Abstract
African swine fever virus (ASFV) is the causative pathogen of the recent African swine fever epidemic, with devastating impacts on economy. A recent study by Wang et al. reveals the multilayer structural details of ASFV at near-atomic resolution, which provides interesting insights about giant virus assembly and paves the way for vaccine development.
In the last two decades, many giant DNA viruses have been discovered, some of which are larger than small bacteria in their genomes and physical sizes, defying the traditional definition of viruses as small and filterable infectious agents. Intensive genomic analyses have been carried out to answer the fundamental evolutionary question about where these giant viruses come from. Cellular genes found in all three kingdoms of life are present in giant virus genomes, leading to controversial debates about how to place them into the current evolution of life.
Most of these giant DNA viruses were isolated in the setting of environmental studies, such as in aquatic and terrestrial systems. Though the majority of them have very limited economic impact to society, one of a few exceptions is the African swine fever virus (ASFV). This is a lethal pathogen to domestic and wild pigs, causing highly contagious hemorrhagic fever with high mortality rates. African swine fever (ASF) was first described in the 1920s in Kenya and gradually spread all over the world. A recent epidemic of ASF from 2018 to 2019 in China [1] has caused hundreds of millions of pigs to either die from the disease or be slaughtered to control the spread of the virus because no vaccines or medicines are currently available. As the largest pork producer and consumer in the world, this recent outbreak of ASFV has posed significant impacts on the economy of China and threatened its food security.
Due to their physical size and structural complexity, the structural studies of the giant viruses have posed significant challenges to current techniques. Cryogenic-electronic microscopy (cryo-EM) is one of the few powerful tools that can be used. Recently, the breakthrough in detectors, combined with other advances in cryo-EM, has made it the dominating tool to determine high resolution structures for biological molecules. However, the challenge of determining high-resolution structures of giant viruses remains because certain assumptions that built the current cryo-EM theory were derived from small molecules embedded in thin ice, which are no longer true for giant viruses frozen in much thicker ice. Exciting breakthroughs occurred in the field between 2018 and 2019. A new method was developed in which the huge map of a giant virus was broken into small blocks and reconstructed separately with localized parameters [2]. This method has been used to study Paramecium bursaria Chlorella virus 1 (PBCV-1), resulting in the first near-atomic structure of a giant virus [3]. A similar method has been applied to ASFV, providing the first urgently needed high-resolution view of the virus. This work was accomplished by a multi-institutional team in China and published in Science [4].
The most interesting feature of ASFV revealed by the study is its unique multiple-layer structure (Figure 1). While most giant icosahedral DNA viruses that have protein shells (capsids) as their exposed layers, ASFV has an outer membrane probably acquired from the host cellular membrane during the budding process. Beneath the membrane is the typical icosahedral protein capsid assembled from capsomers with a triangulation number T of 277. Under this capsid lies an inner lipid membrane, which is commonly seen in many other giant icosahedral DNA viruses [3,5]. However, ASFV has an icosahedral core shell just below the inner membrane, unlike the usual spherical nucleocapsids or genomic sacks of other giant viruses. Within this T = 19 icosahedral core shell is the nucleoid containing the viral DNA (Figure 1). ASFV is closely related to faustovirus [6] and both viruses have similar icosahedral core shell and share the same T number for the outer capsid. However, faustovirus does not have the outer and inner lipid membranes. In addition, the inner core shell of faustovirus has a T number of 64 instead of 19 for ASFV [4,6].
Figure 1. The Multilayer Architecture of African Swine Fever Virus (ASFV).
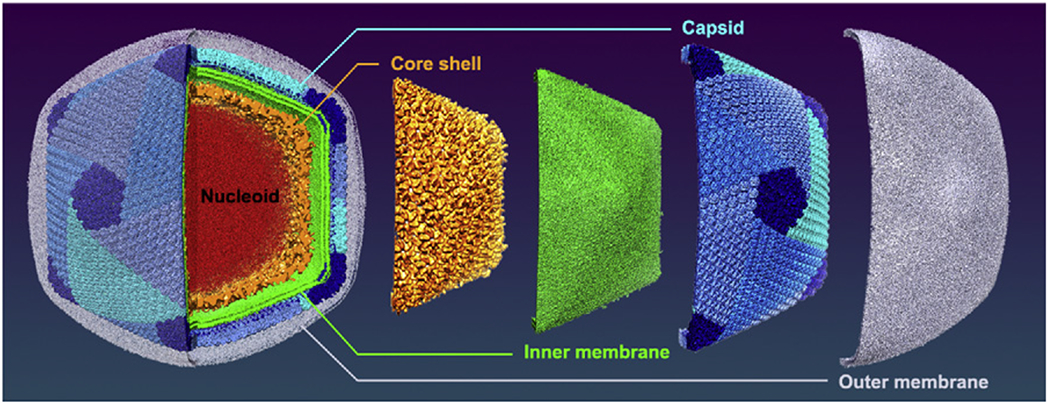
The five layers, including the genome core, nucleocapsid, inner membrane, capsid, and outer membrane, are colored in red, orange, green, blue, and grey, respectively. For a better observation on the capsid, the outer membrane has been depicted with 70% transparency. The capsid surface was colored by pentasymmetrons (dark blue) and trisymmetrons (lighter blue colors including cyan, dodger blue, and cornflower blue).
The most structural details of ASFV revealed in this study lie on its capsid layer, mainly consisting of 2760 pseudo-hexameric capsomers and 12 pentameric capsomers [4]. The capsid of ASFV has a similar capsomer arrangement as other giant DNA viruses [3,5,6]. Patches of capsomers called symmetrons are observed in the ASFV capsid (Figure 1), of which, those around the 12 icosahedral five-fold vertices are called pentasymmetrons and those that cover the 20 faces of the icosahedral capsid are called trisymmetrons. All giant icosahedral viruses have a similar pentasymmetron size of 31 capsomers but different trisymmetron size. The ASFV major capsid protein (MCP) that forms the pseudo-hexameric capsomers shares the same double-jelly-roll fold as seen in other giant DNA viruses. However, the surface loops of ASFV MCP have unique intertwining structures, which may offer targets for vaccine development. Three different interaction modes were observed among the capsomers, consistent with a previous study [7]. The similar arrangement of the capsomers among all giant icosahedral DNA viruses indicates that they might have evolved from a common ancestor and share the same assembly mechanism.
Compared with the 19 minor capsid proteins (mCPs) identified in PBCV-1 [3], only three were found in ASFV [4]. Among these mCPs is the interesting elongated tape-measure protein that was also found in PBCV-1 [3]. The lower number of mCPs of ASFV suggests that the additional icosahedral core shell of ASFV might stabilize the capsid better, or alternatively, could be due to the relatively lower resolution of the map. Based on the structure, the authors of the study also proposed a new assembly hypothesis that is different to previous ones in other giant viruses [3,5].
In summary, the study provided the first high-resolution structure of ASFV, revealing its unique multilayer architecture. It is noteworthy that two more cryo-EM studies of the whole ASFV virus [8,9] and one for its MCP were also published [10] in 2019, making it a very fruitful year for ASFV structural studies. Future structural studies of ASFV will reveal more information on its mCPs and the unique core shell, and nonicosahedrally averaged cryo-EM reconstruction can show whether ASFV has a unique portal as seen in other giant viruses. The resultant structures from all these studies will not only deepen our understanding of the life cycle of ASFV, but also pave the way for vaccine and drug development to fight the disease that is devastating the world pork industry.
Acknowledgments
We really appreciate that the primary authors of the paper on ASFV published in Science rendered the high-resolution images for us to be used in our figure. Authors of this publication were supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM129525. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Zhou L (2019) African swine fever epidemic in China. Vet. Rec 184, 713. [DOI] [PubMed] [Google Scholar]
- 2.Zhu D et al. (2018) Pushing the resolution limit by correcting the Ewald sphere effect in single-particle cryo-EM reconstructions. Nat. Commun 9, 1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fang Q et al. (2019) Near-atomic structure of a giant virus. Nat. Commun 10, 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang N et al. (2019) Architecture of African swine fever virus and implications for viral assembly. Science 366, 640–644 [DOI] [PubMed] [Google Scholar]
- 5.Xiao C et al. (2017) Cryo-EM reconstruction of the Cafeteria roenbergensis virus capsid suggests novel assembly pathway for giant viruses. Sci. Rep 7, 5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klose T et al. (2016) Structure of faustovirus, a large dsDNA virus. Proc. Natl. Acad. Sci. U. S. A 113, 6206–6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xian Y et al. (2019) The roles of electrostatic interactions in capsid assembly mechanisms of giant viruses. Int. J. Mol. Sci 20, E1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S et al. (2019) Cryo-EM Structure of the African swine fever virus. Cell Host Microbe 26, 836–843 [DOI] [PubMed] [Google Scholar]
- 9.Andres G et al. (2019) The cryo-EM structure of African swine fever virus unravels a unique architecture comprising two icosahedral protein capsids and two lipoprotein membranes. J. Biol. Chem 295, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q et al. (2019) Structure of the African swine fever virus major capsid protein p72. Cell Res. 29, 953–955 [DOI] [PMC free article] [PubMed] [Google Scholar]


