Abstract
Various mesenchymal stem cells as easily accessible and multipotent cells can share different essential signaling pathways related to their stemness ability. Understanding the mechanism of stemness ability can be useful for controlling the stem cells for regenerative medicine targets. In this context, OMICs studies can analyze the mechanism of different stem cell properties or stemness ability via a broad range of current high-throughput techniques. This field is fundamentally directed toward the analysis of whole genome (genomics), mRNAs (transcriptomics), proteins (proteomics) and metabolites (metabolomics) in biological samples. According to several studies, metabolomics is more effective than other OMICs ّfor various system biology concerns. Metabolomics can elucidate the biological mechanisms of various mesenchymal stem cell function by measuring their metabolites such as their secretome components. Analyzing the metabolic alteration of mesenchymal stem cells can be useful to promote their regenerative medicine application.
Key Words: Mesenchymal stem cells, metabolic pathways, metabolomics, systems biology
Two main properties of stem cells are including prolonged self- renewal and multi-potent differentiation capacity which make them ideal candidate for cell therapy and regenerative medicine (1-5). Related to these properties, stem cells share several essential genes and signaling pathways (i.e. Hedgehog, Wnt, Notch, phosphatidylinositol 3-kinase/ phosphatase, and nuclear factor-κB signaling pathways) as stemness ability (6-8). In other word, stem cells can preserve their lineage, interaction with the environment, and cross-talk with adjacent cells to keep a balance between repose, proliferation, and restoration, through stemness ability (9-11). However, understanding the mechanism of stemness ability is challenging (9). According to several studies, stable, safe, and more accessible stem cells are considered as an excellent choice for regenerative medicine. In this context, mesenchymal stem cells (MSCs) (as easily accessible, self-renewable, and multipotent cells with few consideration ethics) have significant efficacy in regenerative medicine. (12-26). Furthermore, recent development in OMICs approaches (technologies for understanding the whole activity of cells, tissues, and organs at the molecular level) specifically metabolomics approaches (extensive analysis of metabolites in cells, tissues, and organs) can increase our understanding about the self-renewal and differentiation mechanisms. On the other hand, analysis of chemical alterations related to natural processes of living cells including growth, environmental adaptation, and differentiation can be provided by metabolomics methods (27-29).
OMICs - based stem cell monitoring
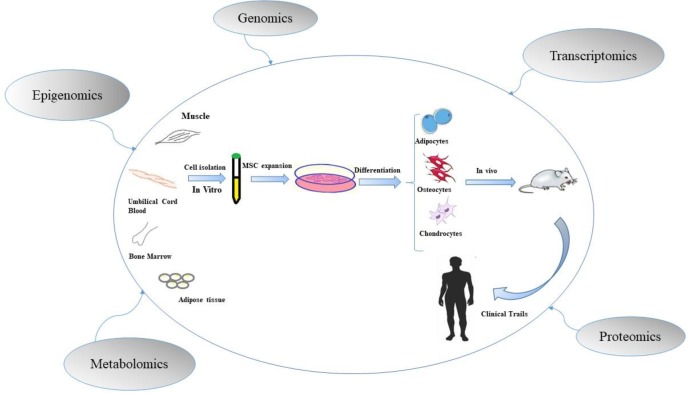
Multi- OMICs approaches including geno-mics, epigenomics, transcriptomics, proteomics, and metabolomics are functional methods to study stem cell biology and its therapeutic application (Fig.1) (30-32).
Fig. 1.
Based stem cell monitoring. Multi- OMICs approaches are functional methods to study stem cell biology and its therapeutic application through evaluation of molecular mechanisms of stem cells properties and quantification of cellular products (33).
At first, human genome project has led to the advancement of genome sequencing and study on DNA by analysis of single nucleotide polymorphisms (SNPs), variation copies, and mutations (34-36). Nowadays, genomics as the most mature approache of OMICs and next generation sequencing (NGS) as the latest technology in this field are used for high-throughput detection and cost effective analysis of biological data (37-40). On the other hand, epigenetic modifications (e.g. methylation and histone acetylation) have an important role in differentiation and development of stem cells (41, 42). The study of heritable modifications (not sequence changes) of DNA is called epigenomics (43, 44). Additionally, qualitative and quantitative transcriptomics can facilitate the investigation of RNAs in stem cells, via molecular and cellular methods such as micro-array and RNA-sequencing (45, 46). It also has a vital role in analyzing key genes and pathways that participate in self-renewal, proliferation, and differentiation of stem cells (47-49). Some transcription factors (related to non-coding RNAs) such as octamer-binding transcription factor 4 (OCT 4) and NANOG can regulate pluripotency feature of stem cells (50, 51). Proteomics tries to evaluate the qualitative and quantitative changes in proteins and identify new markers in stem cell development stages (52, 53). Finally, metabolomics measures and demonstrates the products of metabolism such as amino-acids and fatty-acids. In this respect, metabolomics is an accurate approach to recognize metabolite biomarkers in biological samples (54, 55). Although, application of OMICs, especially metabolomics, for monitoring of stem cell in researches and therapies is in its infancy period, it can be useful to understand different features of cell-based therapy (1, 56).
Stem cells metabolomics
Because of the self-renewal and differentiation properties of stem cells, they can be applied for regenerative medicine, drug screening, toxicity testing, and evaluation of disease phenotypes (57-59). Although they are metabolically inactive population in quiescent state, their metabolic activity increases during differentiation (60). Stem cells niche can preserve them in a quiescent state to maintain their self-renewal ability (61, 62). In other words, morphogens and growth factors in the niche of stem cells can change the regulation of stem cells through numerous metabolic pathways (1, 63, 64). Moreover, molecular mechanisms can regulate differentiation and reprogramming, and also they can control the energy of metabolism in stem cells throughout glycolytic or oxidative phosphorylation (OXPHOS) reactions (1, 65, 66). In other words, changes in glycolysis and OXPHOS have impact on differentiation or reprogramming of stem cells (66-68). Glycolysis and OXPHOS changes can alter the metabolite levels and reduction–oxidation (redox) state (69-71). Subsequently, hypoxia, glycolysis and redox states can affect the homeostasis and regeneration of stem cells (67, 72, 73). For instance, hypoxia has a key role in maintaining undifferentiated state of stem cells by reducing redox state (74-76). For preparing a balance between self-renewal and differentiation ability, the role of redox state can be important (77, 78). Moreover, the increase of reactive oxygen species (ROS) can promote cell differentiation (74, 79). Herein, understanding the mechanism of stem cells (e.g. MSCs) function is momentous for in vitro and in vivo studies and also the stem cells application in cell therapy.
Metabolomics- based comparison of mesenchy-mal stem cells
MSCs as multi-potent stem cells can be extracted from different sources. Their intrinsic properties have drawn the attention for developing more comprehensive studies (13, 14). Moreover, realizing the biological mechanisms of their function can be helpful for developing stem cell researches. Accordingly, metabolomics as a valuable tool for stem cell monitoring can clarify the biological mechanisms of MSCs function through assaying metabolites. Metabolites of MSCs are involved in metabolic or signaling pathways (80-82). Metabolic pathways produce vital signals for the self-renewal, differentiation and other properties of MSCs. On the other hand, undifferentiated state and differentiated state of MSCs can be distinguished via their metabolic profile. Accordingly, in undifferentiated state, mitochondrial OXPHOS is maintained at a low level, while the glycolytic function is maintained at a high level (81, 83). Additionally, in the early phase of MSCs differentiation, down-regulation of some pluripotent genes, up-regulation of terminal genes, and changing the subsets of metabolic enzymes can redirect the new fate of cells. Furthermore, in normoxic states, the proliferation and colony-forming abilities of MSCs are considerably increased (84, 85). In other words, hypoxic condition restricts MSC proliferation to maintain long-term self-renewal capacity. Generally, metabolomics can analyze the rapid kinetics and dynamics of metabolic reactions in different MSCs (86-88). Different types of MSCs share various properties due to their gene expression profile. Additionally, MSCs from various sources have also various secretome and metabolic profile (89, 90).
Metabolomics analysis of mesenchymal stem cells secretome
MSCs have demonstrated a pivotal and therapeutic impact on several diseases by producing a broad spectrum of autocrine and paracrine secretion factors (secretome) (15, 81, 91). The characterization of the MSCs secretome can elucidate their activation mechanism (92). Accordingly, metabolomics analyses can decipher the mechanism of secretome component functions (93). MSCs conditioned media (MSCs-CM) and extracellular vesicles (EVs) are two main MSC-sourced secretome.
Metabolomics study of mesenchymal stem cells
conditioned media
MSCs-CM encompasses multiple growth factors (GFs), metabolites, and cytokines. It can be prepared through 4 steps including isolation and characterization of cells, culture of cells in a proper culture medium, cell expansion, and CM collection (94, 95). Additionally, it has been shown that MSCs-CM can improve various pathophysiology hallmarks of diseases e.g. lung injury, skin wound, Alzheimer’s disease, and Parkinson’s disease. For instance, there are some anti-inflammatory cytokines in MSC-CM (i.e. ciliary neurotrophic factor (CNTF), transforming growth factor 1 (TGF1), neurotrophin 3 (NT-3) factor, interleukin (IL) 13, IL18 binding protein (IL18BP), IL10, IL17E, IL27 or IL1 receptor antagonist (IL1RA)), and also some pro-inflammatory cytokines (including IL1b, IL6, IL8, and IL9) (95, 96). The equilibrium between these two types of cytokines can mediate the anti-inflammatory impact of MSC-CM. On the other hand, MSC-CM has anti-apoptotic activity via reducing the pro-apoptotic factors and increasing the expression of pro-angiogenic factors. Metabolomics can support quantification of MSC-CM metabolites by different targeted and non-targeted methods (91).
Metabolomics profiling of mesenchymal stem cells derived extracellular vesicles
EVs including exosomes and micro -vesicles can be secreted by cells which have an important role in intercellular signaling pathways (15, 97). It has been confirmed that MSC-EVs specifically MSCs-derived exosomes (MSC-Exo) can imitate their origin MSCs therapeutic effects in improvement of different disorders. MSC-EVs carry lipids, genetic materials (mRNA and non-coding RNA), and proteins. Moreover, they can be characterized by some surface markers such as CD29, CD73, CD44, and CD105. On the other hand, it is remarkable that MSCs- EVs from different MSC sources have also different composition (98). Namely, menstrual fluid derived MSCs -Exo has greater neurite outgrowth response than bone marrow (BM), chorion, and umbilical cord-derived MSCs. Metabolomics techniques can be used to analyze the mechanism of different MSC-EVs activity based on their different metabolic profile (99).
Analytical techniques in metabolomics analysis
Metabolomics can assay the metabolite compositions of cells and biological fluids through various targeted and non- targeted techniques (100, 101). A broad range of analytical methods containing capillary electrophoresis (CE) (the separation method in which metabolites are separated based on their migration in the electrical field of the capillary tube), gas chromatography (GC) (a method for separating volatile matters), ultra-performance liquid chromatography (UPLC) (as a modern liquid chromatography method can be used for particles less than 2 µl in diameter), and high performance or high-pressure liquid chromatography (HPLC) (the highly advanced form of column chromatography which pumps the sample of metabolites in mobile phase at high
pressure within a column or the stationary phase) linked to high-throughput techniques including nuclear magnetic resonance (NMR) (a spectroscopic procedure to follow local strong stationary magnetic fields around atomic nuclei which is for absorbing very high-frequency radio waves) and mass spectrometry (MS) (an analytical manner to ionizing chemical samples to identity unknown composites and chemical features of different molecules based on their mass-to-charge ratio) can be used for separation, examination, and quantification of the cellular metabolites composition as metabolomics approaches (107, 112-114). Each of the metabolo-mics approaches has some advantages and disadvantages (Table 1).
Table 1.
Advantages and disadvantages of metabolomics techniques
| Method | Advantages | Disadvantages | References |
|---|---|---|---|
| NMR | - Simple sample preparation -Excellent reproducibility -Quantify a wide-range of organic compounds in the micro-molar range |
-Low sensitivity compared with MS methods - Suitable for quantification of metabolites present in relatively high concentration |
(102, 103) |
| GC-MS | - High separation efficiency - The oldest and a robust tool for qualitative metabolic profiling |
-Non-volatile matrices require additional preparation - Some gases are challenging (CO2, N2, O2, Ar, CO, H2O) |
(104, 105) |
| LC-MS | - High separation efficiency - No derivatization is needed for the analysis of polar or high molecular weight metabolites - Quick analysis of small samples |
- Ion suppression | (103, 106) |
| CE-MS | -Suitable for the separation of polar and charged compounds - Powerful for charged metabolites -High-analyte resolution – providing information mainly on polar or ionic compounds -Short analysis time -Very small sample requirement |
- Poor concentration sensitivity | (107, 108) |
| HPLC-MS | -Robustness -Ease of use - Good selectivity -Adjustable sensitivity |
-Lack of efficiency due to low diffusion coefficients in liquid phase | (109, 110) |
| UPLC-MS | -Powerful technique in biomolecular research - Covers a number of polar metabolites and enlarges the number of detected analytes -Better efficiency with speedy analysis |
Less time life of columns | (107, 111) |
Acknowledgment
The authors would like to thank Dr. Mohsen Khorshidi and Shokouh Salimi for their considera-ble assistance.
Conclusion and future perspectives
Metabolomics is an impressive research area, which can be used for screening the metabolic modifications during the stem cells reprogramming, proliferation, and differentiation (56, 115). Indeed, screening the metabolic modifications of stem cells (e.g. MSCs) can facilitate their application for regenerative medicine purposes via increasing the man control over in vitro manipulation of stem cells including tissue-specific stem cells activation, and promote stem cells for migration to the side of tissue injury. Based on researches, some important metabolic elements can be used to dedifferentiate stem cells toward organ-specific somatic cells (116). Accordingly, in the coming future it seems that the application of generated knowledge on metabolic key methods can be useful for therapeutic targets without the necessity of genetic manipulation. On the other hand, combination of metabolomics technology with other technologies (i.e. genomics, proteomics, structural biology and imaging) can increase its performance to identify novel biological pathways in mechanism of stem cell function, and also to identify disease mechanism (39, 117). Additionally, progress in the development of metabolite databases and in silico fragmentation tools can pave the way for large-scale metabolomics analysis (118, 119).
Conflict of interest
Authors declare no conflict of interest.
References
- 1.Shyh-Chang N, Ng HH. The metabolic programming of stem cells. Genes Dev. 2017;31:336–46. doi: 10.1101/gad.293167.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biehl JK, Russell B. Introduction to stem cell therapy. J Cardiovasc Nurs. 2009;24:98–103; quiz 4-5. doi: 10.1097/JCN.0b013e318197a6a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahim S, Rahim F, Shirbandi K, et al. Sports Injuries: Diagnosis, Prevention, Stem Cell Therapy, and Medical Sport Strategy. Adv Exp Med Biol. 2018 doi: 10.1007/5584_2018_298. [DOI] [PubMed] [Google Scholar]
- 4.Goodarzi P, Falahzadeh K, Aghayan H, et al. Therapeutic abortion and ectopic pregnancy: alternative sources for fetal stem cell research and therapy in Iran as an Islamic country. Cell Tissue Bank. 2019;20:11–24. doi: 10.1007/s10561-018-9741-y. [DOI] [PubMed] [Google Scholar]
- 5.Rahim F, Arjmand B, Shirbandi K, et al. Stem cell therapy for patients with diabetes: a systematic review and meta-analysis of metabolomics-based risks and benefits. Stem Cell Investig. 2018;5:40. doi: 10.21037/sci.2018.11.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsui WH. Cancer stem cell signaling pathways. Medicine (Baltimore) 2016;95:S8–S19. doi: 10.1097/MD.0000000000004765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nwabo Kamdje AH, Takam Kamga P, Tagne Simo R, et al. Developmental pathways associated with cancer metastasis: Notch, Wnt, and Hedgehog. Cancer Biol Med. 2017;14:109–20. doi: 10.20892/j.issn.2095-3941.2016.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCubrey JA, Rakus D, Gizak A. Effects of mutations in Wnt/β-catenin, hedgehog, Notch and PI3K pathways on GSK-3 activity—Diverse effects on cell growth, metabolism and cancer. Biochim Biophys Acta Mol Cell Res. 2016;1863:2942–76. doi: 10.1016/j.bbamcr.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Aponte PM, Caicedo A. Stemness in Cancer: Stem Cells, Cancer Stem Cells, and Their Microenvironment. Stem Cells Int. 2017;2017:5619472. doi: 10.1155/2017/5619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225–38. doi: 10.1016/j.stem.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chosa N, Ishisaki A. Two novel mechanisms for maintenance of stemness in mesenchymal stem cells: SCRG1/BST1 axis and cell-cell adhesion through N-cadherin. Jpn Dent Sci Rev. 2018;54:37–44. doi: 10.1016/j.jdsr.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ullah I, Subbarao RB, Rho GJ. Human mesenchymal stem cells - current trends and future prospective. Biosci Rep. 2015:35. doi: 10.1042/BSR20150025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schafer R, Spohn G, Baer PC. Mesenchymal Stem/Stromal Cells in Regenerative Medicine: Can Preconditioning Strategies Improve Therapeutic Efficacy? Transfus Med Hemother. 2016;43:256–67. doi: 10.1159/000447458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodarzi P, Larijani B, Alavi-Moghadam S, et al. Mesenchymal Stem Cells-Derived Exosomes for Wound Regeneration. Adv Exp Med Biol. 2018;1119:119–31. doi: 10.1007/5584_2018_251. [DOI] [PubMed] [Google Scholar]
- 15.Frese L, Dijkman PE, Hoerstrup SP. Adipose Tissue-Derived Stem Cells in Regenerative Medicine. Transfus Med Hemother. 2016;43:268–74. doi: 10.1159/000448180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payab M, Goodarzi P, Foroughi Heravani N, et al. Stem Cell and Obesity: Current State and Future Perspective. Adv Exp Med Biol. 2018;1089:1–22. doi: 10.1007/5584_2018_227. [DOI] [PubMed] [Google Scholar]
- 17.Goodarzi P, Alavi-Moghadam S, Sarvari M, et al. Adipose Tissue-Derived Stromal Cells for Wound Healing. Adv Exp Med Biol. 2018;1119:133–49. doi: 10.1007/5584_2018_220. [DOI] [PubMed] [Google Scholar]
- 18.Derakhshanrad N, Saberi H, Tayebi Meybodi K, et al. Case Report: Combination Therapy with Mesenchymal Stem Cells and Granulocyte-Colony Stimulating Factor in a Case of Spinal Cord Injury. Basic Clin Neurosci. 2015;6:299–305. [PMC free article] [PubMed] [Google Scholar]
- 19.Larijani B, Aghayan H, Goodarzi P. Clinical Grade Human Adipose Tissue-Derived Mesenchymal Stem Cell Banking. Acta Med Iran. 2015;53:540–6. [PubMed] [Google Scholar]
- 20.Shirian S, Ebrahimi-Barough S, Saberi H, et al. Comparison of Capability of Human Bone Marrow Mesenchymal Stem Cells and Endometrial Stem Cells to Differentiate into Motor Neurons on Electrospun Poly(epsilon-caprolactone) Scaffold. Mol Neurobiol. 2016;53:5278–87. doi: 10.1007/s12035-015-9442-5. [DOI] [PubMed] [Google Scholar]
- 21.Goodarzi P, Aghayan HR, Larijani B, et al. Stem cell-based approach for the treatment of Parkinson's disease. Med J Islam Repub Iran. 2015;29 [PMC free article] [PubMed] [Google Scholar]
- 22.Aghayan HR, Goodarzi P, Arjmand B. GMP-compliant human adipose tissue-derived mesenchymal stem cells for cellular therapy. Methods Mol Biol. 2015;1283:93–107. doi: 10.1007/7651_2014_112. [DOI] [PubMed] [Google Scholar]
- 23.Larijani B, Aghayan HR, Goodarzi P, et al. GMP-grade human fetal liver-derived mesenchymal stem cells for clinical transplantation. Methods Mol Biol. 2015;1283:123–36. doi: 10.1007/7651_2014_101. [DOI] [PubMed] [Google Scholar]
- 24.Arjmand B, Aghayan HR. Cell manufacturing for clinical applications. Stem Cells. 2014;32:2557–8. doi: 10.1002/stem.1751. [DOI] [PubMed] [Google Scholar]
- 25.Goodarzi P, Falahzadeh K, Nematizadeh M, et al. Tissue Engineered Skin Substitutes. Adv Exp Med Biol. 2018;1107:143–88. doi: 10.1007/5584_2018_226. [DOI] [PubMed] [Google Scholar]
- 26.Larijani B, Arjmand B, Ahmadbeigi N, et al. A simple and cost-effective method for isolation and expansion of human fetal pancreas derived mesenchymal stem cells. Arch Iran Med. 2015;18:770–5. [PubMed] [Google Scholar]
- 27.Panopoulos AD, Yanes O, Ruiz S, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–77. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng B, Li H, Peng XX. Functional metabolomics: from biomarker discovery to metabolome reprogramming. Protein Cell. 2015;6:628–37. doi: 10.1007/s13238-015-0185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Izpisua Belmonte JC. Stem Cells: A Renaissance in Human Biology Research. Cell. 2016;165:1572–85. doi: 10.1016/j.cell.2016.05.043. [DOI] [PubMed] [Google Scholar]
- 30.Singh H. Multi-omics approach to stem cell studies. Minerva Biotecno. 2017;29:169–73. [Google Scholar]
- 31.Suravajhala P, Kogelman LJA, Kadarmideen HN. Multi-omic data integration and analysis using systems genomics approaches: methods and applications in animal production, health and welfare. Genet Select Evol. 2016;48:38. doi: 10.1186/s12711-016-0217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler J, Sotiriadou I, Chen S. The potential of embryonic stem cells combined with-omics technologies as model systems for toxicology. Curr Med Chem. 2009;16:4814–27. doi: 10.2174/092986709789909657. [DOI] [PubMed] [Google Scholar]
- 33.Naidoo N, Pawitan Y, Soong R, et al. Human genetics and genomics a decade after the release of the draft sequence of the human genome. Hum Genomics. 2011;5:577–622. doi: 10.1186/1479-7364-5-6-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simonti CN, Capra JA. The evolution of the human genome. Curr Opin Genet Dev. 2015;35:9–15. doi: 10.1016/j.gde.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson MW, Schrijver I. Next generation DNA sequencing and the future of genomic medicine. Genes (Basel) 2010;1:38–69. doi: 10.3390/genes1010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Churko JM, Mantalas GL, Snyder MP, et al. Overview of high throughput sequencing technologies to elucidate molecular pathways in cardiovascular diseases. Circ Res. 2013;112:1613–23. doi: 10.1161/CIRCRESAHA.113.300939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohashi H, Hasegawa M, Wakimoto K, et al. Next-generation technologies for multiomics approaches including interactome sequencing. Biomed Res Int. 2015;2015:104209. doi: 10.1155/2015/104209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaitankar V, Karakulah G, Ratnapriya R, et al. Next generation sequencing technology and genomewide data analysis: Perspectives for retinal research. Prog Retin Eye Res. 2016;55:1–31. doi: 10.1016/j.preteyeres.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Podobinska M, Szablowska-Gadomska I, Augustyniak J, et al. Epigenetic Modulation of Stem Cells in Neurodevelopment: The Role of Methylation and Acetylation. Front Cell Neurosci. 2017;11:23. doi: 10.3389/fncel.2017.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boland MJ, Nazor KL, Loring JF. Epigenetic regulation of pluripotency and differentiation. Circ Res. 2014;115:311–24. doi: 10.1161/CIRCRESAHA.115.301517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123:2145–56. doi: 10.1161/CIRCULATIONAHA.110.956839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trerotola M, Relli V, Simeone P, et al. Epigenetic inheritance and the missing heritability. Hum Genomics. 2015;9:17. doi: 10.1186/s40246-015-0041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roson-Burgo B, Sanchez-Guijo F, Del Canizo C, et al. Transcriptomic portrait of human Mesenchymal Stromal/Stem Cells isolated from bone marrow and placenta. BMC Genomics. 2014;15:910. doi: 10.1186/1471-2164-15-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miura S, Himaki T, Takahashi J, et al. THE ROLE OF TRANSCRIPTOMICS: PHYSIOLOGICAL EQUIVALENCE BASED ON GENE EXPRESSION PROFILES. Reviews in Agricultural Science. 2017;5:21–35. [Google Scholar]
- 46.Yeo JC, Ng HH. Transcriptomic analysis of pluripotent stem cells: insights into health and disease. Genome Med. 2011;3:68. doi: 10.1186/gm284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Churko JM, Lee J, Ameen M, et al. Transcriptomic and epigenomic differences in human induced pluripotent stem cells generated from six reprogramming methods. Nat Biomed Eng. 2017;1:826–37. doi: 10.1038/s41551-017-0141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chin CJ, Li S, Corselli M, et al. Transcriptionally and Functionally Distinct Mesenchymal Subpopulations Are Generated from Human Pluripotent Stem Cells. Stem Cell Reports. 2018;10:436–46. doi: 10.1016/j.stemcr.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huo JS, Zambidis ET. Pivots of pluripotency: the roles of non-coding RNA in regulating embryonic and induced pluripotent stem cells. Biochim Biophys Acta. 2013;1830:2385–94. doi: 10.1016/j.bbagen.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakamoto N, Honma R, Sekino Y, et al. Non-coding RNAs are promising targets for stem cell-based cancer therapy. Noncoding RNA Res. 2017;2:83–7. doi: 10.1016/j.ncrna.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Hoof D, Krijgsveld J, Mummery C. Proteomic analysis of stem cell differentiation and early development. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a008177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pripuzova NS, Getie-Kebtie M, Grunseich C, et al. Development of a protein marker panel for characterization of human induced pluripotent stem cells (hiPSCs) using global quantitative proteome analysis. Stem Cell Res. 2015;14:323–38. doi: 10.1016/j.scr.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17:451–9. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deidda M, Piras C, Bassareo PP. Metabolomics, a promising approach to translational research in cardiology. IJC Metabolic & Endocrine. 2015;9:31–8. [Google Scholar]
- 55.Bhute VJ, Bao X, Palecek SP. Advances in Applications of Metabolomics in Pluripotent Stem Cell Research. Curr Opin Chem Eng. 2017;15:36–43. doi: 10.1016/j.coche.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peffers MJ, Collins J, Fang Y, et al. Age-related changes in mesenchymal stem cells identified using a multi-omics approach. Eur Cell Mater. 2016;31:136–59. doi: 10.22203/ecm.v031a10. [DOI] [PubMed] [Google Scholar]
- 57.Mahla RS. Stem Cells Applications in Regenerative Medicine and Disease Therapeutics. Int J Cell Biol. 2016;2016:6940283. doi: 10.1155/2016/6940283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh VK, Kalsan M, Kumar N, et al. Induced pluripotent stem cells: applications in regenerative medicine, disease modeling, and drug discovery. Front Cell Dev Biol. 2015;3 doi: 10.3389/fcell.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ilic D, Polak JM. Stem cells in regenerative medicine: introduction. Br Med Bull. 2011;98:117–26. doi: 10.1093/bmb/ldr012. [DOI] [PubMed] [Google Scholar]
- 60.Rafalski VA, Mancini E, Brunet A. Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. J Cell Sci. 2012;125:5597–608. doi: 10.1242/jcs.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L, Bhatia R. Stem cell quiescence. Clin Cancer Res. 2011;17:4936–41. doi: 10.1158/1078-0432.CCR-10-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. 2014;20:833–46. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferraro F, Celso CL, Scadden D. Adult stem cels and their niches. Adv Exp Med Biol. 2010;695:155–68. doi: 10.1007/978-1-4419-7037-4_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reinwald Y, Bratt J, El Haj A. Pluripotent stem cells and their dynamic niche. 2016 [Google Scholar]
- 65.Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism in tissue development and aging. Development. 2013;140:2535–47. doi: 10.1242/dev.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathieu J, Ruohola-Baker H. Metabolic remodeling during the loss and acquisition of pluripotency. Development. 2017;144:541–51. doi: 10.1242/dev.128389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burgess RJ, Agathocleous M, Morrison SJ. Metabolic regulation of stem cell function. J Intern Med. 2014;276:12–24. doi: 10.1111/joim.12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Varum S, Rodrigues AS, Moura MB, et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang J, Nuebel E, Daley GQ, et al. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11:589–95. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryall JG, Cliff T, Dalton S, et al. Metabolic Reprogramming of Stem Cell Epigenetics. Cell Stem Cell. 2015;17:651–62. doi: 10.1016/j.stem.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perales-Clemente E, Folmes CD, Terzic A. Metabolic regulation of redox status in stem cells. Antioxid Redox Signal. 2014;21:1648–59. doi: 10.1089/ars.2014.6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pala F, Di Girolamo D, Mella S, et al. Distinct metabolic states govern skeletal muscle stem cell fates during prenatal and postnatal myogenesis. J Cell Sci. 2018:131. doi: 10.1242/jcs.212977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bigarella CL, Liang R, Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141:4206–18. doi: 10.1242/dev.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yun Z, Lin Q. Hypoxia and regulation of cancer cell stemness. Adv Exp Med Biol. 2014;772:41–53. doi: 10.1007/978-1-4614-5915-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sart S, Song L, Li Y. Controlling Redox Status for Stem Cell Survival, Expansion, and Differentiation. Oxid Med Cell Longev. 2015;2015:14 pages. doi: 10.1155/2015/105135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–61. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 77.Iqbal MA, Eftekharpour E. Regulatory role of redox balance in determination of neural precursor cell fate. Stem Cells Int. 2017;2017:13 pages. doi: 10.1155/2017/9209127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang K, Zhang T, Dong Q, et al. Redox homeostasis: the linchpin in stem cell self-renewal and differentiation. Cell Death Dis. 2013;4:e537. doi: 10.1038/cddis.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ji AR, Ku SY, Cho MS, et al. Reactive oxygen species enhance differentiation of human embryonic stem cells into mesendodermal lineage. Exp Mol Med. 2010;42:175–86. doi: 10.3858/emm.2010.42.3.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klontzas ME, Vernardis SI, Heliotis M, et al. Metabolomics Analysis of the Osteogenic Differentiation of Umbilical Cord Blood Mesenchymal Stem Cells Reveals Differential Sensitivity to Osteogenic Agents. Stem Cells Dev. 2017;26:723–33. doi: 10.1089/scd.2016.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ivanova G, Pereira T, Caseiro AR. Metabolomic and Proteomic Analysis of the Mesenchymal Stem Cells’ Secretome Metabolomics-Fundamentals and Applications. InTech. 2016 [Google Scholar]
- 82.Lee SJ, Yi T, Ahn SH, et al. Comparative study on metabolite level in tissue-specific human mesenchymal stem cells by an ultra-performance liquid chromatography quadrupole time of flight mass spectrometry. Anal Chim Acta. 2018;1024:112–22. doi: 10.1016/j.aca.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 83.Ito K, Ito K. Metabolism and the Control of Cell Fate Decisions and Stem Cell Renewal. Annu Rev Cell Dev Biol. 2016;32:399–409. doi: 10.1146/annurev-cellbio-111315-125134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu C, Fan L, Cen P, et al. Energy Metabolism Plays a Critical Role in Stem Cell Maintenance and Differentiation. Int J Mol Sci. 2016;17:253. doi: 10.3390/ijms17020253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Boyette LB, Creasey OA, Guzik L, et al. Human bone marrow-derived mesenchymal stem cells display enhanced clonogenicity but impaired differentiation with hypoxic preconditioning. Stem Cells Transl Med. 2014;3:241–54. doi: 10.5966/sctm.2013-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15:243–56. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fujisawa K, Takami T, Okada S, et al. Analysis of Metabolomic Changes in Mesenchymal Stem Cells on Treatment with Desferrioxamine as a Hypoxia Mimetic Compared with Hypoxic Conditions. Stem Cells. 2018;36:1226–36. doi: 10.1002/stem.2826. [DOI] [PubMed] [Google Scholar]
- 88.Pattappa G, Heywood HK, de Bruijn JD, et al. The metabolism of human mesenchymal stem cells during proliferation and differentiation. J Cell Physiol. 2011;226:2562–70. doi: 10.1002/jcp.22605. [DOI] [PubMed] [Google Scholar]
- 89.Phelps J, Sanati-Nezhad A, Ungrin M. Bioprocessing of Mesenchymal Stem Cells and Their Derivatives: Toward Cell-Free Therapeutics. Stem cells int. 2018;2018:23 pages. doi: 10.1155/2018/9415367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Flower T, Pulsipher V, Moreno A. A new tool in regenerative medicine: mesenchymal stem cell secretome. J Stem Cell Res Ther. 2015;1:1–3. [Google Scholar]
- 91.Vizoso FJ, Eiro N, Cid S, et al. Mesenchymal Stem Cell Secretome: Toward Cell-Free Therapeutic Strategies in Regenerative Medicine. Int J Mol Sci. 2017:18. doi: 10.3390/ijms18091852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cunningham CJ, Redondo-Castro E, Allan SM. The therapeutic potential of the mesenchymal stem cell secretome in ischaemic stroke. J Cereb Blood Flow Metab. 2018;38:1276–92. doi: 10.1177/0271678X18776802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mukherjee P, Mani S. Methodologies to decipher the cell secretome. Biochim Biophys Acta. 2013;1834:2226–32. doi: 10.1016/j.bbapap.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park CW, Kim KS, Bae S, et al. Cytokine secretion profiling of human mesenchymal stem cells by antibody array. Int J Stem Cells. 2009;2:59–68. doi: 10.15283/ijsc.2009.2.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hwang JH, Shim SS, Seok OS, et al. Comparison of cytokine expression in mesenchymal stem cells from human placenta, cord blood, and bone marrow. J Korean Med Sci. 2009;24:547–54. doi: 10.3346/jkms.2009.24.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kwon HM, Hur SM, Park KY, et al. Multiple paracrine factors secreted by mesenchymal stem cells contribute to angiogenesis. Vascul Pharmacol. 2014;63:19–28. doi: 10.1016/j.vph.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 97.Boilard E. Extracellular vesicles and their content in bioactive lipid mediators: more than a sack of microRNA. J Lipid Res. 2018;59:2037–46. doi: 10.1194/jlr.R084640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li Y, Cheng Q, Hu G, et al. Extracellular vesicles in mesenchymal stromal cells: A novel therapeutic strategy for stroke. Exp Ther Med. 2018;15:4067–79. doi: 10.3892/etm.2018.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rani S, Ryan AE, Griffin MD, et al. Mesenchymal Stem Cell-derived Extracellular Vesicles: Toward Cell-free Therapeutic Applications. Mol Ther. 2015;23:812–23. doi: 10.1038/mt.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126:1110–20. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang A, Sun H, Wang P, et al. Modern analytical techniques in metabolomics analysis. Analyst. 2012;137:293–300. doi: 10.1039/c1an15605e. [DOI] [PubMed] [Google Scholar]
- 102.Xiao JF, Zhou B, Ressom HW. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. Trends Analyt Chem. 2012;32:1–14. doi: 10.1016/j.trac.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Klein MS, Shearer J. Metabolomics and type 2 diabetes: translating basic research into clinical application. J Diabetes Res. 2016;2016:10 pages. doi: 10.1155/2016/3898502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Iwasaki Y, Sawada T, Hatayama K, et al. Separation technique for the determination of highly polar metabolites in biological samples. Metabolites. 2012;2:496–515. doi: 10.3390/metabo2030496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang A, Sun H, Xu H, et al. Cell metabolomics. OMICS. 2013;17:495–501. doi: 10.1089/omi.2012.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dunn WB, Broadhurst DI, Atherton HJ, et al. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev. 2011;40:387–426. doi: 10.1039/b906712b. [DOI] [PubMed] [Google Scholar]
- 107.Preet A, Karve TM, Rizk N. Metabolomics: approaches and applications to diabetes research. J Diabetes Metab. 2012;6:S:6. [Google Scholar]
- 108.Stanczyk FZ, Clarke NJ. Advantages and challenges of mass spectrometry assays for steroid hormones. J Steroid Biochem Mol Biol. 2010;121:491–5. doi: 10.1016/j.jsbmb.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 109.Lu J, Xie G, Jia W, et al. Metabolomics in human type 2 diabetes research. Front Med. 2013;7:4–13. doi: 10.1007/s11684-013-0248-4. [DOI] [PubMed] [Google Scholar]
- 110.Scalbert A, Brennan L, Fiehn O, et al. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009;5:435–58. doi: 10.1007/s11306-009-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ramautar R, Mayboroda OA, Somsen GW, et al. CE-MS for metabolomics: Developments and applications in the period 2008-2010. Electrophoresis. 2011;32:52–65. doi: 10.1002/elps.201000378. [DOI] [PubMed] [Google Scholar]
- 112.Kushnir MM, Rockwood AL, Bergquist J. Liquid chromatography-tandem mass spectrometry applications in endocrinology. Mass Spectrom Rev. 2010;29:480–502. doi: 10.1002/mas.20264. [DOI] [PubMed] [Google Scholar]
- 113.Pang B, Zhu Y, Lu L. The Applications and Features of Liquid Chromatography-Mass Spectrometry in the Analysis of Traditional Chinese Medicine. Evid Based Complement Alternat Med. 2016;2016:7 pages. doi: 10.1155/2016/3837270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Plumb R, Castro-Perez J, Granger J, et al. Ultra-performance liquid chromatography coupled to quadrupole-orthogonal time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2004;18:2331–7. doi: 10.1002/rcm.1627. [DOI] [PubMed] [Google Scholar]
- 115.Meissen JK, Yuen BT, Kind T, et al. Induced pluripotent stem cells show metabolomic differences to embryonic stem cells in polyunsaturated phosphatidylcholines and primary metabolism. PLoS One. 2012;7:e46770. doi: 10.1371/journal.pone.0046770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gaspar JA, Doss MX, Hengstler JG, et al. Unique metabolic features of stem cells, cardiomyocytes, and their progenitors. Circ Res. 2014;114:1346–60. doi: 10.1161/CIRCRESAHA.113.302021. [DOI] [PubMed] [Google Scholar]
- 117.Ramalingam A, Kudapa H, Pazhamala LT, et al. Proteomics and Metabolomics: Two Emerging Areas for Legume Improvement. Front Plant Sci. 2015;6:1116. doi: 10.3389/fpls.2015.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fukushima A, Kusano M. Recent progress in the development of metabolome databases for plant systems biology. Front Plant Sci. 2013;4:73. doi: 10.3389/fpls.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wolf S, Schmidt S, Muller-Hannemann M, et al. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinformatics. 2010;11:148. doi: 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]