Abstract
RNA interference (RNAi) is a process through which double‐stranded RNA induces the activation of cellular pathways, leading to potent and selective silencing of genes with homology to the double strand. Much excitement surrounding small interfering RNA (siRNA)‐mediated therapeutics arises from the fact that this approach overcomes many of the shortcomings previously experienced with approaches such as antibodies, antisense oligonucleotides and pharmacological inhibitors. Induction of RNAi through administration of siRNA has been successfully used in treatment of hepatitis, viral infections, and cancer. In this review we will present a brief history of RNAi, methods of inducing RNAi, application of RNAi in the therapeutic setting, and the possibilities of using this highly promising approach in the context of transplantation.
Keywords: Gene silencing, gene therapy, RNA interference, transplantation
Introduction
Methods for manipulating biological systems have included pharmacological drugs, antisense oligonucleotides (AO), ribozymes, and antibodies. In transplantation, all of these approaches have been applied with varying degrees of success. The revolutionary discovery that the endogenous cellular process of RNA interference (RNAi) can be artificially manipulated for inducing gene‐specific silencing through administration of small interfering RNA (siRNA) has led to an explosion of interest in this technique. The attractiveness of RNAi in contrast to other methods of manipulation arises from its extremely high inhibitory activity, the fact that the inhibition is very specific, and the ease with which various methods of inducing RNAi can be applied. Owing to the explosion of interest in siRNA, several others have reviewed this field concentrating on genetic mechanisms of RNAi (1), delivery methodologies (2), and effects of global gene silencing (3). In this paper we will review the therapeutic aspects of siRNA and apply them to transplant research.
RNA Interference
RNA interference is an endogenous cellular defence mechanism against viruses and transposable elements in the genome (4). Upon recognition of these ‘dangerous’ double‐stranded RNA (dsRNA), enzymatic complexes degrade any mRNA transcripts with homology to the dsRNA. This implies that artificial induction of RNAi can be useful for silencing pathological genes in a therapeutic manner. The fact that RNAi is a natural defence mechanism suggests that manipulation of this phenomenon for intervention would be a more biological approach to induce genetic alteration, as compared with other methods such as AO or chemical inhibitors of enzymes.
The initial suggestion of RNAi came from work in petunia flowers in which overexpression of the gene responsible for purple pigmentation actually caused the flowers to lose their endogenous colour (5). This phenomenon was termed ‘cosuppression’, as both the inserted gene transcript and the endogenous transcript were suppressed. The mechanism remained unclear until 1998 when Fire et al. found that the combined sense and antisense RNA led to more potent suppression of gene expression than sense or antisense used individually. The dsRNA seemed to be inducing inhibition through a pathway distinct from classical antisense inhibition, as the suppressive effect observed using the dsRNA was more potent than ever seen before. Approximately 1–3 molecules of duplexed RNA per cell were effective at knocking down gene expression. This seminal paper was the first to describe RNAi and to coin its name accordingly (6).
Despite the potency of gene inhibition, at the initial description, RNAi could not be used for any therapeutic purposes in mammalian cells owing to long dsRNA (>25 nucleotides) activating a ‘panic’ response in eukaryotic cells, part of which includes nonspecific inhibition of gene transcription and production of interferon‐α (7). The enzymes PKR and 2′5′ oligosynthetase interact with dsRNA and trigger this nonspecific response. However, these problems were overcome with the discovery of the effector mechanism of gene‐inhibition for dsRNA in 2001. It was demonstrated that after a long dsRNA duplex enters the cytoplasm, a ribonuclease III‐type enzyme, termed ‘DICER’, cleaves the duplex into smaller 21–23 base‐pairs (Figure 1A). It is these small duplexes, called ‘siRNA’, that are active in silencing endogenously produced mRNA transcripts. When siRNA with homology to a mRNA transcript enters a cell, it mobilizes a self‐aggregating complex called RNA‐induced silencing complex (RISC) that then unwinds the siRNA, hybridizes with the mRNA, induces cleavage of the mRNA, and then subsequently continues performing the same process, but without the ‘panic’ response. As depicted in Figure 1 (B), administration of preformed siRNA duplexes bypasses the nonspecific activation of PKR and 2′5′ oligosynthetase while allowing for only gene‐specific silencing through the stimulation of RISC.
Figure 1.
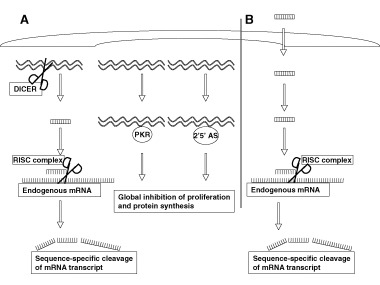
Induction of RNA interference. (A) Natural gene silencing by long double‐stranded RNA (dsRNA). Upon viral infection or the presence of other dsRNA transducted intracellularly, an innate defense system is activated that causes the sequential degradation of the dsRNA by the type III endonuclease DICER. This endonuclease subsequently cleaves the dsRNA into 21 nucleotide double‐stranded fragments. These fragments then associate with the RNA‐induced silencing complex (RISC) complex and induce cleavage of endogenous mRNA transcripts in a sequence‐ and length‐ specific manner. However, long dsRNA also activates 2′5 oligosynthetase which induces nonspecific interferon response and global shutdown of protein synthesis. (B) Artificial gene silencing through siRNA. To take advantage of the gene‐silencing effect while circumventing nonspecific cellular effects, synthetic dsRNA of 21 nucleotides are transfected into the cell. This small interfering RNA (siRNA) is not recognized by DICER or 2′5 oligosynthetase, but instead directly binds the RISC complex that subsequently induces selective silencing of endogenous transcripts.
Methods of Inducing RNAi
The discovery that siRNA is the effector mechanism of endogenous RNAi prompted investigation into the ability to utilize exogenously administered siRNA, or vectors inducing the expression of siRNA, for gene‐specific silencing. Genetic manipulation using such a strategy raises several issues that need to be addressed: (1) Stability of siRNA; (2) ability to constitutively express the siRNA; (3) possibility of tissue‐specific delivery; and (4) finding the best method for identifying effective silencing sites on the mRNA transcript. In order to answer these questions, various versions of siRNA have been developed.
Chemically synthesized siRNA
Chemical synthesis of oligonucleotides is a readily used procedure in molecular biology. Owing to the short size, 21–23 nucleotides in length of siRNA, typical nucleotide synthesis techniques can be used. However, the production of siRNA requires several additional steps including generation of the two homologous strands, annealing of the strands in vitro, addition of chemical entities to increase stability, and ensuring that 2‐nucleotide overhangs are present. The purpose of these overhangs is to activate the RNAi‐inducing enzymatic complex (RISC). In addition, the siRNA duplex requires a‐3′ hydroxyl group and a 5′ phosphate group for functional activity (8). Commercial synthesis of customized siRNA is presently available on a widespread level. Despite the ease of generating chemically synthesized siRNA, a key consideration is choosing the appropriate sequence of the duplex that would most effectively silence the mRNA transcript whose inhibition is desired. It is known that efficacy of silencing varies with segments of the transcripts that are targeted (9). At present no clear‐cut rules exist for choosing the best segment to silence, however, it is suggested that the target region should be at a least 70–100 nucleotides away from the translational initiation site of the transcript and that the AU: GC content should be as close to 50% as possible (10). Additionally, the siRNA should target coding sequences, as the process of RNAi occurs only in the cytoplasm. Using these suggestions, as well as empirical testing, a variety of experiments have been performed with chemically presynthesized siRNA.
The first utilization of chemically synthesized siRNA duplexes demonstrated effective silencing of the cytoskeletal proteins lamin A/C, lamin B1, nuclear mitotic apparatus protein (NuMA) and vimentin in human cell lines (11). Suppression was specific to the target transcript, and was detected both at the mRNA and at the protein level. Importantly, cellular viability and function was not affected by the silencing procedure. Subsequently, chemically synthesized siRNA was used as a substitute for ‘knockout’ animals or for chemical inhibitors of enzymatic pathways (12).
Of particular relevance to the field of transplantation is the paper by Hara et al. (13) elucidating the importance of the ‘raptor’ protein in mediating signal transduction induced by rapamycin through the target of rapamycin (TOR) protein. When siRNA specific to raptor was added to TOR‐overexpressing cells, the downstream signalling of this protein was abolished. In addition to the inhibition of biological pathways, chemically synthesized siRNA has been extensively used for other therapeutic and in vivo experiments, which will be described in later sections.
Enzymatically synthesized siRNA
A problem with using chemically synthesized siRNA is that the most effective target sequence on the transcript is unpredictable. However, simultaneously targeting different segments of the same transcript leads to more effective gene silencing. For example, targeting the HIV entry coreceptor CXCR‐4 using several siRNA duplexes against different segments of the mRNA transcript resulted in higher silencing activity than using a siRNA against a single target (14). An alternative to simultaneously using several chemically synthesized siRNA duplexes was recently presented, in which siRNA duplexes to every mRNA target site are generated by enzymatically cleaving long dsRNA homologous to the target gene in vitro using RNAse III extracted from Escherichia coli. Generation of long, double‐stranded siRNA is typically performed by in vitro transcription of the target gene, both in sense and antisense using the T7 RNA polymerase (15). This promoter possesses the advantage of early termination and has been widely used for in vitro transcription of RNA for almost two decades (16). This ‘multiple siRNA’ approach has the advantage of using long dsRNA for silencing without inducing the classical nonspecific ‘panic’ response (17). Although interferon production has been reported by one group using enzymatically generated siRNA (18). Despite this potential drawback, enzymatically generated siRNA was demonstrated to induce specific and potent gene silencing in a recent study where targeting of both exogenous puromycin‐resistance gene and endogenous expression of H‐ras, c‐jun and c‐fos was performed (19). The advantage of such an enzymatic approach is the rapid and effortless identification of the optimal siRNA species for silencing the desired biological function.
Comparison between chemically and enzymatically synthesized siRNA
While chemically synthesized siRNA possesses the advantage being easy to manufacture, it is very costly. Additionally, sequence selection is difficult owing to variability in targeting efficacy owing to the positional effect described earlier. The ability to enzymatically generate siRNA allows for cheaper production and more effective gene‐silencing ability, as the enzymatically generated siRNA can correspond to sequences overlapping the entire gene. Indeed, higher silencing ability using the enzymatically generated siRNA has been reported (20). A drawback of this approach is that spin columns or other methods of purification must be used to separate out the generated siRNA from contaminating uncleaved RNA duplexes, or residual nucleic acids. Despite this, it is the opinion of the authors that enzymatically synthesized siRNA is a more effective and convenient method of inducing gene‐silencing than the chemically synthesized forms.
siRNA‐expressing vectors
Several variations on a theme have been made in order to improve utility of the siRNA‐mediated gene‐silencing technique. Owing to the double‐stranded feature of siRNA, if a partially palindromic hairpin loop mRNA is expressed from a plasmid, the stalk portion of the loop would hypothetically be recognized as dsRNA and cleaved into siRNA by DICER. Such an approach possesses several advantages compared with administration of chemically synthesized siRNA: (1) The siRNA could be constitutively expressed, allowing for a higher level of silencing; (2) regulatory elements could be added to the promoter region of the plasmid such that tissue‐specific silencing occurs with a systemically administered plasmid; and (3) permanent gene ‘knock‐down’ cell lines can be established for in vitro work, or for generation of ‘knock‐down’ animals through cloning.
As both RNA pol III promoters U6 and H1 cause termination after the second uridine, the transcript formed mimics the siRNA that is naturally formed after cleavage of long dsRNA by DICER. This siRNA contains two symmetrical 3′ overhanging T or U nucleotides (nt) that are necessary for gene‐specific silencing (21). Comparisons between the efficacy of tandem and hairpin loop expressed siRNA suggest a stronger in vivo silencing efficacy using the hairpin approach. In a study by Kobayashi et al. Small interfering RNA specific to green fluorescent protein (GFP) plasmid was administered to mice using the hydrodynamic method of transfection. A superior silencing efficacy and longer inhibitory effect was observed using hairpin expressed siRNA compared with the tandem approach (22).
A novel advancement in siRNA expression is the ability to selectively activate silencing through administration of exogenous agents. Inducible siRNA‐promoters were subsequently optimized through combining RNA pol III‐elements with various commonly used repressor systems. For example a tetracycline‐inducible plasmid expressing siRNA‐silenced PI‐3 kinase in an in vivo model of prostate cancer (23). It is anticipated that the future development of tissue‐specific promoters to drive siRNA expression will occur. Such an approach would allow the systemic administration of siRNA‐expressing plasmids with activity only in the desired target tissue.
siRNA‐expression cassettes
Owing to the time‐consuming process of cloning siRNA into plasmid‐expressing constructs and the need for verification of the cloned sequence, an easier approach to screening sequences was developed. This method involves production of ‘siRNA‐expression cassettes’ (SECs). Basically, SECs consist of a PCR product, which, once transcribed, forms a RNA hairpin loop which is intracellularly cleaved into siRNA. Gene‐specific SECs are generated through a series of PCR reactions. The end result is a PCR product that contains a Pol III promoter, a DNA sequence that, once transcribed, forms hairpin siRNA and a terminator sequence (24). As the SEC can be designed with restriction sites, it is possible to clone effective SEC sequences into expression plasmids in order to raise large quantities of SECs. While the SEC technique does not allow permanent transfection of cells with siRNA, the expediency and low cost of this procedure lends itself to mass screening of siRNA libraries as well as identification of siRNA target sites.
A recent modification of the SEC method has been reported, which involves generation of a PCR product with tandem promoters to drive siRNA hairpin loop formation (25). Utilizing both a human H1 and murine U6 promoters, the sense and antisense nucleotides are transcribed in opposing directions but on the same template to generate duplex siRNA. This ‘dual promoter’ vector has successfully been used for high‐throughput screening of cDNA libraries. These types of approaches will be useful for identifying novel functional aspects of genes without a priori knowledge of the specific gene.
Delivery Strategies for siRNA
Various delivery methods have been developed for in vitro and in vivo gene silencing. The originally developed transfection protocols for siRNA used liposomal‐based reagents. Such reagents typically allow greater than 90% transfection efficacy (26). Unfortunately, they are costly and toxic in vivo. Several methods of inducing siRNA entry into cells in order to overcome conventional drawbacks are described later.
Direct administration of siRNA
The first direct delivery of siRNA in vivo was performed using the ‘hydrodynamic’ technique of administering short duplexes of naked siRNA in a large volume of saline through the tail vein (27). Successful inhibition of GFP and hepatitis surface antigen B was achieved. The observation that transfection of the siRNA in vivo does not require a liposomally based transfection reagent suggested that naked siRNA may have an endocytic pathway of entry into cells. It was demonstrated that intranasal administration of naked siRNA targeting the organ‐protecting enzyme heme oxygenase‐1 led to effective gene silencing and consequently an increase in ischemia‐reperfusion injury (28).
Infectious delivery of siRNA by viral vectors
Stable transfection of siRNA‐expressing constructs has been performed using various types of viral vector approaches. Applicability of retroviral transfection with p53‐targeting siRNA was successful in both cell lines and primary fibroblasts (29). Another viral approach involves using adenoviruses. Adenoviral delivery of siRNA was effective at decreasing formation of pathological polyglutamine‐mediated cellular aggregation in a murine model (30). A drawback of these viral approaches is that incorporation is dependent on the proliferation of target cells. In contrast, lentiviral vectors can incorporate with great efficacy in nondividing cells. In vitro silencing of GFP using lentiviral‐delivered siRNA was highly effective and long lasting (>25 days) in culture (31). Furthermore, lentiviral delivery of siRNA was capable of inhibiting HIV production from primary human T cells (32) and macrophages (33) in vitro. In vivo administration of siRNA using lentiviral vectors has demonstrated gene silencing in transgenic mice (12). Although previous to this paper, transgenic mice and rats were generated by microinjection of a DNA construct expressing siRNA (34).
siRNA Therapy
Application of siRNA as a ‘drug’ was demonstrated in recent studies (35, 36, 37, 38). The therapeutic promise of siRNA has to fulfill the following conditions: (1) Bioavailability, (2) lack of toxicity, (3) specificity of silencing effects, and (4) efficacy in vivo. It appears that siRNA meets all of these criteria. The original therapeutic indications for siRNA were performed in vivo using viral and cancer models. Now, this approach has been applied to the treatment of various diseases (Table 1).
Table 1.
In vivo therapeutic utilization of small interfering RNA
| Disease | Target gene | siRNA Type | Delivery | Reference |
|---|---|---|---|---|
| Con‐A hepatitis | Fas | Pre‐siRNA | IV | 40 |
| Fas‐induced hepatitis | Caspase‐8 | Pre‐siRNA | IV | 77 |
| Pathologic ocular angiogenesis | VEGF | Pre‐siRNA | SR | 35 |
| LPS‐sepsis | TNF | Pre‐siRNA | IP | 36 |
| Polyglutamine‐mediated | ||||
| neuro‐degeneration | Polyglutamine repeats | Retroviral | IV | 30 |
| Hepatitis B | HbsAg | Pre‐siRNA | IV | 37 |
| HIV | HIV‐REV | Lentiviral | IV | 65 |
| Colon cancer | Beta‐catenin | Pre‐siRNA | IV | 51 |
IV = intravenous, SR = subretinal, IP = intraperitoneal.
Viral infection
Small interfering RNA has been successful in treatment of viral diseases such as HIV, hepatitis (39, 40), and even the severe acute respiratory syndrome (SARS)‐associated coronavirus (41). In the case of HIV, effective silencing of both the primary HIV receptor, CD4, and the HIV coreceptor, CXCR‐4 (14), has been successfully accomplished by siRNA, resulting in the prevention of viral entry into target cells. A practical utilization of blocking HIV entry into cells could be transfecting hematopoietic stem cells with siRNA‐expressing constructs so that progeny cells are not susceptible to infection. This approach was effective in rendering monocytes derived from transfected progenitors resistant to HIV infection (42).
Induction of RNAi to target hepatitis viruses was performed in virally infected cell lines. Addition of siRNA to silence various portions of the hepatitis C virus genome led to a 98% reduction in a detectable virally infected cell (43). The in vivo applicability of siRNA was demonstrated using a systemic siRNA‐administration approach in mice expressing the hepatitis B genome in the liver. That study demonstrated reduction in viral mRNA, viral antigens, and viral genomic DNA in both liver and sera of siRNA recipients (37).
Additionally, researchers have begun exploring the in vitro utility of siRNA against a wide variety of viruses. For example, targeting the E6 gene from human papilloma virus it was possible to induce apoptosis in primary patient tumor samples (44). Plasmid‐driven siRNA specific to influenza and West Nile Virus were effective in suppressing viral transcripts and infectious virion production in virally infected cell lines (45). Using cytotoxicity of Vero cells as a surrogate marker of SARS‐virus infection, it was demonstrated that transfection with siRNA was able to effectively inhibit replication of this coronavirus subtype (41). Therefore the field of antiviral siRNA therapeutics is a very aggressively studied and newly developing area of investigation.
Cancers
As siRNA mediates very precise silencing activity, its use in blocking expression of aberrantly expressed or mutated proteins is very appealing. This concept lends itself well to therapy for tumors that possess well‐known and common oncogenic features. Initial in vitro studies have demonstrated effective silencing of a wide variety of mutated oncogenes such as K‐Ras (46), mutated p53 (47), Her2/neu (48), and bcr‐abl (49). The appeal of siRNA‐targeting, compared with conventional cytostatic drugs, is the promise of cancer‐specific killing in the absence of collateral non‐neoplastic cell damage. This concept is demonstrated by the observation that even the ‘specific’ chemically generated bcr‐abl inhibitor Gleevec also possesses inhibitory effects on nonleukemic hematopoietic stem cells (50). Such nonspecific effects are not anticipated with siRNA‐targeting.
In vivo utilization of siRNA was effectively performed by targeting the colorectal cancer‐associated gene beta‐catenin. Subsequent to siRNA‐mediated silencing of beta‐catenin, in human colon cancer cells, decreased proliferation and diminished invasiveness was observed. Additionally, when these treated cancer cells where placed in a nude mouse, prolonged survival was seen compared with mice receiving unmanipulated tumors (51). Similarly, silencing of the oncogene H‐Ras led to inhibition of in vivo tumor growth of human ovarian cancer in a SCID mouse model (52). A class of molecular targets that are very attractive in the field of oncology are the antiapoptotic family of proteins. Small interfering RNA inhibition of bcl‐2 family members is associated with increased susceptibility of prostate cancer to chemotherapeutic intervention (53). Thus, owing to the overwhelming amount of cancer‐specific genes identified on an almost daily basis, the utilization of siRNA for elucidating gene function, and perhaps for therapeutic intervention, becomes increasingly important.
Drawbacks of siRNA therapeutics
The discovery that siRNA can induce gene‐specific silencing in absence of interferon response and global protein inhibition has now come under some attack. The original paper by Elshabir et al. demonstrated that administration of chemically synthesized siRNA did not lead to interferon production or nonspecific gene inhibition (11). Indeed, in the author's hands, administration of siRNA did not elicit such effects on dendritic cells (DCs), one of the most sensitive cell types to interferon (54). Despite this, Sledz et al. (55) recently reported that administration of siRNA can induce interferon production through a PKR‐dependent mechanism. As siRNA has been administered for a variety of therapeutic usages in vivo (Table 1) and no report of nonspecific interferon induction or toxicity was published, the biological relevance of these findings should be evaluated. Another group reported that certain vectors utilized for delivery of siRNA can lead to a similar induction of interferon responses (56). This is a more plausible scenario, as plasmid DNA may cause formation of long hairpin RNA duplexes that would induce PKR. One possible explanation of the induction of the interferon response could be chemical modifications at the 3′ end of siRNA. Such a scenario was proposed by Kim et al. (18) who demonstrated that ‐3′ triphosphates on the duplex play a critical role in the activation of PKR. In any case, development of siRNA therapeutics will have to ensure that the duplex does not evoke a clinically meaningful interferon response, such a response could hypothetically lead to a wide variety of toxicities in human studies.
Utilization of siRNA in Immunology and Transplantation
The process of immune modulation offers a plethora of molecular targets for siRNA silencing such as (1) molecules on lymphocytes associated with activation; (2) molecules on antigen presenting cells (APCs) which stimulate lymphocytes; (3) soluble molecular signals such as cytokines; (4) molecules associated with lymphocyte extravasation and homing; and (5) effector molecules of immunity such as complement, perforin, or granzymes. In the context of transplantation, even more molecular targets arise, such as genes associated with ischemia/reperfusion damage and genes causing apoptosis of transplanted organs.
Immunological applications of siRNA
One of the most devastating immune‐mediated pathologies is bacterial sepsis mediated by systemic release of TNF‐α. The utilization of siRNA to silence this gene has been successfully accomplished in a murine model of sepsis (36). In addition, the ability of siRNA to modify immunological parameters was recently demonstrated in a study of leukocyte adhesion under flow conditions over TNF‐α‐activated human umbilical cord endothelial cells (HUVECs). Silencing of E‐selectin on the activated HUVECs by siRNA was effective as witnessed by lack of mRNA transcripts and inhibition of E‐selectin protein. Most importantly, leukocytes flowing over the activated HUVECs did not adhere after siRNA treatment (57). DCs silenced for IL‐12p35 exhibited higher IL‐10 production and could modulate immune responses from Th1 to Th2 in an antigen‐specific manner both in vitro and in vivo (54). Small interfering RNA‐mediated silencing of the NF‐kB p50 subunit was used to generate DCs that possessed a reduced expression of IL‐12 but still maintained maturation ability (58). The feasibility of inhibiting transcription factors in DCs was further illustrated in a study where silencing of the CIITA transcription factor not only inhibited MHC expression but also blocked the production of plexin, a structural protein that endows DCs with dendritic processes (59).
Manipulation of macrophages, peripheral blood mononuclear cells, and T cells was successfully accomplished with siRNA. These findings, combined with the demonstrated pharmacological activity of siRNA, raises the prospect of using siRNA as an immune suppressant. Previous approaches to suppressing T‐cell responses included administration of drugs (e.g. cyclosporine), antibodies (e.g. anti‐CD154), or fusion proteins (e.g. CTLA4‐Ig). Unfortunately, these strategies all possess significant drawbacks such as organotoxicity, lack of specificity, increased thromboembolisms, and poor pharmacokinetics (60, 61, 62). Based on the previous therapeutic utilization of siRNA to accomplish a wide variety of gene silencing therapeutically, we anticipate that silencing of immunological genes in T cells will be a feasible and practical alternative to traditional immune suppressants. Although an earlier study raised concern that inhibitory effects of RNAi may be diluted in proliferating T cells after administration of duplexed siRNA (63), more recent studies using plasmid‐driven (64) or lentiviral‐delivered (65) siRNA have not suffered this drawback.
siRNA gene targets in transplantation
Immunological attack of the grafted organ is initially mediated by T‐cell responses. Inhibition of this cellular target would require identification of ‘master regulator’ genes that control a plethora of downstream biological cascades. Molecular targeting of T cells is limited in that antibodies can only inhibit extracellular proteins, whereas pharmacological inhibitors often possess lack of specificity. Targeting of specific receptor subunits, something difficult to perform with antibodies, can be performed with siRNA. One such target would be the cytokine receptor common gamma chain. Knockout mice whose T cells are deficient in this protein allow for permanent survival of islet allografts (66). Another class of targets that would be particularly attractive in transplantation are transcription factors associated with T‐cell inflammatory responses. For example, the signal transducer and activator of transcription (STAT)‐4 is a DNA‐binding protein that is implicated in activation of Th1 inflammatory T‐cell responses (67). The relevance of targeting such a protein in contrast to specific cytokines can be seen in experiments where IFN‐γ knockout recipients possess similar or accelerated rates of allograft rejection as wild‐type mice (68), whereas STAT‐4 knockout recipients have a significant decrease in graft pathology and prolonged allograft acceptance (69). The added attractiveness of targeting STAT‐4 would be the endowment of T cells with an increased predisposition to induction of tolerance, as was elegantly demonstrated by Zhou et al. in STAT‐4 knockout mice (70). T‐bet is another inflammatory‐associated transcription factor whose absence results in deficient Th1 development (71). Silencing of this gene may yield results comparable to STAT‐4 inhibition in transplantation.
Small interfering RNA may also be used for gene‐silencing on the APC side of the immune response. Previous reports using AO have demonstrated that inhibition of the costimulatory molecules CD80 and CD86 on DCs prolongs allograft survival (72). Additionally, administration of CD40− DCs induces antigen‐specific tolerance through the generation of Treg cells (73). Therefore targeting such costimulatory molecules on APCs using siRNA appears to be potentially fruitful approach. As stated previously, the transcription factor NF‐κB possesses potent immune stimulatory activity through its ability to activate several signalling pathways in APCs leading to robust T‐cell stimulation. Feasibility of silencing NF‐κB subunits was already demonstrated to result in generation of Th2‐promoting DCs (58). Owing to the potent tolerogenic activity of other NF‐κB inhibitors, we anticipate siRNA silencing of this target would be a useful approach to inducing APC‐mediated tolerance. Cytokines elaborated by APCs involved in induction of naïve T‐cell differentiation would also be a promising target. We have previously demonstrated that silencing of IL‐12 p35 in DCs induces Th2 deviation in vitro and in vivo (54). Future studies will evaluate more potent Th1 inducers such as IL‐18 (74) and IL‐23 (75).
Clinical applicability of siRNA
The promise of siRNA‐therapeutics is held back by the question of delivery. Although viral vectors are promising in preclinical models, the fatality reported in a clinical trial of gene therapy using such a vector has placed a significant roadblock in the implementation of viral approaches (76). Murine studies have indicated that siRNA can be administered through the ‘hydrodynamic approach’; however, such a strategy would clearly not be ethical clinically (37, 77). One attractive method is through delivery of siRNA using cell‐specific immunoliposomes. Immunoliposomes are artificial model membranes with specific antibodies attached to the outer lipid leaflet thus enabling specific discharge of liposomal contents into cells expressing the surface antigen recognized by the respective antibody. The attractiveness of this approach is that unique cellular specificity can be achieved. The utility of liposomal techniques has been demonstrated clinically in studies where liposomal drugs allow for much lower administration of the said drug without losing the desired therapeutic effect (78, 79). This approach has been successfully used to target chemotherapeutic agents against tumors using antibodies to the oncogenic protein HER‐2 (80). The ability of immunoliposomes to deliver nucleic acids to specific target cells was recently demonstrated (81). Additionally, in vivo delivery of siRNA has been previously reported using liposomes (82). A type of liposome currently used for cell‐specific targeting is the polyethelene glycol (PEG)‐immunoliposome, in which nucleic acids are entrapped in the fluid phase of the liposome and the antibodies are coupled to PEG and are anchored in the lipid bilayer (83). Such immunoliposomes have been demonstrated to be innocuous in a variety of in vivo toxicological models (84). Specifically, for transplantation, one could use immunoliposomes to specifically target siRNA to Th cells via CD4 (85); to APCs such as DCs using CD11c (86) or macrophages using STEALTH liposomes (87).
Another area of siRNA application is in the form of organ‐storage solutions. Donor organs are subjected to flushing and storage in hypothermic conditions (4 °C) in specially formulated solutions (organ storage solutions) in order to wash out debris and to decrease damage during transportation (88). A variety of groups have performed modifications to typical perfusion solutions to attain better graft function (89, 90, 91, 92). Despite these modifications, little work has been performed on altering organ and tissue immunogenicity, which is directly related to graft rejection. Chen et al. (93) transfected kidneys with naked antisense DNA in order to suppress expression of intracellular adhesion molecule‐1 (ICAM‐1). Successful prevention of reperfusion injury was noted. Similarly, suppression of NF‐κB activation was reported in rat hearts by administration of decoy oligonucleotides to the organ in the perfusion solution (94). Previous attempts to modify direct antigen presentation by donor DCs using monoclonal antibodies have failed owing to the inability of the perfusion process to deliver the antibodies to a sufficient number of target cells (95). Addition of siRNA‐targeting genes associated with immune rejection, endothelial activation, and apoptosis to the organ storage solution is a potentially useful avenue of ex vivo administration of siRNA. In support of this approach are studies demonstrating efficient gene‐silencing through siRNA administration into whole kidney cultures (96). The observation that intranasal delivery of siRNA leads to pulmonary gene silencing (28) also strengthens the notion that siRNA can be used for localized gene silencing in isolated organs.
Conclusions
The discovery of RNAi opens the door for gene‐specific manipulation in a safe and physiologically useful manner. Silencing gene expression through siRNA is superior to conventional gene‐ or antibody‐blocking approaches owing to the following: (1) Blocking efficacy is more potent (97); (2) targeting of gene expression is more specific (98); (3) inhibitory effects can be passed on for multiple generations (99); (4) in vitro transfection efficacy is higher and can be expressed in a stable manner (100); (5) in vivo use is more practical and safer owing to the lower concentration needed for a therapeutic effect; (6) tissue‐ or cell‐specific gene targeting is possible using a specific promoter vector (101, 102) or specific antibody conjugated liposomes; and (7) simultaneous targeting multiple genes or multiple exons is possible for increasing efficacy (103). We anticipate that the discovery of new physiological targets will be matched by specific and potent siRNA strategies, which will lead to overall improved graft survival in recipients of organ transplants.
Acknowledgments
We thank Dr Gill Strejan, University of Western Ontario, for critical review and comments. This study was partially supported by Roche Organ Transplant Research Foundation, Heart and Stroke Foundation of Canada, Kidney Foundation of Canada, and a research grant from the Multi‐Organ Transplant Program, London Health Sciences Centre and Lawson Health Research Institute.
References
- 1. Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA Interference. biology, mechanism, and applications. Microbiol Mol Biol Rev 2003; 67: 657–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davidson BL. Hepatic diseases – hitting the target with inhibitory RNAs. N Engl J Med 2003; 349: 2357–2359. [DOI] [PubMed] [Google Scholar]
- 3. Wall NR, Shi Y. Small RNA: can RNA interference be exploited for therapy Lancet 2003; 362: 1401–1403. [DOI] [PubMed] [Google Scholar]
- 4. Ullu E, Djikeng A, Shi H, Tschudi C. RNA interference: advances and questions. Philos Trans R Soc Lond B Biol Sci 2002; 357: 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jorgensen RA, Cluster PD, English J, Que Q, Napoli CA. Chalcone synthase cosuppression phenotypes in petunia flowers: comparison of sense vs. antisense constructs and single‐copy vs. complex T‐DNA sequences. Plant Mol Biol 1996; 31: 957–973. [DOI] [PubMed] [Google Scholar]
- 6. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans. Nature 1998; 391: 806–811. [DOI] [PubMed] [Google Scholar]
- 7. Proud CG. PKR: a new name and new roles. Trends Biochem Sci 1995; 20: 241–246. [DOI] [PubMed] [Google Scholar]
- 8. Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol 2003; 4: 457–467. [DOI] [PubMed] [Google Scholar]
- 9. Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res 2002; 30: 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Caplen NJ, Mousses S. Short interfering RNA (siRNA)‐mediated RNA interference (RNAi) in human cells. Ann N Y Acad Sci 2003; 1002: 56–62. [DOI] [PubMed] [Google Scholar]
- 11. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21‐nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001; 411: 494–498. [DOI] [PubMed] [Google Scholar]
- 12. Tiscornia G, Singer O, Ikawa M, Verma IM. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc Natl Acad Sci U S A 2003; 100: 1844–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hara K, Maruki Y, Long X et al Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002; 110: 177. [DOI] [PubMed] [Google Scholar]
- 14. Ji J, Wernli M, Klimkait T, Erb P. Enhanced gene silencing by the application of multiple specific small interfering RNAs. FEBS Lett 2003; 552: 247–252. [DOI] [PubMed] [Google Scholar]
- 15. Katoh T, Susa M, Suzuki T, Umeda N, Watanabe K. Simple and rapid synthesis of siRNA derived from in vitro transcribed shRNA. Nucleic Acids Res Suppl 2003: 249–250. [DOI] [PubMed] [Google Scholar]
- 16. Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 1987; 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amarasinghe AK, Calin‐Jageman I, Harmouch A, Sun W, Nicholson AW. Escherichia coli ribonuclease III. affinity purification of hexahistidine‐tagged enzyme and assays for substrate binding and cleavage. Methods Enzymol 2001; 342: 143–158. [DOI] [PubMed] [Google Scholar]
- 18. Kim DH, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol 2004; 22: 321–325. [DOI] [PubMed] [Google Scholar]
- 19. Kawasaki H, Suyama E, Iyo M, Taira K. siRNAs generated by recombinant human Dicer induce specific and significant but target site‐independent gene silencing in human cells. Nucleic Acids Res 2003; 31: 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Myers JW, Jones JT, Meyer T, Ferrell JE Jr. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat Biotechnol 2003; 21: 324–328. [DOI] [PubMed] [Google Scholar]
- 21. Tuschl T. Expanding small RNA interference. Nat Biotechnol 2002; 20: 446–448. [DOI] [PubMed] [Google Scholar]
- 22. Kobayashi N, Matsui Y, Kawase A et al Vector‐based in vivo RNA interference: dose‐ and time‐dependent suppression of transgene expression. J Pharmacol Exp Ther 2003; 308: 688–693. [DOI] [PubMed] [Google Scholar]
- 23. Czauderna F, Santel A, Hinz M et al Inducible shRNA expression for application in a prostate cancer mouse model. Nucleic Acids Res 2003; 31: e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Castanotto D, Li H, Rossi JJ. Functional siRNA expression from transfected PCR products. RNA 2002; 8: 1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zheng L, Liu J, Batalov S et al An approach to genomewide screens of expressed small interfering RNAs in mammalian cells. Proc Natl Acad Sci U S A 2004; 101: 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Surowiak P. Evaluation of transfection effectiveness using fluorescein‐labelled oligonucleotides and various liposomes. Folia Morphol (Warsz) 2003; 62: 397. [PubMed] [Google Scholar]
- 27. McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature 2002; 418: 38–39. [DOI] [PubMed] [Google Scholar]
- 28. Zhang X, Shan P, Jiang D et al Small interfering RNA targeting heme oxygenase‐1 enhances ischemia‐reperfusion‐induced lung apoptosis. J Biol Chem 2003; 279: 10677–10684. [DOI] [PubMed] [Google Scholar]
- 29. Barton GM, Medzhitov R. Retroviral delivery of small interfering RNA into primary cells. Proc Natl Acad Sci U S A 2002; 99: 14943–14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xia H, Mao Q, Paulson HL, Davidson BL. siRNA‐mediated gene silencing in vitro and in vivo. Nat Biotechnol 2002; 20: 1006–1010. [DOI] [PubMed] [Google Scholar]
- 31. Abbas‐Terki T, Blanco‐Bose W, Deglon N, Pralong W, Aebischer P. Lentiviral‐mediated RNA interference. Hum Gene Ther 2002; 13: 2197–2201. [DOI] [PubMed] [Google Scholar]
- 32. Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV‐1 infection in human T cells by lentiviral‐mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A 2003; 100: 183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee MT, Coburn GA, McClure MO, Cullen BR. Inhibition of human immunodeficiency virus type 1 replication in primary macrophages by using Tat‐ or CCR5‐specific small interfering RNAs expressed from a lentivirus vector. J Virol 2003; 77: 11964–11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hasuwa H, Kaseda K, Einarsdottir T, Okabe M. Small interfering RNA and gene silencing in transgenic mice and rats. FEBS Lett 2002; 532: 227–230. [DOI] [PubMed] [Google Scholar]
- 35. Reich SJ, Fosnot J, Kuroki A et al Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis 2003; 9: 210–216. [PubMed] [Google Scholar]
- 36. Sorensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J Mol Biol 2003; 327: 761–766. [DOI] [PubMed] [Google Scholar]
- 37. Giladi H, Ketzinel‐Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther 2003; 8: 769–776. [DOI] [PubMed] [Google Scholar]
- 38. Novina CD, Murray MF, Dykxhoorn DM et al siRNA‐directed inhibition of HIV‐1 infection. Nat Med 2002; 8: 681–686. [DOI] [PubMed] [Google Scholar]
- 39. Wilson JA, Jayasena S, Khvorova A et al RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci U S A 2003; 100: 2783–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song E, Lee SK, Wang J et al RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med 2003; 9: 347–351. [DOI] [PubMed] [Google Scholar]
- 41. Zhang R, Guo Z, Lu J et al Inhibiting severe acute respiratory syndrome‐associated coronavirus by small interfering RNA. Chin Med J (Engl) 2003; 116: 1262–1264. [PubMed] [Google Scholar]
- 42. Li MJ, Bauer G, Michienzi A et al Inhibition of HIV‐1 infection by lentiviral vectors expressing Pol III‐promoted anti‐HIV RNAs. Mol Ther 2003; 8: 196–206. [DOI] [PubMed] [Google Scholar]
- 43. Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci U S A 2003; 100: 235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Butz K, Ristriani T, Hengstermann A, Denk C, Scheffner M, Hoppe‐Seyler F. siRNA targeting of the viral E6 oncogene efficiently kills human papillomavirus‐positive cancer cells. Oncogene 2003; 22: 5938–5945. [DOI] [PubMed] [Google Scholar]
- 45. McCown M, Diamond MS, Pekosz A. The utility of siRNA transcripts produced by RNA polymerase i in down regulating viral gene expression and replication of negative‐ and positive‐strand RNA viruses. Virology 2003; 313: 514–524. [DOI] [PubMed] [Google Scholar]
- 46. Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNA (Val) promoter significantly induce RNAi‐mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res 2003; 31: 700–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martinez LA, Naguibneva I, Lehrmann H et al Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways. Proc Natl Acad Sci U S A 2002; 99: 14849–14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Choudhury A, Charo J, Parapuram SK et al Small interfering RNA (siRNA) inhibits the expression of the Her2/neu gene, upregulates HLA class I and induces apoptosis of Her2/neu positive tumor cell lines. Int J Cancer 2004; 108: 71–77. [DOI] [PubMed] [Google Scholar]
- 49. Scherr M, Battmer K, Winkler T, Heidenreich O, Ganser A, Eder M. Specific inhibition of bcr‐abl gene expression by small interfering RNA. Blood 2003; 101: 1566–1569. [DOI] [PubMed] [Google Scholar]
- 50. Dewar AL, Domaschenz RM, Doherty KV, Hughes TP, Lyons AB. Imatinib inhibits the in vitro development of the monocyte/macrophage lineage from normal human bone marrow progenitors. Leukemia 2003; 17: 1713–1721. [DOI] [PubMed] [Google Scholar]
- 51. Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB. Small interfering RNAs directed against beta‐catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin Cancer Res 2003; 9: 1291–1300. [PubMed] [Google Scholar]
- 52. Yang G, Thompson JA, Fang B, Liu J. Silencing of H‐ras gene expression by retrovirus‐mediated siRNA decreases transformation efficiency and tumor growth in a model of human ovarian cancer. Oncogene 2003; 22: 5694–5701. [DOI] [PubMed] [Google Scholar]
- 53. Ray S, Almasan A. Apoptosis induction in prostate cancer cells and xenografts by combined treatment with Apo2 ligand/tumor necrosis factor‐related apoptosis‐inducing ligand and CPT‐11. Cancer Res 2003; 63: 4713–4723. [PubMed] [Google Scholar]
- 54. Hill JA, Ichim TE, Kusznieruk KP et al Immune modulation by silencing IL‐12 production in dendritic cells using small interfering RNA. J Immunol 2003; 171: 691–696. [DOI] [PubMed] [Google Scholar]
- 55. Sledz CA, Holko M, De Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short‐interfering RNAs. Nat Cell Biol 2003; 5: 834–839. [DOI] [PubMed] [Google Scholar]
- 56. Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet 2003; 34: 263–264. [DOI] [PubMed] [Google Scholar]
- 57. Nishiwaki Y, Yokota T, Hiraoka M et al Introduction of short interfering RNA to silence endogenous E‐selectin in vascular endothelium leads to successful inhibition of leukocyte adhesion. Biochem Biophys Res Commun 2003; 310: 1062–1066. [DOI] [PubMed] [Google Scholar]
- 58. Laderach D, Compagno D, Danos O, Vainchenker W, Galy A. RNA interference shows critical requirement for NF‐kappa B p50 in the production of IL‐12 by human dendritic cells. J Immunol 2003; 171: 1750–1757. [DOI] [PubMed] [Google Scholar]
- 59. Wong AW, Brickey WJ, Taxman DJ et al CIITA‐regulated plexin‐A1 affects T‐cell–dendritic cell interactions. Nat Immunol 2003; 4: 891–898. [DOI] [PubMed] [Google Scholar]
- 60. Dumont FJ. Treatment of transplant rejection: are the traditional immunosuppressants good enough Curr Opin Investig Drugs 2001; 2: 357–363. [PubMed] [Google Scholar]
- 61. Billaud EM. Clinical pharmacology of immunosuppressive drugs: year 2000 – time for alternatives. Therapie 2000; 55: 177–183. [PubMed] [Google Scholar]
- 62. Wijdenes J, Roy C, Morel‐Fourrier B, Racadot E. Monoclonal antibodies in human organ transplantation and auto‐immune diseases. Therapie 1992; 47: 283–287. [PubMed] [Google Scholar]
- 63. McManus MT, Haines BB, Dillon CP et al Small interfering RNA‐mediated gene silencing in T lymphocytes. J Immunol 2002; 169: 5754–5760. [DOI] [PubMed] [Google Scholar]
- 64. Kojima S, Vignjevic D, Borisy GG. Improved silencing vector co‐expressing GFP and small hairpin RNA. Biotechniques 2004; 36: 74–79. [DOI] [PubMed] [Google Scholar]
- 65. Banerjea A, Li MJ, Bauer G et al Inhibition of HIV‐1 by lentiviral vector‐transduced siRNAs in T lymphocytes differentiated in SCID‐hu mice and CD34+ progenitor cell‐derived macrophages. Mol Ther 2003; 8: 62–71. [DOI] [PubMed] [Google Scholar]
- 66. Li XC, Ima A, Li Y, Zheng XX, Malek TR, Strom TB. Blocking the common gamma‐chain of cytokine receptors induces T cell apoptosis and long‐term islet allograft survival. J Immunol 2000; 164: 1193–1199. [DOI] [PubMed] [Google Scholar]
- 67. Nishikomori R, Usui T, Wu CY, Morinobu A, O'Shea JJ, Strober W. Activated STAT4 has an essential role in Th1 differentiation and proliferation that is independent of its role in the maintenance of IL‐12R beta 2 chain expression and signaling. J Immunol 2002; 169: 4388–4398. [DOI] [PubMed] [Google Scholar]
- 68. Bishop DK, Chan Wood S, Eichwald EJ, Orosz CG. Immunobiology of allograft rejection in the absence of IFN‐gamma: CD8+ effector cells develop independently of CD4+ cells and CD40–CD40 ligand interactions. J Immunol 2001; 166: 3248–3255. [DOI] [PubMed] [Google Scholar]
- 69. Koglin J, Glysing‐Jensen T, Gadiraju S, Russell ME. Attenuated cardiac allograft vasculopathy in mice with targeted deletion of the transcription factor STAT4. Circulation 2000; 101: 1034–1039. [DOI] [PubMed] [Google Scholar]
- 70. Zhou P, Szot GL, Guo Z et al Role of STAT4 and STAT6 signaling in allograft rejection and CTLA4‐Ig‐mediated tolerance. J Immunol 2000; 165: 5580–5587. [DOI] [PubMed] [Google Scholar]
- 71. Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T‐bet in TH1 lineage commitment and IFN‐gamma production in CD4 and CD8 T cells. Science 2002; 295: 338–342. [DOI] [PubMed] [Google Scholar]
- 72. Liang X, Lu L, Chen Z et al Administration of dendritic cells transduced with antisense oligodeoxyribonucleotides targeting CD80 or CD86 prolongs allograft survival. Transplantation 2003; 76: 721–729. [DOI] [PubMed] [Google Scholar]
- 73. Martin E, O'Sullivan B, Low P, Thomas R. Antigen‐specific suppression of a primed immune response by dendritic cells mediated by regulatory T cells secreting interleukin‐10. Immunity 2003; 18: 155–167. [DOI] [PubMed] [Google Scholar]
- 74. Stoll S, Jonuleit H, Schmitt E et al Production of functional IL‐18 by different subtypes of murine and human dendritic cells (DC): DC‐derived IL‐18 enhances IL‐12‐dependent Th1 development. Eur J Immunol 1998; 28: 3231–3239. [DOI] [PubMed] [Google Scholar]
- 75. Puccetti P, Belladonna ML, Grohmann U. Effects of IL‐12 and IL‐23 on antigen‐presenting cells at the interface between innate and adaptive immunity. Crit Rev Immunol 2002; 22: 373–390. [PubMed] [Google Scholar]
- 76. Grilley BJ, Gee AP. Gene transfer: regulatory issues and their impact on the clinical investigator and the good manufacturing production facility. Cytotherapy 2003; 5: 197–207. [DOI] [PubMed] [Google Scholar]
- 77. Zender L, Hutker S, Liedtke C et al Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc Natl Acad Sci U S A 2003; 100: 7797–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Vail DM, Kurzman ID, Glawe PC et al STEALTH liposome‐encapsulated cisplatin (SPI‐77) versus carboplatin as adjuvant therapy for spontaneously arising osteosarcoma (OSA) in the dog: a randomized multicenter clinical trial. Cancer Chemother Pharmacol 2002; 50: 131–136. [DOI] [PubMed] [Google Scholar]
- 79. Rosenthal DI, Yom SS, Liu L et al A phase I study of SPI‐077 (Stealth liposomal cisplatin) concurrent with radiation therapy for locally advanced head and neck cancer. Invest New Drugs 2002; 20: 343–349. [DOI] [PubMed] [Google Scholar]
- 80. Park JW, Hong K, Kirpotin DB et al Anti‐HER2 immunoliposomes: enhanced efficacy attributable to targeted delivery. Clin Cancer Res 2002; 8: 1172–1181. [PubMed] [Google Scholar]
- 81. Rodriguez M, Coma S, Noe V, Ciudad CJ. Development and effects of immunoliposomes carrying an antisense oligonucleotide against DHFR RNA and directed toward human breast cancer cells overexpressing HER2. Antisense Nucleic Acid Drug Dev 2002; 12: 311–325. [DOI] [PubMed] [Google Scholar]
- 82. Sioud M, Sorensen DR. Cationic liposome‐mediated delivery of siRNAs in adult mice. Biochem Biophys Res Commun 2003; 312: 1220–1225. [DOI] [PubMed] [Google Scholar]
- 83. Mercadal M, Domingo JC, Petriz J, Garcia J, De Madariaga MA. Preparation of immunoliposomes bearing poly (ethylene glycol) – coupled monoclonal antibody linked via a cleavable disulfide bond for ex vivo applications. Biochim Biophys Acta 2000; 1509: 299–310. [DOI] [PubMed] [Google Scholar]
- 84. Zhang YF, Boado RJ, Pardridge WM. Absence of toxicity of chronic weekly intravenous gene therapy with pegylated immunoliposomes. Pharm Res 2003; 20: 1779–1785. [DOI] [PubMed] [Google Scholar]
- 85. Phillips NC, Gagne L, Tsoukas C, Dahman J. Immunoliposome targeting to murine CD4+ leucocytes is dependent on immune status. J Immunol 1994; 152: 3168–3174. [PubMed] [Google Scholar]
- 86. Steinman RM, Pack M, Inaba K. Dendritic cells in the T‐cell areas of lymphoid organs. Immunol Rev 1997; 156: 25–37. [DOI] [PubMed] [Google Scholar]
- 87. Vertut‐Doi A, Ishiwata H, Miyajima K. Binding and uptake of liposomes containing a poly (ethylene glycol) derivative of cholesterol (stealth liposomes) by the macrophage cell line J774. influence of PEG content and its molecular weight. Biochim Biophys Acta 1996; 1278: 19–28. [DOI] [PubMed] [Google Scholar]
- 88. Menasche P, Termignon JL, Pradier F et al Experimental evaluation of Celsior, a new heart preservation solution. Eur J Cardiothorac Surg 1994; 8: 207–213. [DOI] [PubMed] [Google Scholar]
- 89. Baker CJ, Longoria J, Gade PV, Starnes VA, Barr ML. Addition of a water‐soluble alpha‐tocopherol analogue to University of Wisconsin solution improves endothelial viability and decreases lung reperfusion injury. J Surg Res 1999; 86: 145–149. [DOI] [PubMed] [Google Scholar]
- 90. McAnulty JF, Huang XQ. The effect of simple hypothermic preservation with Trolox and ascorbate on lipid peroxidation in dog kidneys. Cryobiology 1996; 33: 217–225. [DOI] [PubMed] [Google Scholar]
- 91. Natori S, Higuchi H, Contreras P, Gores GJ. The caspase inhibitor IDN‐6556 prevents caspase activation and apoptosis in sinusoidal endothelial cells during liver preservation injury. Liver Transpl 2003; 9: 278–284. [DOI] [PubMed] [Google Scholar]
- 92. Randsbaek F, Kimose HH, Hansen SB, Jacobsen B, Botker HE, Nielsen TT. Captopril improves oxygen and glucose extraction in pig hearts during reperfusion after cold cardioplegic storage. Scand Cardiovasc J 2000; 34: 201–208. [DOI] [PubMed] [Google Scholar]
- 93. Chen W, Bennett CF, Wang ME et al Perfusion of kidneys with unformulated ‘naked’ intercellular adhesion molecule‐1 antisense oligodeoxynucleotides prevents ischemic/reperfusion injury. Transplantation 1999; 68: 880–887. [DOI] [PubMed] [Google Scholar]
- 94. Vos IH, Govers R, Grone HJ et al NF kappa B decoy oligodeoxynucleotides reduce monocyte infiltration in renal allografts. FASEB J 2000; 14: 815–822. [DOI] [PubMed] [Google Scholar]
- 95. Goldberg LC. Pretreatment of kidney allografts with monoclonal antibodies to CD45: results of a multicentre study. CD45 Study Group. Transpl Int 1994; 7 (Suppl 1): S252–S254. [DOI] [PubMed] [Google Scholar]
- 96. Davies JA, Ladomery M, Hohenstein P et al Development of an siRNA‐based method for repressing specific genes in renal organ culture and its use to show that the Wt1 tumour suppressor is required for nephron differentiation. Hum Mol Genet 2004; 13: 235–246. [DOI] [PubMed] [Google Scholar]
- 97. Bertrand J, Pottier M, Vekris A, Opolon P, Maksimenko A, Malvy C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem Biophys Res Commun 2002; 296: 1000–1004. [DOI] [PubMed] [Google Scholar]
- 98. Celotto AM, Graveley BR. Exon‐specific RNAi: a tool for dissecting the functional relevance of alternative splicing. RNA 2002; 8: 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science 2000; 287: 2494–2497. [DOI] [PubMed] [Google Scholar]
- 100. Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002; 296: 550–553. [DOI] [PubMed] [Google Scholar]
- 101. Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat Biotechnol 2002; 20: 505–508. [DOI] [PubMed] [Google Scholar]
- 102. Devroe E, Silver PA. Retrovirus‐delivered siRNA. BMC Biotechnol 2002; 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Yang D, Buchholz F, Huang Z et al Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci U S A 2002; 99: 9942–9947. [DOI] [PMC free article] [PubMed] [Google Scholar]


