways involved in cell migration, proliferation, and differentiation (1,2). Integrins are heterodimeric proteins composed of α and β subunits that associate noncovalently. Each subunit consists of a large extracellular ectodomain, a single-pass transmembrane helix (TM), and a short cytoplasmic tail (CT) (Fig. 1 A; (3,4)). There are 24 different combinations of integrin α- and β-subunits. Integrin αIIbβ3 is one of the well-known types that is only expressed in platelets and is necessary for the hemostatic function of platelets (5,6). Integrins mediate bidirectional, inside-out, and outside-in signaling across the membrane. Inside-out signaling involves the interaction of intracellular proteins with the integrin CT, which modulates integrin’s affinity for extracellular ligands (7, 8, 9).
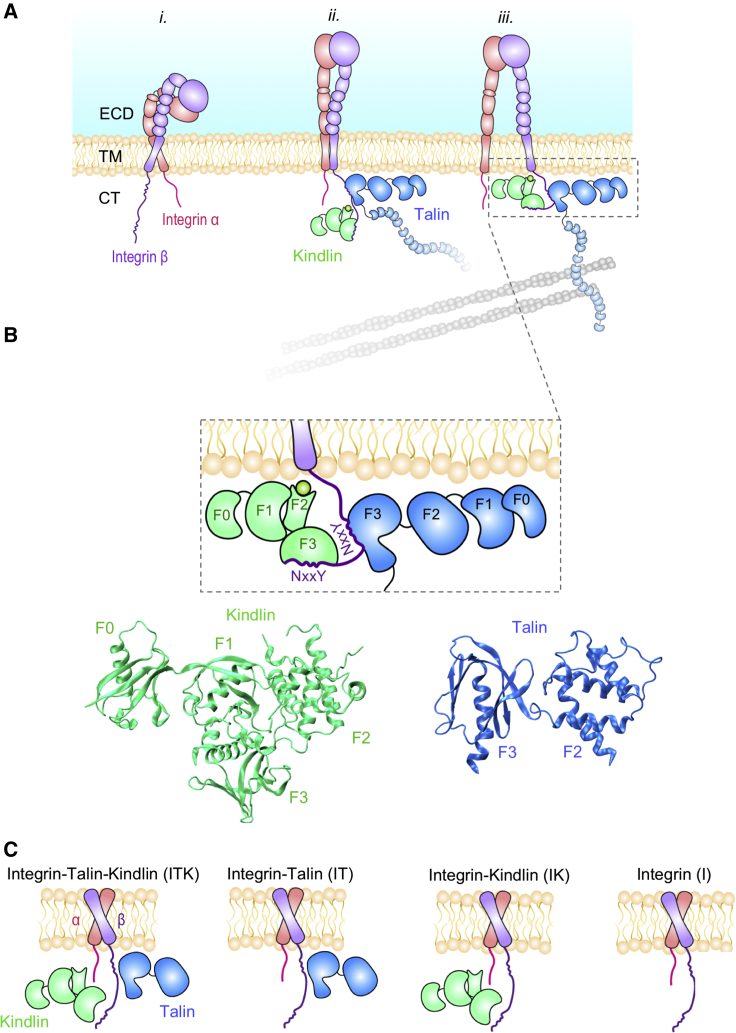
Figure 1.
Cytoplasmic interactions with the integrin β tail mediate inside-out signaling. (A) Schematic representations of the three conformational states of integrin are shown: i) bent closed (inactive), ii) extended closed, and iii) extended open (activated). Talin and kindlin are both involved in integrin activation by binding to the cytoplasmic domain of β. Talin association to the actin cytoskeleton provides a linkage between the extracellular environment and the cytoskeleton. (B) The F3 subdomain of talin and kindlin bind to the membrane proximal and membrane distal regions of β-integrin, respectively (top). The crystal structure of kindlin2 (F0–F3 subdomains) and talin1 (F2–F3 domains) are shown here (bottom). (C) Schematic representations of the four molecular models used in our simulations are shown: integrin in complex with talin and kindlin (ITK), integrin in complex with talin (IT), integrin in complex with kindlin (IK), and integrin only (I). To see this figure in color, go online.
Three distinct conformational states of integrin have been determined: bent closed or resting state, extended closed, and extended open (Fig. 1 A; (6)). Integrin activation entails a conformational transition of integrin from a low-affinity (bent-closed) to a high-affinity state (extended-open) (Fig. 1 A; (10,11)). It was previously believed that talin was both necessary and sufficient for inducing integrin activation (12,13), whereas recent studies contradict this notion (14, 15, 16) and suggest that kindlin is also needed as a coactivator of integrin (14,17,18). For example, Moser et al. (19) showed that the knockout of kindlin3 results in inactivated integrins despite the presence of talin. Moreover, kindlin-associated diseases like leukocyte adhesion disease III, reflecting deficiencies of integrin function, show the necessity of kindlin in integrin activation (5).
Both talin (talin-1 and 2) and kindlin (kindlin-1, 2, and 3) isoforms include a 4.1 protein, ezrin, radixin, moesin (FERM) domain, which is composed of four structural subdomains F0–F3 (20). Kindlin also features a pleckstrin homology domain inserted into its F2 subdomain, which has been shown to directly interact with the lipid membrane (Fig. 1 B; (21)). Talin and kindlin both bind to the CT domain of integrin β through their F3 subdomains. Specifically, talin binds to the NxxY motif at the membrane proximal region, whereas kindlin associates with this motif at the membrane distal part of the CT, suggesting that kindlin and talin can simultaneously bind to the integrin tail and trigger activation (22). The molecular mechanisms of the interplay among talin and kindlin for inducing integrin activation are not yet well understood.
Recent experimental and computational studies have revealed important molecular details of integrin inside-out activation. These studies have suggested that the inner membrane clasp (IMC) between the CT domains of integrin subunits is critical for maintaining the closed state of integrin. In addition, these studies have shown that the unclasping of the IMC induced by talin or other activators triggers integrin activation (23, 24, 25) and ultimately results in an increase in the crossing angle between α and β monomers (26, 27, 28). However, all computational studies on integrin-mediated inside-out signaling to date have overlooked the role of kindlin, largely because of the lack of sufficient structural information (7,29, 30, 31). In this study, we use the recently solved crystal structure of kindlin (32) to investigate the molecular mechanism of integrin activation through simultaneous binding of talin and kindlin.
Previous studies from our group have also looked at the molecular interactions between focal adhesion proteins using molecular dynamics (MD) simulations (33, 34, 35, 36). For example, Golji et al. (34,35) investigated the interaction of talin with two different states of vinculin, i.e., autoinhibited and activated. This study showed that talin could completely bind to activated vinculin. MD is a powerful technique to shed light into molecular details of interactions between proteins, especially when the resolved protein structures are available. To identify these critical interactions, different simulation scenarios can be designed in which the effect of different molecules can be exclusively and/or simultaneously be studied. We use all-atomic microsecond-scale MD simulations of integrin αIIbβ3 TM/CT structure in an explicit lipid-water environment (I) and under the following three conditions: in complex with talin1 F2–F3 subdomains (IT), with kindlin2 (IK), and with both talin1 and kindlin2 (ITK) to uncover the role of kindlin in integrin activation (Fig. 1 C). Specifically, we explore how talin and kindlin binding affect the allosteric transition of forces across the integrin molecule, i.e., from the CT to the TM and, eventually, to the ectodomain. Our results show that kindlin2 cooperates with talin1 to facilitate integrin αIIbβ3 activation by enhancing talin1 interaction with the membrane proximal (MP) region of β3-integrin.
Materials and Methods
System setups
The crystal structure of TM/CT domains of integrin αIIbβ3 (Protein Data Bank, PDB: 2KNC (37)), talin1 (PDB: 3IVF (21)), and kindlin2 FERM domain (PDB: 5XPY (32)) were taken from the PDB. Because the talin FERM domain is both necessary and sufficient for integrin activation (38,39), the talin rod domain was excluded from our models to reduce the system size and minimize the computational overhead. Two regions of the kindlin2 FERM domain in F1 and F2 subdomains are missing in the current resolved crystal structure. These regions are connected to the current crystal structure via flexible linkers, and the motions of these missing regions are independent of other parts of the molecule (32). Also, because there are no known interactions between the kindlin missing residues and talin or integrin molecules, no homology modeling techniques were used because they would introduce uncertainties.
Four different models were built using VMD: integrin αIIbβ3 (I), integrin αIIbβ3 in complex with either talin1 (IT) or kindlin2 (IK), or integrin αIIbβ3 in complex with both talin1 and kindlin2 (ITK). First, the TM domains of integrin were embedded in a patch of POPC lipid bilayer with dimensions of 200 × 250 Å2 using VMD. Also, the lipid chains overlapping with the integrin were removed. Then, the structures of integrin in complex with talin (IT) (PDB: 3G9W (40)) and kindlin (IK) (PDB: 5XQ1 (32)) were used as templates for finding the best orientation of talin and kindlin molecules with respect to integrin and the membrane. Two different orientations were tested for IT and ITK scenarios (IT, IT_o, ITK, and ITK_o). Additionally, two different trial runs were conducted for ITK (ITK and ITK_2).
All systems were solvated using the TIP3P explicit water model in nanoscale MD (41), and water molecules inside the lipid membrane were removed. Then, all systems were neutralized and ionized with 150 mM of KCl. The number of atoms reached 744,946 in ITK and ITK_2; 556,876 in IT; 625,742 in IK; 425,820 in I; 745,559 in ITK_o; and 556,511 in IT_o.
MD simulations
We performed all-atom MD simulations to investigate the conformational changes of integrin αIIbβ3 upon simultaneous interaction with talin1 and kindlin2 (ITK, ITK_2, and ITK_o) and compared them to the control simulations (I, IT, IT_o, and IK). MD simulations were carried out using NAMD and CHARMM36 force field (42). Molecular visualization and analysis were performed using the VMD package (43). Periodic boundary conditions were used in all three dimensions, and a 2-fs time step was used in all simulations. The Langevin piston, the Nosé-Hoover pressure control algorithm, and the Langevin damping thermostat for temperature control were used (42). Pressure was maintained at 1 bar and temperature at 310 K with a damping coefficient of 5/ps.
Each trial was initially minimized for 100,000 timesteps using the conjugate gradient and line search algorithm to relax the structures and remove all bad contacts. After the minimization process, each configuration was equilibrated for 25 ns or longer until equilibrium was reached. Fully equilibrated structures were then used in the final production simulations that ran for 1000 ns for ITK and IT, 760 ns for IK, 625 ns for I, and 400 ns for ITK_o, IT_o, and ITK_2.
To provide a side-by-side comparison between different simulation scenarios, we presented density plots that show the distribution of combined data obtained from various orientations and repetitions for each ITK and IT conditions. The density plots of the talin-integrin analyses for ITK_a (i.e., ITK, ITK_2, and ITK_o) and IT_a (i.e., IT and IT_o) data are overlaid to facilitate the comparison.
Hbond calculations
The VMD hbonds plug-in (version 1.2) (43) was used to calculate the number of hydrogen bonds (hbonds) between regions of interest. The cutoff distance and angle were set to 4 and 20 Å, respectively. The density- and time-dependent plots were all prepared in R matrixStats and gplot (44,45).
Principal component analysis
Principal component analysis (PCA) was used to examine the time evolution of the integrin structure along all trajectories and understand the conformational differences induced by the cytoplasmic interactions. The principal components were defined as the orthogonal axes of maximal variance and were identified by superpositioning the structures on the invariant core and calculating the variance using the tools introduced by the Bio3D package (46).
Residue cross correlation
To examine how atomic fluctuations and displacements within the integrin heterodimer are correlated throughout our simulations, we performed pairwise residue cross correlation analysis using the cross correlation function (“dccm.xyz”) in the Bio3D package (46). The matrix of all pairwise cross correlations between residues was visualized using a dynamical cross correlation heatmap.
Cross corelation function
The cross correlation function (CCF) in R was used to understand the relationship between two time series representing different features along the trajectories. The first argument was treated as the predictor (cause) of the second argument. The lag period indicates when the effect of a change in one feature is reflected in the other feature (46).
Solvent-accessible surface area
The solvent-accessible surface area (SASA) of each molecule was calculated using the “measure SASA” command in VMD (43).
Force distribution analysis
To monitor changes in the internal forces of the integrin heterodimer upon cytoplasmic interactions, we performed time-resolved force distribution analysis (TRFDA) implemented in the GROMACS software package (47). Atomic pairwise forces were calculated for all residues of both integrin subunits and included only the nonbonded electrostatic interactions acting on each residue. The comparison between force propagation along the transmembrane domains of integrins indicated the allosteric signal propagation across the membrane. The punctual stress measured for α- and β-subunits is the sum of absolute values of scalar pairwise forces exerted on each atom.
Structural alignment
To determine the changes in the conformation of kindlin and talin upon interactions with integrin, we compared the initial structures of these molecules with the final structures after ∼1000 ns of simulation. The final structures of talin in the ITK or IT simulations were aligned to the initial structure of talin using the “struct.aln” function in the Bio3D package in R (46). Similarly, the final structures of kindlin in the ITK or IK simulations were aligned to the initial kindlin structure. To quantify the distances between residues in the two aligned structures, a difference vector was calculated between the two structures using the “dist.xyz” function in R (46).
Results
To explore the role of kindlin in the activation of integrin as part of the integrin-mediated inside-out signaling, we developed all-atomic microsecond-scale MD simulations of αllbβ3 using an explicit lipid-water environment under four distinct scenarios: integrin alone (I) and in complex with talin1 F2–F3 subdomains (IT), with kindlin2 (IK), and with both talin1 and kindlin2 (ITK).
The interactions of talin1 and kindlin2 with the CT of integrin β3 regulate its conformation
The mechanism of interactions of talin1 and kindlin2 with integrin β3 can be characterized based on the conformational transitions of the integrin heterodimer. These conformational changes were determined by comparing the first and last frames of our microsecond MD simulations, featuring an identical starting conformation of integrin for all scenarios (Fig. 2 A). The crossing angle between the transmembrane regions of integrin monomers was notably increased in IT simulation, changing the shape to open scissors (see below for further details). Unexpectedly, the final shape of integrin in the ITK simulation was changed to closed scissors (Fig. 2 A). No significant change was observed in the crossing angle in the IK simulation, and only a small decrease in this angle was seen in the I simulation.
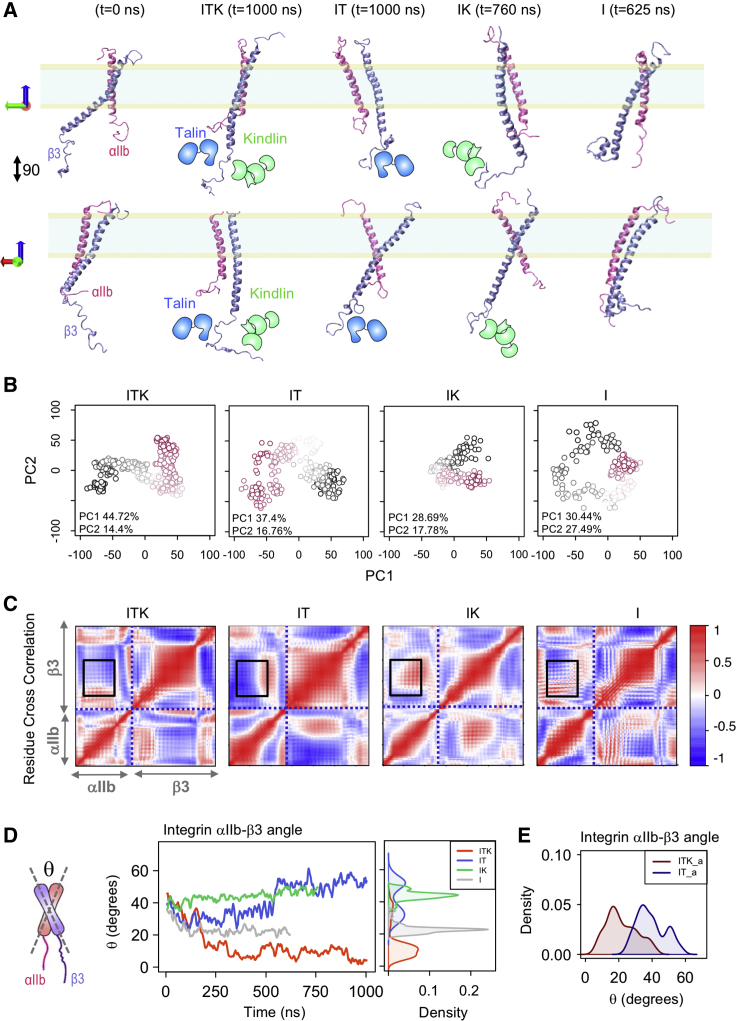
Figure 2.
Conformational changes of integrin αIIbβ3. (A) The schematics of talin and kindlin are overlaid with the cartoon representations of the integrin αIIb (mauve) β3 (ice blue) heterodimer in two different views (90°). Snapshots of the trajectories of the integrin αIIbβ3 dimer for the last simulation frame of the ITK (t = 1000 ns), IT (t = 1000 ns), IK (t = 760 ns), and I (t = 625 ns) simulations are shown. (B) The PCA of all trajectories is shown. Each point is a structural state in the PC1-PC2 space. The points corresponding to the beginning of all trajectories are shown in red, the middle frames are represented by a smooth color change to white, and the end frames are illustrated in black. (C) Residue cross correlation heatmaps of the integrin αIIb and β3 helices averaged over simulation time are shown here. The αIIb and β3 regions are indicated on the heatmaps. Black boxes indicate the TM regions of integrin dimer. A representative heatmap is shown for each of the four ITK, IT, IK, and I simulations. (D) A schematic representation of the integrin dimer showing angle θ between the αIIb and β3 helices (right) is given. The time and density plots of angle θ for the ITK, IT, IK, and I simulations are shown in red, blue, green, and gray, respectively. (E) The density plots of the θ angle are shown for the combined ITK simulations (ITK_a) and combined IT simulations (IT_a) in dark red and dark blue, respectively. To see this figure in color, go online.
Conformational transitions of the integrin dimer in response to cytoplasmic interactions with talin1 and/or kindlin2 were quantified via PCA. For PCA calculations, the invariant core of the structure in each simulation was identified. The distribution of structures was extracted from each trajectory and conformational differences were quantified for equivalent residues. The first two principal components accounted for over 45% of the variance in all simulations. Because the contribution of all other principal components was relatively minimal, the structural distribution was projected in PC1-PC2 space as shown in Figs. 2 B and S1 A. The color gradient from red to black represents the time evolution of simulation trajectories in the PCA plots. For IT, ITK, IT_o, ITK_o, and ITK_2, the final state of the system falls far from the initial state in the principlal component (PC) space, indicating a remarkable conformational transition; however, this is not notable in either IK or I.
Residue cross correlation analyses between all residue pairs of the integrin αIIb and β3 in ITK, ITK_2, ITK_o IT, IT_o, IK, and I were performed to examine how the fluctuations within the integrin dimer are correlated (Figs. 2 C and S1 B). Interestingly, we observed a correlation and hence a coupling between the transmembrane regions of integrin αIIb and β3 (black boxes in Figs. 2 C and S1 B). This coupling was dominantly observed in ITK and IT scenarios. The level of residue correlations was notably lower in IK and I simulations (Fig. 2 C).
In addition, we calculated the crossing angle between αIIb and β3 helices (θ) as a function of time (Fig. 2 D) to quantify the conformational changes of integrins. Specifically, we calculated the angle between the line crossing residues 967–979 in αIIb and the line crossing residues 697–709 in β3. As expected, an increase in crossing angle of β3 in IT simulations was observed. Interestingly, in ITK, the θ angle starts from 40° and then approaches zero after 250 ns of simulation and αIIb and β3 helices become almost parallel to each other. In the case of binding of kindlin2 without talin1 (IK), our results showed a small angle change (10°) in θ, which is clearly in contrast with the cases of binding of talin1 that resulted in a noticeable change of θ (i.e., 30° change in IT and 40° change in ITK). Also, the crossing angle immediately drops in the first 150 ns of the simulation, in which we only considered integrin (I), and stabilizes to ∼20° after 150 ns.
Moreover, we added an overlaid density plot that shows the distribution of the combined data obtained from various orientations and repetitions for ITK and IT scenarios. Basically, the density plots of the angle between integrin monomers for all the ITK (i.e., ITK, ITK_o, and ITK_2) and IT (i.e., IT and IT_o) data are overlaid (Fig. 2 E). The distinction between the peak of the combined ITK density plots (i.e., ITK_a) and the peak of the combined IT density plots (i.e., IT_a) clearly shows higher angle values for IT compared to ITK in each orientation and each repetition.
Interactions between integrin αIIb and β3 upon binding of talin1 and kindlin2
To determine the conformational transition in integrin β3 upon binding of talin1 and kindlin2, we next analyzed the interaction energy between the αIIb and β3 helices. Two interaction interfaces on the integrin αIIbβ3 TM/CT domains are known to stabilize the resting conformation of the dimer: the inner membrane clasp (IMC) and the outer membrane clasp (OMC). The IMC is characterized by the interactions between the highly conserved 991GFFKR995 motif of αIIb and W715, K716, and I719 residues of β3 and a salt bridge between residue R995 on αIIb and D723 on β3 (Fig. 3 A). The R995-E726 salt bridge is also known to be involved in maintaining integrin in an inactive conformation. OMC is defined as an interaction network between 972GXXXG976 on αIIb and V700, M701, and I704 residues on β3, as well as associations between αIIb 979LL980 and β3 705LXXG708 residues (Fig. 3 A). Mutations in the residues involved in IMC or OMC result in disruption of interactions between αIIb and β3 helices, leading to integrin activation (6,20,29,48,49).
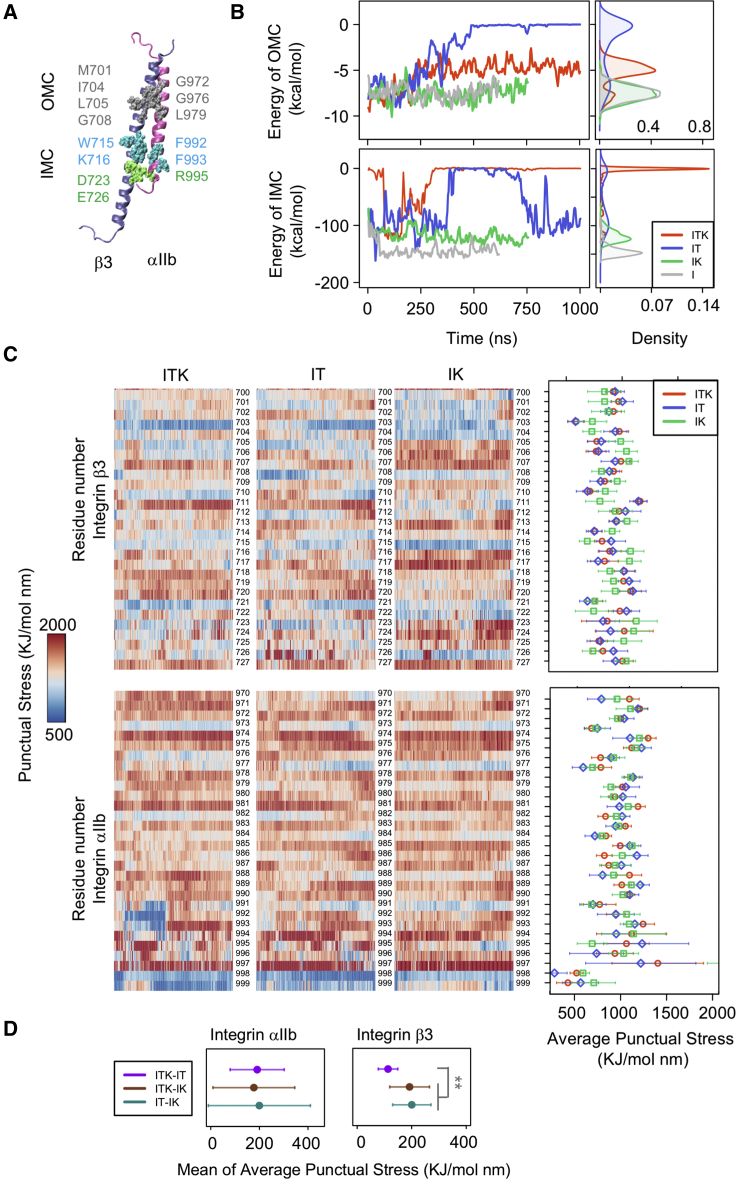
Figure 3.
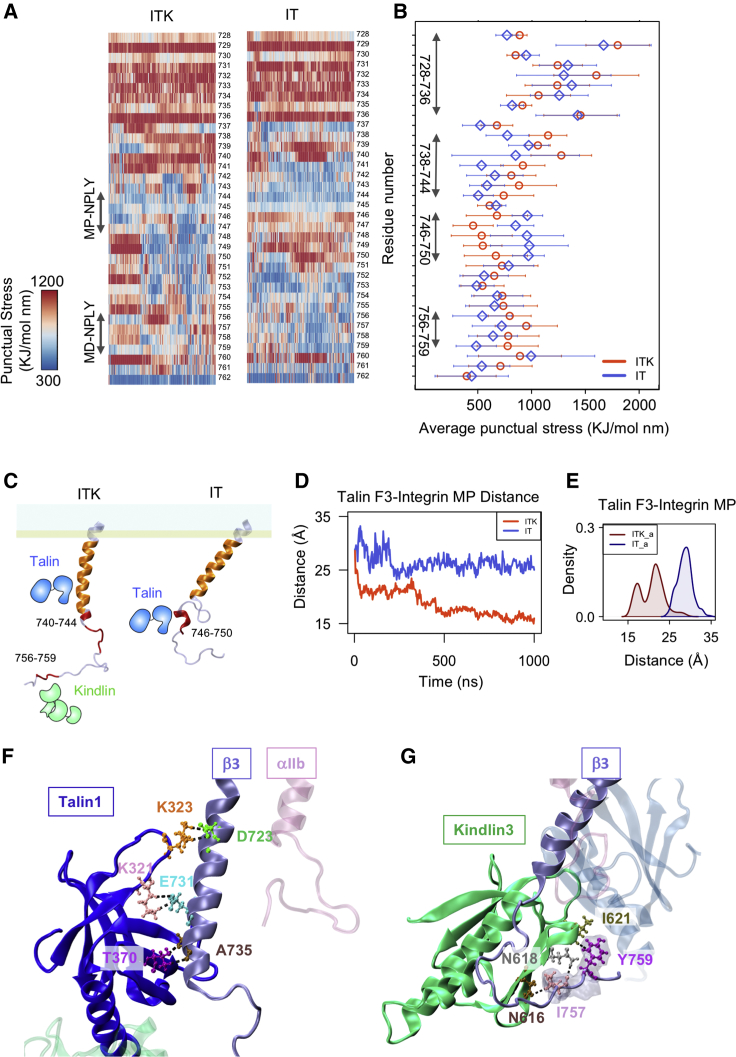
Interactions between αIIb and β3 dimer. (A) The structure of the αIIb (mauve) β3 (ice blue) heterodimer is shown. Interacting residues in the OMC region are shown in silver, interacting residues in the IMC region are shown in blue, and the αIIb R995-β3 D723/D726 salt bridges are shown in green. (B) The time and density plots of energy of the interactions between OMC (top) and IMC (bottom) residues are shown for the ITK, IT, IK, and I simulations in red, blue, green, and gray respectively. (C) The TRFDA and time-averaged punctual stresses (for the last 300 ns of the simulations) within the integrin αIIb and β3 dimer were calculated. Representative heatmaps of per-residue punctual stress values over simulation time for the integrin β3 residues 700–727 and αIIb residues 971–996 are shown. A representative heatmap is shown for each of the three ITK, IT, and IK simulations. (D) The average values and standard deviations are shown for the mean of the time average of punctual stresses of ITK-IT, ITK-IK, and IT-IK. The mean of time average of ITK-IT is significantly lower than ITK-IT and IT-IK. p-value <1e −8. To see this figure in color, go online.
To examine whether the binding of kindlin2 or talin1 could disrupt the IMC and OMC interactions in our simulations, we calculated the nonbonded interaction energies between IMC and OMC regions over the course of simulation time (Fig. 3 B). Our results showed that the interaction energies of IMC and OMC decrease upon simultaneous binding of talin1 and kindlin2 (ITK simulation) and stabilize after 250 ns to approximately zero in IMC and −4 kcal/mol in OMC. In IT simulation, the nonbonded interaction energy in OMC approaches zero and remains zero after 500 ns. On the other hand, IMC interactions in IT simulations switch between zero and −100 kcal/mol. In IK and I simulations, there is no change in the OMC interactions, and a stable nonbonded interaction energy of ∼−7 kcal/mol is maintained throughout the simulation. The IMC level is also stable but slightly higher in I (∼−150 kcal/mol) compared to IK (∼−120 kcal/mol) because of an initial increase in the IMC level in I. This initial IMC increase in I simulation can possibly result in the initial decrease in the θ angle as shown in Fig. 2 D.
These results indicate that the cooperation of kindlin2 and talin1 can destabilize integrin αIIbβ3 dimer at the IMC region and weaken their association in the OMC region. Our results also suggest that kindlin2 alone is not sufficient to disrupt the IMC or OMC interactions. Also, the stable interactions (IMC and OMC) in the I simulation confirm that the disruption of these interactions in IT and ITK are not random and are a result of the binding of kindlin2 and talin1 to the cytoplasmic regions of integrin. Moreover, significant conformational changes in the TM of integrin in ITK and IT compared to IK and I (Fig. 2) show a correlation between OMC disruption and the open/close structure of integrin. Basically, the disruption of interactions in OMC region causes separation of αIIb and β3 helices, leading to the conformational changes in the TM region of integrin, as discussed in more detail in the following section.
Mechanical signal transmission entails dynamic force redistribution across the integrin αIIbβ3 molecule. To investigate how cytoplasmic interaction(s) with talin1 and/or kindlin2 allosterically impact the stress distribution within the TM, especially near the IMC and OMC, we performed TRFDA for the binding simulations as shown in Fig. 3 C. For this purpose, we calculated punctual stresses for residues of integrin αIIb and β3 helices (see Materials and Methods) and showed the results over time or time averaged of the last 300 ns of the simulations. The time average of punctual stress of all integrin residues was used as a measure to compare force distribution patterns between simulations as shown in Fig. 3 C on the right side, the bars indicating standard deviation. Moreover, to see the differences between the punctual stresses of the simulations, the time average of the punctual stresses was calculated for the differences between any pair of two out of the three states of ITK, IT, and IK simulations for each residue (ITK-IT, ITK-IK, and IT-IK) (Fig. S2).
Significant differences in the punctual stresses of integrin β3 were almost exclusively observed in IK compared to ITK and IT simulations. Basically, more similarities in the time-averaged punctual stresses can be seen between ITK and IT simulations (Figs. 3, C and D and S2). Specifically, time-averaged punctual stresses experienced in residues 704 and 705 (part of IMC); 706, 711, and 715 (part of OMC); and 717, 722, 723, and 724 were remarkably different in IK, although they were almost the same in IT and ITK simulations. Notably, residue 711 showed a most significantly different punctual stress within the IK simulation.
Force distribution analysis (FDA) results show the punctual stress changes within the TM and cytoplasmic regions of integrin αIIbβ3 are imposed by talin1 and/or kindlin2 binding to the integrin cytoplasmic region, resulting in IMC and OMC disruption and separation of αIIb and β3 helices in ITK and IT.
αIIb-β3 angle change (θ) is correlated with OMC
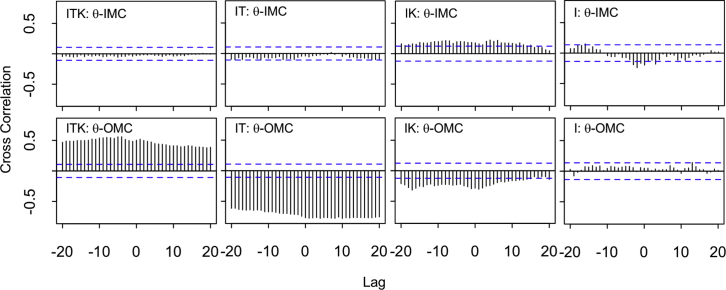
To understand whether changes in the angle (θ) between αIIb and β3 (Fig. 2, D and E) were correlated with the interaction energies of IMC and OMC (Fig. 3 B), we performed a cross correlation analysis using the CCF for time series in R, as described in Materials and Methods (Figs. 4 and S3). This function computes the covariance between two time series up to a defined lag. Based on the results shown in Figs. 4 and S3 (top), the θ angle was not a significant predictor of IMC, most likely because the IMC was either abruptly disrupted or remained stable throughout the simulation. This suggests that IMC dissociation is directly regulated by cytoplasmic interactions and not through angle change. In other words, interaction at the cytoplasmic region exerts tension that is transmitted by both bonded and nonbonded interactions across integrin residues toward the membrane, which eventually breaks the IMC and allows angle change. Conversely, dynamics of OMC interactions were highly correlated with θ in ITK and IT simulations (Fig. 4, bottom). The reverse correlations of ITK and IT reflects opposite directions of angle change. OMC interactions were also correlated with θ in ITK_2, ITK_o, and IT_o simulations (Fig. S3, bottom). Because there is no significant change in angle (θ) in the IK and only a small rapid decrease in θ in I simulations, we see a minimal θ-OMC correlation in IK and I compared to ITK and IT simulations (Fig. 4, bottom).
Figure 4.
θ angle correlation with IMC and OMC. Cross correlation of the IMC and angle changes of αIIb and β3 (top) and the cross correlation of the OMC and angle changes of αIIb and β3 (bottom) are shown. The horizontal blue lines are ∼95% confidence interval. A cross correlation plot is shown for each of the four ITK, IT, IK, and I simulations. High correlations exist between θ and OMC in ITK and IT, whereas the other correlations are negligible. To see this figure in color, go online.
Talin1 binds more effectively and strongly to integrin αIIbβ3 in the presence of kindlin2
Integrin β3 tail contains 47 residues and two NxxY motifs. The first NPLY motif (membrane proximal) is the binding site for talin1, and the second NITY motif (membrane distal) is the kindlin2 binding site. Talin1 and kindlin2 can simultaneously bind to β3 through their F3 domains and initiate inside-out signaling (3). To understand how this inside-out signaling may be initiated by the cooperation between kindlin2 and talin1, we investigated the molecular mechanisms of the simultaneous binding of talin1 and kindlin2 to the short CT of β3 and compared them with the individual binding of integrin with talin1 and kindlin2 in our simulations. Specifically, we calculated nonbonded interaction energies between integrin β3 and kindlin2 or talin1, as well as SASAs of integrin β3 CT, talin1, and kindlin2.
We first calculated the nonbonded interaction energies between the CT of integrin β3 and talin1 or kindlin2 F3 domains (Fig. 5, A and B). Specifically, we calculated the interaction between residues 720–762 of β3 and residues 569–680 of talin1 or residues 560–680 of kindlin2.
Figure 5.
Integrin β3-talin1 or -kindlin2 binding. (A) The time and density plots of interaction energies between integrin β3 CT and talin1 F3 domain (top) and integrin β3 CT and kindlin2 F3 domain (bottom) are shown in three simulations of ITK, IT, and IK. (B) The density plots of interaction energies are shown for ITK_a and IT_a in dark red and dark blue, respectively (top); the density plots of interaction energies are shown for ITK_a and IK in dark red and green, respectively (bottom). (C) Density plot representations of the number of hbonds between the integrin β3 CT and talin1 F3 domain (top) are shown for each of the ITK and IT simulations. The same plot is shown for kindlin2 (bottom) for each of the ITK and IK simulations. (D) The solvent-accessible surface area (SASA) of talin1 (subdomain that binds integrin) in IT or ITK simulations (top), the SASA of kindlin2 (subdomain that binds integrin) in IK or ITK simulations (middle), and the SASA of the CT of integrin in IT, IK, or ITK simulations (bottom) are shown. (E) CCF between binding energy of talin1-integrin β3 and θ in ITK and IT simulations is shown. (F) Cross correlation between binding energy of kindlin2-integrin β3 and θ in ITK and IK is shown (bottom). To see this figure in color, go online.
In the IT simulation, talin1 and β3 maintain an average interaction energy of 105 kcal/mol (Fig. 5 A, top). However, talin1 is able to bind much more strongly to integrin β3 over time in the ITK simulation. In this simulation, the nonbonded interaction energy between talin1 and β3 increases from 100 to 400 kcal/mol with an average energy of 222 ± 94 kcal/mol. Similarly, the overlaid density plots of the talin-integrin interaction for the combined ITK (ITK_a) and combined IT (IT_a) data show higher energies in ITK_a compared to IT_a (Fig. 5 B, top), confirming the stronger talin-integrin interaction in the presence of kindlin. Kindlin2, on the other hand (Fig. 5 A, bottom), binds more strongly to integrin β3 in the IK simulation with an interaction energy of 258 ± 61 kcal/mol, compared with 124 ± 57 kcal/mol in the ITK simulation. The overlaid density plots of kindlin-integrin interactions for ITK_a and IK data show lower peak value in ITK_a compared with IK (Fig. 5 B, bottom).
The crystal structures of the kindlin2-integrinβ3 and talin1-integrinβ3 complexes revealed the hbond network between the molecules (32). To determine the stability of kindlin2 or talin1-integrin complex, we calculated the number of hbonds between integrin CT-kindlin F3 and integrin CT-talin F3 (Fig. 5 C). Our results show that talin1 forms a slightly higher number of hbonds with the integrin β3 tail in the presence of kindlin2. However, kindlin2 can form a larger number of hbonds with the integrin β3 tail in the absence of talin1.
Next, we calculated the SASA of the integrin-binding F3 subdomains of talin1 and kindlin2 and the integrin β3 CT (see Materials and Methods; Fig. 5 D). Our results show that the SASA of talin1 is lower in ITK simulations (5929 ± 210 Å2) compared with the IT simulation (6034 ± 163 Å2) (Fig. 5 A, top). The SASA of kindlin2 immediately drops in the first 250 ns of simulation in IK simulations. However, when talin1 is present, the SASA of kindlin2 fluctuates much more slowly and is much higher in value, as shown in the density plots (Fig. 5 D, middle). The SASA of integrin CT is the lowest in ITK simulations compared to IT or IK simulations. The average values of 2575 ± 189 for ITK, 2698 ± 221 for IT, and 2805 ± 158 for IK suggest that both can bind to various regions on integrin and reduce its SASA (Fig. 5 D, bottom). Also, in the IT simulation, the SASA of integrin CT peaks at a slightly lower value than in the IK simulation, indicating a closer contact between integrin and talin1, compared with integrin and kindlin2 in conditions in which only one of them is present. Based on the interaction energies and SASA results, it is evident that talin1 binds more effectively in the presence of kindlin2, and kindlin2 can bind to integrin much more strongly in the absence of talin1.
Finally, to determine whether the interaction energies between integrin αIIbβ3 CT and talin1 or kindlin2 were directly correlated with the changes observed in the crossing angle between αIIb-β3 (θ), we also calculated the cross correlation between these parameters (Fig. 5, E and F). The interaction between talin1 and integrin β3 was promptly formed in both IT and ITK, whereas kindlin2 binding was relatively gradual. In both IT and ITK, talin1 binding did not show a major correlation with the angle change (Fig. 5 E), mainly because talin1 interaction was stable throughout the simulations. This suggests that a stable binding with talin1 is necessary for the subsequent changes in the integrin conformation. However, kindlin2 binding lagged the angle change in both IK and ITK (Fig. 5 F). This shows that the role of kindlin2 is more complex because it may both directly and indirectly modulate integrin activation.
FDA of integrin β3 CT
We performed TRFDA to monitor the dynamics of stress redistribution within the β3 cytoplasmic region upon simultaneous binding to talin1 and kindlin2. We have calculated the punctual stresses, scalar pairwise forces exerted on each atom, for residues 728–762 of the integrin β3 cytoplasmic domain over time for both IT and ITK simulations (Fig. 6 A). It can be seen from the heatmap that the punctual stress fluctuates in most of the residues, except in residues 729 and 736, in which the punctual stress remains high all the time both in IT and ITK.
Figure 6.
FDA of integrin αIIbβ3 CT. (A) TRFDA of integrin αIIb and β3 subunits are shown. Representative heatmaps of per-residue punctual stress values over simulation time for the integrin β3 cytoplasmic regions are shown. A representative plot is shown for each of the three ITK, IT, and IK simulations. (B) Time-averaged punctual stress for the last 300 ns of the IT and ITK simulations for integrin β3 CT residues is shown. Different distinct regions can be identified based on whether the punctual stress of IT or ITK dominates. (C) Schematic representations of the final structure of the integrin β3 CT in ITK and IT simulations are given. Regions with high punctual stress values are shown in red and orange on the β3 CT. (D) The time plot of distance between the center of mass of residues 720–736 in the integrin β3 and talin1 F3 subdomain is shown for the ITK and IT simulations in red and blue, respectively. (E) The density plots of the distance between the center of mass of residues 720–736 in integrin β3 and talin1 F3 subdomain are shown for the combined ITK simulations (ITK_a) and combined IT simulations (IT_a) in dark red and dark blue, respectively. (F) Strong interactions between the talin1 F3 subdomain and integrin β3 MP region in ITK simulation, mainly between residues K321, K323, and T370 on talin1 and residues D723, E731, and A735 on β3, are shown. hbonds are shown with dashed black lines. (G) Interactions between kindlin2 F3 subdomain and integrin β3 NITY motif in ITK simulation, mainly between residues N616, N618, and TI621 on kindlin2 and residues I757 and Y759 on β3, are shown. Hbonds are shown with dashed black lines. To see this figure in color, go online.
Also, we have calculated the time-averaged punctual stresses over the last 300 ns of the simulations (Fig. 6 B). Different distinct regions can be identified on Fig. 6, B and C based on whether the punctual stress of IT or ITK dominates. Time-averaged punctual stress is higher in residues 746–750, which overlaps with the talin1 binding site (NPLY), in IT compared to ITK. Conversely, the kindlin2 binding site (NITY) consisting of residues 756–759 shows higher time-averaged punctual stress values in ITK compared to IT. This most likely indicates that direct binding of integrin with kindlin can locally increase stress levels. However, the region consisting of residues 738 to 744 exhibits higher average stress values in ITK compared to IT. We should note that in ITK, the stress value suddenly declines in residues 748 and 749 while at the same time increasing in residues 737 and 739.
Moreover, we see similar high-stress values in the region consisting of residues 728–736 in both IT and ITK. Our analysis of binding interactions in this region revealed that these residues interact strongly with the membrane in both IT and ITK. On the other hand, the distance between the talin1 F3 subdomain and integrin β3 residues 728–736 decreases more significantly when kindlin2 is present (Fig. 6 D). Interestingly, the SASA of the MP (residues 720–736) region of the integrin CT and the SASA of the talin1 F3 S1–S2 loop (residues 320–327) also decrease dramatically after 400 ns in ITK, although it only slightly fluctuates in IT (Fig. S4). Also, the distinct peak of the combined ITK density plot (i.e., ITK_a) from the peak of the combined IT density plot (i.e., IT_a) in talin-MP region distance and the SASA of the MP region clearly show the enhancement of talin-integrin binding mediated by kindlin in all various orientations and repetitions of ITK simulations (Figs. 6 E and S4 B). Therefore, the presence of kindlin2 resulted in significantly stronger interaction of talin1 with the integrin β3 MP region in ITK simulations compared to IT simulations. This is most probably an important factor in integrin activation because it can disrupt the IMC.
Furthermore, we studied the interactions of talin1 and kindlin2 with high-stress regions of integrin β3 over the course of the ITK simulation. We observed three important interactions between the F3 subdomain of talin1 and the MP region of integrin β3 (residues 723–736), namely 1) talin K323-β3 D723, 2) talin K321-β3 E731, and 3) talin T370-β3 A735 (Fig. 6 F). In addition, interactions formed between the F3 subdomain of kindlin2 and the NITY motif of integrin β3 (residues 756–759). Specifically, residues N616/N618 and I621 of kindlin2 interacted with I757 and Y759 of integrin β3, respectively, which remained stable for the last 50 ns of the ITK simulation (Fig. 6 G).
Distinct conformational changes of kindlin2 and talin1 in presence of each other
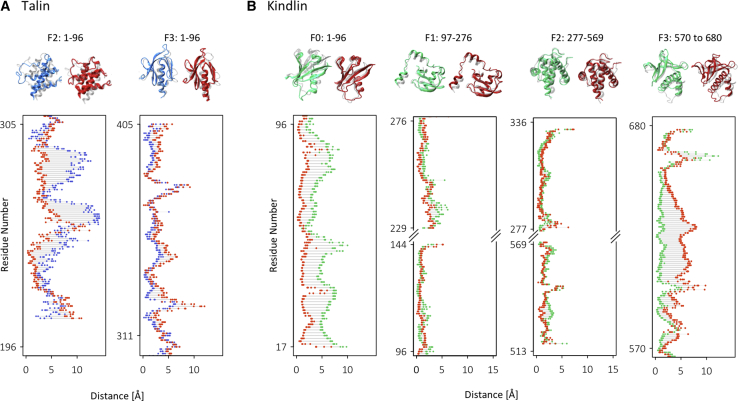
To determine whether any conformational changes were induced in kindlin2 or talin1 by their mutual interactions with integrin αIIbβ3, we performed structural alignments between the final structures of kindlin2 (at t = 760 ns) and talin1 (at t = 1000 ns) in each simulation, and their initial crystal structures. The structural alignments were performed using all subdomains that were simulated (i.e., F2–F3 for talin1 and F0–F3 for kindlin2). However, for clarity, the alignments are shown separately for each subdomain (Fig. 7). To quantify the deviation of each residue from its original position, the distance between residues of the initial and final structures were calculated after structural alignment was performed (Fig. 7). Our results show that the conformational changes observed in the F2 subdomain (residues 196–305) of talin1 are much higher in the IT simulation compared with the ITK simulation (Fig. 7 A). These conformational changes are mainly observed in two α-helices (∼residues 224–294) in the four-helix bundle of the F2 subdomain in the IT simulation. The conformational changes observed in the F3 subdomain of talin1 were similar between the IT and ITK conditions and mostly limited to the loop regions, as expected. The movement limitation imposed by simultaneous binding of the talin1 F3 domain with the integrin CT and the membrane (Fig. S5, A and C, left) can account for lower conformational changes of F3 compared to F2 in IT and ITK.
Figure 7.
Kindlin2 and talin1 structural alignment. (A) The final frames of talin1 in IT (blue) and ITK (red) were aligned to the initial PDB structure of talin1 (transparent). The distance shows the three-dimensional distance between each residue in the two structures after alignment. (B) The final frames of kindlin2 in IK (green) and ITK (red) were aligned to the initial PDB structure of kindlin2 (transparent) (by root mean-square deviation minimization methods). The distance shows the three-dimensional distance between each residue in the two structures after structural alignment. To see this figure in color, go online.
The structural alignment of kindlin2 showed significant conformational changes in the F0 subdomain in IK simulations and F3 in ITK simulations, whereas the F1 and F2 subdomains showed very small changes in both simulations. Previous work showed that the F0 subdomain of kindlin2 is highly dynamic and moves independently of F1–F3 subdomains. Also, the kindlin F0 subdomain is interacting with the membrane during our simulations and has a stronger and more stable interaction in ITK compared to IT in the last 200 ns of the simulations (Fig. S5 B, bottom and C, right). This stronger lipid interaction can account for lower conformational changes of F0 in ITK compared to IK. On the other hand, the conformational changes in F3 were seen in integrin-binding regions (i.e., residues 590–656) and are likely attributable to the distinct interactions of kindlin2 with integrin β3 in IK versus ITK simulations. These results suggest that conformational changes are induced in talin1 and kindlin2 upon interactions with integrin β3 and that these changes are distinct when both molecules are present.
Discussion
Integrin activation is central to a variety of cell functions, including cell-matrix adhesion, cell migration, and apoptosis. Kindlin and talin are both cytoplasmic proteins that directly interact with the integrin tail and induce a high-affinity state of integrin for extracellular ligands (3). Although talin can induce conformational changes in integrin, it has been shown that talin alone is not sufficient for the activation of integrin, and kindlin also plays a key role in this process (15,18). Although molecular mechanisms of talin-mediated integrin activation have been extensively studied, it is not yet clear how kindlin interferes with this process. Kindlin dysfunction has been associated with a number of human diseases, which are likely linked to deficiency in integrin activation; thus, knowing the molecular mechanism of kindlin-mediated integrin activation has potential impacts in therapeutic purposes. Here, we investigated the interplay between talin and kindlin in integrin activation using microsecond-long MD simulations (Fig. 1).
Previous studies have shown that kindlin binding to integrin is not sufficient but is required for triggering integrin activation (6,14,16). Moreover, Ma et al. (15) showed that simultaneous transfection of kindlin2 with the head domain of talin enhances integrin activation and identified kindlin2 as a coactivator of talin. However, the mechanisms of kindlin’s involvement remain elusive; in particular, it is still not known whether kindlin can induce conformational changes to integrin heterodimer. To address these questions and provide molecular details of the interaction among kindlin and talin with integrin, we developed all-atom microsecond-scale MD simulations of αIIbβ3 using an explicit lipid-water environment under three distinct scenarios: integrin in complex with talin1 (IT), kindlin2 (IK), and both talin1 and kindlin2 (ITK). We also performed a control simulation (I), in which only integrin was considered. This control simulation helps with better understanding of the effect of talin and kindlin interactions in integrin conformational changes (Fig. 8 A). Our IK simulation featuring integrin αIIbβ3 and kindlin2 showed that the main interactions that keep integrin in an inactive state, namely the IMC and OMC, remained stable over the course of 760 ns of the simulation when only kindlin is present (Figs. 3 B and 8 B). This further confirmed that kindlin binding is insufficient for unclasping the cytoplasm and transmembrane regions of αIIbβ3.
Figure 8.
Schematic representation of the proposed kindlin-mediated integrin activation mechanism. (A) Integrin remains in a closed conformation when it is not interacting with talin or kindlin (with stable and strong interactions in the IMC and OMC regions). (B) Kindlin alone is unable to disrupt the IMC or OMC interactions and, therefore, unable to induce conformational changes (i.e., angle changes between integrin α- and β-helices) in the transmembrane domain of integrin. (C) Talin binding to the integrin CT causes the complete disruption of OMC and destabilizes IMC interactions. Also, the angle between integrin α and β helices increases significantly upon talin binding. (D) Kindlin interaction with the membrane distal NPXY motif of integrin indirectly pushes talin toward the integrin MP region, resulting in a stronger interaction between talin and the MP motif and a complete disruption of IMC, OMC, and the parallelization of the transmembrane α- and β-helices. To see this figure in color, go online.
Interestingly, our simulations showed that kindlin2 binds to the integrin β3 tail more strongly in the absence of talin1, whereas talin1 binding to integrin is reinforced by kindlin2 (Fig. 5). Talin1 interaction was particularly enhanced with the MP region of integrin β3 (residues 720–736), although its interaction with the membrane proximal NPXY motif was gradually weakened over time. It should be noted that talin1 can still bind to the integrin CT in IT, and kindlin2 is not necessary for this interaction (Fig. 5, A and B). However, this interaction is mostly with the NPLY motif of integrin, and the interaction with the integrin MP region is very minimal (Fig. 6, D and E). Therefore, when kindlin2 is present, it indirectly pushes talin1 toward the membrane, resulting in a strong interaction with the MP region of integrin as well—thus enhancing the talin-integrin interaction—and a more effective disruption of the IMC in ITK compared to IT. This way, kindlin2 modifies the activation pathway by positioning talin1 more upward and closer to the membrane, resulting in stronger binding and a complete IMC disruption (Fig. 8 D). Also, kindlin2 decreased the intradomain conformational changes of talin1, especially in the F2 subdomain, which promoted both talin1-lipid and talin1-integrin β3 CT interactions (Fig. 7 A). Therefore, we suggest that the strength of kindlin2 binding to integrin does not necessarily correlate with activation, whereas stronger talin1 association can expedite the activation process. This further confirms the indirect role of kindlin2 as a cooperative partner for talin1 rather than a direct activator of integrin αIIbβ3. Although we did not observe a strong interaction between talin1 and the MP region of β3 in IT simulations, it is quite conceivable that even without kindlin2’s assistance, talin1 can eventually bind to the proximal region of integrin tail in a longer timescale.
On the other hand, the stronger binding of kindlin2 to integrin in the absence of talin1 observed in our simulations may have implications in talin-independent roles of kindlin in the formation and maturation of focal adhesions. Theodosiou et al. (16) showed that subsequent to the cooperative activation of integrin by talin and kindlin, kindlin initiates a talin-independent signaling pathway at new adhesion sites by binding to other molecules such as paxillin to induce cell spreading. Based on previous studies and our results, we propose that kindlin2 cooperates with talin1 to activate integrin by enhancing talin1’s interactions with the CT of integrin. On the other hand, kindlin2 can bind more strongly to integrin in the absence of talin1 to perform talin1-independent signaling subsequent to integrin activation (16).
It is now widely accepted that the IMC and OMC between integrin subunits maintain the resting state of integrin, and unclasping them triggers the inside-out signaling (6). In the IMC, R995 of β-integrin forms two salt bridges with D723 and E726 on α-integrin; however, it has been shown that disrupting the R995-D723 interaction is most critical for integrin activation (6). Our simulations indicated that talin1 disrupts R995-D723 interaction in both the presence and absence of kindlin2; however, kindlin2 is also required for dissociating R995-E726 interaction within 1 μs of simulation (Fig. 3 B).
Conversely, the OMC was destabilized in both cases (IT and ITK), causing significant conformational changes in the transmembrane region of integrin αIIbβ3 (Figs. 2, 3, and 8, C and D). The destabilization of OMC is highly correlated with the dramatic angle change between αIIb and β3 (θ) in both IT and ITK (Figs. 4 and S3). We noticed that the angle change in ITK was in reverse direction relative to that in IT, i.e., decreasing the angle in ITK and increasing the angle in IT (Fig. 2). Previous studies suggested that talin-mediated integrin activation involved an increase in the αIIb-β3 crossing angle and subsequent separation of the transmembrane domains (7,28). However, recent studies showed that transmembrane domains of integrin subunits were close to each other in all conformational snapshots, including active, inactive, and intermediate states, contradicting the notion of chain separation as an indicator of integrin activation (29,50). Because the angle change in ITK led to disruption of the IMC and OMC interactions, we propose that the degree of angle change is more pivotal in integrin activation than the direction of such change. This is most likely because talin1 and kindlin2 eliminate the inhibitory effects of the IMC and OMC, which allows for mechanical signals to be transmitted across the membrane without complete separation of the transmembrane regions. This is also demonstrated by changes in the force distribution patterns of the transmembrane domains of integrin in both IT and ITK simulations (Figs. 3 C and S2). Furthermore, we showed previously that residue A711 in the β3 helix is key for signal transmission across the transmembrane domain of integrin, and an A711P mutation can disrupt integrin activation (33). Our FDA analysis also indicated that A711 bears high forces in both IT and ITK, but not in IK (Fig. 3 C), implying that signal transmission is activated upon IMC disruption. Overall, our study suggests that not only are both talin1 and kindlin2 essential for efficiently initiating the activation process, but kindlin2 may also modify the molecular mechanism of inside-out signaling at the integrin β3 tail, which eventually leads to the opening of the integrin ectodomain.
Although not studied in this work, it is important to note that kindlin may have several other roles in integrin activation and the formation of focal adhesions. For example, one possible role of kindlin in integrin activation may be preventing the association of cytoplasmic inhibitors of integrin activation such as filamin or α-actinin to the integrin tail. Also, kindlin may be involved in integrin clustering through dimerization (32). Moreover, kindlin binding to actin can enhance cytoskeletal coupling to the membrane. Therefore, simultaneous binding of kindlin and talin to the integrin β increases the overall force applied to the integrin tail, which may facilitate focal adhesion formation and maturation.
Although our results focus on the integrin β3, the CTs of integrins are more than 60% similar to each other (Fig. S6). This high level of similarities with the conserved active sites between integrins shows the potential of having the same activation mechanisms and suggests that activation of different integrin receptors may at least be initiated in a similar manner. However, further computational studies are needed to uncover the mechanism of kindlin-mediated integrin activation in other members of integrin subfamilies.
Taken together, our results shed light on the molecular mechanisms by which kindlin2 cooperates with talin1 in integrin αIIbβ3 activation. This is the first computational study, to the best of our knowledge, that models the interplay between kindlin2 with talin1 in promoting inside-out signaling through integrin binding.
Author Contributions
All authors contributed to the design and implementation of the project and the computational experiments. All authors provided critical feedback in all steps of the project, which shaped the final research. Z.H. conducted the MD simulations with inputs from H.S. and Z.J. All authors were involved in postprocessing and analysis of the data and writing of the manuscript. M.R.K.M. supervised the project and contributed materials and analysis tools.
Acknowledgments
We thank Mehrdad Mehrbod for fruitful conversations in the early stages of this research.
This research was supported by the National Science Foundation through grant CMMI-1538707. Additionally, this research used resources of the Extreme Science and Engineering Discovery Environment (XSEDE) supercomputing facilities, supported by the National Science Foundation (NSF) grant No. ACI-1053575.
Editor: Guy Genin.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.02.023.
Supporting Material
References
- 1.Humphries J.D., Chastney M.R., Humphries M.J. Signal transduction via integrin adhesion complexes. Curr. Opin. Cell Biol. 2019;56:14–21. doi: 10.1016/j.ceb.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Kim M., Carman C.V., Springer T.A. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 3.Luo B.-H., Carman C.V., Springer T.A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong J.-P., Stehle T., Arnaout M.A. New insights into the structural basis of integrin activation. Blood. 2003;102:1155–1159. doi: 10.1182/blood-2003-01-0334. [DOI] [PubMed] [Google Scholar]
- 5.Nieswandt B., Varga-Szabo D., Elvers M. Integrins in platelet activation. J. Thromb. Haemost. 2009;7(Suppl 1):206–209. doi: 10.1111/j.1538-7836.2009.03370.x. [DOI] [PubMed] [Google Scholar]
- 6.Bledzka K., Qin J., Plow E.F. Integrin αIIbβ3. Platelets. 2019:227–241. [Google Scholar]
- 7.Mehrbod M., Trisno S., Mofrad M.R.K. On the activation of integrin αIIbβ3: outside-in and inside-out pathways. Biophys. J. 2013;105:1304–1315. doi: 10.1016/j.bpj.2013.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jahed Z., Shams H., Mofrad M.R.K. Mechanotransduction pathways linking the extracellular matrix to the nucleus. Int. Rev. Cell Mol. Biol. 2014;310:171–220. doi: 10.1016/B978-0-12-800180-6.00005-0. [DOI] [PubMed] [Google Scholar]
- 9.Cheng B., Wan W., Huang G., Li Y., Genin G., Mofrad M.R.K., Lu T.J., Xu F., Lin M. Nanoscale integrin cluster dynamics controls cellular mechanosensing via FAK Y397 phosphorylation. Science Advance. 2020;6 doi: 10.1126/sciadv.aax1909. eaax1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ye F., Kim C., Ginsberg M.H. Reconstruction of integrin activation. Blood. 2012;119:26–33. doi: 10.1182/blood-2011-04-292128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Z., Costell M., Fässler R. Integrin activation by talin, kindlin and mechanical forces. Nat. Cell Biol. 2019;21:25–31. doi: 10.1038/s41556-018-0234-9. [DOI] [PubMed] [Google Scholar]
- 12.Ye F., Hu G., Ginsberg M.H. Recreation of the terminal events in physiological integrin activation. J. Cell Biol. 2010;188:157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shattil S.J., Kim C., Ginsberg M.H. The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol. 2010;11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plow E.F., Qin J. The Kindlin Family of Adapter Proteins. Circ. Res. 2019;124:202–204. doi: 10.1161/CIRCRESAHA.118.314362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Y.-Q., Qin J., Plow E.F. Kindlin-2 (Mig-2): a co-activator of beta3 integrins. J. Cell Biol. 2008;181:439–446. doi: 10.1083/jcb.200710196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theodosiou M., Widmaier M., Fässler R. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife. 2016;5:e10130. doi: 10.7554/eLife.10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kammerer P., Aretz J., Fässler R. Lucky kindlin: a cloverleaf at the integrin tail. Proc. Natl. Acad. Sci. USA. 2017;114:9234–9236. doi: 10.1073/pnas.1712471114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plow E.F., Qin J., Byzova T. Kindling the flame of integrin activation and function with kindlins. Curr. Opin. Hematol. 2009;16:323–328. doi: 10.1097/MOH.0b013e32832ea389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moser M., Nieswandt B., Fässler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat. Med. 2008;14:325–330. doi: 10.1038/nm1722. [DOI] [PubMed] [Google Scholar]
- 20.Ye F., Snider A.K., Ginsberg M.H. Talin and kindlin: the one-two punch in integrin activation. Front. Med. 2014;8:6–16. doi: 10.1007/s11684-014-0317-3. [DOI] [PubMed] [Google Scholar]
- 21.Elliott P.R., Goult B.T., Barsukov I.L. The structure of the talin head reveals a novel extended conformation of the FERM domain. Structure. 2010;18:1289–1299. doi: 10.1016/j.str.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bledzka K., Liu J., Plow E.F. Spatial coordination of kindlin-2 with talin head domain in interaction with integrin β cytoplasmic tails. J. Biol. Chem. 2012;287:24585–24594. doi: 10.1074/jbc.M111.336743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalli A.C., Campbell I.D., Sansom M.S.P. Conformational changes in talin on binding to anionic phospholipid membranes facilitate signaling by integrin transmembrane helices. PLoS Comput. Biol. 2013;9:e1003316. doi: 10.1371/journal.pcbi.1003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau T.L., Kim C., Ulmer T.S. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009;28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calderwood D.A., Campbell I.D., Critchley D.R. Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 2013;14:503–517. doi: 10.1038/nrm3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurtz L., Kao L., Zhu Q. Integrin αIIbβ3 inside-out activation: an in situ conformational analysis reveals a new mechanism. J. Biol. Chem. 2012;287:23255–23265. doi: 10.1074/jbc.M112.360966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehrbod M., Mofrad M.R.K. Localized lipid packing of transmembrane domains impedes integrin clustering. PLoS Computational Biology. 2013;9:e1002948. doi: 10.1371/journal.pcbi.1002948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim C., Ye F., Ginsberg M.H. Talin activates integrins by altering the topology of the β transmembrane domain. J. Cell Biol. 2012;197:605–611. doi: 10.1083/jcb.201112141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jamali Y., Jamali T., Mofrad M.R.K. An agent based model of integrin clustering: exploring the role of ligand clustering, integrin homo-oligomerization, integrin-ligand affinity, membrane crowdedness and ligand mobility. J. Comput. Phys. 2013;244:264–278. [Google Scholar]
- 30.Truong T., Shams H., Mofrad M.R.K. Mechanisms of integrin and filamin binding and their interplay with talin during early focal adhesion formation. Integr. Biol. 2015;7:1285–1296. doi: 10.1039/c5ib00133a. [DOI] [PubMed] [Google Scholar]
- 31.Kalli A.C., Wegener K.L., Sansom M.S.P. The structure of the talin/integrin complex at a lipid bilayer: an NMR and MD simulation study. Structure. 2010;18:1280–1288. doi: 10.1016/j.str.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Deng Y., Yu C. Structural basis of kindlin-mediated integrin recognition and activation. Proc. Natl. Acad. Sci. USA. 2017;114:9349–9354. doi: 10.1073/pnas.1703064114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shams H., Mofrad M.R.K. α-Actinin induces a kink in the transmembrane domain of β3-integrin and impairs activation via talin. Biophys. J. 2017;113:948–956. doi: 10.1016/j.bpj.2017.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Golji J., Mofrad M.R.K. The interaction of vinculin with actin. PLoS Comput. Biol. 2013;9:e1002995. doi: 10.1371/journal.pcbi.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golji J., Lam J., Mofrad M.R.K. Vinculin activation is necessary for complete talin binding. Biophys. J. 2011;100:332–340. doi: 10.1016/j.bpj.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S.E., Chunsrivirot S., Mofrad M.R.K. Molecular dynamics study of talin-vinculin binding. Biophys. J. 2008;95:2027–2036. doi: 10.1529/biophysj.107.124487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weljie A.M., Hwang P.M., Vogel H.J. Solution structures of the cytoplasmic tail complex from platelet integrin alpha IIb- and beta 3-subunits. Proc. Natl. Acad. Sci. USA. 2002;99:5878–5883. doi: 10.1073/pnas.092515799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calderwood D.A., Yan B., Ginsberg M.H. The phosphotyrosine binding-like domain of talin activates integrins. J. Biol. Chem. 2002;277:21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 39.Moes M., Rodius S., Kieffer N. The integrin binding site 2 (IBS2) in the talin rod domain is essential for linking integrin β subunits to the cytoskeleton. J. Biol. Chem. 2007;282:17280–17288. doi: 10.1074/jbc.M611846200. [DOI] [PubMed] [Google Scholar]
- 40.Anthis N.J., Wegener K.L., Campbell I.D. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 2009;28:3623–3632. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 42.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 44.Bengtsson H., Bravo H.C., Montgomery B. 2019. Functions that apply to rows and columns of matrices (and to vectors) [Google Scholar]
- 45.Package ‘gplots’ Title Various R Programming Tools for Plotting Data. 2019.
- 46.Grant B.J., Rodrigues A.P.C., Caves L.S.D. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 47.Costescu B.I., Gräter F. Time-resolved force distribution analysis. BMC Biophys. 2013;6:5. doi: 10.1186/2046-1682-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J., Wang Z., Zhu J. The dual structural roles of the membrane distal region of the α-integrin cytoplasmic tail during integrin inside-out activation. J. Cell Sci. 2015;128:1718–1731. doi: 10.1242/jcs.160663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo J., Zhang Y., Xu C. Intramembrane ionic protein-lipid interaction regulates integrin structure and function. PLoS Biol. 2018;16:e2006525. doi: 10.1371/journal.pbio.2006525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eng E.T., Smagghe B.J., Springer T.A. Intact alphaIIbbeta3 integrin is extended after activation as measured by solution X-ray scattering and electron microscopy. J. Biol. Chem. 2011;286:35218–35226. doi: 10.1074/jbc.M111.275107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.