Abstract
Objectives
To investigate the different CT characteristics which may distinguish influenza from 2019 coronavirus disease (COVID-19).
Methods
A total of 13 confirmed patients with COVID-19 were enrolled from January 16, 2020, to February 25, 2020. Furthermore, 92 CT scans of confirmed patients with influenza pneumonia, including 76 with influenza A and 16 with influenza B, scanned between January 1, 2019, to February 25, 2020, were retrospectively reviewed. Pulmonary lesion distributions, number, attenuation, lobe predomination, margin, contour, ground-glass opacity involvement pattern, bronchial wall thickening, air bronchogram, tree-in-bud sign, interlobular septal thickening, intralobular septal thickening, and pleural effusion were evaluated in COVID-19 and influenza pneumonia cohorts.
Results
Peripheral and non-specific distributions in COVID-19 showed a markedly higher frequency compared with the influenza group (p < 0.05). Most lesions in COVID-19 showed balanced lobe localization, while in influenza pneumonia they were predominantly located in the inferior lobe (p < 0.05). COVID-19 presented a clear lesion margin and a shrinking contour compared with influenza pneumonia (p < 0.05). COVID-19 had a patchy or combination of GGO and consolidation opacities, while a cluster-like pattern and bronchial wall thickening were more frequently seen in influenza pneumonia (p < 0.05). The lesion number and attenuation, air bronchogram, tree-in-bud sign, interlobular septal thickening, and intralobular septal thickening were not significantly different between the two groups (all p > 0.05).
Conclusions
Though viral pneumonias generally show similar imaging features, there are some characteristic CT findings which may help differentiating COVID-19 from influenza pneumonia.
Key Points
• CT can play an early warning role in the diagnosis of COVID-19 in the case of no epidemic exposure.
• CT could be used for the differential diagnosis of influenza and COVID-19 with satisfactory accuracy.
• COVID-19 had a patchy or combination of GGO and consolidation opacities with peripheral distribution and balanced lobe predomination.
Keywords: Coronavirus; Influenza; Pneumonia; Virus; Tomography, X-ray
Introduction
In December 2019, a cluster of pneumonia cases of unknown etiology emerged, now known as novel coronavirus disease (COVID-19), in Wuhan, Hubei, China [1–3]. In a recent study [4], five patients in a family cluster who presented with unknown-cause pneumonia after returning to Shenzhen, Guangdong Province, China, from Wuhan and members of another family who did not have any travel or residence history were finally diagnosed with COVID-19, indicating person-to-person transmission of novel coronavirus pneumonia. Another study based on the reproductive number (R0) also suggested rapid person-to-person transmission of COVID-19 [2]. In their study, R0 of COVID-19 was 2.2, indicating disease transmission from each patient to 2.2 other individuals. As of January 30, 2020, the World Health Organization (WHO) has designated the outbreak as a global health emergency. By February 25, 2019, 77,779 confirmed cases and 2666 deaths were reported in China, while 2459 confirmed cases and 34 deaths were reported in 33 different countries outside of China.
Viruses are the most common cause of respiratory infection [5]. Influenza viruses are important human respiratory pathogens that cause seasonal upper respiratory tract infections in the community, endemic infections, and periodic, unpredictable pandemics [5, 6]. Influenza viruses are single-stranded ribonucleic acid (RNA) viruses belonging to the family Orthomyxoviridae. Influenza type A is the most important of all respiratory viruses in terms of morbidity and mortality in the general population. Influenza A virus is transmitted from person to person through aerosolized or respiratory droplets. In the USA, more than 35,000 deaths and 200,000 hospitalizations due to influenza occur annually, and the number continues to increase [6]. Coronaviruses (CoVs) are a large family of viruses that cause illnesses ranging from the common cold to more severe diseases such as the Middle East respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS). The 2002–2003 pandemic of SARS and the ongoing emergence of MERS have demonstrated that CoVs posed significant threats to public health [7, 8]. A novel coronavirus is a new strain that has not been previously identified in humans [2, 9, 10]. The outbreak of COVID-19 has caused a wide concern worldwide.
The clinical manifestations of viral pneumonia are diverse and overlap [6]. The clinical symptoms such as fever, cough, and fatigue are found in both influenza pneumonia and COVID-19. It is difficult to predict etiology based on clinical manifestations. Patients with real-time polymerase chain reaction (RT-PCR) revealed positive detection of COVID-19 in throat swabs or lower respiratory tract of patients were admitted to Shanghai Public Health Center for further treatment [7, 11]. False-negative COVID-19 cases were reported in recent investigations [12, 13]. In our center, RT-PCR was performed the first three times in one case of 13 confirmed patients with COVID-19, revealing a negative detection of COVID-19. Another case who did not have epidemic exposure accompanied by dry cough was finally confirmed as COVID-19. However, CT images of the two cases showed lung lesions. Additionally, the chest CT findings of viral pneumonia were non-specific and difficult to differentiate from those of other infections [6]. However, a similar pathogenesis of pneumonia caused by the same viral family often shows similar imaging characteristics. Although not all cases manifest with typical patterns, most typical imaging patterns of viral pneumonia can be classified according to viral families [5, 6]. Recent studies have demonstrated that CT can play a critical role in the early identification of pneumonia and help in accurate diagnosis [7, 8, 10, 11, 14]. Some cases with CT characteristics, such as ground-grass opacity (GGO) lesions following peripheral and crazy-paving patterns, combined with the history of epidemic exposure can be highly suspected of having novel coronavirus pneumonia. In this season, influenza is more common than COVID-19. An accurate diagnosis in the case of no epidemic exposure and no fever symptom is worth exploration. Recent studies [15, 16] have shown that the early identification, isolation, and treatment of COVID-19 were very important. However, direct comparisons of the imaging features of the two different forms of viral pneumonia have not been performed. This study aimed to investigate the differential diagnostic CT findings between influenza and COVID-19.
Materials and methods
Patients
The retrospective study was approved by the Institutional Ethics Committee of the hospital. Patients with viral pneumonia were divided into COVID-19 and influenza groups.
COVID-19 group: A total of 13 patients with COVID-19 confirmed by RT-PCR assay were enrolled from January 16, 2020, to February 25, 2020. The first confirmed patient was diagnosed on January 16, 2020. All patients with COVID-19 underwent chest CT.
Influenza pneumonia group: A total of 4225 patients confirmed with influenza A and 1533 with influenza B by the colloidal gold method were enrolled for further evaluation from January 1, 2019, to February 25, 2020. Among these patients, 203 confirmed patients with influenza A and 65 confirmed patients with influenza B underwent chest CT. In the influenza A group, 127 patients were excluded. Of these, 87 patients showed negative CT manifestation, 5 had breathing motion artifact, 3 had tuberculosis, 3 had pneumonedema, 1 had lung cancer, 4 had a mucous plug combined with atelectasis, and 24 had bronchitis. In the influenza B group, 49 patients were excluded. Of these, 46 have negative CT manifestation, 1 had tuberculosis, 1 had bronchiectasis combined with infection, and 1 had lung cancer. A total of 123 (133/268, 49.6%) patients had negative CT manifestation. The two subgroups were combined into one influenza pneumonia group. A total of 92 patients with influenza pneumonia, including 76 confirmed with influenza A and 16 confirmed with influenza B, were included in the final study. The time duration from onset of symptoms to the CT examination (time duration of CT) was also evaluated.
CT image acquisition
All CT scans were implemented with a 64-section scanner (SOMATOM Definition, Siemens). The CT parameters were as follows: voltage, 120 kV; tube current, 140 mA; section thickness, 7 mm and breath-hold at full inspiration. All images were obtained with a 1.25-mm reconstruction section thickness and sent to the picture archiving and communication system (PACS, DJ Health Union Systems Corporation).
Two radiologists (H. W. and B. S.) with 15 and 25 years of experience, respectively, in thoracic imaging diagnosis reviewed the images of all patients using PACS independently. Each radiologist was blinded to laboratory assay results and demographic information including date of CT examination and patient name. The discrepancies were resolved by discussion.
CT images were evaluated for distribution, lobe predomination (superior lobe, inferior lobe, middle lobe, and balanced predomination), lesion number, lesion attenuation (GGO, consolidation, and GGO with consolidation), lesion margin (clear, vague), and GGO lesion involvement pattern. The distribution of lung abnormalities was recorded as central (involving mainly the central regions of the lung without subpleural involvement), peripheral (involving mainly the peripheral area of the lung), diffuse (continuous involvement without respect to lung segments), or non-specific (without predilection for subpleural or central regions) (Fig. 1). GGO was defined as a hazy increase in lung attenuation with no obscuration of the underlying vessels [6, 17]. GGO lesion involvement pattern was defined as patchy GGO, cluster-like GGO, combination of GGO and consolidation opacities, whole consolidation, which represented the lesion attenuation and involvement regions (Fig. 2). The other parameters were lesion contour (shrinking, non-shrinking, Fig. 3), bronchial wall thickening (Fig. 4), air bronchogram, tree-in-bud sign, interlobular septal thickening, intralobular septal thickening, and pleural effusion.
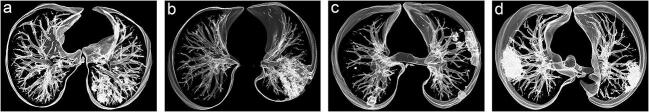
Fig. 1.
Distribution of pulmonary lesions. Volume rendering reconstruction was implemented into images a–d. a Central distribution. b Diffuse distribution. c Non-specific distribution. d Peripheral distribution
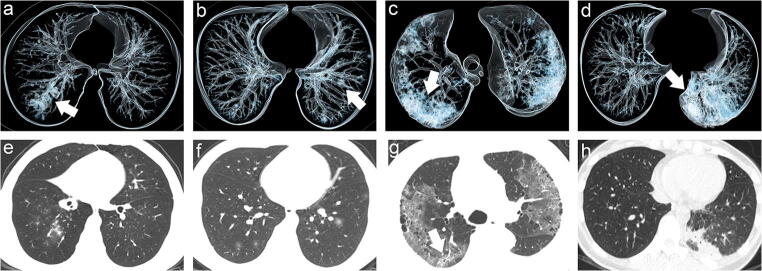
Fig. 2.
GGO lesion involvement pattern. Volume rendering reconstruction was implemented into images a–d. And e–g were the original thin-slice images. a Cluster-like involvement pattern (white arrow). b Patchy involvement pattern (white arrow). c Combination of GGO and consolidation opacities pattern (white arrow). d Whole consolidation pattern (white arrow). e Image e showed cluster-like GGO along the bronchial tree in the right inferior lobe. f Image f showed multifocal patchy GGO located in bilateral lung inferior lobe. g Image g showed combination of GGO and consolidation opacities (arrow). h Image h showed whole consolidation
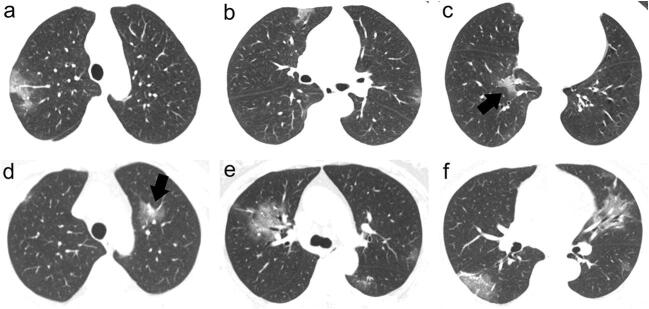
Fig. 3.
a–c A 60-year-old female patient with COVID-19 showing patchy GGO, shrinking GGO contour (black arrow), and peripheral distribution in bilateral lungs. d–f A 69-year-old female patient with COVID-19 showing combination of GGO and consolidation opacities, shrinking GGO contour (black arrow), and non-specific distribution
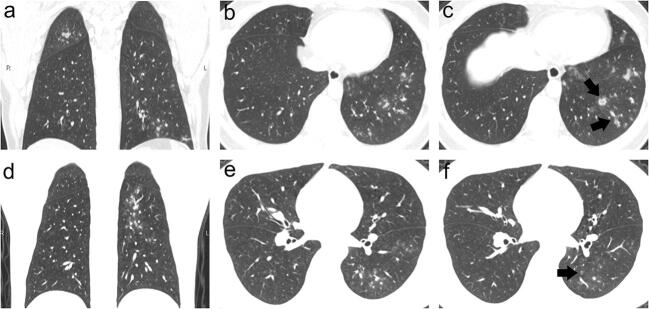
Fig. 4.
a–c A 33-year-old male patient with influenza A pneumonia showing diffuse cluster-like GGO along the bronchial tree and bronchial wall thickening (black arrow) with central distribution. d–f A 38-year-old male patient with influenza B pneumonia showing bronchial wall thickening (black arrow) and vague cluster-like GGO along the bronchial tree located in the inferior lobe
Laboratory assay
All patients underwent blood routine and influenza A and B virus antigen detection (the colloidal gold method) in our center. All confirmed patients with COVID-19 underwent RT-PCR assay in Shanghai Center for Disease Control of China.
Statistical analysis
Categorical variables were presented as a percentage and assessed using the χ2 test and Fisher’s exact test. Continuous parameters were evaluated using the Kolmogorov–Smirnov test. Continuous data with normal distribution were expressed as mean ± standard deviation and assessed using the Student t test. Two-sided p < 0.05 indicated statistical significance. Interobserver agreement between the two readers was evaluated according to the Cohen κ coefficient.
Results
Demographics
The demographic data are shown in Table 1.
Table 1.
Demographic and clinical characteristics in 13 patients with COVID-19 and 92 with influenza pneumonia
| Parameters | Virus | p* | |||
|---|---|---|---|---|---|
| COVID-19 | Influenza | ||||
| Total | Influenza A | Influenza B | |||
| Age | 49.54 ± 17.419 | 53.91 ± 20.311 | 57.29 ± 19.977 | 37.88 ± 13.236 | 0.371 |
| Sex | 1 | ||||
| Male | 7 (53.8%) | 51 (55.4%) | 43 (56.6%) | 8 (50%) | |
| Female | 6 (46.2%) | 41 (44.6%) | 33 (43.4%) | 8 (50%) | |
| Signs and symptoms | |||||
| Fever | 12 (92.3%) | 73 (79.3%) | 59 (77.6%) | 14 (87.5%) | 0.454 |
| Cough | 4 (30.8%) | 76 (82.6%) | 61 (80.3%) | 15 (93.8%) | 0.000 |
| Myalgia | 2 (15.4%) | 19 (20.7%) | 16 (21.1%) | 3 (18.8%) | 1 |
| Laboratory assay results | |||||
| White blood cell count (× 109/l) | 5.46 ± 1.377 | 6.76 ± 2.667 | 6.84 ± 2.629 | 6.33 ± 2.893 | 0.076 |
| Neutrophil count (× 109/l) | 3.58 ± 1.139 | 5.13 ± 2.498 | 5.19 ± 2.495 | 4.86 ± 2.580 | 0.061 |
| Lymphocyte count (× 109/l) | 1.29 ± 0.565 | 1.06 ± 0.529 | 1.10 ± 0.557 | 0.89 ± 0.333 | 0.515 |
| Monocyte count (× 109/l) | 0.40 ± 0.219 | 0.50 ± 0.261 | 0.49 ± 0.257 | 0.54 ± 0.285 | 0.548 |
| Time duration (days)# | 1.54 ± 0.946 | 4.42 ± 3.557 | 4.41 ± 3.636 | 4.44 ± 3.265 | 0.019 |
*p < 0.05 indicates a significant difference
#Time duration: The time duration from onset of symptoms to the CT examination
A total of 13 confirmed patients with COVID-19 were in the novel coronavirus pneumonia group, including 6 (46.2%) women and 7 (53.8%) men; their age ranged from 20 to 72 years (49.54 ± 17.419). Further, 33 (43.4%) women and 43 (56.6%) men were diagnosed with influenza A pneumonia, with age ranging from 18 to 92 years (57.29 ± 19.977). Also, 8 (50%) women and 8 (50%) men were diagnosed with influenza B, with age ranging from 18 to 70 years (37.88 ± 13.236). The most clinical symptoms were fever (12/13, 92.3% in the COVID-19 group; 73/92, 79.3% in the influenza group), cough (4/13, 30.8% in the COVID-19 group, 76/92, 82.6% in the influenza group), and myalgia (2/13, 15.4% in the COVID-19 group; 19/92, 20.7% in the influenza group). Other symptoms included fatigue, diarrhea, and chest pain.
Further, 11 of 13 (84.6%) patients with COVID-19 and 78 of 92 (84.8%) patients with influenza pneumonia showed a normal white blood cell count. No significant difference of white blood cell count was found in the two groups (p > 0.05). The time duration of CT in COVID-19 (1.54 ± 0.946) was significantly shorter than influenza pneumonia (4.42 ± 3.557) (p < 0.05).
CT image findings
The CT imaging data are summarized in Table 2.
Table 2.
CT imaging findings in 13 patients with COVID-19 and 92 with influenza pneumonia
| CT findings | COVID-19 | Influenza pneumonia | p* | Cohen κ# |
|---|---|---|---|---|
| Lesion distribution | 0.000 | 0.945 | ||
| Central | 1 (7.7%) | 69 (75.0%) | ||
| Peripheral | 5 (38.5%) | 3 (3.3%) | ||
| Diffuse | 0 | 20 (21.7%) | ||
| Non-specific | 7 (53.8%) | 0 | ||
| Lobe predomination | 0.001 | 0.927 | ||
| Superior lobe | 3 (23.1%) | 22 (23.9%) | ||
| Inferior lobe | 2 (15.4%) | 53 (57.6%) | ||
| Middle lobe | 1 (7.7%) | 7 (7.6%) | ||
| Balanced predomination | 7 (53.8%) | 10 (10.9%) | ||
| Lesion number | 0.285 | 0.914 | ||
| Single lesion | 2 (15.4%) | 12 (13.0%) | ||
| Multiple lesions in one lobe | 2 (15.4%) | 28 (30.4%) | ||
| Multiple lesions in one lung | 0 | 12 (13.0%) | ||
| Multiple lesions in bilateral lungs | 9 (69.2%) | 40 (43.5% | ||
| Lesion attenuation | 0.908 | 0.851 | ||
| GGO | 4 (30.8%) | 35 (38.0%) | ||
| Consolidation | 1 (7.7%) | 10 (10.9%) | ||
| GGO with consolidation | 8 (61.5%) | 47 (51.1%) | ||
| Lesion margin | 0.004 | 0.819 | ||
| Clear | 6 (46.2%) | 10 (10.9%) | ||
| Vague | 7 (53.8%) | 82 (89.1%) | ||
| GGO involvement pattern | 0.000 | 0.887 | ||
| Patchy GGO | 5 (38.5%) | 5 (5.4%) | ||
| Cluster-like GGO | 1 (7.7%) | 71 (77.2%) | ||
| Combination of GGO and consolidation opacities | 6 (46.2%) | 6 (6.5%) | ||
| Whole consolidation | 1 (7.7%) | 10 (10.9%) | ||
| Lesion contour | 0.000 | 0.807 | ||
| Shrinking | 9 (69.2%) | 1 (1.1%) | ||
| Non-shrinking | 4 (30.8%) | 91 (98.9%) | ||
| Bronchial wall thickening | 0 | 30 (32.6%) | 0.018 | 0.838 |
| Air bronchogram | 6 (46.2%) | 25 (27.2%) | 0.197 | 0.841 |
| Tree-in-bud sign | 0 | 22 (23.9%) | 0.065 | 0.885 |
| Interlobular septal thickening | 0 | 6 (6.5%) | 1 | 0.904 |
| Intralobular septal thickening | 2 (15.4%) | 5 (5.4%) | 0.208 | 0.941 |
| Pleura effusion | 0 | 5 (5.4%) | 1 | 1 |
GGO, ground-glass opacity. *p < 0.05 indicates a significant difference
#Cohen κ coefficient when using Kappa test
A satisfactory interobserver agreement was obtained between the two radiologists (all Cohen κ coefficient > 0.8) (Table 2). Peripheral and non-specific distributions in the COVID-19 group showed a markedly higher frequency compared with the influenza group (p < 0.05). Most lesions were located in the inferior lobe in influenza pneumonia, while COVID-19 showed a balanced lobe predomination (p < 0.05). Both COVID-19 and influenza pneumonia presented GGO and GGO with consolidation. The lesion number and attenuation showed no significant difference in the two groups (p > 0.05). More lesions had a vague margin in the influenza pneumonia group, while almost half of COVID-19 CT images presented a clear margin (p < 0.05). In terms of GGO lesion involvement pattern, patients with COVID-19 had patchy or combination of GGO and consolidation opacities, while patients with influenza pneumonia had cluster-like involvement (p < 0.05). The lesion contour showed significant differences between the groups (p < 0.05). Shrinking lesion contour was more frequently found in the COVID-19 group. Furthermore, 30 of 92 patients with influenza pneumonia showed bronchial wall thickening, while this characteristic was not found in any of the patients with COVID-19 (p < 0.05). No significant differences in CT findings, such as air bronchogram, tree-in-bud sign, interlobular septal thickening, intralobular septal thickening, and pleura effusion, were found between the two groups (all p > 0.05).
Discussion
On February 28, the WHO adjusted the global risk assessment of COVID-19 as very high. The outbreak of COVID-19 has threatened public health worldwide [18]. Early detection, isolation, and effective treatment are critical to block the spread of COVID-19, which has been declared an emergency by the WHO emergency committee [15]. Additionally, influenza has been a very common cause of respiratory tract infection in this period in China. Influenza leads to significant morbidity and mortality worldwide every year [19]. Influenza pneumonia and COVID-19 usually show similar clinical manifestations, such as cough, fever, and myalgia. The two different forms of viral pneumonia can cause a decrease in white blood cell and lymphocyte counts. These similar clinical characteristics can lead to confusion among physicians in differentiating between the two kinds of disease. It is hard for clinicians to provide an accurate diagnosis in the case of no epidemic exposure. However, treatment and clinical outcomes are completely different. Our study showed shorter time duration from onset of symptoms to the CT examination in COVID-19, because in some sites, CT was used to diagnose COVID-19 before the PCR test, and perhaps this is the reason for the short period when compared with influenza. Recently, some published studies have analyzed CT images of COVID-19 and reported many helpful CT findings. In this study, the CT findings of COVID-19 were analyzed and compared with those of influenza pneumonia. This study showed that CT could supply some positive hints in distinguishing COVID-19 from influenza pneumonia.
Peripheral and non-specific distributions showed a markedly higher frequency in the COVID-19 group (92.3%) compared with the influenza group (3.3%) in this study. A recent study including 51 confirmed patients with COVID-19 demonstrated that peripheral distribution was a characteristic distribution pattern [7]. Most pulmonary lesions in patients with COVID-19 involved bilateral lungs with multiple lung lobes and a balanced predominant distribution in the peripheral part of the lungs in this study. Previous studies reported that influenza pneumonia had lower lung lobe involvement or normal CT images [5, 20]. Most pulmonary lesions were located in the inferior lobe in influenza pneumonia (57.6%), while COVID-19 showed a balanced lung lobe predomination (53.8%). The lung lobe predomination was found to be significant in the two groups, and the results were consistent with other recent findings [5, 7].
Both COVID-19 and influenza presented GGO or GGO with consolidation in this study, and no significant difference was found in the two groups. GGO and consolidation are non-specific and common CT findings in viral pneumonia [5, 6, 20, 21]. However, COVID-19 presented a high frequency of clear lesion margin (46.2%) and shrinking contour (69.2%) compared with influenza pneumonia (10.9% and 1.1%, respectively). Additionally, more patients with influenza pneumonia showed a cluster-like pattern along the bronchial tree (77.2%), while patchy (38.5%) and combination of GGO and consolidation opacity pattern (46.2%) were seen in most patients with COVID-19. The clinicopathological features of influenza pneumonia include bronchial and surrounding alveolar hyperemia, inflammatory exudation, and formation of a transparent membrane [22], contributing to a clustered GGO lesion involvement pattern and bronchial wall thickening. A previous study reported that pulmonary lesions in SARS included bilateral extensive consolidation; localized hemorrhage and necrosis; desquamative pulmonary alveolitis and bronchitis; proliferation and desquamation of alveolar epithelial cells; exudation of proteins, monocytes, lymphocytes, and plasma cells in alveoli; and hyaline membrane formation [23]. A recent report demonstrated that the pathological features of COVID-19 greatly resembled those seen in SARS coronavirus infection, leading to similar CT manifestations such as GGO, combination of GGO and consolidation opacities, consolidation, and peripheral distribution.
Additionally, although interlobular and intralobular septal thickening was a typical characteristic of COVID-19 in recent published studies [7, 8, 11, 24], it was rarely found in this study. Only 2 of 13 (15.4%) patients with COVID-19 presented intralobular septal thickening, while none showed interlobular septal thickening. Similarly, a few patients with influenza pneumonia showed interlobular (6.5%) and intralobular septal thickening (5.4%) in this study. This was probably because most of the enrolled patients in this study were the early stage of COVID-19 (mean time from onset of symptoms to the CT examination was only 1.5 days). Multifocal lesions, GGO, and consolidation were the common CT findings of viral pneumonia; therefore, the findings including lesion number and attenuation were not significantly different between the two groups. Additionally, air bronchogram, tree-in-bud sign, and pleura effusion were more found in bacterial pneumonia, which performed not significantly different between the two groups.
This study had several limitations. First, the sample size was modest, especially for the COVID-19 cohort, limiting the differentiated performance of this study. Nevertheless, CT manifestations in COVID-19 in this study were consistent with those in previous studies. Second, this study was retrospective in design, especially for the influenza pneumonia cohort. Most patients did not undergo CT scanning, leading to a selection bias and therefore undermining the validity of the results. Third, influenza A viruses can be classified into subtypes according to a combination of 16 known hemagglutinins (H1–H16) and 9 neuraminidases (N1–N9) [25]. In this study the influenza A viruses were not divided into subgroups according to this standard. Fourth, only 13 confirmed patients with COVID-19 were enrolled in this study. They were not divided into different types according to different pathological processes, which usually present diverse CT manifestations. Finally, the findings were not compared between the influenza A and influenza B subgroups because the treatment and clinical outcomes usually were similar. Nevertheless, the results were encouraging and should be used to generate hypotheses for future large-scale multicenter studies using CT to distinguish COVID-19 from other forms of viral pneumonia.
In summary, CT can play an early warning role in the diagnosis of COVID-19 in the case of no epidemic exposure. Additionally, the results indicated that CT images, a convenient and non-invasive method, could be used for the differential diagnosis of influenza and COVID-19 with satisfactory accuracy. GGO and GGO with consolidation were the common CT findings of viral pneumonia. COVID-19 had a patchy or combination of GGO and consolidation opacities with peripheral distribution and balanced lobe predomination. However, influenza pneumonia tended to have a cluster-like pattern along the bronchial tree with central distribution, bronchial wall thickening, and inferior lobe predomination.
Acknowledgments
We thank the doctors (Chenyang Wang, Yimin Zhu, Qian Zhao, Yuanyuan Chen, Yu Kong) for searching data in the Department of Radiology, Minhang Hospital, Fudan University. We also thank Lili Gu (Department of Administration) for invaluable help in manuscript preparation.
Abbreviations
- COVID-19
2019 novel coronavirus disease
- CoVs
Coronaviruses
- CT
Computed tomography
- GGO
Ground-glass opacity
- MERS
Middle East respiratory syndrome
- PACS
Picture archiving and communication system
- R0
Reproductive number
- RNA
Ribonucleic acid
- RT-PCR
Real-time polymerase chain reaction
- SARS
Severe acute respiratory syndrome
- WHO
World Health Organization
Funding information
The authors state that this work has not received any funding.
Compliance with ethical standards
Guarantor
The scientific guarantor of this publication is Bin Song.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic study
• performed at one institution
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hao Wang and Ran Wei contributed equally to this work and share first authorship.
Contributor Information
Jie Zhu, Email: zhujie_1982@163.com.
Bin Song, Email: songbin@fudan.edu.cn.
References
- 1.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang D, Hu B, Hu C et al (2020) Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed]
- 3.Zhou Z, Guo D, Li C et al (2020) Coronavirus disease 2019: initial chest CT findings. Eur Radiol. 10.1007/s00330-020-06816-7 [DOI] [PMC free article] [PubMed]
- 4.Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koo HJ, Lim S, Choe J, Choi SH, Sung H, Do KH. Radiographic and CT features of viral pneumonia. Radiographics. 2018;38:719–739. doi: 10.1148/rg.2018170048. [DOI] [PubMed] [Google Scholar]
- 6.Franquet T. Imaging of pulmonary viral pneumonia. Radiology. 2011;260:18–39. doi: 10.1148/radiol.11092149. [DOI] [PubMed] [Google Scholar]
- 7.Song F, Shi N, Shan F et al (2020) Emerging coronavirus 2019-nCoV pneumonia. Radiology. 10.1148/radiol.2020200274:200274
- 8.Pan Y, Guan H, Zhou S et al (2020) Initial CT findings and temporal changes in patients with the novel coronavirus pneumonia (2019-nCoV): a study of 63 patients in Wuhan, China. Eur Radiol. 10.1007/s00330-020-06731-x [DOI] [PMC free article] [PubMed]
- 9.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung M, Bernheim A, Mei X et al (2020) CT imaging features of 2019 novel coronavirus (2019-nCoV). Radiology. 10.1148/radiol.2020200230:200230 [DOI] [PMC free article] [PubMed]
- 11.Pan F, Ye T, Sun P et al (2020) Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 10.1148/radiol.2020200370:200370
- 12.Hao W, Li M (2020) Clinical features of atypical 2019 novel coronavirus pneumonia with an initially negative RT-PCR assay. J Infect. 10.1016/j.jinf.2020.02.008 [DOI] [PMC free article] [PubMed]
- 13.Huang P, Liu T, Huang L et al (2020) Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 10.1148/radiol.2020200330:200330 [DOI] [PMC free article] [PubMed]
- 14.Fang Y, Zhang H, Xu Y, Xie J, Pang P, Ji W (2020) CT manifestations of two cases of 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 10.1148/radiol.2020200280:200280 [DOI] [PMC free article] [PubMed]
- 15.Sohrabi C, Alsafi Z, O'Neill N et al (2020) World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19). Int J Surg. 10.1016/j.ijsu.2020.02.034 [DOI] [PMC free article] [PubMed]
- 16.Wilder-Smith A, Freedman DO (2020) Isolation, quarantine, social distancing and community containment: pivotal role for old-style public health measures in the novel coronavirus (2019-nCoV) outbreak. J Travel Med. 10.1093/jtm/taaa020 [DOI] [PMC free article] [PubMed]
- 17.Ye Z, Zhang Y, Wang Y, Huang Z, Song B (2020) Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur Radiol. 10.1007/s00330-020-06801-0 [DOI] [PMC free article] [PubMed]
- 18.Li K, Fang Y, Li W et al (2020) CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19). Eur Radiol. 10.1007/s00330-020-06817-6 [DOI] [PMC free article] [PubMed]
- 19.Reich NG, McGowan CJ, Yamana TK, et al. Accuracy of real-time multi-model ensemble forecasts for seasonal influenza in the U.S. PLoS Comput Biol. 2019;15:e1007486. doi: 10.1371/journal.pcbi.1007486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller WT, Jr, Mickus TJ, Barbosa E, Jr, Mullin C, Van Deerlin VM, Shiley KT. CT of viral lower respiratory tract infections in adults: comparison among viral organisms and between viral and bacterial infections. AJR Am J Roentgenol. 2011;197:1088–1095. doi: 10.2214/AJR.11.6501. [DOI] [PubMed] [Google Scholar]
- 21.Yuan Y, Tao XF, Shi YX, Liu SY, Chen JQ. Initial HRCT findings of novel influenza A (H1N1) infection. Influenza Other Respir Viruses. 2012;6:e114–e119. doi: 10.1111/j.1750-2659.2012.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita J, Ohtsuki Y, Higa H, et al. Clinicopathological findings of four cases of pure influenza virus A pneumonia. Intern Med. 2014;53:1333–1342. doi: 10.2169/internalmedicine.53.1174. [DOI] [PubMed] [Google Scholar]
- 23.Ding Y, Wang H, Shen H, et al. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J Pathol. 2003;200:282–289. doi: 10.1002/path.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi H, Han X, Zheng C (2020) Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019-nCoV) pneumonia in Wuhan, China. Radiology. 10.1148/radiol.2020200269:200269 [DOI] [PMC free article] [PubMed]
- 25.Wang Q, Zhang Z, Shi Y, Jiang Y. Emerging H7N9 influenza A (novel reassortant avian-origin) pneumonia: radiologic findings. Radiology. 2013;268:882–889. doi: 10.1148/radiol.13130988. [DOI] [PubMed] [Google Scholar]