Abstract
Many membrane proteins are thought to function as dimers or higher oligomers, but measuring membrane protein oligomerization in lipid membranes is particularly challenging. Förster resonance energy transfer (FRET) and fluorescence cross-correlation spectroscopy are noninvasive, optical methods of choice that have been applied to the analysis of dimerization of single-spanning membrane proteins. However, the effects inherent to such two-dimensional systems, such as the excluded volume of polytopic transmembrane proteins, proximity FRET, and rotational diffusion of fluorophore dipoles, complicate interpretation of FRET data and have not been typically accounted for. Here, using FRET and fluorescence cross-correlation spectroscopy, we introduce a method to measure surface protein density and to estimate the apparent Förster radius, and we use Monte Carlo simulations of the FRET data to account for the proximity FRET effect occurring in confined two-dimensional environments. We then use FRET to analyze the dimerization of human rhomboid protease RHBDL2 in giant plasma membrane vesicles. We find no evidence for stable oligomers of RHBDL2 in giant plasma membrane vesicles of human cells even at concentrations that highly exceed endogenous expression levels. This indicates that the rhomboid transmembrane core is intrinsically monomeric. Our findings will find use in the application of FRET and fluorescence correlation spectroscopy for the analysis of oligomerization of transmembrane proteins in cell-derived lipid membranes.
Significance
Membrane proteins often function as dimers or higher oligomers. Measuring membrane protein oligomerization in native lipid membranes is thus very important, but it is also particularly challenging. Here, we address key problems and develop a method to measure surface protein density and to estimate the apparent Förster radius, and we use Monte Carlo simulations of the Förster resonance energy transfer (FRET) data to account for the proximity FRET effect occurring in confined two-dimensional environments. Using this method, we find that rhomboid protease RHBDL2 is monomeric in lipid membranes. Our findings will find use in the application of FRET and fluorescence correlation spectroscopy for the analysis of oligomerization of transmembrane proteins in native cellular lipid membranes.
Introduction
Membrane proteins frequently form functionally important homo- or hetero-oligomeric complexes, and their surrounding milieu, i.e., lipid membrane, can shape their properties and interactions profoundly (1). It is thus important to study membrane protein interactions directly in their native membranes in a minimally invasive way. Optical, fluorescence-based methods appear ideal for this purpose. Förster resonance energy transfer (FRET) is a powerful technique for measuring distances at the nanometer scale, and thus, it can potentially address protein oligomerization. FRET reports on the vicinity of protein that carries an energy donor in the presence of an acceptor, typically provided by fluorescent protein reporters, fused genetically to the protein of interest. In a three-dimensional (3D) space (in solution), FRET response is observed almost exclusively when the proteins of interest are indeed physically interacting in oligomers. However, in a two-dimensional (2D) confinement of membranes, protein density can be elevated to such a level that even if the two proteins do not physically interact in a complex, energy can be transferred to multiple acceptor molecules that are in close proximity of the donor (2). Studying protein oligomers within cellular lipid membranes by FRET is therefore ideal for situations of low protein density, i.e., the interaction has to be rather strong. Measuring dissociation constants of weaker interactions requires a higher protein surface density, which results in increased FRET “background” arising merely from the proximity of noninteracting fluorescence acceptors (3, 4, 5, 6). We aimed at analyzing and compensating these and other limitations to allow usage of fluorescence techniques for the study of oligomerization of polytopic membrane proteins spanning a wide range of dissociation constants in live-cell-derived lipid membranes.
Already under mild overexpression conditions, common in cell-biological experiments, the “proximity-induced FRET” effect mentioned above may become significant, and it has to be properly taken into account to avoid spurious results. This problem can be ameliorated by the use of FRET controls, i.e., validated known interacting and noninteracting protein pairs. In addition to that, protein density level in the membrane has to be evaluated, and only samples with similar surface densities of donor- and acceptor-labeled proteins should be mutually compared. In most membrane structures of living cells, direct protein surface density determination is not feasible because the membrane area from which the fluorescence signal is collected cannot be measured. Therefore, the fluorescence intensity of the donor and acceptor is used instead for relative comparison (7). Even such a comparison, however, requires careful interpretation because the level of FRET is influenced not only by oligomerization and density-induced proximity but also possibly by the length of the linker between the protein of interest and the fluorescent probe. In addition, it is affected by the excluded area of studied proteins (8). Importantly, global protein surface density at the membrane can be determined in spherical giant plasma membrane vesicles (GPMVs) formed from cells expressing proteins of interest (9, 10, 11, 12). Finally, recent work analyzing the FRET efficiency signatures of noninteracting membrane proteins both experimentally and by Monte Carlo (MC) simulations indicates how the overall FRET efficiency should be corrected for the “proximity-induced FRET” effect mentioned above (2).
We show that if FRET is combined with quantification of lateral concentrations of the proteins and combined with MC simulations, the true interaction can be distinguished from the proximity effect, and the quantification of oligomerization directly in the membrane becomes possible. Understanding the interaction quantitatively helps evaluate its importance, i.e., the value of dissociation constants refers to the protein concentration at which the oligomerization occurs. Knowing this, we can decide whether the interaction of endogenous proteins is their intrinsic property or whether other physical forces may need to be involved to explain oligomerization if it occurs in cells, such as other proteins, the cytoskeleton, or lipid arrangement.
Here, we focus on intramembrane proteases of the rhomboid family, proteins formed by six or seven α-helical transmembrane segments (reviewed in 13). These enzymes occur widely across evolution and regulate EGF receptor signaling, mitochondrial dynamics, mitophagy, apoptosis, or invasion of apicomplexan parasites and are relevant for a growing number of diseases, including malaria, cancer, and Parkinson’s disease (reviewed in 14). Based largely on in vitro experiments in the detergent-solubilized state of three bacterial rhomboid proteases, these enzymes have been proposed to form stable dimers and thereby become allosterically activated (15). If indeed rhomboid protease activity is regulated by dimerization, this could be significant for the interpretation of biological mechanisms involving rhomboids. Thus, to clarify this issue, here we zoom in on the well-characterized human rhomboid protease RHBDL2 (16, 17, 18, 19, 20, 21), which is localized to the plasma membrane and consists of seven transmembrane helices, and analyze its oligomeric status in cell-derived lipid membranes.
Strong interactions are detectable by single-molecule approaches such as fluorescence cross-correlation spectroscopy (FCCS) or photobleaching, but to observe weaker interactions in the native membranes, higher protein densities are required. To investigate the oligomeric status of RHBDL2 as a model transmembrane protein, we use human cells expressing fluorescently labeled RHBDL2 to create GPMVs, which have a simple geometry and show homogeneous distribution of the observed molecules. In addition to FCCS, we then employ FRET in individual vesicles in combination with MC simulations to investigate the oligomeric state of RHBDL2. In doing so, we build on the MC-FRET approach introduced by Johansson et al. (22,23) and later applied to lipid clustering by others (24,25). Within this context, we develop and implement several novel considerations that are essential when addressing large membrane inclusions (such as RHBDL2) linked to fluorescent proteins. We focus on the direct determination of protein surface density in the 2D environment of GPMVs, we address the issue of not being in the isotropic dynamic limit in these conditions, and we show that the commonly used Förster radius (R0) cannot be satisfactorily used in FRET quantification. Finally, we use MC simulations to model the FRET measurements in GPMVs and thus account for the lack of a priori knowledge of the dissociation constant and the geometry of the putative dimer. Comparing the simulation readout with the measured data then allows drawing conclusions on the dimerization of the membrane protein.
In summary, we address several key limitations of using FRET for oligomerization studies of polytopic membrane proteins in live-cell-derived membranes and demonstrate their use by showing the lack of dimerization of rhomboid protease RHBDL2, which we confirm independently also by cell-biological “relocalization” experiments. Our results can find use in the application of FRET and FCCS for the analysis of oligomerization of transmembrane proteins in lipid membranes.
Materials and Methods
Materials
Oligonucleotides were from Sigma-Aldrich (St. Louis, MO), and restriction endonucleases and other enzymes for DNA cloning were from New England Biolabs (Ipswich, MA). All other chemicals were from Sigma-Aldrich unless stated otherwise.
Cloning and constructs
The fluorescence reporters eGFP and mCherry were cloned as fusions to the N-terminus of human RHBDL2 (GenBank: NP_060291.2) in the pEGFP vector (18). For relocalization experiments, the DNA sequence encoding the peptide KDEL preceded by a (GS)3 linker was cloned at the 3′ end of the RHBDL2 gene in the eGFP/mCherry-RHBDL2 constructs. The constructs encoding fluorescently tagged human glutamyl carboxypeptidase II (GCPII) were generated by fusing eGFP or mCherry and a 17-amino acid linker to the N-terminus of human GCPII (GenBank: NP_004467.1) harboring mutations L4A/L5A (26,27) that impair endocytosis. The control constructs for RHBDL2 encoding its N-terminal cytosolic domain (R2Ncyto) of 72 amino acids fused to eGFP or mCherry and equipped with an His tag (eGFP-R2Ncyto-His6 and mCherry-R2Ncyto-His6) were cloned into the pET25b + vector for bacterial expression. The constructs encoding His6-eGFP or His6-mCherry were kind gifts of Evzen Bouřa and had been generated by cloning eGFP- or mCherry-encoding fragments into the pHis2 vector downstream of an His6 purification tag and TEV protease cleavage site (28), yielding a spacer of 18 amino acids between the His tag and the fluorescent protein. All constructs were verified by Sanger sequencing.
Protein expression and purification
The His-tagged fusions eGFP-R2Ncyto-His6, mCherry-R2Ncyto-His6, eGFP-GCPIINcyto-His6, and mCherry-GCPIINcyto-His6 were expressed from T7-driven vectors in Escherichia coli BL21(DE3) at 20°C, induced by 0.5 mM IPTG for 12 h. Cells were broken by three passages through an Emulsiflex C3 (Avestin, Ottawa, Canada) in the presence of 1 mM serine protease inhibitor phenylmethylsulfonylfluoride, and the insoluble fraction was removed by centrifugation at 15,000 × g for 30 min at 4°C. The His-tagged proteins were purified from the supernatant by metal-chelate affinity chromatography using NiNTA agarose (Qiagen, Hilden, Germany) and eluted into 20 mM HEPES (pH 8.0), 300 mM NaCl, and 10% (w/v) glycerol using 250 mM imidazole, which was immediately removed by desalting into phosphate-buffered saline (pH 7.4) using a PD-10 desalting column (GE Healthcare, Chicago, IL). Protein concentration was determined from absorbance at 280 nm, and purified proteins were flash frozen into liquid nitrogen and stored at −80°C. The identity of purified proteins was further validated by quantitative amino acid analysis, mass spectrometry, and sodium dodecyl sulfate–polyacrylamide gel electrophoresis with fluorescence scanning.
Cell culture
HeLa cells (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco’s Modified Eagle Medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum (Thermo Fisher Scientific, Waltham, MA) at 37°C and 10% CO2. For transfection, 2 × 105 cells were seeded per well of a four-chamber dish (catalogue no. D35C4-20-1.5-N; Cellvis, Mountain View, CA) and transfected by FuGene6 (Promega, Madison, WI). The eGFP-RHBDL2 and mCherry-RHBDL2 mutant fusion construct plasmids were transfected at 50:50 ng and50:500 ng DNA per well for donor-acceptor ratios of ∼1:1 and 1:10, respectively, for fluorescence lifetime imaging (FLIM)-FRET experiments, and 50:50 ng (donor-acceptor ratio of∼1:1) for FCCS experiments. The eGFP-GCPII and mCherry-GCPII mutant fusion construct plasmids were transfected at 250:250, 250:750, and 75:750 ng DNA per well for donor-acceptor ratios of ∼1:1, 1:3, and 1:10, respectively, for FLIM-FRET experiments and 250:250 ng (donor-acceptor ratio of ∼1:1) for FCCS experiments. The transfection mixtures were complemented with empty vector pcDNA3.1 to 1 μg of total DNA mass. Natural heterogeneity in expression levels within the cell population was exploited to choose cells with a range of fluorescence intensities of expressed reporters. For the relocalization experiments, 50 ng of FP-RHBDL2 vector with corresponding 50 ng FP-RHBDL2-KDEL fusion construct plasmids was transfected.
Spontaneously immortalized human keratinocytes HaCaT (29) (item no. 300493; Cell Lines Service, Eppelheim, Germany) were cultured in Dulbecco’s Modified Eagle Medium (Invitrogen). To deplete endogenous RHBDL2, HaCaT cells were transduced with a recombinant lentivirus expressing the short hairpin RNA #01 targeting RHBDL2 (17) as described (16) and selected for puromycin resistance.
Immunoblotting
The integrity of the expressed fusion proteins and the expression levels of RHBDL2 were examined by immunoblotting. Cell lysates were separated by 4–20% gradient Tris-Glycine SDS-PAGE (Bio-Rad Laboratories, Hercules, California), electroblotted onto a polyvinylidene fluoride membrane (Immobilon-FL; Millipore, Burlington, MA), optionally stained by the revert protein stain (Li-Cor Biosciences, Lincoln, NE), scanned using infrared fluorescence scanner Odyssey CLx (Li-Cor Biosciences), destained, blocked in Casein Blocker solution (Thermo Fisher Scientific) for 1 h at 25°C, and exposed to primary antibodies over night at 4°C. Primary antibodies were from rabbit (with the exception of αGCPII-02 (30), which was from mouse), and were used at the following concentrations: α-RHBDL2 (catalogue no. 12467-1-AP; Proteintech, Rosemont, IL) at 1:500, α-eGFP (catalogue no. 2956; Cell Signaling Technology, Danvers, MA) at 1:1000, α-RFP (catalogue no. R10367; Thermo Fisher Scientific) at 1:3000, αGCPII-02 at 1:5000, and secondary antibody donkey α-rabbit IgG (H + L) or donkey α-mouse IgG (H + L) cross-adsorbed and conjugated to DyLight 800 (catalogue no. SA5-10044; Thermo Fisher Scientific) at 1:10,000. Secondary antibody fluorescence was visualized using near-infrared fluorescence scanner Odyssey CLx (Li-Cor Biosciences) and optionally quantified using Image Studio Lite (Li-Cor Biosciences).
GPMV preparation
GPMVs were prepared as described elsewhere (31). Briefly, HeLa cells transiently expressing eGFP and mCherry fusions to human RHBDL2 were washed by the GPMV buffer (10 mM HEPES, 150 mM NaCl, 2 mM CaCl2 (pH 7.4)). Then, a 2 mM solution of N-ethylmaleimide in the GPMV buffer was added, and the cells were incubated for a minimum of 1 h at 37°C. Once GPMVs were formed, they were immediately used for microscopy experiments in situ. GPMVs containing GCPII constructs were produced using the chloride salt method as described (10).
Giant unilamellar vesicle preparation
Giant unilamellar vesicles (GUVs) were prepared by electroformation (32). Chloroform-lipid mixture was prepared so that total lipid concentration was 5 μg/μL, containing 75 mol % of POPC (1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), 20 mol % cholesterol, and 5 mol % DGS-NTA(Ni) (all lipids purchased from Avanti Polar Lipids, Alabaster, AL). 10 μL of the mixture was spread on two ITO-coated glass electrodes each. The electrodes were dried under vacuum overnight and then assembled in parallel into a homemade Teflon holder containing 5 mL of 600 mM sucrose solution. For the electroformation, 10 Hz harmonically oscillating voltage of 1 V peak value was applied to the electrodes for 1 h in an incubator set to 60°C. For the imaging, bovine serum albumin-coated four-chamber glass bottom dishes (Cellvis) were used, and 100 μL of GUVs was mixed with 100 μL of isoosmotic buffer (25 mM Tris (pH 8), 10 mM MgCl2, 20 mM imidazole, 261.5 mM NaCl, 2 mM β-mercaptoethanol) containing the His-tagged fluorescent proteins.
For the experiments that were carried out to validate the FCCS-based lateral concentration measurement, we prepared GUVs consisting of POPC and ATTO488- and ATTO647N-DOPE (ATTO-TEC, Siegen, Germany). The amount of the acceptor-labeled lipid was varied so that it was sufficient for the single-molecule measurement as well as for the FRET, i.e., from 0.01 to 10%. The amount of donor was 0.01%.
Microscopy
All the microscopy images were acquired on an LSM 780 confocal microscope (Zeiss, Jena, Germany) using a 40×/1.2 water objective. For the FLIM-FRET and FCCS measurements, the external tau-single photon avalanche diode (SPAD) detectors equipped with time-correlated single-photon counting (TCSPC) electronics (Picoquant, Berlin, Germany). For FCCS experiments, eGFP and ATTO488 were excited with the 490-nm line of the Intune laser (Zeiss) pulsing at a 40-MHz repetition frequency, and mCherry and ATTO647N were excited continuously at 561 and 633 nm, respectively. The excitation light was focused on the apical membrane of giant liposomes. The precise positioning was checked by maximizing fluorescence intensity and the apparent molecular brightness (33). Fluorescence intensity, collected by the same objective lens, was refocused on the pinhole (1 airy unit), and the recollimated light behind the pinhole was split on the external tau-SPADs, in front of which emission band pass filters 520/45, 600/52, and 679/41 for eGFP and ATTO488, mCherry and ATTO647N signal, respectively, were placed. The intensity of the excitation light at the back aperture of the objective was 2, 6, and 0.5 μW for 490-, 561-, and 633-nm laser lines, respectively. The collected data were correlated by home-written script in MATLAB (The MathWorks, Natick, MA) according to a described algorithm (34,35).
To avoid detector cross talk, i.e., the bleedthrough from the GFP channel to the mCherry channel, the red channel fluorescence signal was split according to its TCSPC pattern (exponential for the signal generated by the pulsed Intune laser and flat for the 561- or 633-nm continuous wave laser) into two contributions, and only the signal assigned to the flat TCSPC profile was correlated (Fig. S1). The data were processed as described (35,36). The FLIM-FRET data were acquired in the equatorial plane of giant liposomes. The equatorial plane was chosen because it is well defined and easy to find, and the measurement is not affected by the small drifts of focus that are possible during FRET measurements. The data were accumulated during fast repetitive frame scans. Framed sequentially, the acceptor signal was collected. The mean fluorescence signal of the donor and acceptor was used to obtain information on their concentrations by extrapolation of the calibration measurement as described in the Results. The excitation intensity of the 561- and 633-nm laser was decreased to 0.5 and 0.05 μW, respectively, to minimize photobleaching of the acceptor.
MC simulations
For each FLIM measurement carried out on a single GPMV/GUV, an MC simulation was executed. The simulation algorithm is illustrated step by step in Fig. S2. The input parameters for the simulation were the surface concentrations of donor- and acceptor-labeled proteins that were obtained directly during FLIM acquisition with the help of the calibration measurement (see further). The simulations were run for the following variable parameters: 1) Förster radius (R0app) or 2) dissociation constant, KD, and excluded radius, L0, when oligomerization was addressed.
MC simulations were performed on the grid of 10,000 × 10,000 pixels. The size of the pixel was set to be 0.01 × R0. First, the total amount of proteins in the simulated field was calculated. Based on the assumed KD, overall numbers of monomers and dimers were calculated. In our planar system, the dissociation constant KD is defined as follows:
| (1) |
where cD and cM are the surface concentrations of dimer and monomer, respectively.
All the monomers and one of the partners from each dimer were placed to the field so that they did not overlap (each occupies a circular area of a diameter of an excluded distance), i.e., a random position was generated, and if it was occupied, it was rejected and another one was generated. Subsequently, the partners in the dimers were located, a random, already localized molecule was selected, and a new one was placed directly next to it (so that the distance of their centers equals the excluded distance) at a random angle. Once the molecule was located, a random number was generated that distinguished whether the molecule was a donor or an acceptor based on the experimentally known ratio between those. The system uses periodic boundary.
Once the molecules were localized in the field, a random donor was selected, and the overall FRET rate Ωi was determined according to the formula
| (2) |
where Rij is the distance between the selected donor and all acceptors closer than the cutoff distance RC (10 × R0), τD is the lifetime of the donor in the absence of acceptor, and C2 is the number of the acceptors inside the circle of the Förster radius (22,23,37).
For the FRET efficiency calculations, 100 configurations of donors and acceptors within the simulated field are generated. For each configuration, 10 random donors are selected, and their FRET rates are generated according to Eq. 2. For the overall FRET efficiency, it holds ), where the sum goes over all selected donors in all configurations.
For the fluorescence decay calculations, a random time Δti at which the energy transfer occurs was generated: , where α is a random number from the interval of 0–1. For each situation, i.e., each GPMV/GUV, 100 configurations of donors and acceptors were generated, and at each configuration, 100 random excitations were performed. First, the probability that the donor does not transfer its energy to an acceptor is calculated by cumulative histogramming of Δti. Second, this probability is multiplied by the acceptor-free donor decay (fit of the eGFP decay), and third, the resulting function is convolved with the instrumental response function.
The simulated efficiencies as well as the fluorescence decays for the given acceptor/donor surface densities were compared with the measured data by calculating the mean-square deviation (MSD).
Experimental values of FRET efficiencies were obtained from the fits of experimental decays from the amplitude-weighted average of the decay times:
| (3) |
where Ai and τi stand for amplitudes and characteristic times of biexponential fits of the experimental decays of donor in the presence and absence of acceptors.
FRET-based determination of surface acceptor concentration
According to Baumann and Fayer (37), the donor fluorescence decay F(t) consists of two contributions:
| (4) |
1) the decay of the donor in the absence of acceptors characterized by the amplitudes αi and the corresponding decay times τi and 2) the probability Gs(t) that a donor does not transfer its energy to an acceptor from the moment of having been excited until time t. Gs(t) is a product of an intra- and intermembrane leaflet energy transfer
| (5) |
and
| (6) |
Here, C2 is the number of acceptors within in a single leaflet in the circular area of R0 in radius, d is the membrane thickness, Г is the gamma function, μ = 3t(R0/d)6/2τ, and s = 2μcos6θr/3, where θr is the angle between the bilayer normal and a vector that connects positions of donor and acceptor dipoles.
We used the model to determine C2 for a 2D membrane system, where ATTO488 and ATTO647N-DOPE served as donor and acceptor, respectively. First, we have determined the amplitudes αi and the decay times τi of the donor in the absence of acceptors by two-exponential fitting of the donor fluorescence decay. Second, we have fitted the donor decay in the presence of acceptors with Eq. 4. The fitting involved convolution with the experimental instrument response function and was implemented in MATLAB.
Results
Investigating dimerization of human rhomboid protease RHBDL2 by FCCS
The joint motion of proteins of interest provides the most convincing evidence of their physical interaction, and single-molecule techniques based on tracking individual fluorescently labeled proteins thus represent straightforward tools to address protein-protein oligomerization. Because single-molecule-based approaches require protein concentrations low enough to distinguish individual molecules, they are applicable only to strongly interacting species (i.e., those that form significant fraction of oligomers at low total concentrations).
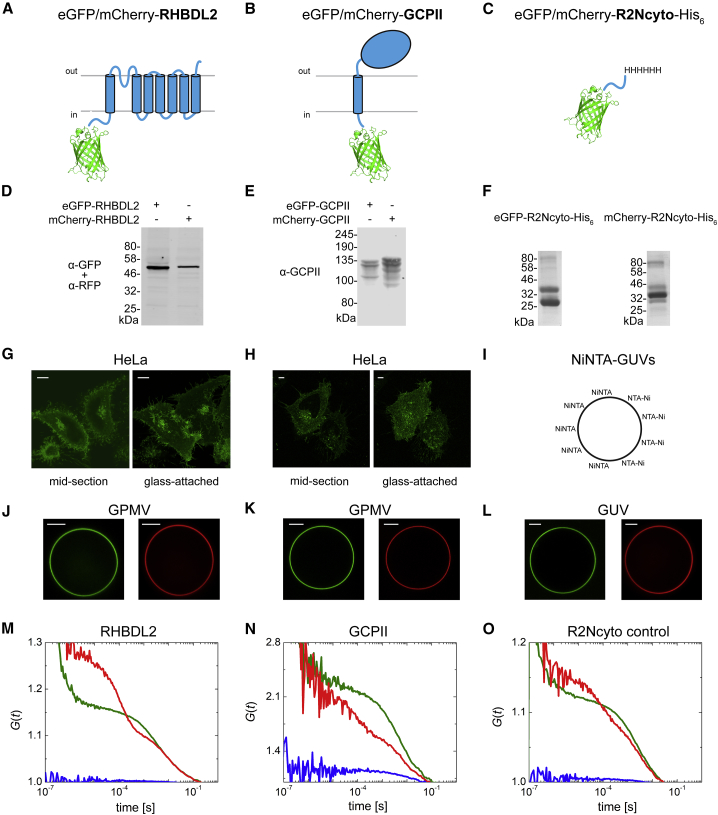
We have first employed FCCS (38) to investigate the dimerization status of human RHBDL2 in live-cell-derived membranes of GPMVs (31). The FCCS technique analyzes fluorescence intensity fluctuations arising from individual molecular transits through the diffraction-limited laser focus, and the temporal cross correlation between the signal of fluorescence-reporter-labeled membrane proteins suggests their joint motion. We have performed the experiment in the apical membrane of GPMVs (Fig. 1 J) derived from HeLa cells coexpressing eGFP-RHBDL2 and mCherry-RHBDL2. Both fusion proteins localized largely to the plasma membrane, were mostly full length (Fig. 1, A, D, and G), and exhibited normal protease activity of RHBDL2 against a well-established model substrate (Fig. S3). Cross-correlation functions (Fig. 1 M) did not suggest any interaction of RHBDL2 monomers at this concentration range. The same experiment was performed with murine GCPII (Fig. 1, B, E, H, K, and N), which is known to form dimers and localize to the plasma membrane (39,40). As expected, the coexpressed eGFP and mCherry fusions of GCPII exhibited a positive cross-correlation amplitude (Fig. 1 N), indicative of a dimer. In contrast, eGFP and mCherry fused to the His-tagged N-terminal cytosolic domain of RHBDL2 (R2Ncyto) attached to the membrane via Ni-NTA-lipid-containing GUVs (Fig. 1, C, F, I, and L) did not show any cross-correlation (Fig. 1 O), serving as negative, monomeric controls. More details on fits and the analysis of correlation amplitudes are given in Fig. S4 and Table S1. Taken together, these results indicated that if RHBDL2 dimers exist at all, their interaction is relatively weak, and their analysis thus requires methods that can access higher concentration ranges. The use of FCCS has been shown to be subject solely to brightness of molecules and can also principally address higher concentrations, provided that detectors can handle high fluorescence countrates and that effects of laser fluctuations can be corrected (41). This approach represents a great potential in addressing protein-protein interactions but cannot be achieved with most of the common microscope setups at the moment, including ours. Our detectors can work linearly to ∼0.2 MHz, which means that we can observe a maximum of ∼100 fluorophores (with brightness of 2–3 kHz per molecule, as is common for eGFP or mCherry) in focus (which corresponds to ∼0.4 × 10−3 fluorophores per nm2), and corrections of fluctuations of laser power and fluctuation of 2D system, such as thermal motion of the membrane, have not been addressed.
Figure 1.
The experimental system used and fluorescence cross-correlation analysis of dimerization of RHBDL2 and GCPII. (A–C) The construct schemes for eGFP-RHBDL2 and mCherry-RHBDL2 (A), eGFP-GCPII and mCherry-GCPII (B), and eGFP-R2Ncyto-His6 and mCherry-R2Ncyto-His6 (C) are shown. (D and E) Western blots showing expression and integrity of eGFP-RHBDL2 and mCherry-RHBDL2 (D) and eGFP-GCPII and mCherry-GCPII (E) are shown. (F) An image of the Coomassie-stained SDS-PAGE showing the integrity of recombinant eGFP-R2Ncyto-His6 and mCherry-R2Ncyto-His6 is shown. (G and H) Images of live-cell green fluorescence protein showing subcellular localization of eGFP-RHBDL2 (G) and eGFP-GCPII (H) expressed in HeLa cells are shown. Scale bars, 10 μm. (I) A scheme of GUVs spiked with DGS-NTA(Ni) is shown. (J–L) Fluorescence images illustrating GPMVs containing eGFP-RHBDL2 and mCherry-RHBDL2 (J) (scale bar, 2 μm), eGFP-GCPII and mCherry-GCPII (K) (scale bar, 2 μm), and NTA-decorated GUVs containing surface-bound eGFP-R2Ncyto-His6 or mCherry-R2Ncyto-His6 (L) (scale bar, 5 μm) are shown. (M–O) Auto- and cross-correlation functions of eGFP-RHBDL2 (green) and mCherry-RHBDL2 (red) in GPMVs (M), eGFP-GCPII (green) and mCherry-GCPII (red) in GPMVs (N), and R2Ncyto-His6 (green) and mCherry-R2Ncyto-His6 (red) bound to DGS-NTA(Ni)-spiked GUVs (O) are shown. Blue lines denote the cross-correlation functions. Representative measurements are displayed. A number of GPMVs/GUVs were measured for each construct pair, with detailed statistics of the FCCS experiment available in Fig. S4.
Investigating weakly dimerizing polytopic membrane proteins by FRET
From a biophysical point of view, quantifying weaker protein-protein interactions by fluorescence techniques requires working at concentrations that are close to the dissociation constant, which may exceed the maximal levels used in single-molecule experiments (typically fewer than ∼0.4 × 10−3 fluorophores per nm2). Therefore, spectroscopic techniques probing the vicinity of fluorescent probes are required, such as FRET. Working at high protein densities with membrane-embedded entities of a nonnegligible excluded volume that are potentially forming dimers of unknown spatial orientation, however, brings specific problems such as proximity-induced FRET and the need to know the surface density of donors and acceptors at the site of measurement. To accommodate all the specifics of dealing with weakly interacting polytopic transmembrane proteins fused to fluorescent protein reporters, we discuss several underlying physical and methodological considerations below.
Determination of protein surface densities in the membrane of giant liposomes
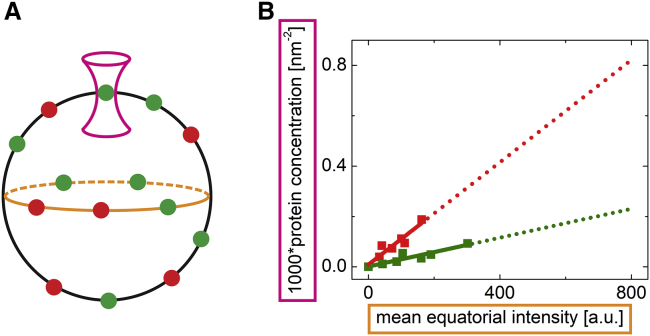
Quantitative determination of dissociation constants requires simulations of FRET efficiencies by the MC approach (24), and for this, knowledge of surface densities of donors and acceptors is essential. Because of the large differences in geometry of 3D and 2D systems, calibration of fluorescence intensity versus known protein concentration feasible in 3D is not transferrable to a 2D system. Therefore, we employed a single-molecule technique of fluorescence correlation spectroscopy (FCS) that enables counting fluorescent molecules within a diffraction-limited spot. The full width in half maximum, represented as FWHM, of the spot light profile is given as , where λ is the excitation wavelength and NA stands for the numerical aperture of the objective lens. We used FCS to measure fluctuations of fluorescence intensity in the upper membrane of a liposome (its apical surface) (Fig. 2 A; (42)), and an analysis of the intensity trace yielded the absolute number of fluorophores within this focal spot. Because the FRET measurement, for which we need to know the surface densities of donors and acceptors, is made in the equatorial plane of a spherical liposome (GPMV) (Fig. 2 A) at a lower excitation power, the relationship between the mean equatorial intensity at the excitation power used for FRET and the apical surface density needs to be established. FRET protein densities are much higher than the single-molecule experiment requires; therefore, the dependence between the equatorial signal and the apical protein density is linearly extrapolated from the FCS data (Fig. 2 B).
Figure 2.
Determination of protein surface densities in the membrane of giant liposomes. (A) A scheme of the calibration experiment is shown: FCCS determining the protein concentration was acquired in the apical membrane of the GUVs (magenta laser light profile), and the FRET experiment requiring the equatorial intensity of the fluorescent proteins was carried out in the middle section of the GUVs (orange ellipse). (B) The linear dependence of the fluorescent protein surface concentration on the mean equatorial intensity is shown; green and red lines denote His6-eGFP and His6-mCherry, respectively. The dotted lines show the extrapolation to the concentrations for which FCCS could not be carried out.
To validate our approach, we decided to compare the results obtained by the FCS/extrapolation approach with an FRET-based approach. In a planar system of donors and acceptors that are in the dynamic isotropic regime such as ATTO488 and ATTO647N headgroup-labeled lipids, the fluorescence decay of donors in the “sea” of acceptors has been theoretically described by Baumann and Fayer (37). For details, see Materials and Methods. This analysis (Fig. S5) shows a reasonably good agreement between the two methods of acceptor surface density determination, indicating that our approach is a valid way of determination of protein surface densities in giant liposomes.
Determination of the apparent Förster radius
Förster radius represents the main characteristics of an FRET pair. It equals the distance in which FRET efficiency drops to 50%, and it thus refers to the distances that can be addressed by the pair. Förster radius (R0) can be determined as follows:
| (7) |
where NA stays for the Avogadro’s number, n is the refractive index, QD is the quantum yield of the donor in the absence of the acceptor, J is the spectral overlap integral, and κ2 is the orientation factor kappa squared, which equals 2/3, provided that rotational depolarization upon excitation occurs much faster than transfer of energy and all orientations of the fluorophores are equally distributed (i.e., dynamic isotropic limit). Fluorescent proteins attached to membranes via fusions to integral membrane proteins are thought to rotate slowly compared to the donor fluorescence lifetime, and thus the dynamic limit may no longer hold for them. In addition, membranes are naturally anisotropic, which prevents the fluorophores from sampling the entire rotational space, and these factors together make correct mathematical treatment of such a system exceedingly complicated. In these cases, the distribution of acceptor transition dipole orientations becomes distance dependent as a direct consequence of the motion restrictions and system anisotropy. Therefore, the appropriate physical description of the system would require knowledge of not only a mean value of the orientation factor (if we had a chance to access it) but of its entire donor-acceptor distance distribution. In single-molecule studies, the orientation factors were determined for some single donor-acceptor pairs (43,44). In our laterally crowded system, however, a donor can transfer its energy to several acceptors with different probabilities. Experimentally, in such laterally crowded systems, an individual energy transfer cannot be separated from transfers to other neighboring acceptors, and this has to be taken into account.
These considerations imply that the Förster radius obtained for isotropic dynamic limit cannot be automatically used for quantitative description of membrane attached fluorescent proteins. Moreover, even the simple equations that are commonly used when characterizing FRET are no longer valid because of the need for quantum yield averaging. Despite that, the established formalism for FRET efficiency η can be kept as
| (8) |
where QDA and QD are quantum yields of donor in the presence and absence of the acceptor, respectively. For their ratio, it holds
| (9) |
where summing over i holds for all acceptors in the vicinity of a selected donor and summing over M stays for all donors in the system in all possible configurations. We introduce the apparent Förster radius, R0app, which formally behaves as a common Förster radius but is not only specific for the given donor-acceptor pair but for the entire situation, i.e., angular distribution of all transition dipole moments at given protein densities. It has to be emphasized that R0app is an empirical constant that is principally influenced by the linker length between the transmembrane protein and the fluorescent protein, as well as to the given donor-acceptor surface density or heterogeneities in lateral protein organization.
To characterize protein-protein interaction by means of dissociation constant, R0app has to be determined in a reference system that experimentally best resembles the system of our interest (which are GPMVs with transmembrane proteins fused to fluorescent proteins). In our approach, we decided on a reference system that consists of the N-terminal cytosolic extramembrane part of the studied transmembrane protein RHBDL2 (R2Ncyto), fused to the fluorescent protein and attached to the membrane of GUVs via His-tag and an NTA-lipid anchor. Geometrically, both the reference system and the studied system represent planes with green- and red-fluorescing protein barrels separated by the cytosolic domain from the plane, i.e., the rotational motion of the barrels and their mutual geometry would be comparable. Therefore, we assume that the donor-acceptor-distance-dependent κ2 factor distribution would be similar to that in GPMVs containing polytopic membrane proteins fused to fluorescence proteins. The use of this approach would be restricted only to the systems in which this assumption holds, i.e., those with similar rotational dynamics and geometry. It has to be pointed out that in any FRET experiments using established interacting or noninteracting membrane proteins as positive or negative controls, this assumption is tacitly expected to hold because otherwise the differences in FRET efficiencies could be caused by a different κ2 distribution rather than by interaction between the proteins of interest.
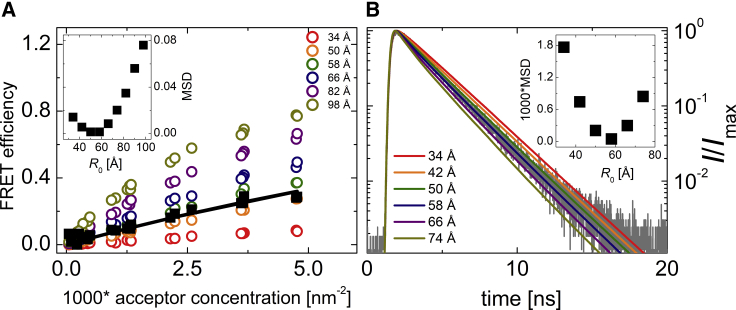
To estimate the R0app for two fluorescent proteins attached to the membrane, we made use of the discussed proximity FRET effect. Knowing that the distribution of His6-R2Ncyto-eGFP and His6-R2Ncyto-mCherry attached to the membrane of GUVs spiked with DGS-NTA(Ni) is homogeneous and that no significant interaction between the two proteins occurs, the combination of FRET with MC simulations allows for the estimation of the apparent Förster radius of this fluorophore pair. At given surface densities of donors and acceptors and with knowledge of the excluded areas of the protein barrels, the apparent Förster radius is the only variable parameter required for the determination of FRET efficiency. We have thus prepared GUVs containing DGS-NTA(Ni) and measured FRET efficiency for various amounts of His-tagged fluorescent proteins added to the GUVs. For every GUV, the mean equatorial intensity was evaluated, the surface protein density of donors and acceptors was calculated from the extrapolation of the calibration measurement described above, and FRET efficiency based on FLIM was determined (Fig. 3 A).
Figure 3.
Determination of the R0app for two fluorescent proteins attached to the membrane. (A) FRET efficiency as a function of acceptor concentration is shown; black squares denote the measured data; circles denote the simulated data with increasing apparent Förster radius. The best agreement between the measured and the simulated data was found for the apparent Förster radius equalling 50–58 Å. The black line is a fit of experimental data by a numerical model introduced by Snyder and Freire (8), giving the value of R0app = 54 Å. (B) MC simulation of the fluorescence decay (gray line) obtained for a selected GPMV is shown here; colored lines are obtained from the simulation with increasing apparent Förster radius. The best agreement between the data and the simulation was obtained for R0app = 58 Å. The insets show the dependence of MSD, calculated from the measured data and each of the simulated dependences, on the apparent Förster radius. Table S2 lists all of the concentrations of donor and acceptors.
The surface densities of donors and acceptors obtained for each GUV were taken as input parameters for MC simulations that were employed to calculate a theoretical level of FRET efficiency for the given situation including the proximity FRET phenomenon. Varying the value of the apparent Förster radius (R0app) and assuming that the donor (eGFP) and acceptor (mCherry) cannot come closer to each other than 30 Å (4), we have obtained the best agreement between our experimental data and the MC simulation for R0app ranging between 50 and 58 Å (Fig. 3 A, inset). Also, when fitting our experimental data with the numerical model established by Snyder and Freire (8), we obtained a value of R0app of 54 Å (for a fixed value of the excluded distance of 30 Å). Both for the MC simulations and for the Snyder-Freire model, we used the closest donor-acceptor distance of 30 Å. It is of note that we do not precisely know the distance, but based on the protein geometry, it has to fall within the range of 25–35 Å. The numerical model gives corresponding values of R0app 52 and 56 Å, which means that the error in the closest protein-protein distance estimate is much smaller than the range of R0app obtained from our simulation.
Both the values we obtained from the FRET efficiency simulations as well as those we obtained from the numerical model are very close to the value of R0 for the eGFP/mCherry pair at dynamic isotropic limit, 52 Å. In addition, we have performed the reference experiment also with a shorter linker of 18 amino acids instead of the whole 72-amino acid R2Ncyto and obtained very similar values of FRET efficiency, suggesting that the effect of the linker length is negligible (see Fig. S6). Collectively, this suggests that despite our concerns, the FRET behavior of the fluorescence proteins barrels resembles the dynamic isotropic regime in our system.
Our MC simulations were not only adopted for the FRET efficiency calculations but also for modeling of fluorescence decays. The advantage of FLIM combined with the decay simulations over the efficiencies is that the whole decay bears significantly more complex information on the energy transfer. Therefore, even from a single GUV measurement, the information on Förster radius or interaction parameters (see later) can be drawn. To prove this, we have chosen a few single-GUV measurements and analyzed fluorescence decay data gathered during imaging of the GUVs in their equatorial plane. Comparing the measured decay with the simulated data revealed the best agreement for R0app of 58 Å (see Fig. 3 B), which is remarkably close to the values resulting from the analysis of FRET efficiencies (Fig. 3 A).
Dimerization of RHBDL2 and GCPII
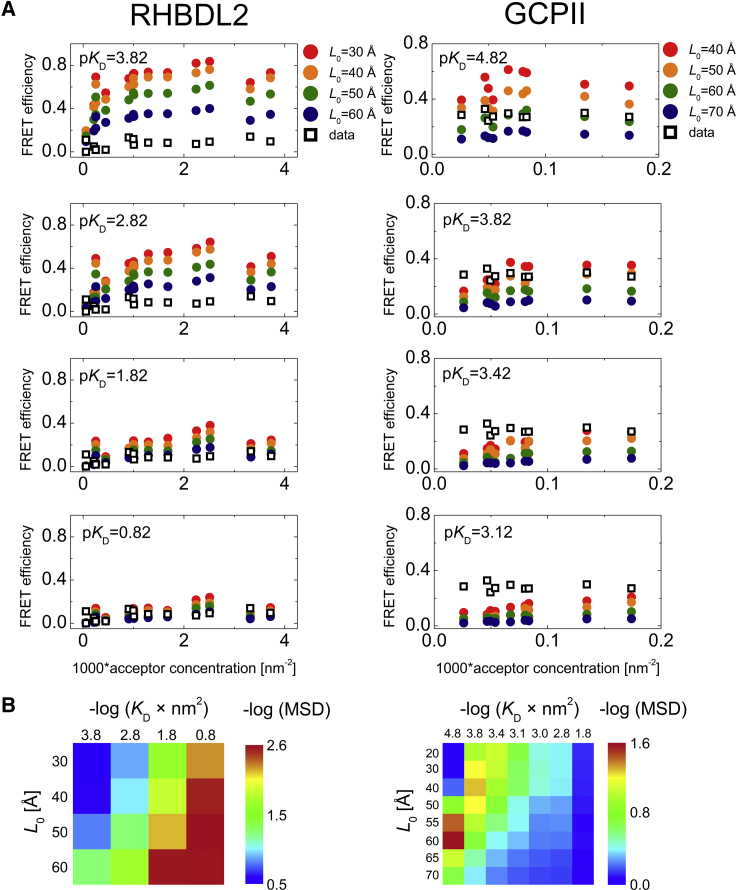
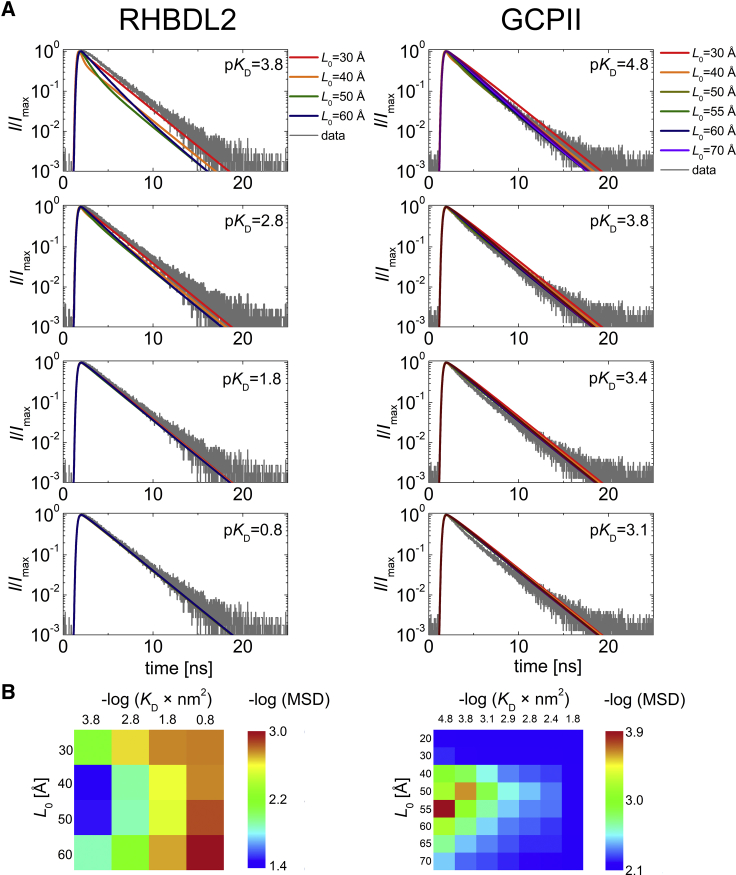
To address the thermodynamic propensity of the human rhomboid intramembrane protease RHBDL2 to form dimers in the natural lipid environment and at high protein densities, we have measured FRET in GPMVs prepared from HeLa cells coexpressing eGFP and mCherry fusions to RHBDL2, using GCPII (also known as NAALADase) as a dimeric positive control (Fig. 1; (39,40)). The use of the spherical GPMVs was crucial for quantification of protein surface densities in the area from which the signal was collected. In living cells, membrane proteins are synthetized at the endoplasmic reticulum (ER) membranes and are then trafficked to the plasma membrane. Because plasma membrane surface is not simply planar but is complicated by ruffles and numerous filopodia-like protrusions (Fig. 1, G and H), plenty of signal intensity heterogeneities are visible when focusing on the plasma membrane adhering to the glass (Fig. 1, G and H). This could also be due to numerous ER-plasma membrane contact sites. As a consequence, the overall area of the plasma membrane cannot be easily measured in live cells. Thus, GPMVs with low fluorescence signal were used for the calibration of surface protein density by FCS, as described above, and protein density in the apical surface of the highly fluorescent GPMVs used for FRET measurements was calculated from the mean equatorial fluorescence. These surface protein densities were used as input parameters for MC simulations in which the value of KD and the closest donor-acceptor distance (excluded radius) were varied, and the results were compared with the experimental values of FRET efficiency (Fig. 4).
Figure 4.
Analysis of dimerization of RHBDL2 and GCPII in GPMVs derived from HeLa cells by FRET measurements and MC simulations. (A) A comparison of acceptor-concentration-dependent FRET efficiencies obtained experimentally (black squares) and from MC simulations (circles) is shown. The MC simulations were carried out for RHBDL2 and GCPII at increasing KD-values and for excluded radii ranging from 30 to 60 Å and from 20 to 70 Å for RHBDL2 and GCPII, respectively. Donor concentrations are not depicted for the sake of simplicity, but for every analyzed GPMV, they were used as input parameters for the MC simulations. (B) Heat maps of −log (MSD) visualizing the agreement between the measured and the simulated data for various values of KD and the excluded radii are shown. Table S2 lists all concentrations of donor and acceptors. pKD = −log(KD); MSD, mean-square deviation.
The results for RHBDL2 show that independently of the excluded radius of the protein (L0 in Fig. 4 B), experimental data match the simulations only at relatively high KD-values (low −log(KD × nm2)) of ∼0.16 nm−2 (corresponding to ∼4.5 mol %, assuming lipid headgroup size is 60 Å2). This is one or two orders of magnitude higher than the highest acceptor concentration achieved in our experiment and a few more orders of magnitude higher than the concentration of the endogenous RHBDL2 in HeLa cells (Fig. S7). It is thus highly unlikely that the rhomboid protease scaffold of RHBDL2 is intrinsically dimeric in lipid membranes at physiological concentrations. For the positive control, GCPII, the simulation predicts an apparent KD-value of 1.5 × 10−5–1.5 × 10−4 nm−2 (corresponding to 0.00045–0.0045 mol %, respectively, provided the lipid headgroup size is 60 Å2), which is three to four orders of magnitude lower than that of RHBDL2 and in agreement with the known dimeric character of this protein (39,40).
For both RHBDL2 and GCPII, we have also employed the analysis of fluorescence donor decays. We have selected several GPMVs with a higher acceptor density to ensure a high enough level of FRET. We have simulated the decay for the selected GPMV characterized by the surface concentrations of donors and acceptors. Similar to efficiency simulations, we have executed the simulations for increasing KD-values and radii of excluded volumes (L0). Eventually, the simulated data were compared with the experimental decay curve, which shows that the KD-values as well as the excluded radii obtained from the decay analysis are in a good agreement with those obtained from the analysis of efficiencies (Fig. 5).
Figure 5.
Analysis of dimerization of RHBDL2 and GCPII in GPMVs derived from HeLa cells by FLIM-FRET measurements and MC simulations. (A) A comparison donor fluorescence decay (gray line) obtained at given lateral concentration of donors and acceptors with decay curves obtained from MC simulations (colored lines) is shown. The MC simulations were carried out for RHBDL2 and GCPII, at increasing KD- and L0-values. (B) Heat maps of −log (MSD) visualizing the agreement between the measured and the simulated data for various values of KD and the excluded radii L0 are shown. pKD = −log(KD); MSD, mean-square deviation.
It is worth noticing that the levels of FRET observed for RHBDL2 are comparable to those obtained with R2Ncyto, the negative control, for similar levels of acceptor density (the donor density was small in both situations; see Table S2). This means that the observed FRET efficiency can be attributed entirely to the proximity phenomena in case of RHBDL2 and R2Ncyto. In contrast, in the case of GCPII, large FRET efficiencies (around 30%) were observed already at protein densities almost an order of magnitude lower than the highest densities used with RHBDL2 and R2Ncyto. At the concentrations used for GCPII, the level of proximity FRET for a GCPII monomer would not exceed 5%; for dimer, it would be even lower (2). Most of the FRET efficiency in GCPII experiments, thus, arises from the strong interaction between GCPII monomers.
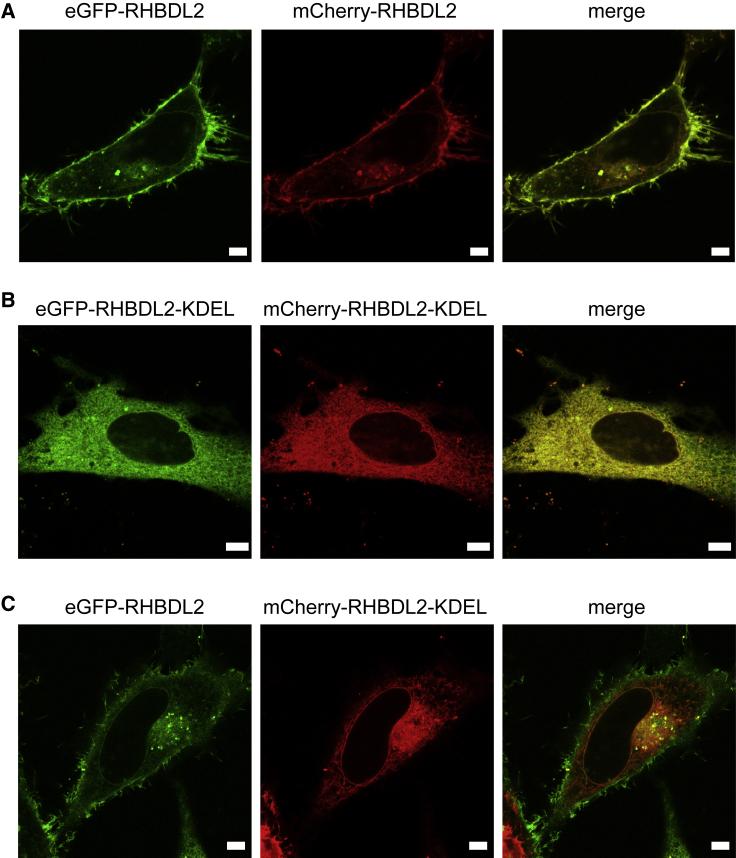
Our analysis indicates that RHBDL2 is not dimeric to any significant degree in cell-membrane-derived vesicles at expression levels far exceeding the endogenous ones (17- to 56-fold higher; Fig. S7). We, however, could not exclude that in other intracellular compartments such as the ER or Golgi, which have a different lipid composition and hydrophobic thickness, RHBDL2 may dimerize. Because interactions within the intracellular compartments of the secretory pathway are not easily directly addressable by FRET or FCCS, we resorted to a cell-biological approach exploiting the cellular mechanism of retrieving proteins back to the ER from early Golgi via the KDEL tag (45) and membrane-bound KDEL receptor (46,47). In fact, KDEL tagging was shown to exert a strong dominant negative effect on the secretion of dimeric proteins such as TGFβ (48), documenting the feasibility of this approach. When eGFP-RHBDL2 and mCherry-RHBDL2 are coexpressed in HeLa cells, both constructs show predominantly plasma membrane localization, notably labeling filamentous extrusions of the cell surface (Fig. 6 A; Fig. S8 A). When both of these constructs are equipped with a C-terminal, luminal KDEL tag, both predominantly show an ER localization with a complete overlap and complete loss of filopodia-like labeling (Fig. 6 B; Fig. S8 B). When, however, mCherry-RHBDL2-KDEL and eGFP-RHBDL2 are coexpressed, only mCherry-RHBDL2-KDEL relocalizes to the ER, whereas the localization of eGFP-RHBDL2 is barely affected and the fluorescence of the two reporters overlaps only minimally (Fig. 6 C; Fig. S8 C). The same is true when eGFP-RHBDL2 is KDEL tagged and mCherry-RHBDL2 is not (data not shown). These independent qualitative data strongly indicate the absence of stable dimers of RHBDL2 that could traffic together. In other words, RHBDL2 appears monomeric in all major membrane compartments of the secretory pathway where it normally resides.
Figure 6.
Relocalization analysis of RHBDL2 in live cells. Fluorescent constructs of human RHBDL2 fused to either eGFP or mCherry with or without the ER-retaining KDEL signal fused to the very C-terminus of each protein were coexpressed in HeLa cells, and live cell fluorescence was recorded 20–24 h after transfection. (A) shows eGFP-RHBDL2 coexpressed with mCherry-RHBDL2, (B) shows eGFP-RHBDL2-KDEL coexpressed with mCherry-RHBDL2-KDEL, and (C) shows eGFP-RHBDL2 coexpressed with mCherry-RHBDL2-KDEL. Note that although both fusions show strong plasma membrane localization including filopodia (A), KDEL tagging effectively relocalizes both fusions to the ER (B), whereas KDEL tagging of only one of the fusion proteins does not relocalize the other coexpressed one (C), meaning that the two fusion proteins do not stably interact with one another within the cell. Scale bars, 5 μm. Images of more cells from (A)–(C) are shown in Fig. S8.
Discussion
For addressing weak protein oligomerization in cellular membranes, FRET combined with FLIM represents one of few applicable optical noninvasive techniques. It has been shown that the expression levels of fluorescent protein acceptor fusion have to be kept under control because they may contribute to the overall FRET efficiency by the proximity effect (2). Having full control of lateral protein density does not seem to be possible in living cells, but it is possible in GPMVs derived from the cells of interest using an external calibration (4). To separate the impact of the proximity effect from the impact caused by protein oligomerization on the overall FRET efficiency, we have utilized MC simulations.
A large fluorescent protein attached to a membrane-residing protein of interest, however, cannot freely rotate in all directions, and it rotates at timescales much slower than the donor fluorescence lifetime. Such fluorescent fusion membrane proteins hence do not fulfill the requirements for the dynamic isotropic limit, which predicts the value of the orientation factor κ2 to be 2/3. Knowledge of κ2 and, consequently, of the Förster radius is, however, essential for meaningful quantification of FRET. In situations that are neither isotropic nor dynamic, estimation of κ2 is very difficult and usually requires understanding the dynamics of the involved proteins. Moreover, to calculate FRET efficiency under the static or intermediate conditions, the overall dependence of the κ2 factor on the donor-acceptor distance has to be known (49). Here, we propose using an apparent value of R0 (R0app) that would allow us to compare systems resembling each other well as regards rotational dynamics and geometry of the fluorophores. In this sense, we consider our system, consisting of transmembrane proteins fused to fluorescence proteins, comparable to the eGFP/mCherry donor-acceptor pair attached by a linker and His-tag to the membrane of GUVs. Our approach then simulates FRET efficiencies at experimentally obtained protein densities and compares those with the measured FRET efficiency values. The only optimized parameter in the simulations is the apparent Förster radius. Our results clearly indicate that the apparent Förster radius for eGFP and mCherry in our system falls into the range of 50–58 Å, which is in very good agreement with the value reported in literature for the dynamic isotropic limit, i.e., 52 Å (50). This result, however, does not prove that the pair is in the dynamic isotropic regime. Whether it is close to it or whether it is just a coincidence cannot be distinguished from our data. Our results, however, imply that a pair of similar barrel-like fluorescent proteins attached to a membrane plane via a transmembrane protein may be treated as if they were in dynamic isotropic regime because their geometrical situation is similar to our system. Being aware that the rotational rigidity caused by the close proximity of the fluorescent protein barrel and the lipid membrane can be significantly decreased by the presence of a flexible linker, we tested two different linker lengths: 18 and 72 amino acids. Because we did not observe any difference between those two, we can conclude that 18 residues probably already allow maximal mobility of the fluorescence protein barrel.
The quantitative determination of the dissociation constant by the MC simulation can be generally applied also for any higher-order protein-protein interaction. It is, however, limited by the a priori knowledge of the stoichiometry of the protein complexes. When it is known, it can be straightforwardly implemented into the MC simulation. When stoichiometry is unknown, single-molecule or other biochemical approaches can be employed to acquire this information.
Apart from FRET, we have also applied FCCS to address protein dimerization. In our hands, the two techniques are more or less complementary. Although FCCS is used for lower surface concentrations of proteins (typically up to ∼0.4 × 10−3 protein fluorophores per nm2), FRET, even though it can be applied generally in all concentration ranges, is beneficial especially in ranges at which FCCS cannot be easily used in practice (typically above ∼0.4 × 10−3 protein fluorophores per nm2). Although the concentration range for which FCCS can be applied can generally be much broader (theoretically unlimited (41)), its limits are in practice set by the technical features of the used microscope, specifically by the dynamic range of the detectors or by the ability to correct for the laser fluctuations. In addition, the membrane thermally fluctuates, which would probably also add a noise component to the correlation curve. This may be negligible at low protein densities because its amplitude is very small, but it could become significant at high protein concentrations. The actual donor concentrations in experiments depicted in Fig. 4 are reported in Table S2.
To validate our method, we have applied it to the dimeric membrane protein GCPII. The data indicated that GCPII was dimeric in GPMVs, and the values of the dissociation constant of GCPII were three to four orders of magnitude lower than those of the rhomboid intramembrane protease RHBDL2, which was previously proposed to be dimeric (15). Interestingly, recent single-molecule photobleaching analysis of several rhomboid proteases and pseudoprotease by Kreutzberger and Urban (51) also found no evidence for dimerization in live cells. This study was, however, limited to low concentrations of the species compatible with single-molecule studies. Our study additionally provides data from the high concentration range of rhomboid and is testing the possibility of weak interactions between rhomboid monomers. Because our study also finds no evidence for rhomboid dimerization, these two studies together strongly indicate that the rhomboid domain is intrinsically monomeric in membranes. This is particularly relevant for the related rhomboid pseudoproteases iRhoms (52) and Derlins (53), which are involved in inflammatory signaling (54) and ER-associated degradation (55), respectively. Derlins were proposed to be dimeric based on detergent solubilization and pull-down experiments (56), but it is highly likely that Derlins themselves do not dimerize in membranes. Indeed, dimerization of the rhomboid-family proteins could occur if there was an additional force that drove the partners to one another, such as dimerization of their extramembrane domains (57), interaction via a third partner, segregation to some specific lipid pools, or binding to juxtamembrane structures.
Conclusions
Here, we adapt the usage of FRET for the analysis of dimerization of polytopic transmembrane proteins by taking into account proximity-induced FRET, careful estimation of the apparent Förster radius that can principally deviate from the value commonly used in isotropic conditions, and employing MC simulations to interpret FRET results. In addition, we have developed and validated a method for lateral concentration determination that is crucial for having distribution of donors and acceptors under the control. Using these methods, our biophysical and cell-biological experiments do not provide any evidence for the dimerization of human rhomboid protease RHBDL2 in lipid membranes of live cells, which suggests that the transmembrane core of rhomboid protease is intrinsically monomeric, unlike proposed previously (15).
Acknowledgments
We thank Evzen Bouřa for the His6-eGFP- and His6-mCherry-encoding plasmids, Pavel Šácha for GCPII constructs, Cyril Bařinka for consultations, and Marek Cebecauer and Radek Šachl for critical reading of the manuscript.
K.S. and J.H. acknowledge support from Czech Science Foundation (project no. 18-09556S and 19-18917S), Ministry of Education, Youth and Sports of the Czech Republic (project no. LO1302), and Operational Programme European Regional Development Fund (no. CZ.02.1.01/0.0/0.0/16_019/0000729).
Footnotes
J. Škerle and J. Humpolíčková contributed equally to this work.
Editor: Dimitrios Stamou.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.03.011.
Contributor Information
Jana Humpolíčková, Email: jana.humpolickova@uochb.cas.cz.
Kvido Strisovsky, Email: kvido.strisovsky@uochb.cas.cz.
Author Contributions
K.S., J.H., and J.Š. designed research. J.Š., J.H., P.R., N.J., D.J., A.S., E.P., M.F., and J.D. performed research. J.H., J.Š., D.J., N.J., and K.S. analyzed data. J.H. and K.S. wrote the manuscript with input from co-authors.
Supporting Material
References
- 1.Gupta K., Donlan J.A.C., Robinson C.V. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature. 2017;541:421–424. doi: 10.1038/nature20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King C., Sarabipour S., Hristova K. The FRET signatures of noninteracting proteins in membranes: simulations and experiments. Biophys. J. 2014;106:1309–1317. doi: 10.1016/j.bpj.2014.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King C., Raicu V., Hristova K. Understanding the FRET signatures of interacting membrane proteins. J. Biol. Chem. 2017;292:5291–5310. doi: 10.1074/jbc.M116.764282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L., Novicky L., Hristova K. Measuring the energetics of membrane protein dimerization in mammalian membranes. J. Am. Chem. Soc. 2010;132:3628–3635. doi: 10.1021/ja910692u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer B.H., Segura J.M., Vogel H. FRET imaging reveals that functional neurokinin-1 receptors are monomeric and reside in membrane microdomains of live cells. Proc. Natl. Acad. Sci. USA. 2006;103:2138–2143. doi: 10.1073/pnas.0507686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakraborty H., Chattopadhyay A. Excitements and challenges in GPCR oligomerization: molecular insight from FRET. ACS Chem. Neurosci. 2015;6:199–206. doi: 10.1021/cn500231d. [DOI] [PubMed] [Google Scholar]
- 7.Strachotová D., Holoubek A., Palková Z. Ato protein interactions in yeast plasma membrane revealed by fluorescence lifetime imaging (FLIM) Biochim. Biophys. Acta. 2012;1818:2126–2134. doi: 10.1016/j.bbamem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Snyder B., Freire E. Fluorescence energy transfer in two dimensions. A numeric solution for random and nonrandom distributions. Biophys. J. 1982;40:137–148. doi: 10.1016/S0006-3495(82)84468-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarabipour S., Del Piccolo N., Hristova K. Characterization of membrane protein interactions in plasma membrane derived vesicles with quantitative imaging Förster resonance energy transfer. Acc. Chem. Res. 2015;48:2262–2269. doi: 10.1021/acs.accounts.5b00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Piccolo N., Placone J., Hristova K. Production of plasma membrane vesicles with chloride salts and their utility as a cell membrane mimetic for biophysical characterization of membrane protein interactions. Anal. Chem. 2012;84:8650–8655. doi: 10.1021/ac301776j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen S., Ushiro H., Chinkers M. A native 170,000 epidermal growth factor receptor-kinase complex from shed plasma membrane vesicles. J. Biol. Chem. 1982;257:1523–1531. [PubMed] [Google Scholar]
- 12.Scott R.E., Perkins R.G., Maercklein P.B. Plasma membrane vesiculation in 3T3 and SV3T3 cells. I. Morphological and biochemical characterization. J. Cell Sci. 1979;35:229–243. doi: 10.1242/jcs.35.1.229. [DOI] [PubMed] [Google Scholar]
- 13.Strisovsky K. Structural and mechanistic principles of intramembrane proteolysis--lessons from rhomboids. FEBS J. 2013;280:1579–1603. doi: 10.1111/febs.12199. [DOI] [PubMed] [Google Scholar]
- 14.Düsterhöft S., Künzel U., Freeman M. Rhomboid proteases in human disease: mechanisms and future prospects. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:2200–2209. doi: 10.1016/j.bbamcr.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 15.Arutyunova E., Panwar P., Lemieux M.J. Allosteric regulation of rhomboid intramembrane proteolysis. EMBO J. 2014;33:1869–1881. doi: 10.15252/embj.201488149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson N., Březinová J., Strisovsky K. Quantitative proteomics screen identifies a substrate repertoire of rhomboid protease RHBDL2 in human cells and implicates it in epithelial homeostasis. Sci. Rep. 2017;7:7283. doi: 10.1038/s41598-017-07556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adrain C., Strisovsky K., Freeman M. Mammalian EGF receptor activation by the rhomboid protease RHBDL2. EMBO Rep. 2011;12:421–427. doi: 10.1038/embor.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohi O., Urban S., Freeman M. Diverse substrate recognition mechanisms for rhomboids; thrombomodulin is cleaved by Mammalian rhomboids. Curr. Biol. 2004;14:236–241. doi: 10.1016/j.cub.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Noy P.J., Swain R.K., Bicknell R. Sprouting angiogenesis is regulated by shedding of the C-type lectin family 14, member A (CLEC14A) ectodomain, catalyzed by rhomboid-like 2 protein (RHBDL2) FASEB J. 2016;30:2311–2323. doi: 10.1096/fj.201500122R. [DOI] [PubMed] [Google Scholar]
- 20.Cheng T.L., Wu Y.T., Wu H.L. Functions of rhomboid family protease RHBDL2 and thrombomodulin in wound healing. J. Invest. Dermatol. 2011;131:2486–2494. doi: 10.1038/jid.2011.230. [DOI] [PubMed] [Google Scholar]
- 21.Pascall J.C., Brown K.D. Intramembrane cleavage of ephrinB3 by the human rhomboid family protease, RHBDL2. Biochem. Biophys. Res. Commun. 2004;317:244–252. doi: 10.1016/j.bbrc.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 22.Johansson L.B.Å., Engström S., Lindberg M. Electronic energy transfer in anisotropic systems. III. Monte Carlo simulations of energy migration in membranes. J. Chem. Phys. 1992;96:3844–3856. [Google Scholar]
- 23.Engström S., Lindberg M., Johansson L.B.Å. Monte Carlo simulations of electronic energy transfer in three-dimensional systems: a comparison with analytical theories. J. Chem. Phys. 1988;89:204–213. [Google Scholar]
- 24.Štefl M., Šachl R., Hof M. Dynamics and size of cross-linking-induced lipid nanodomains in model membranes. Biophys. J. 2012;102:2104–2113. doi: 10.1016/j.bpj.2012.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Šachl R., Amaro M., Hof M. On multivalent receptor activity of GM1 in cholesterol containing membranes. Biochim. Biophys. Acta. 2015;1853:850–857. doi: 10.1016/j.bbamcr.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Mlcochová P., Barinka C., Konvalinka J. Prostate-specific membrane antigen and its truncated form PSM’. Prostate. 2009;69:471–479. doi: 10.1002/pros.20894. [DOI] [PubMed] [Google Scholar]
- 27.Rajasekaran S.A., Anilkumar G., Rajasekaran A.K. A novel cytoplasmic tail MXXXL motif mediates the internalization of prostate-specific membrane antigen. Mol. Biol. Cell. 2003;14:4835–4845. doi: 10.1091/mbc.E02-11-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubankova A., Humpolickova J., Boura E. Negative charge and membrane-tethered viral 3B cooperate to recruit viral RNA dependent RNA polymerase 3D pol. Sci. Rep. 2017;7:17309. doi: 10.1038/s41598-017-17621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boukamp P., Petrussevska R.T., Fusenig N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barinka C., Mlcochová P., Konvalinka J. Amino acids at the N- and C-termini of human glutamate carboxypeptidase II are required for enzymatic activity and proper folding. Eur. J. Biochem. 2004;271:2782–2790. doi: 10.1111/j.1432-1033.2004.04209.x. [DOI] [PubMed] [Google Scholar]
- 31.Sezgin E., Kaiser H.J., Levental I. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat. Protoc. 2012;7:1042–1051. doi: 10.1038/nprot.2012.059. [DOI] [PubMed] [Google Scholar]
- 32.Humpolickova J., Mejdrová I., Boura E. Fluorescent inhibitors as tools to characterize enzymes: case study of the lipid kinase phosphatidylinositol 4-kinase IIIβ (PI4KB) J. Med. Chem. 2017;60:119–127. doi: 10.1021/acs.jmedchem.6b01466. [DOI] [PubMed] [Google Scholar]
- 33.Humpolícková J., Gielen E., Engelborghs Y. Probing diffusion laws within cellular membranes by Z-scan fluorescence correlation spectroscopy. Biophys. J. 2006;91:L23–L25. doi: 10.1529/biophysj.106.089474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wahl M., Gregor I., Enderlein J. Fast calculation of fluorescence correlation data with asynchronous time-correlated single-photon counting. Opt. Express. 2003;11:3583–3591. doi: 10.1364/oe.11.003583. [DOI] [PubMed] [Google Scholar]
- 35.Gregor I., Enderlein J. Time-resolved methods in biophysics. 3. Fluorescence lifetime correlation spectroscopy. Photochem. Photobiol. Sci. 2007;6:13–18. doi: 10.1039/b610310c. [DOI] [PubMed] [Google Scholar]
- 36.Kapusta P., Macháň R., Hof M. Fluorescence lifetime correlation spectroscopy (FLCS): concepts, applications and outlook. Int. J. Mol. Sci. 2012;13:12890–12910. doi: 10.3390/ijms131012890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumann J., Fayer M.D. Excitation transfer in disordered two-dimensional and anisotropic three-dimensional systems: effects of spatial geometry on time-resolved observables. J. Chem. Phys. 1986;85:4087–4107. [Google Scholar]
- 38.Schwille P., Meyer-Almes F.J., Rigler R. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys. J. 1997;72:1878–1886. doi: 10.1016/S0006-3495(97)78833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ptacek J., Nedvedova J., Barinka C. The calcium-binding site of human glutamate carboxypeptidase II is critical for dimerization, thermal stability, and enzymatic activity. Protein Sci. 2018;27:1575–1584. doi: 10.1002/pro.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schülke N., Varlamova O.A., Olson W.C. The homodimer of prostate-specific membrane antigen is a functional target for cancer therapy. Proc. Natl. Acad. Sci. USA. 2003;100:12590–12595. doi: 10.1073/pnas.1735443100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laurence T.A., Ly S., Coleman M.A. Fluorescence correlation spectroscopy at micromolar concentrations without optical nanoconfinement. J. Phys. Chem. B. 2014;118:9662–9667. doi: 10.1021/jp505881z. [DOI] [PubMed] [Google Scholar]
- 42.Magde D., Elson E., Webb W.W. Thermodynamic fluctuations in a reacting system - measurement by fluorescence correlation spectroscopy. Phys. Rev. Lett. 1972;29:705–708. [Google Scholar]
- 43.Wozniak A.K., Schröder G.F., Oesterhelt F. Single-molecule FRET measures bends and kinks in DNA. Proc. Natl. Acad. Sci. USA. 2008;105:18337–18342. doi: 10.1073/pnas.0800977105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diez M., Zimmermann B., Gräber P. Proton-powered subunit rotation in single membrane-bound F0F1-ATP synthase. Nat. Struct. Mol. Biol. 2004;11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- 45.Munro S., Pelham H.R. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 46.Lewis M.J., Sweet D.J., Pelham H.R. The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell. 1990;61:1359–1363. doi: 10.1016/0092-8674(90)90699-f. [DOI] [PubMed] [Google Scholar]
- 47.Lewis M.J., Pelham H.R. A human homologue of the yeast HDEL receptor. Nature. 1990;348:162–163. doi: 10.1038/348162a0. [DOI] [PubMed] [Google Scholar]
- 48.Matsukawa S., Moriyama Y., Kuroda H. KDEL tagging: a method for generating dominant-negative inhibitors of the secretion of TGF-beta superfamily proteins. Int. J. Dev. Biol. 2012;56:351–356. doi: 10.1387/ijdb.123514sm. [DOI] [PubMed] [Google Scholar]
- 49.Dale R.E., Eisinger J., Blumberg W.E. The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys. J. 1979;26:161–193. doi: 10.1016/S0006-3495(79)85243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akrap N., Seidel T., Barisas B.G. Förster distances for fluorescence resonant energy transfer between mCherry and other visible fluorescent proteins. Anal. Biochem. 2010;402:105–106. doi: 10.1016/j.ab.2010.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kreutzberger A.J.B., Urban S. Single-molecule analyses reveal rhomboid proteins are strict and functional monomers in the membrane. Biophys. J. 2018;115:1755–1761. doi: 10.1016/j.bpj.2018.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zettl M., Adrain C., Freeman M. Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell. 2011;145:79–91. doi: 10.1016/j.cell.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greenblatt E.J., Olzmann J.A., Kopito R.R. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat. Struct. Mol. Biol. 2011;18:1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adrain C., Zettl M., Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335:225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neal S., Jaeger P.A., Hampton R.Y. The Dfm1 derlin is required for ERAD retrotranslocation of integral membrane proteins. Mol. Cell. 2018;69:306–320.e4. doi: 10.1016/j.molcel.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goder V., Carvalho P., Rapoport T.A. The ER-associated degradation component Der1p and its homolog Dfm1p are contained in complexes with distinct cofactors of the ATPase Cdc48p. FEBS Lett. 2008;582:1575–1580. doi: 10.1016/j.febslet.2008.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lazareno-Saez C., Arutyunova E., Lemieux M.J. Domain swapping in the cytoplasmic domain of the Escherichia coli rhomboid protease. J. Mol. Biol. 2013;425:1127–1142. doi: 10.1016/j.jmb.2013.01.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.