Abstract
Mealybugs (Hemiptera: Pseudococcidae) are economically significant agricultural pests on many different crops. Because of their small size and lack of easily visible characters for identification, determination of their taxonomic status is difficult and requires technical competency to prepare a slide-mounted specimen. The standard mounting technique does not allow for analysis of the genome of the specimen. Conversely, preparatory techniques for genetic analysis of mealybugs cause either loss of the entire individual or physical damage that can make morphology-based identification difficult. This study describes a simple protocol that does not impact physical integrity of the specimen for fixation and microscopic examination yet enables simultaneous DNA extraction for DNA-based identification of four mealybug species. All species prepared yielded high quality slide mounts, identified as Planococcus citri Risso, Pseudococcus viburni Signoret, Rhizoecus kondonis Kuwana, or Rhizoecus californicus Ferris. DNA extracted in this manner had higher purity and yield in the final eluate than in samples extracted using standard methods. DNA extracted was successfully amplified by polymerase chain reaction using primers for the cytochrome oxidase I gene and subsequently sequenced for all specimens. This protocol is likely to be applicable to other Hemiptera taxa that are preserved by slide mounting, allowing for both the preparation of a high-quality voucher specimen for morphological identification and simultaneous analysis of DNA for the same specimen. The methods used are technically less challenging than current standard procedures.
Keywords: mealybug, taxonomy, identification, slide-mount, DNA extraction
Mealybugs (Hemiptera: Pseudococcidae) are the second largest family of scale insects, with approximately 2,000 described species in more than 270 genera ( Ben-Dov et al. 2003 ). The family Pseudococcidae has a worldwide distribution but is more common in the subtropics and tropics ( Ben-Dov 1994 ). Their name derives from a white, waxy secretion found on the bodies of adult females and nymphs of most species. Many members of this family are pests of a wide variety of crops grown in tropical, subtropical, and temperate regions, with some species causing significant impact on yield and quality. Economic crop losses occur either from large populations of mealybugs and excessive production of honeydew that serves as a substrate for the growth of sooty molds ( Geiger and Daane 2001 ) or from transmission of viruses that can drastically reduce crop yields ( Golino et al. 2002 ).
Mealybug taxonomy has generally been based on morphological characters of adult females ( Downie and Gullan 2004 ), with relatively few studies focused on adult males ( Beardsley 1960 , Beardsley 1962 , Afifi 1968 , Hodgson 2012 ). This is likely due to the ephemeral nature of adult males and difficulty in capturing them. Furthermore, some species of mealybug are parthenogenic ( Lloyd 1952 , Gullan et al. 2010 ), thus eliminating the possibility of using males to identify those species and creating inconsistencies in phylogenetic studies. Because of this, use of adult females remains the standard for taxonomy, phylogenetic analysis, and species descriptions in the family Pseudococcidae. Recent advances in genetics have made DNA barcoding a convenient means to classify the members of Pseudococcidae and compare against the traditional morphology-based techniques. Multiple studies have begun studying molecular variation and using sequence data to construct phylogenies for the Pseudococcidae ( Downie and Gullan 2004 , Hardy et al. 2008 , Malausa et al. 2011 ). Only recently have phylogenetic studies integrated morphological data with DNA sequence data ( Hardy et al. 2008 , Malausa et al. 2011 ).
One issue with the preparation of mealybugs for slide-mounting and identification is the physical manipulation of the specimen to adequately clear the body contents, so that important morphological features are recognizable. The Systematic Entomology Laboratory of the United States Department of Agriculture (USDA) has a document that provides instructions for slide-mounting scales and mealybugs ( http://www.ars.usda.gov/SP2UserFiles/Place/12754100/IDService/scaleslides.pdf ), with variations of this standard protocol mentioned in many additional publications ( Gullan 2000 , Downie and Gullan 2004 , Triplehorn and Johnson 2005 , Malausa et al. 2011 ). Although this technique yields high-quality slide mounts, it damages the specimen, requires manipulation to clear body contents that takes a high level of skill, and results in the loss of genetic material. Herein we describe a novel and easy-to-use technique to extract DNA from adult female mealybugs while maintaining a fully intact specimen that can be slide mounted without suffering external morphological damage that is suitable for serving as a physical voucher for the exact sequence data to support research or extension activities requiring mealybug identification to species. The technique developed will hereafter be termed the “EPED protocol” (extended proteinase and extended detergent).
Materials and Methods
Sample Collection
Four mealybug species were selected for this study. Planococcus citri Risso and Pseudococcus viburni Signoret were collected from various plants in greenhouses at the University of California, Davis, CA. Specimens of Rhizoecus kondonis Kuwana and Rhizoecus californicus Ferris were collected from soil samples and were observed feeding on grass roots in a vineyard from Oakville, CA. These species were tentatively identified on-site.
DNA Extraction and Clearing of Specimens
For both P. citri and Ps. viburni, 10 adult females were used for extraction of DNA and preparation of voucher specimens using the EPED protocol. An additional 10 specimens for each P. citri and Ps. viburni had DNA extracted from them using the standard protocol, where the insects were individually macerated and DNA was extracted using the DNeasy Blood and Tissue Kit (Qiagen, Carol Stream, IL) following manufacturer’s instructions to compare DNA yield (ng/µl) and DNA purity (absorbance ratio 260/280) between the Qiagen protocol and the EPED protocol. For R. kondonis, two adult females were used for the EPED protocol, and due to scarcity of specimens, no individuals could be spared for use in the standard protocol. For R. californicus , four adult females were prepared using the EPED protocol, and due to scarcity of specimens no individuals were processed using the standard protocol. For the EPED protocol, adult female mealybugs were placed in a 1.5 ml microcentrifuge tube with 180 μl of buffer ATL and 20 μl of Proteinase K provided in the DNeasy Blood and Tissue kit and lysed at 56°C from 8 h to 3 d, depending on the size of the mealybug. Once the specimen became transparent, the 200 μl of supernatant was transferred to a new 1.5 ml microcentrifuge tube, and the DNA extraction protocol provided in the kit was followed on the supernatant. Next, 200 μl of buffer ATL was added to the tube with the mealybug exoskeleton followed by 200 μl buffer AL. At no stage was the tube with the exoskeleton vortexed. Once the buffer AL was added, the tube was placed at 96°C until the fat bodies and wax completely dissolved, a period ranging from 1 d to 1 wk. Fat bodies and wax appeared as a small, clear sphere within the exoskeleton at 96°C, but if the specimen was cooled to room temperature before the fat bodies were dissolved, the sphere became white. DNA was extracted from 10 individuals of P. citri and Ps. viburni using the DNeasy Blood and Tissue Kit as per the manufacturer’s instructions. DNA quantification was performed on final extract for each specimen of each species using a NANODROP 1000 Spectrophotometer (Thermo Scientific, Waltham, MA).
Slide-Mounting and Photography
The specimen was washed from the microcentrifuge tube with 85% ethanol onto a Kimwipe sheet (Kimberly-Clark Professional, Roswell, GA) placed over a Petri dish. Next, the specimen was transferred with a paintbrush to a Petri dish containing double stain (BioQuip, Rancho Dominguez, CA) and left at room temperature for 2 h. The specimen was then transferred to a Petri dish containing 85% ethanol and gently washed until the desired level of stain was achieved, then transferred with a paintbrush to a Petri dish containing clove oil and left at room temperature for a minimum of 12 h. The specimens were subsequently placed on a dry microscope slide and about 100 μl of Euparal Mounting Medium (BioQuip, Rancho Dominguez, CA) was placed over the specimen, and a coverslip was added. The slides were then dried on a slide warmer at 40°C for 1 wk. Once dried, the specimens were identified to genus and then to species using the key provided by McKenzie (1967) . Slide-mounted specimens were identified and photographed on a Leica DM5000B using the image builder option in LAS V43 (Leica Microsystems, Wetzlat, Germany). The standard protocol referred to for slide preparation is that of the USDA protocol ( http://www.ars.usda.gov/SP2UserFiles/Place/12754100/IDService/scaleslides.pdf ) and was used as a basis of comparison.
Polymerase Chain Reaction, Sequence Analysis, and Data Analysis
DNA extracted from mounted specimens and specimens prepared with the Qiagen protocol was subjected to polymerase chain reaction (PCR) using primers specific to the cytochrome oxidase I (COI) gene in the mitochondrial genome. The forward primer sequence was 5’-TTG ATT TTT TGG TCA TCC AGA AGT-3’ and the reverse primer sequence was 5’-TCC AAT GCA CTA ATC TGC CAT ATT A-3’ ( Simon et al. 1994 ). PCRs were performed in 25 μl reactions with 0.5 µM of each primer, 200 µM dNTPs, 5X Green GoTaq Flexi Buffer, 25 mM MgCl 2, and 1.5 U GoTaq Flexi DNA Polymerase with the remaining volume made up with DEPC treated H 2 O. Two microliters of DNA template were used for each reaction. Thermal cycling conditions were an initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 56°C, extension for 2 min at 72°C followed by a final extension for 5 min at 72°C. Product was visualized for successful amplification on a 2% agarose gel. PCR product was purified using the QIAquick PCR Purification Kit (Qiagen, Carol Stream, IL) as per manufacturer’s instructions. DNA sequencing was done at the DNA Sequencing Facility at the University of California, Davis (Davis, CA). Contiguous files were created using Vector NTI v11.5 (Life Technologies, Benicia, CA), aligned using MEGA 5.2 ( Tamura et al. 2011 ), and were BLAST searched against the GenBank database for comparison.
DNA yield and purity were compared between the two protocols for P. citri and Ps. viburni using a standard, one-way analysis of variance in SAS 9.4.
Results and Discussion
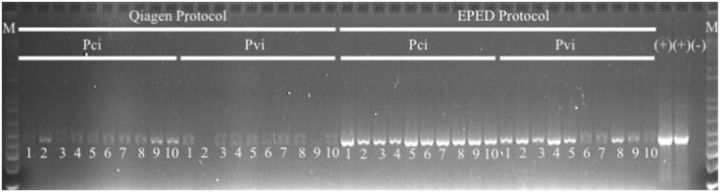
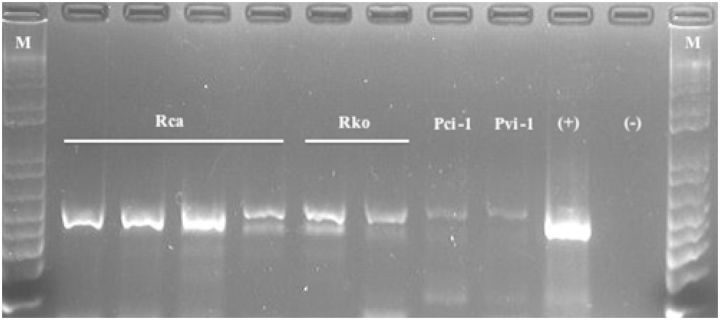
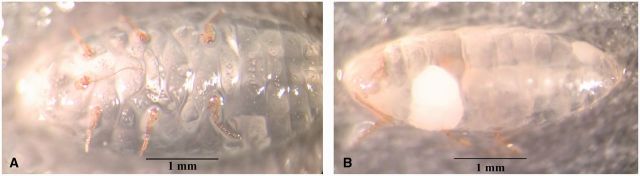
DNA yield data for samples of P. citri, Ps. viburni extracted using the standard protocol, that required complete maceration of samples and for the EPED protocol are presented in Table 1 . DNA purity data for samples of P. citri, Ps. viburni extracted using the standard protocol, that required complete maceration of samples and for the EPED protocol are presented in Table 2 . There was a statistically significant difference between the two extraction protocols for both DNA yield between P. citri and Ps. viburni and DNA purity in the final extract for both P. citri and Ps. viburni . On average, extractions conducted using the EPED protocol yielded 10.6 ng/μl more DNA for P. citri and 16.8 ng/μl more DNA for Ps. viburni than the Qiagen protocol and more pure eluate for both species. Although the difference in actual DNA yield was higher in the EPED protocol, the values obtained from the Qiagen protocol are high enough for amplification by PCR and suitable for research purposes. The higher purity of DNA in the final extract produced by this protocol than that of the standard protocol is highly significant and appears to produce higher quality PCR results ( Fig. 1 ). The 260/280 ratio measurement is an evaluation of the contaminants present in the eluate with accepted ratios to be between 1.8 and 2.0 for DNA (T009-Technical Bulletin, Thermo Scientific, Waltham, MA). The high values presented in Table 1 for both species are likely due to residual cellular components that were not completely cleaned out and, possibly, from minute fragments of exoskeleton present from maceration that do not exist in the eluate produced from the EPED protocol. Regardless of the nature of the contaminants, they likely contributed to lower quality PCR that included weaker signals and nonspecific bands in many of the samples ( Fig. 1 ), whereas samples amplified from eluate produced from the EPED protocol produced relatively strong symbols and produced only one band of the expected size ( Fig. 1 ). Although the extraction methods could not be compared for the two species of Rhizoecus examined in this study, sufficient DNA of adequate purity was produced by the EPED protocol to allow for successful amplification for these specimens ( Fig. 2 ). All specimens of R. californicus and R. kondonis yielded substantially lower quantities of DNA per μl of eluate than did extractions of P. citri and Ps. viburni but much stronger signals were produced in the PCR reactions when compared with the positive controls of P. citri and Ps. viburni that were extracted with the Qiagen protocol ( Fig. 2 ). This is also an indicator that a high degree of DNA purity in the final eluate is essential for efficient and consistent PCR reactions. Complete clearing of an individual after lysis and dissolving of wax and fat bodies is shown in Fig. 3 .
Table 1.
DNA yield data obtained from specimens of P. citri , Ps. viburni , R. californicus, and R. kondonis using Qiagen DNeasy blood and tissue kit
| Species |
Qiagen protocol
|
EPED protocol
|
F test | P | ||||
|---|---|---|---|---|---|---|---|---|
| n | Range | Mean ± SE | n | Range | Mean ± SE | |||
| P. Citri | 10 | 7.88–26.27 | 17.19 ± 1.9 | 10 | 18.1–40.04 | 27.7 ± 3.2 | 8.22 | 0.010 |
| Ps. viburni | 10 | 9.19–40.50 | 21.3 ± 4.2 | 10 | 15.03–56.44 | 38.1 ± 4.7 | 7.14 | 0.016 |
| R. californicus | 0 | NA | NA | 4 | 3.11–5.01 | 4.16 ± 0.41 | NA | NA |
| R. kondonis | 0 | NA | NA | 2 | 6.21–10.02 | 8.12 ± 1.91 | NA | NA |
Extractions used the whole body of adult females. Values represent ng/µl in final eluate.
Table 2.
DNA purity data obtained from specimens of P. citri , Ps. viburni , R. californicus, and R. kondonis using Qiagen DNeasy blood and tissue kit
| Species |
Qiagen protocol
|
EPED protocol
|
F test | P | ||||
|---|---|---|---|---|---|---|---|---|
| n | Range | Mean ± SE | n | Range | Mean ± SE | |||
| P. citri | 10 | 3.99 to 36.36 | 7.70 ± 3.24 | 10 | 1.73 to 2.05 | 1.90 ± 0.03 | 32.078 | <0.0001 |
| Ps. viburni | 10 | −0.99 to 11.36 | 3.87 ± 1.04 | 10 | 1.84 to 2.11 | 1.98 ± 0.03 | 32.912 | <0.0001 |
| R. californicus | 0 | NA | NA | 4 | 1.54 to 2.20 | 1.87 ± 0.14 | NA | NA |
| R. kondonis | 0 | NA | NA | 2 | 1.51 to 1.74 | 1.63 ± 0.12 | NA | NA |
Extractions used the whole body of adult females.
Fig. 1.
Gel electrophoresis of amplicons produced from DNA extracted from mealybugs using standard DNA extraction protocol and the EPED protocol; samples present under EPED correspond to voucher specimens in Figs. 4 and 5 , first (+) =plasmid with COI region of P. citri , second (+) = plasmid with COI region of Ps. viburni , (−) = water control, M = 1 kb + ladder.
Fig. 2.
Gel electrophoresis of amplicons produced from DNA extracted from R. californicus (Rca) and R. kondonis (Rko) using protocol described herein. Pci, amplicon from P. citri using Qiagen protocol; Pvi, amplicon from Ps. viburni using Qiagen protocol; (+), Ps. viburni COI amplicon in plasmid; (−), water control; M, 1 kb + ladder.
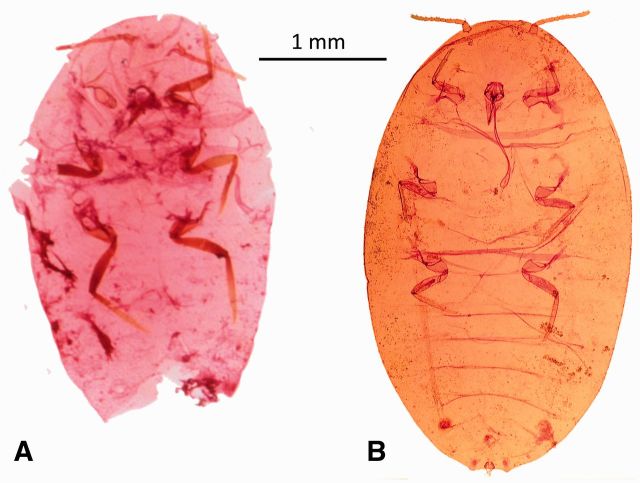
Fig. 3.
Exoskeleton of Ps. viburni specimen after being properly cleared (A) and with remnant wax and fat bodies (B).
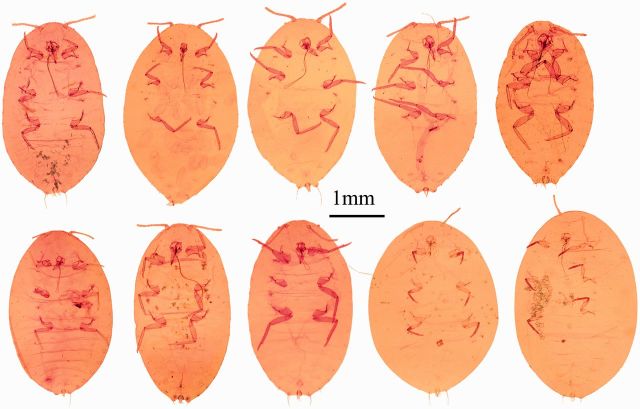
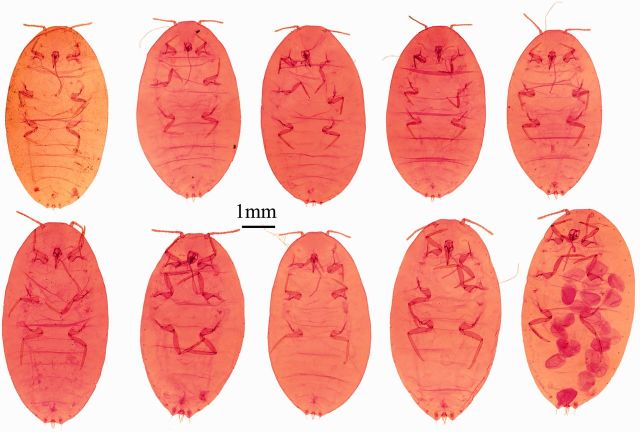
For all specimens prepared with the EPED protocol, a series of slide-mounted vouchers was obtained for P. citri ( Fig. 4 ), Ps. viburni ( Fig. 5 ), R. californicus ( Fig. 6 ), and R. kondonis ( Fig. 7 ) with all relevant morphological characters necessary for identification by visible keys and terminology provided by McKenzie (1967) ( Fig. 8 ). The two species collected from greenhouses were identified as P. citri and Ps. Viburni, and the specimens collected from soil samples were identified as R. californicus and R . kondonis . Morphological identification made on these voucher specimens matched identifications based on comparison of COI sequence data for two of the four species examined in this study. Sequence data obtained from vouchers identified as P. citri (KR014243) shared 99% identity with P. citri sequences available on GenBank (JF714160.1). Sequence data obtained from vouchers identified as Ps. viburni (KR014244) shared 100% identity with Ps. viburni sequences available on GenBank (JF714166.1). The two species where sequence data could not be compared were R. californicus and R. kondonis because sequence data for the COI gene for these species were not present in GenBank, but they were found to share 88% identity with Ps. viburni (KJ530622.1) for the same region examined. Sequence data for R. kondonis (KR014242) in this study and R. californicus for this region of COI represent new barcodes.
Fig. 4.
Image plate for corresponding voucher specimens created for P. citri from Fig. 2 .
Fig. 5.
Image plate for corresponding voucher specimens created for Ps. viburni from Fig. 2 .
Fig. 6.
Image plate for corresponding voucher specimens created for R. californicus from Fig. 3 .
Fig. 7.
Image plate for corresponding voucher specimens created for R. kondonis from Fig. 3 .
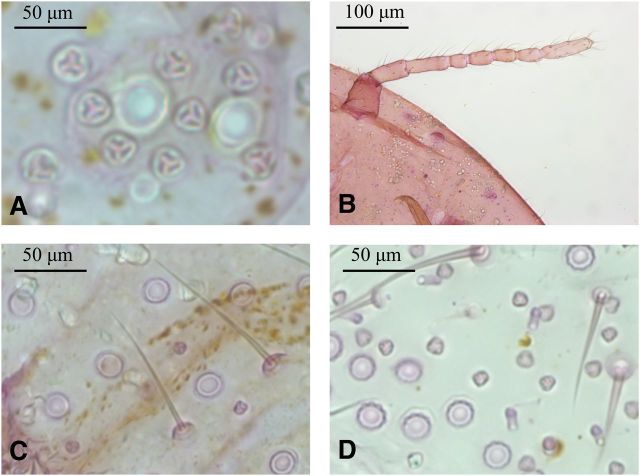
Fig. 8.
Common morphological characters used in mealybug identification: triocular pores (A) antennal morphology (B) ocular pores and setae, Ps. viburni (C) ocular pores, triocular pores, and oral-collar tubular ducts, P. citri (D).
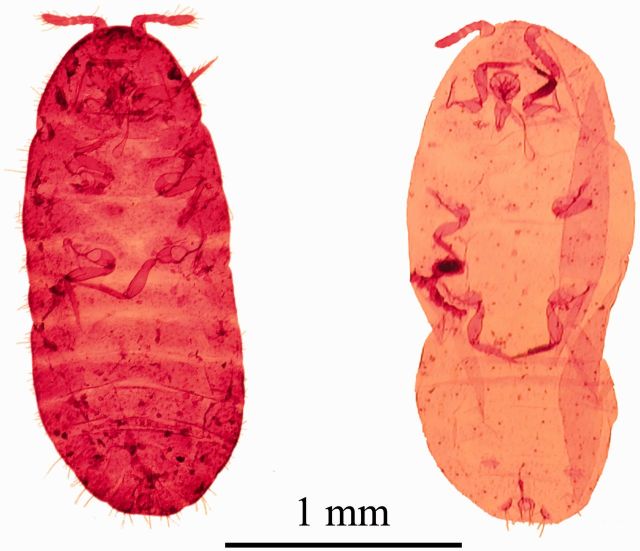
Specimens prepared using the EPED protocol yielded similar or higher quality vouchers than specimens prepared using the methods described by the USDA Systematic Entomology Laboratory ( Fig. 9 ). This EPED protocol yields a voucher specimen in about a week that is available for photography and identification and required substantially less time for smaller specimens. Indeed, the actual handling time for a specimen is only about 30 min using the EPED protocol. Once the initial lysis is complete, DNA extraction and PCR can be completed within a day and depending on access to sequencing facilities DNA data can be obtained within a few days of beginning the procedure, allowing for molecular identification to occur before the production of a voucher specimen assuming sequence data is available for the species in question. Table 3 presents a timeline for completing the EPED protocol and the actual time spent handling a specimen during that period. Experienced specialists can likely produce a slide-mounted specimen of equal quality to those presented here as well as provide a visual identification in less time than is necessary with the EPED protocol. Such professionals are not present at most research stations where identification is needed for research or extension purposes. The technical skills needed to produce high-quality specimens using the standard protocol takes a substantial amount of time to reach perfection, whereas the EPED protocol requires little technical skill and yields consistently high-quality specimens with accompanying sequence data. While a trained professional could produce a specimen and identify it in less time than that presented in the EPED protocol, when considering shipment of specimens to a specialist for preparation and identification and associated communication of results, using the EPED protocol would likely be suitable for research purposes requiring molecular research, with the additional benefit of maintaining a voucher specimen and rapid dissemination of information.
Fig. 9.
Specimens of Ps. viburni prepared using standard USDA Systematic Entomology Laboratory protocol (A) and protocol described in this article (B).
Table 3.
Average time required to complete each step for all specimens of each species included in study
| Species | Lysis | Clearing | Mounting | Handling |
|---|---|---|---|---|
| P. citri | 36.4 ± 0.9 h | 72.1 ± 0.2 h | 2.63 ± 0.12 min | 30.3 ± 0.63 min |
| Ps. viburni | 70.3 ± 0.7 h | 95.4 ± 0.1 h | 1.21 ± 0.05 min | 26.2 ± 0.21 min |
| R. kondonis | 8.5 ± 0.5 h | 2.25 ± 0.25 h | 2.90 ± 0.23 min | 35.1 ± 0.41 min |
| R. californicus | 9.2 ± 0.5 h | 2.40 ± 0.11 h | 2.88 ± 0.15 min | 32.5 ± 0.22 min |
While the EPED protocol in this article was developed using Pseudococcidae, this technique could easily be modified to accommodate other soft-bodied Hemipteran taxa, such as aphids, whiteflies, phylloxera, and scale insects. Other studies have used whole-body DNA extraction to study armored scales ( Morse and Normark 2006 ) and aphids ( Sunnucks and Hales 1996 ); however, individuals in these studies were crushed to obtain genetic material and it is unclear if crushed individuals were then mounted for identification or if separate specimens were prepared for slide-mounts while some were crushed for genetic analysis. Malausa et al. (2011) described a technique where the specimen was placed intact into a tube with the lysis time extended to between 5 and 8 h before being cleared using standard practices to prepare a slide-mounted specimen. While this technique also allows for the production of a voucher specimen with its corresponding sequence data, the level of physical manipulation and delicate handling of the specimen necessitates a high degree of technical skill.
The EPED protocol makes is possible for entomologists with little experience in mealybug taxonomy or technical skills to prepare a slide-mounted specimen to obtain an exact match between a genome and a physical voucher specimen thereby saving time and money needed to obtain species identification from a specialist. We hope that this technique can facilitate research that will increase understanding of this important and fascinating taxon.
Acknowledgments
Funding for this research was provided by the United States Department of Agriculture - Agricultural Research Service Current Research Information System project number 2032-22000-015-00D and 2023-22000-015-29S as well as the Fruit Tree, Nut Tree and Grapevine Improvement Advisory Board of the California Department of Food and Agriculture. The authors thank Trent Lawler, Ashley Li, Meredith Castillo, and Marion Eng for technical assistance in the laboratory and Dr. Sudeep Bag for assistance with sequence data.
References Cited
- Afifi S. A. 1968. . Morphology and taxonomy of the adult males of the families Pseudococcidae and Eriococcidae (Homoptera: Coccoidea) . Bull. Br. Museum (Nat. Hist.) Entomol. Suppl . 13 : 1 – 210 . [Google Scholar]
- Beardsley J. W. 1960. . A preliminary study of the males of some Hawaiian mealybugs (Homoptera: Pseudococcidae) . Proc. Hawaiian Entomol. Soc. 17 : 199 – 243 . [Google Scholar]
- Beardsley J. W. 1962. . Descriptions and notes on male mealybugs (Homoptera: Pseudococcidae) . Proc. Hawaiian Entomol. Soc. 18 : 81 – 98 . [Google Scholar]
- Ben-Dov Y. 1994. . A systematic catalogue of the mealybugs of the World (Insecta: Homoptera: Coccoidea: Pseudococcidae and Putoidae) with data on geographical distribution, host plants, biology, and economic importance. Intercept, Andover, MA . [Google Scholar]
- Ben-Dov Y., Miller D. R., Gibson G.A.P. . 2003. . ScaleNet: a database of the scale insects of the world. ( http://www.sel.barc.usda.gov/scalenet/scalenet.htm ) . [Google Scholar]
- Downie D. A., Gullan P. J. . 2004. . Phylogenetic analysis of mealybugs (Hemiptera: Coccoidea: Pseudococcidae) based on DNA sequences from three nuclear genes, and a review of the higher classification . Syst. Entomol. 29 : 238 – 259 . [Google Scholar]
- Geiger C. A., Daane K. M. . 2001. . Seasonal movement and distribution of the grape mealybug (Homoptera: Pseudococcidae): developing a sampling program for San Joaquin Valley vineyards . J. Econ. Entomol. 94 : 291 – 301 . [DOI] [PubMed] [Google Scholar]
- Golino D. A., Sim S. T., Gill R., Rowhani A. . 2002. . California mealybugs can spread grapevine leafroll disease . Calif. Agric. 56 : 196 – 201 . [Google Scholar]
- Gullan P. J. 2000. . Identification of the immature instars of mealybugs (Hemiptera: Pseudococcidae) found on citrus in Australia . Aust. J. Entomol. 39 : 160 – 166 . [Google Scholar]
- Gullan P. J., Bora Kaydan M., Hardy N. B. . 2010. . Molecular phylogeny and species recognition in the mealybug genus Ferrisia Fullaway (Hemiptera: Pseudococcidae) . Syst. Entomol. 35 : 329 – 339 . [Google Scholar]
- Hardy N. B., Gullan P. J., Hodgson C. J. . 2008. . A subfamily-level classification of mealybugs (Hemiptera: Pseudococcidae) based on integrated molecular and morphological data . Syst. Entomol. 33 : 51 – 71 . [Google Scholar]
- Hodgson C. 2012. . Comparison of the morphology of the adult males of the rhizoecine, phenacoccine, and pseudococcine mealybugs (Hemiptera: Sternorrhyncha: Coccoidea), with the recognition of the family Rhizoecidae Williams . Zootaxa 3291 : 1 – 79 . [Google Scholar]
- Lloyd D. C. 1952. . Parthenogenesis in the mealybug, Phenacoccus solani Ferris . Can. Entomol. 84 : 308 – 310 . [Google Scholar]
- Malausa T., Fenis A., Warot S., Germain J.-F., Ris N., Prado E., Botton M., Vanlerberghe-Masutti F., Sfroza R., Cruaud C., et al. . 2011. . DNA markers to disentangle complexes of cryptic taxa in mealybugs (Hemiptera: Pseudoccocidae) . J. Appl. Entomol. 135 : 142 – 155 . [Google Scholar]
- McKenzie H. L. 1967. . Mealybugs of California, 525 pp . University of California Press; , Berkeley, CA: . [Google Scholar]
- Morse G. E., Normark B. B. . 2006. . A molecular phylogenetic study of armoured scale insects (Hemiptera: Diaspididae) . Syst. Entomol. 31 : 338 – 349 . [Google Scholar]
- Simon C., Frati F., Beckenbach A., Crespi B., Liu H., Flook P. . 1994. . Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers . Ann. Entomol. Soc. Am. 87 : 651 – 701 . [Google Scholar]
- Sunnucks P., Hales D. F. . 1996. . Numerous transposed sequences of mitochondrial cytochrome oxidase I-II in aphids of the genus Sitobion (Hemiptera: Aphididae) . Mol. Biol. Evol. 13 : 510 – 524 . [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. . 2011. . MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods . Mol. Biol. Evol. 28 : 2731 – 2739 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplehorn C. A., Johnson N. F. . 2005. . Borror and DeLong’s introduction to the study of insects . Cengage Learning; , Boston, MA: . [Google Scholar]