Abstract
We report new lipid-based, high-density, environmentally sensitive (HIDE) probes that accurately and selectively image endo-lysosomes and their dynamics at super-resolution for extended times. Treatment of live cells with the small molecules DiIC16TCO or DiIC16’TCO followed by in situ tetrazine ligation reaction with the silicon-rhodamine dye SiR-Tz generates the HIDE probes DiIC16-SiR and DiIC16’-SiR in the endo-lysosomal membrane. These new probes support the acquisition of super-resolution videos of organelle dynamics in primary cells for more than 7 minutes with no detectable change in endosome structure or function. Using DiIC16-SiR and DiIC16’-SiR, we describe the first direct evidence of endosome motility defects in cells from patients with Niemann-Pick Type-C disease. In wild-type fibroblasts, the probes reveal distinct but rare inter-endosome kiss-and-run events that cannot be observed using confocal methods. Our results shed new light on the role of NPC1 in organelle motility and cholesterol trafficking.
Introduction
Endosomes are challenging to visualize because they are small – typically less than 200 nm in diameter5 – and dynamic. As a result, observing endosomes as discrete objects demands super-resolution methods that are compatible with live cells, such as structured illumination (SIM)6, stochastic optical reconstruction (STORM)7, and stimulated emission depletion (STED)8 microscopy. Visualizing endosome dynamics using either STORM or STED for more than 3 minutes is currently not possible, as even highly resistant fluorophores photo-bleach rapidly under the conditions required for these experiments6. SIM has been used to image endosomes for over 3 minutes, but the resolution possible with this technique (> 100 nm)9 is insufficient to reliably differentiate individual endosomes. Quantifying endosome dynamics is even more challenging, as doing so requires the combination of high temporal resolution (> 1 frame/sec)10, deep cell penetration to visualize the critical perinuclear region11, and high spatial resolution to differentiate individual endosomes from within a densely packed array6. While each of these essential elements has been demonstrated individually to varying degrees of success6, 10, 11, all three are needed simultaneously to capture and rigorously quantify endosome motility and understand its connection to normal cell function and disease.
A large number of neurodegenerative endo-lysosomal storage disorders, including Niemann-Pick, Gauchers, and Tay-Sachs disease12,13 are characterized by aberrant endosome motility. Lysosomal storage disorders typically result from inborn errors of metabolism that delete, inactivate, or deplete an enzyme, often one involved in lipid processing, that is critical for late endo-lysosomal function. The end result is impaired lipid trafficking and the toxic accumulation of lipids, including cholesterol and glycosphingolipids, which manifests as neurologic disease13. In the case of Niemann-Pick C disease (NPD), loss of function of the lysosomal transmembrane proteins NPC1 or NPC2 cause cholesterol to accumulate in the late endosomal lumen14. Although the precise mechanism is debated15–18, the absence of cholesterol trafficking by NPC1 leads eventually to neurodegeneration, hepatosplenomegaly, and premature death.
NPC1 is a large protein (1251 residues) and over 380 NPC1 mutations induce cholesterol accumulation and present as Niemann-Pick C disease in patients (https://ghr.nlm.nih.gov/gene/NPC1#resources). These cellular effects of these mutations are typically characterized using conjugated polyene macrolides known as filipins, which bind cholesterol and provide a fluorescent read-out of cholesterol accumulation19. Some NPC1 mutations (such as I1061T) lead to cholesterol accumulation by inducing NPC1 mis-folding in the ER and subsequent degradation so that little intact protein reaches the endo-lysosome20. Others, such as R404Q, affect the interaction of NPC1 with NPC2 and thus the export of LDL-derived cholesterol21. Still other mutations (such as 1920delG) have no obvious effects on disease22. It has been impossible thus far to directly correlate cholesterol accumulation with endo-lysosome motility and disease severity in NPC1 mutant cells because there were no tools available to quantify organelle dynamics across multiple mutant cell lines. As a result, the cholesterol accumulation phenotype found in NPD patient cells is often induced in cultured cells using small molecule NPC1 inhibitors23 such as U18666A and the endosomes visualized by fusing a late endosome-resident protein to a fluorescent protein such as GFP or YFP24–26. These studies have led to the widely held view that cholesterol accumulation and aberrant endosome motility are directly correlated27, but this relationship has never been tested in cells that express endogenous levels of NPD-relevant proteins.
Previous work has shown that organelles such as the Golgi, ER, mitochondria, and plasma membrane can be visualized at super-resolution for extended times using ‘HIDE’ (High-Density Environmentally sensitive) probes28. HIDE probes consist of an organelle-specific lipid or lipid-like small molecule that is linked in cellulo with a STED- or single molecule switching (SMS)-appropriate silicon rhodamine (SiR) dye29. When the linkage localizes the SiR dye to within an organelle membrane, dye photostability is enhanced by up to 50-fold30. This increase in photostability facilitates the acquisition of exceedingly long time-lapse images at the nanoscale31, 32. In this contribution, we expand the HIDE toolbox29, 30 to include two new lipid-like small molecules, DiIC16TCO (1) and DiIC16’TCO (2), which were modified to carry a reactive trans-cyclooctene moiety (Figure 1a). When conjugated in cellulo with a SiR dye carrying a tetrazine reaction partner (SiR-Tz (3), Figure 1b), the resultant HIDE probes support the acquisition of long time-lapse, super-resolution images of late endo-lysosomes and a quantitative analysis of their dynamics. Unlike previously reported super-resolution probes for late endo-lysosomes that rely on large fusion proteins6, fixed cells33, or that fail to support long-term dynamic imaging due to insufficient photostability8, the HIDE probes reported here are compatible with primary and hard-to-transfect cells, avoid the over-expression of an organelle-specific protein, and engender no detectable change in organelle function. Using these new HIDE probes, we report the first direct evidence of endosome motility defects in cells from patients with Niemann-Pick Type-C disease. We can also visualize distinct kiss-and-run events between endosomes in wild type fibroblasts. These motility defects and examples of inter-organelle communication are opaque when imaged using confocal methods.
Figure 1. DiIC16-SiR and DiIC16’-SiR are new HIDE probes for late endosomal imaging.
(a) Structures of DiIC16TCO and DiIC16’TCO, which (b) react with SiR-Tz in cellulo to generate DiIC16-SiR and DiIC16’-SiR. In a typical labeling reaction, HeLa cells are treated with 5 μM DiIC16TCO or DiIC16’TCO for 1 hour, followed by addition of 2 μM SiR-Tz and a 10 min incubation. Cells are then visualized by confocal and/or STED microscopy.
Results
Design of late endosome HIDE probes.
Previous work has demonstrated that the commercially available and cell-permeant dialkylindocarbocyanine dye DiIC16(3) (4) localizes effectively and selectively in living cells to late endosomes and lysosomes (but not early endosomes), and its fluorescence allows visualization of these organelles using confocal microscopy (Supplementary Figure 1a, 1b, and 1c). Unfortunately, DiIC16(3) photo-bleaches with a half-life of about 50 seconds under the conditions needed to support STED microscopy (Supplementary Figure 1d). To overcome the limitation imposed by low photostability while preserving the targeting activity of DiIC16(3), we designed a pair of DiIC16(3) analogs that carry a TCO moiety appended to either the aromatic core (DiIC16TCO) or the terminus of a single lipid tail (DiIC16’TCO) (Figure 1a). We hypothesized that DiIC16TCO and DiIC16’TCO would remain cell-permeant and localize selectively to late endosomes and lysosomes when added to live cells; addition of SiR-Tz and subsequent tetrazine ligation would generate the HIDE probes DiIC16-SiR and DiIC16’-SiR, respectively (Supplementary Figure 1e) and localize the photostable and red-shifted silicon rhodamine dye within the late endosomal membrane. Previous work suggests that in this environment, DiIC16-SiR and DiIC16’-SiR would support long time-lapse imaging of the late endo-lysosomal membrane by STED microscopy. DiIC16TCO and DiIC16’TCO were synthesized (Supplementary Note) and the rate of their reaction with SiR-Tz studied in vitro (Supplementary Figure 1f); we also characterized the absorption and emission of the resultant HIDE probes DiIC16-SiR and DiIC16’-SiR (Supplementary Figure 1g and 1h). The kinetic studies confirmed that the rate of the tetrazine ligation reaction between DiIC16TCO and SiR-Tz in vitro was extremely rapid, with a reaction half-life of 23 ± 4.5 s (k2 = 605 ± 78 M−1 s−1). This value agrees well with previously reported values for analogous reactions34.
DiIC16-SiR and DiIC16’-SiR localize to endolysosomes.
To test these hypotheses, we first used confocal microscopy to quantify the extent to which DiIC16-SiR and DiIC16’-SiR, assembled in cellulo, would colocalize with one of three endosomal markers: GFP-Rab5 (early endosomal marker), GFP-Rab7 (late endosomal marker), and GFP-Lamp1 (lysosomal marker). HeLa cells were incubated with 5 μM DiIC16TCO or DiIC16’TCO for 1 hour and then with 2 μM SiR-Tz for 10 min to assemble DiIC16-SiR or DiIC16’-SiR; the cells were then visualized using an inverted Zeiss LSM 880 confocal microscope (Figure 2a and Figure 2b). As expected, only low levels of SiR fluorescence were observed in GFP-Rab7-expressing HeLa cells treated with only SiR-Tz (Figure 2a). The average Pearson’s Correlation Coefficient (PCC) representing the colocalization of DiIC16-SiR or DiIC16’-SiR with GFP-Rab5 was significantly lower (PCC < 0.2) than that representing the colocalization of DiIC16-SiR or DiIC16’-SiR with GFP-Rab7 and GFP-Lamp1 (PCC > 0.4) (Figure 2b, Figure 2c). There was no significant difference between the PCC value representing colocalization of DiIC16-SiR or DiIC16’-SiR with GFP-Rab7 and GFP-Lamp1, which is expected. Moreover, the PCC values we calculate for DiIC16-SiR and DiIC16’-SiR are comparable to those representing the colocalization of DiIC16(3) with the same three endosomal markers (Supplementary Figure 1b and 1c). No intracellular fluorescence due to SiR was observed in HeLa cells treated with the pre-assembled (but cell-impermeant) HIDE probes DiIC16-SiR or DiIC16’-SiR (Supplementary Figure 2a). No intracellular fluorescence due to SiR was observed when cells expressing GFP-Rab7 were treated with SiR-Tz and DiIC16(3) (Supplementary Figure 2b), or with either DiIC16TCO or DiIC16’TCO and SiR-COOH (Supplementary Figure 2c). Based on these results, we conclude that the HIDE probes DiIC16-SiR or DiIC16’-SiR localize selectively to late endosomes and lysosomes.
Figure 2. DiIC16-SiR and DiIC16’-SiR selectively label late endosomes.
Laser-scanning confocal microscopy of HeLa cells (a) expressing GFP-Rab7 and treated with only SiR-Tz or (b) expressing GFP-Rab5, GFP-Rab7, or Lamp1-GFP and treated with DiIC16TCO or DiIC16’TCO and SiR-Tz. Scale bars: 20 μm. (c) Average Pearson’s correlation coefficient (PCC) representing the colocalization of DiIC16-SiR or DiIC16’-SiR with endosome markers. Data shown are mean ± s.e.m., n = 7 cells per endosome marker, N = 3 biological replicates, (***p < 0.001 and not significant (ns) for p > 0.05, one-way ANOVA followed by Tukey’s multiple comparisons test).
To further characterize DiIC16-SiR or DiIC16’-SiR, we designed a competition experiment to determine the fractional labeling of DiIC16TCO upon addition to cells and subsequent reaction with SiR-Tz. HeLa cells were treated with 5 μM DiIC16TCO followed by 2 μM SiR-Tz, as described in Figure 1b and the Online Methods. We then added between 0.5 and 2 μM BODIPY Fl-Tz to react with the remaining DiIC16TCO (Supplementary Figure 2d) and measured the fluorescence intensity of both SiR and BODIPY-Fl. We then plotted the ratio of BODIPY Fl-Tz and SiR fluorescence intensity as a function of the concentration of added BODIPY Fl-Tz (Supplementary Figure 2e). We observed that the BODIPY/SiR ratio plateaued at [BODIPY Fl-Tz] > 1.5 μM (Supplementary Figure 2e). This analysis suggests that approximately 1.5 μM DiIC16TCO remains reactive after treatment with 2 μM SiR-Tz, implying that the virtually all of the SiR-Tz has been converted to the HIDE probe DiIC16-SiR.
Late endosome HIDE probes do not affect endosomes.
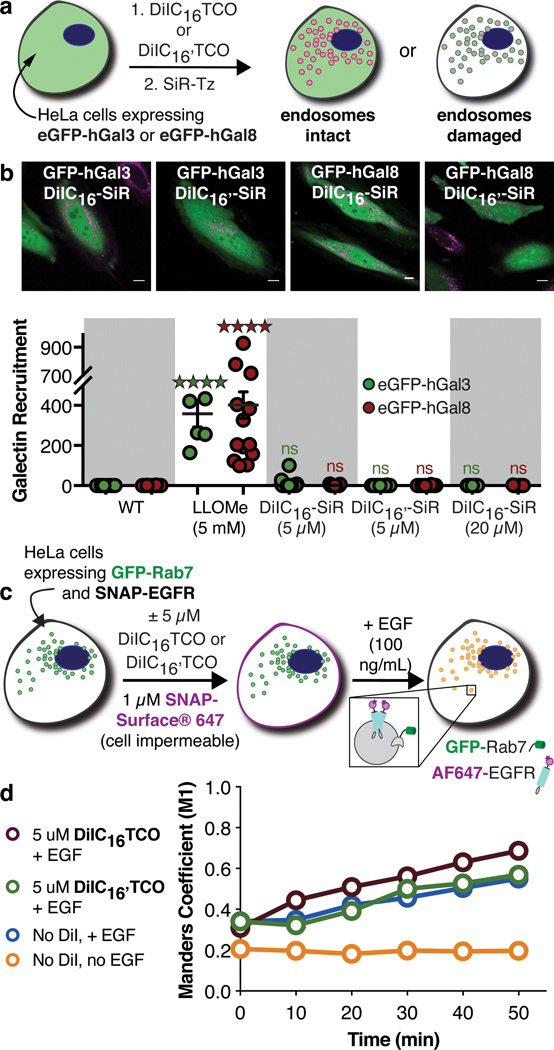
We next sought to verify that the presence of the HIDE probe DiIC16-SiR or DiIC16’-SiR had no measurable effect on endosome integrity and did not induce cell death. First, we utilized a galectin-recruitment assay to detect any potential HIDE probe-induced endosomal damage. This assay exploits the fact that damaged endosomes display β-galactosides on their surface that recruit otherwise cytosolic galectin proteins such as hGal3 and hGal8. When these galectin proteins are fused to eGFP, the recruitment of eGFP-hGal335 and eGFP-hGal836 can be visualized easily using confocal microscopy (Figure 3a). As expected, eGFP-hGal3 and eGFP-hGal8 remained cytosolic in untreated HeLa cells but were recruited extensively to endosomal surfaces when the cells were treated with the known endosomolytic agent LLOMe (Figure 3b and Supplementary Figure 3a). No such recruitment was observed when cells were treated with 5 μM DiIC16TCO or DiIC16’TCO and 2 μM SiR-Tz (Figure 3b), or with higher concentrations of both components, or with DiIC16TCO/DiIC16’TCO alone (Supplementary Figure 3b, 3c and 3d). Additionally, no galectin recruitment was observed either before and after irradiation with the excitation and STED depletion laser (Supplementary Figure 3e).
Figure 3. DiIC16-SiR and DiIC16’-SiR do not damage endosomal membranes or alter endocytic trafficking.
(a) Damage to lysosomes and late endosomes can be detected using eGFP-hGal334 or eGFP-hGal835. (b) Representative laser-scanning confocal microscopy of HeLa cells expressing eGFP-hGal3 or eGFP-hGal8 and labeled with DiIC16-SiR or DiIC16’-SiR (scale bar: 10 μm). Data shown are mean ± s.e.m., n = 9 cells, N = 2 replicates (****p < 0.0001 and not significant (ns) for p > 0.05, one-way ANOVA followed by post-hoc Dunnett’s test). (c) HeLa cells expressing both SNAP-EGFR and GFP-Rab7 were treated with DiIC16TCO or DiIC16’TCO, SNAP-Surface-647, and EGF, and visualized over 50 min using spinning disk confocal microscopy. (d) The average Manders M1 correlation coefficient between AF647-EGFR and GFP-Rab7 upon EGF treatment over time is unchanged in the presence of DiIC16TCO and DiIC16’TCO. Data shown are average from 2 independent experiments, n = 4 cells per condition per experiment; s.e.m. is 0.238 (not shown). Representative microscopy images in Supplementary Figure 4.
Late endosome HIDE probes are benign to EGFR trafficking.
We next sought to verify that the presence of DiIC16-SiR or DiIC16’-SiR had no measurable effect on endosome function, in particular on the rate of ligand-stimulated trafficking of the Epidermal Growth Factor Receptor (EGFR). HeLa cells over-expressing GFP-Rab7 and SNAP-EGFR were labeled with 1 μM of the cell-impermeant SNAP tag substrate SNAP-Surface 647 and either 5 μM DiIC16TCO or DiIC16’TCO to generate AF647-EGFR (Figure 3c, Supplementary Figure 4a); stimulation with Epidermal Growth Factor (EGF) induces trafficking of AF647-EGFR along the degradative endocytic pathway. The fraction of AF647-positive pixels that are also GFP-positive (Manders’ M1 Colocalization Coefficient (MCC)) was used to quantify the trafficking of AF647-EGFR into GFP-Rab7-positive vesicles over 50 minutes (Figure 3d). Without EGF stimulation, the M1 value reflecting the colocalization of AF647-EGFR and GFP-Rab7 remained constant over 50 minutes, indicating negligible trafficking of unstimulated EGFR into Rab7-positive endosomes (Supplementary Figure 4c). By contrast, after EGF treatment, the M1 value increased significantly over the course of 50 minutes and to the same extent regardless of the presence or absence of DiIC16TCO or DiIC16’TCO (Figure 3d, Supplementary Figure 4b). Thus, the presence of DiIC16TCO or DiIC16’TCO at the concentration used for super-resolution imaging (vide infra) does not significantly alter the rate of EGFR trafficking into late endosomes characterized by the expression of Rab7. We additionally investigated the effects of sustained incubation of DiIC16-SiR in HeLa cells expressing GFP-Rab7 (Supplementary Figure 4d) and found no change in co-localization of SiR between DiIC16-SiR and GFP-Rab7 or in the SiR signal over 24 hours (Supplementary Figure 4e).
Late endosome HIDE probes are non-toxic.
We also evaluated HIDE probe toxicity using two different methods. HeLa cells labeled with DiIC16TCO and DiIC16’TCO continued to divide over 20 hours under continuous irradiation (Supplementary Figure 5a). In addition, the nuclear content of HeLa cells treated overnight with DiIC16-SiR, as assessed by flow cytometry, showed little or no change (Supplementary Figure 5b). Taken as a whole, these diverse control experiments suggest that late endosome HIDE probes have little or no measurable effect on endosome integrity and are non-toxic.
DiIC16-SiR enables STED imaging of endosome dynamics.
With the knowledge that DiIC16-SiR and DiIC16’-SiR were non-toxic, induced no detectable endosomal damage, and had no effect on the kinetics of EGFR trafficking, we proceeded to evaluate their ability to support long-term STED nanoscopy of late endosomes in live cells (Figure 4). Here the two probes performed differently; using a Leica SP8 STED microscope, the HIDE probe generated from DiIC16TCO and SiR-Tz could visualize late endosomes for as long as 7 minutes (Supplementary Video 1), whereas images obtained using DiIC16’TCO and SiR-Tz lasted for less than 4 minutes (Supplementary Figure 5c, Supplemental Video 2). These differences are readily apparent in a plot of normalized intensity of SiR emission as a function of time; the time required to deplete half of the initial DiIC16-SiR signal was 250 seconds, whereas this time required to deplete half of the DiIC16’-SiR signal was less than 100 seconds (Figure 4a). Late endosome images obtained using either HIDE probe lasted significantly longer than those generated by reacting Halo-Rab7 with SiR-CA, which bleached within 40 seconds under STED conditions (Supplementary Figure 5c). Furthermore, using DiIC16TCO and SiR-Tz, late endosomes could be visualized with a resolution of 80 nm, significantly better than 400 nm resolution observed in the diffraction-limited image (Figure 4b and 4c). Endosomes in healthy fibroblasts imaged over the course of 7 minutes by simultaneous STED and confocal had smaller radii (median: 0.178 μm) by STED than when imaged by confocal (median: 0.260 μm) (Figure 4d).
Figure 4. DiIC16-SiR enables long-time lapse STED imaging of rare events in live HeLa cells and fibroblasts.
(a) Comparative STED and confocal microscopy of a HeLa cell labeled with DiIC16-SiR (scale bar: 1 μm). (b) Plot of average normalized fluorescence intensity of cells labeled with Halo-Rab7-SiR, DiIC16-SiR, and DiIC16’-SiR when imaged using STED. (Data shown are mean ± s.e.m., n = 4 cells per condition, N = 3 biological replicates). (c) Plot of normalized fluorescence intensity across the line profile shown (teal) in cells labeled with DiIC16-SiR and visualized using STED or confocal microscopy. The dashed teal and grey lines represent the Lorentzian fits derived from the STED and confocal image, respectively. (d) Particle detection by the TrackMate plugin of individual endosomes in the same cell imaged by confocal and STED displayed different distributions of particle radii. Endosomes imaged by STED had smaller radii than the same particles imaged by confocal. The particle distributions were then fit to a normal gaussian distribution (shown with shading). (e) Using STED microscopy and DiIC16-SiR, we visualized multiple inter-endosome “kiss-and-run” events that could not be resolved using confocal methods. Representative images from simultaneous confocal and STED microscopy of endosomes in WT fibroblasts. The experiment was repeated 6 times with similar results. Fission/fusion events are identified by white arrows (scale bar: 1 μm).
Notably, the increase in resolution achieved using STED microscopy and DiIC16-SiR is apparent over the full duration of the video, revealing multiple apparent inter-endosome “kiss-and-run” type events (Figure 4e). “Kiss-and-run” type events provide a portal for transient exchanges between closely juxtaposed endosomes that regulate cargo trafficking; they are exceedingly difficult to visualize due to increased endosomal clustering in the perinuclear region. Indeed, the endosomes visualized using HIDE probes and STED are so close that they appear as a singular blob-like entity when visualized using confocal methods (Figure 4e). Moreover, the dynamic “kiss-and-run” events that we see are rare; they are clearly observed only over the course of long time-lapse (7-minute) STED movies of both HeLa cells and WT fibroblasts (Supplementary Video 1 and Supplementary Video 3).
Visualizing motility defects in NPD patient fibroblasts.
Because HIDE probes are assembled from cell-permeant small molecules, they can visualize organelles in primary cells and cell lines that are difficult to transfect, such as neurons37 and fibroblasts. Probes such as BODIPY-lactosylceramide derivatives that have been used previously to study endosomal trafficking bleach rapidly and are not suitable for live-cell time-lapse microscopy, especially at the nanoscale38. To highlight the versatility of DiIC16-SiR, we used it to characterize differences in endosome motility between normal fibroblasts (GM05399) and those from patients with Niemann-Pick C disease (NPD). We focused on fibroblasts that harbor mutations in NPC1, an integral membrane protein that collaborates with NPC2 to shuttle cholesterol from the late endosome to the endoplasmic reticulum. These fibroblasts include those harboring the common I1061T NPC1 mutation (GM18453, Supplemental Video 4), the heterozygous P237S/I1061T mutation (GM03123, Supplemental Video 5), the R404Q mutation (GM18388, Supplemental Video 6), as well as the silent 1920delG NPC1 mutation (GM23945, Supplemental Video 7). The I1061T and P237S/I1061T mutations both lead to NPC1 misfolding in the ER20, 39, whereas the R404Q mutation leads to reduced binding of properly folded and localized NPC1 with NPC221. All of these mutations result in cholesterol accumulation in endo-lysosomes, as evaluated using filipin staining22, 40.
NPC1 mutations are believed to cause defects in endosome motility24, but these defects have never been evaluated in fibroblasts using either confocal microscopy or STED. Endosome motility may be quantified by the value of Vmax, the maximum velocity exhibited by an endosome over the course of its trajectory24. Using the HIDE probe DiIC16-SiR and simultaneous STED and confocal microscopy, we accurately detected individual endosomes in WT fibroblasts (Figure 5a) and monitored their movement over 7 minutes. In healthy fibroblasts, the number of endosomes detected by STED was 50% greater than the number detected using confocal microscopy (Figure 5b). We then proceeded to use STED, DiIC16-SiR, and the tracking software TrackMate to calculate late endosomal Vmax distributions in WT fibroblasts as well as fibroblasts harboring 1920delG, P237S/I1061T, I1061T, and R404Q mutations in NPC1 (Figure 5c and 5d). We found reduced endosome motility only in cells harboring the mutations P237S/I1061T and I1061T; endosomes in cells harboring the R404Q mutation displayed normal motility (Figure 5d) despite high levels of accumulated cholesterol. As noted previously, P237S/I1061T and I1061T NPC1 mutations result in protein misfolding in the ER and degradation and thus deplete functional NPC1 from the endo-lysosome39, whereas the R404Q mutation leads to reduced binding of NPC1 to NPC241. Both types of mutations result in massive levels of cholesterol accumulation as determined by filipin staining19. Thus, the data implies that cholesterol accumulation per se does not cause reduced endosome motility27, and instead supports the concept that interactions between intact NPC1 and ER-resident proteins (e.g. ORP5) govern endosome-ER contacts42 and endosome motility43. Critically, the differences in endosome motility are not seen by confocal microscopy and are only revealed by super-resolution imaging using the HIDE probe DiIC16-SiR (Figure 5d and Supplementary Figure 6a and 6b).
Figure 5. Endosome Motility defects in Niemann-Pick C disease are only visible with HIDE probes and STED Microscopy.
(a) Representative images from simultaneous confocal and STED microscopy of endosomes in WT fibroblasts. The experiment was repeated 6 times with similar results. Scale bar = 1 μm. (b) There are 53% more tracks detected for endosomes in WT fibroblasts by STED than by confocal. Videos of simultaneous confocal/STED imaging (of various durations) in WT fibroblasts labeled with DiIC16-SiR were analyzed by TrackMate. The number of tracks in both confocal and STED for each cell were plotted against each other, and the positive linear relationship was characterized by least squares fit (n = 6 cells, N = 1 sample). (c) Plots illustrating the distribution of endosome Vmax values over 3 minutes in healthy (grey) and NPC1 1920delG (blue) fibroblasts labeled with DiIC16-SiR and visualized by STED microscopy. (d) Plots illustrating the distribution of endosome Vmax values over 3 minutes in NPC1 P237S/I1061T (pink), NPC1 I1061T (green) and NPC1 R404Q (yellow) fibroblasts (with comparisons to healthy fibroblasts – grey) labeled with DiIC16-SiR and visualized by STED microscopy. Data shown are mean ± s.e.m., n = 10 cells per mutant, N = 5 samples. Comparison made between the median Vmax values from each mutant cell line (***p < 0.001, **p < 0.01, *p < 0.05, and not significant (ns) for p > 0.05, one-way ANOVA followed by Dunnett’s multiple comparisons test).
Discussion
Here we report new lipid-based, high-density environmentally sensitive (HIDE) probes that accurately and selectively image late endosomes and their dynamics at super-resolution for extended times. Treatment of live cells with the small molecules DiIC16TCO or DiIC16’TCO followed by SiR-Tz and an in situ tetrazine ligation reaction generates the HIDE probes DiIC16-SiR and DiIC16’-SiR within the late endosomal membrane. In this environment, the probes support the acquisition of super-resolution movies of late endosomes that last for more than 7 minutes. In contrast, movies of late endosomes visualized using SiR-tagged proteins last for only 20 seconds.
The ability to image endosomes at super-resolution for extended times revealed dynamic exchanges between closely juxtaposed endosomes, events that are often referred to as “kiss-and-run” events4. Kiss-and-run events are rare, transient interactions between a single endosome and a cluster of endocytic vesicles known as a reserve pool and are facilitated by small (< 50 nm) pores present in the target membrane44. Through kiss-and-run fission and fusion events, endosomes direct cellular signaling pathways and exocytosis of various cargo, including catecholamines and neuropeptides45. While exocytic “kiss-and-run” events have been visualized previously using confocal methods4, 46, the role of “kiss-and-run” phenomena in perinuclear endosome maturation and cargo trafficking is poorly characterized. With HIDE probes and STED microscopy, we can now resolve previously unrecognized kiss-and-run events that occur within the perinuclear region between single endosomes and reserve pools, a difficult feat with diffraction-limited microscopy (Figure 4e). This observation suggests that HIDE probes could be used to unravel the factors that dictate kiss-and-run events and illuminate their importance in various lysosome storage disorders like Niemann-Pick disease.
This work also provides new information about the effects of NPC1 mutations on late endosome motility. Previous work on the relationship between endosome motility and NP-D has relied on microscopy experiments performed in model cell lines in which cholesterol accumulation has been induced artificially, either by treating the cells with small molecule NPC1 inhibitors such as U18666A23 or by genetically manipulating cells to express GFP or YFP fusions to late endosome-resident proteins like NPC1, CD6324, Rab726, ORP1L47 and folate receptor48. These fusion proteins are typically over-expressed in cells with inactivating point mutations49 or truncation mutations that reduce cholesterol egress from the late endosome50, as characterized by filipin staining. The dynamics of these NPD-model endosomes are then analyzed with live-cell confocal microscopy.
Here we characterized endosome dynamics using STED microscopy and HIDE probes in a set of genetically distinct NPC1 patient fibroblasts (Figure 5d). These studies revealed previously unrecognized relationships between endosome motility and different types of NPD-relevant mutations that are evident only upon imaging with STED (Supplementary Figure 6a and 6b). Specifically, endosome motility defects are only seen in fibroblasts harboring NPC1 I1061T or NPC1 P237S/I1061T, but not NPC1 R404Q. As discussed previously, the NPC1 mutations I1061T and NPC1 P237S/I1061T to misfolding and ER-assisted protein degradation (ERAD)20. Whereas the R404Q mutation perturbs binding of NPC1 to NPC221, the luminal cholesterol binding protein in the late endosome, with no change in expression or folding. The observation that endosome motility is diminished only when NPC1 is present at low levels suggests a direct role for NPC1 in dictating endosome motility, potentially through endosome-ER interactions mediated by NPC1 and the ER-resident protein ORP5 or other proteins. The implementation of these new tools could facilitate large-scale screening to identify small molecules that alter endosome and inter-endosome dynamics or improve cholesterol export or establish a comprehensive panel of biomarkers for NPC diagnosis and treatment.
Online Methods
DiIC16TCO and DiIC16’TCO Synthesis:
Synthetic procedures are provided in the Supplementary Note.
Materials, reagents, and instrumentation for cell culture and imaging
Cell culture:
HeLa cells (ATCC) and NPC fibroblasts (Coriell Cell Repository) were cultured in Dulbecco’s modified Eagle medium (DMEM) (Gibco) supplemented with 10% FBS (Sigma-Aldrich), penicillin (100 unit/mL) and streptomycin (100 μg/mL), BacMam CellLight GFP-Rab5, GFP-Rab7, GFP-Lamp1 reagents (Life Technologies), DiIC16(3) (Life Technologies) and SNAP-Surface-647 (New England Biolabs). All cells were cultured at 37°C in a CO2/air (5%/95%) incubator. All cells were bona fide lines purchased from ATCC or Coriell and periodically tested for mycoplasma with DNA methods.
Laser-scanning confocal microscopy experiments were performed on an inverted Zeiss LSM 880 laser scanning confocal microscope equipped with a Plan-Apochromat 63x/1.2 NA oil immersion lens and a diode pumped solid-state 488 nm and 561 nm laser as well as a 633 nm argon laser. The day prior to imaging, cells were plated on a 35 mm MatTek dish in DMEM ph(−). For two-component labeling of late endosomes51, HeLa cells plated on 35 mm MatTek dishes were labeled with 500 μL of 5 μM DiIC16TCO or 500 μL of 5 μM DiIC16’TCO in DPBS (Gibco) with 1% BSA for 5 minutes at 37°C. The probe was removed and the cells were incubated in DMEM ph(−) at 37°C for 1 hr. The cells were washed with warm DPBS and incubated with 500 μL of 2 μM SiR-Tz in DPBS with 1% casein hydrolysate for 10 minutes at 37°C52. The dye solution was replaced with DMEM ph(−) and the cells were incubated for 20 minutes at 37°C. The cells were washed with warm DPBS and imaged in DMEM ph(−).
Colocalization with GFP-Rab proteins and Image analysis
Cells were seeded in eight-well Lab-Tek chambers (Nunc, Thermo Fisher Scientific) at a density of 7 × 103 per well. 6–8 hours post seeding, 2 μL of CellLight Reagent for either GFP-Rab5 (C10586), GFP-Rab7 (C10588), or GFP-LAMP1 (C10596) were added to the requisite wells and cells were incubated for 16–20 hours at 37°C51, 53. Cells were washed 3 times with warm DPBS, labeled with DiIC16TCO or DiIC16’TCO and SiR-Tz as described above and imaged by confocal laser scanning microscopy. Specificity for Rab7-positive vesicles54 was determined by overlap of SiR-Tz signal with GFP-Rab proteins using the Pearsons’ Correlation Coefficient (PCC) for individual cells using the JaCOP plugin on the FIJI software (Fiji is just ImageJ).
Galectin-based Assay for Assessing Endosome Rupture
eGFP-hGal3 and -hGal8 lentiviral particles55 were added to HeLa cells (10% FBS in DMEM, no p/s) plated on eight-well Lab-Tek chambers (Nunc, Thermo Fisher Scientific) at a density of 7 × 103 cells/well. After 16 hours, the cells were washed with warm DPBS three times and labeled with DiIC16TCO/SiR-Tz or DiIC16’TCO/SiR-Tz as described preciously and imaged by confocal microscopy. The amount of galectin recruitment was quantified in Fiji; first, a threshold was created that clearly distinguished between punctate fluorescence as ruptured endosome56. The “Analyze Particles” tool was used to quantify the total area of signal that was above the threshold, and this was divided by the total area of the cell to evaluate galectin recruitment. To obtain the Galectin Recruitment Coefficient (GRC), this number was multiplied by 1000.
Spinning-disk confocal microscopy was performed using an Improvision UltraVIEW VoX system (Perkin-Elmer) built around a Nikon Ti-E inverted microscope with an EMCCD Hamamatsu C9100–50 camera, equipped with PlanApo objectives (60Å~ 1.45-NA) and controlled by the Volocity software (Improvision). Green (GFP) and magenta (AF647-EGFR) channels mentioned in the manuscript correspond to the following filters: 527 ± 12 nm, 615 ± 35 nm and 705 ± 45 nm. 3D images with a thickness of 14–15 μm with 1-μm step size between each layer were taken. Merged XY planes shown in Supplementary Figure 4.
SNAP-EGFR Trafficking Assay
HeLa cells were electroporated with SNAP-EGFR57 using the NEPA21 Electroporator (Nepa Gene Co., Ltd), and transduced with GFP-Rab7 (Life Technologies, C10588) two days prior to imaging. Cells expressing SNAP-EGFR58 and GFP-Rab7 were labeled with DiIC16TCO, DiIC16’TCO, or DMSO control for 5 minutes in DPBS with 1% BSA. The probe mixture was replaced with DMEM ph(−) and the cells were allowed to incubate for 15 minutes at 37°C. 1 μM SNAP-Surface-647 (NEB, S9136S) was added as per the manufacturers protocol. The cells were washed 3x with warm DPBS and incubated in DMEM ph(−) at 37°C for 20 minutes prior to imaging. 2 mL fresh DMEM ph(−) was added to the cells, and cells expressing both SNAP-EGFR and GFP-Rab7 were found. One frame was taken before addition of 100 ng/mL EGF in DMEM and all subsequent stacks were taken after the addition of growth factor.
STED Microscopy
HeLa cells were labeled with DiIC16TCO or DiIC16’TCO and SiR-Tz as described above. STED microscopy was performed on a Leica TCS SP8 Gated STED 3x microscope equipped with a tunable pulsed white light laser (460 – 660 nm) for excitation and two HyD detectors for tunable spectral detection. The microscope offers three STED depletion lasers (592 nm, 660 nm, and 775 nm). For all live-cell imaging, the microscope was equipped with a Tokai Hit stage top incubator (model: INUBG2A-GSI) with temperature and CO2 control to maintain an environment of 37°C and 5% CO2. In this work, SiR derivatives were excited at 633 nm (15% power: HeLa cell imaging, Figure 4a; 20% power: fibroblasts, Figure 4e, Figure 5a) and their emission was detected using a HyD detector from 650–737 nm. The 775 nm depletion laser was used for STED microscopy (20% power). Imaging was conducted with a 100x oil immersion objective (HC PL APO 100x/1.40 OIL) at 1000 Hz with 1-line accumulation and bi-directional x-scanning in a 19.38 μm2 field of view (1024 × 1024 pixels at 18.94 nm/pixel). Raw microscopy data were Gaussian blurred (1.0 pixels) in ImageJ. The FWHM value were obtained by fitting line profiles to a Lorentz distribution using Origin 8.2 (www.originlab.com).
Halo-Rab7 Plasmid Construction and Transfection
A Halo PCR amplified fragment from the pFN23K-Halo plasmid (Promega, G2861) was added on to the N-terminus of the Rab7 sequence using Gibson Assembly (NEB, E2611S). The product was transformed into XL-10 Gold Ultracompetent cells (Agilent) and plated on LB/agar plates with 50 mg/mL kanamycin. Colonies were picked and grown overnight, and plasmids were purified using a Miniprep Kit (Qiagen). DNA sequencing confirmed the identity of the Halo-Rab7 product plasmid. HeLa cells were transfected with the Halo-Rab7 plasmid using FuGene HD (Promega), according to the manufacturer’s protocol. The transfected cells were then labeled with 500 μL of 1 μM SiR-CA in DPBS with 1% casein hydrolysate for 30 minutes at 37°C. The cells were washed with warm DBPS 3 times and incubated in DMEM ph(−) with 10% FBS for 2 hours at 37°C. After another washing step with DPBS, the media was replaced with fresh DMEM ph(−) with 10% FBS and imaged by STED.
Labeling and tracking in fibroblasts
Fibroblasts were plated in 35 mm MatTek dishes at 42,000 cells/mL in DMEM ph(−) with 5% LPDS (Alfa Aesar, J65182). Cells were labeled with DiIC16TCO and SiR-Tz as described for HeLa cells51, and imaged on the Leica TCS SP8 STED microscope in DMEM ph(−) with 5% LPDS at 37°C and 5% CO2. Endosome motility was measured using the LoG detector on the TrackMate plugin on Fiji59. An estimated diameter of 0.5 pixels and a threshold of 0.25 was optimal for endosome tracking in the STED videos obtained. A linking max distance and gap-closing max distance of 0.5 pixels was chosen, with a gap-closing max frame gap of 0.
Data Availability Statement
The materials and data reported in this study are available upon reasonable request from the corresponding author.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01GM131372–01, (A.S.), R01GM118486 (D.T.)), and the Wellcome Trust (095927/A/11/Z) and in part by the NIH (S10 OD020142 (Leica SP8)). A.G. was in part supported by the NIH (5T32GM06754 3–12). A.G. thanks Dr. Joe Wolenksi and Dr. Al Mennone for assistance with confocal and STED microscopy.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Maxfield FR; McGraw TE, Endocytic recycling. Nat Rev Mol Cell Bio 2004, 5 (2), 121–132. [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino JS; Neefjes J, Moving and positioning the endolysosomal system. Curr Opin Cell Biol 2017, 47, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadowski L; Pilecka I; Miaczynska M, Signaling from endosomes: Location makes a difference. Exp Cell Res 2009, 315 (9), 1601–1609. [DOI] [PubMed] [Google Scholar]
- 4.Alabi AA; Tsien RW, Perspectives on Kiss-and-Run: Role in Exocytosis, Endocytosis, and Neurotransmission. Annu Rev Physiol 2013, 75, 393–422. [DOI] [PubMed] [Google Scholar]
- 5.Gruenberg J, The endocytic pathway: A mosaic of domains. Nat Rev Mol Cell Bio 2001, 2 (10), 721–730. [DOI] [PubMed] [Google Scholar]
- 6.Li D; Shao L; Chen BC; Zhang X; Zhang MS; Moses B; Milkie DE; Beach JR; Hammer JA; Pasham M; Kirchhausen T; Baird MA; Davidson MW; Xu PY; Betzig E, Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 2015, 349 (6251). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shim SH; Xia CL; Zhong GS; Babcock HP; Vaughan JC; Huang B; Wang X; Xu C; Bi GQ; Zhuang XW, Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. P Natl Acad Sci USA 2012, 109 (35), 13978–13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson DS; Gregor C; Winter FR; Urban NT; Sahl SJ; Willig KI; Hell SW, SRpHi ratiometric pH biosensors for superresolution microscopy. Nat Commun 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wegel E; Gohler A; Lagerholm BC; Wainman A; Uphoff S; Kaufmann R; Dobbie IM, Imaging cellular structures in super-resolution with SIM, STED and Localisation Microscopy: A practical comparison. Sci Rep 2016, 6, 27290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das A; Barroso MM, Endosome-mitochondria interactions are modulated by iron release from transferrin. Mol Biol Cell 2016, 27, 831–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ba Q; Raghavan G; Kiselyov K; Yang G, Whole-Cell Scale Dynamic Organization of Lysosomes Revealed by Spatial Statistical Analysis. Cell Rep 2018, 23 (12), 3591–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Platt FM; Boland B; van der Spoel AC, Lysosomal storage disorders: The cellular impact of lysosomal dysfunction. J Cell Biol 2012, 199 (5), 723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Futerman AH; van Meer G, The cell biology of lysosomal storage disorders. Nat Rev Mol Cell Bio 2004, 5 (7), 554–565. [DOI] [PubMed] [Google Scholar]
- 14.Pfeffer SR, NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes. J Biol Chem 2019, 294 (5), 1706–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim CY; Davis OB; Shin HR; Zhang J; Berdan CA; Jiang XT; Counihan JL; Ory DS; Nomura DK; Zoncu R, ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C. Nat Cell Biol 2019, 21 (10), 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung K; Chakraborty K; Saminathan A; Krishnan Y, A DNA nanomachine chemically resolves lysosomes in live cells. Nat Nanotechnol 2019, 14 (2), 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vacca F; Vossio S; Mercier V; Moreau D; Johnson S; Scott CC; Montoya JP; Moniatte M; Gruenberg J, Cyclodextrin triggers MCOLN1-dependent endo-lysosome secretion in Niemann-Pick type C cells. J Lipid Res 2019, 60 (4), 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vivas O; Tiscione SA; Dixon RE; Ory DS; Dickson EJ, Niemann-Pick Type C Disease Reveals a Link between Lysosomal Cholesterol and PtdIns(4,5)P-2 That Regulates Neuronal Excitability. Cell Reports 2019, 27 (9), 2636–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanier MT; Latour P, Laboratory diagnosis of Niemann-Pick disease type C: The filipin staining test. Method Cell Biol 2015, 126, 357–375. [DOI] [PubMed] [Google Scholar]
- 20.Gelsthorpe ME; Baumann N; Millard E; Gale SE; Langmade SJ; Schaffer JE; Ory DS, Niemann-Pick type C1I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J Biol Chem 2008, 283 (13), 8229–8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deffieu MS; Pfeffer SR, Niemann-Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. P Natl Acad Sci USA 2011, 108 (47), 18932–18936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park WD; O’Brien JF; Lundquist PA; Kraft DL; Vockley CW; Karnes PS; Patterson MC; Snow K, Identification of 58 novel mutations in Niemann-Pick disease Type C: Correlation with biochemical phenotype and importance of PTC1-like domains in NPC1. Hum Mutat 2003, 22 (4), 313–325. [DOI] [PubMed] [Google Scholar]
- 23.Lu F; Liang Q; Abi-Mosleh L; Das A; De Brabander JK; Goldstein JL; Brown MS, Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. Elife 2015, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko DC; Gordon MD; Jin JY; Scott MP, Dynamic movements of organelles containing Niemann-Pick C1 protein: NPC1 involvement in late endocytic events. Mol Biol Cell 2001, 12 (3), 601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millat G; Marcais C; Tomasetto C; Chikh K; Fensom AH; Harzer K; Wenger DA; Ohno K; Vanier MT, Niemann-Pick C1 disease: Correlations between NPC1 mutations, levels of NPC1 protein, and phenotypes emphasize the functional significance of the putative sterol-sensing domain and of the cysteine-rich luminal loop. Am J Hum Genet 2001, 68 (6), 1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lebrand C; Corti M; Goodson H; Cosson P; Cavalli V; Mayran N; Faure J; Gruenberg J, Late endosome motility depends on lipids via the small GTPase Rab7. Embo J 2002, 21 (6), 1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobo K; Le Blanc I; Luyet PP; Fivaz M; Ferguson C; Parton RG; Gruenberg J; van der Goot FG, Late Endosomal Cholesterol Accumulation Leads to Impaired Intra-Endosomal Trafficking. Plos One 2007, 2 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson AD; Bewersdorf J; Toomre D; Schepartz A, HIDE Probes: A New Toolkit for Visualizing Organelle Dynamics, Longer and at Super-Resolution. Biochemistry-Us 2017, 56 (39), 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erdmann RS; Takakura H; Thompson AD; Rivera-Molina F; Allgeyer ES; Bewersdorf J; Toomre D; Schepartz A, Super-Resolution Imaging of the Golgi in Live Cells with a Bioorthogonal Ceramide Probe. Angew Chem Int Edit 2014, 53 (38), 10242–10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takakura H; Zhang YD; Erdmann RS; Thompson AD; Lin Y; McNellis B; Rivera-Molina F; Uno SN; Kamiya M; Urano Y; Rothman JE; Bewersdorf J; Schepartz A; Toomre D, Long time-lapse nanoscopy with spontaneously blinking membrane probes. Nat Biotechnol 2017, 35 (8), 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bottanelli F; Kilian N; Ernst AM; Rivera-Molina F; Schroeder LK; Kromann EB; Lessard MD; Erdmann RS; Schepartz A; Baddeley D; Bewersdorf J; Toomre D; Rothman JE, A novel physiological role for ARF1 in the formation of bidirectional tubules from the Golgi. Molecular Biology of the Cell 2017, 28 (12), 1676–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bottanelli F; Kromann EB; Allgeyer ES; Erdmann RS; Baguley SW; Sirinakis G; Schepartz A; Baddeley D; Toomre DK; Rothman JE; Bewersdorf J, Two-colour live-cell nanoscale imaging of intracellular targets. Nature Communications 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Revelo NH; Kamin D; Truckenbrodt S; Wong AB; Reuter-Jessen K; Reisinger E; Moser T; Rizzoli SO, A new probe for super-resolution imaging of membranes elucidates trafficking pathways. J Cell Biol 2014, 205 (4), 591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackman ML; Royzen M; Fox JM, Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J Am Chem Soc 2008, 130 (41), 13518–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paz I; Sachse M; Dupont N; Mounier J; Cederfur C; Enninga J; Leffler H; Poirier F; Prevost MC; Lafont F; Sansonetti P, Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell Microbiol 2010, 12 (4), 530–544. [DOI] [PubMed] [Google Scholar]
- 36.Thurston TLM; Wandel MP; von Muhlinen N; Foeglein A; Randow F, Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 2012, 482 (7385), 414–U1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson AD; Omar MH; Rivera-Molina F; Xi Z; Koleske AJ; Toomre DK; Schepartz A, Long-Term Live-Cell STED Nanoscopy of Primary and Cultured Cells with the Plasma Membrane HIDE Probe DiI-SiR. Angew Chem Int Ed Engl 2017, 56 (35), 10408–10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun XF; Marks DL; Park WD; Wheatley CL; Puri V; O’Brien JF; Kraft DL; Lundquist PA; Patterson MC; Pagano RE; Snow K, Niemann-Pick C variant detection by altered sphingolipid trafficking and correlation with mutations within a specific domain of NPC1. Am J Hum Genet 2001, 68 (6), 1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salman A; Cougnoux A; Farhat N; Wassif CA; Porter FD, Association of NPC1 Variant p. P237S with a Pathogenic Splice Variant in Two Niemann-Pick Disease Type C1 Patients. Am J Med Genet A 2017, 173 (4), 1038–1040. [DOI] [PubMed] [Google Scholar]
- 40.Pipalia NH; Subramanian K; Mao S; Ralph H; Hutt DM; Scott SM; Balch WE; Maxfield FR, Histone deacetylase inhibitors correct the cholesterol storage defect in most Niemann-Pick C1 mutant cells. J Lipid Res 2017, 58 (4), 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li XC; Saha P; Li J; Blobel G; Pfeffer SR, Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. P Natl Acad Sci USA 2016, 113 (36), 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eden ER, The formation and function of ER-endosome membrane contact sites. Bba-Mol Cell Biol L 2016, 1861 (8), 874–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raiborg C; Wenzel EM; Stenmark H, ER-endosome contact sites: molecular compositions and functions. Embo J 2015, 34 (14), 1848–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe S; Rost BR; Camacho-Perez M; Davis MW; Sohl-Kielczynski B; Rosenmund C; Jorgensen EM, Ultrafast endocytosis at mouse hippocampal synapses. Nature 2013, 504 (7479), 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staal RGW; Mosharov EV; Sulzer D, Dopamine neurons release transmitter via a flickering fusion pore. Nat Neurosci 2004, 7 (4), 341–346. [DOI] [PubMed] [Google Scholar]
- 46.Saffi GT; Botelho RJ, Lysosome Fission: Planning for an Exit. Trends Cell Biol 2019, 29 (8), 635–646. [DOI] [PubMed] [Google Scholar]
- 47.Vihervaara T; Uronen RL; Wohlfahrt G; Bjorkhem I; Ikonen E; Olkkonen VM, Sterol binding by OSBP-related protein 1L (ORP1 L) regulates late endosome motility and function. Cellular and Molecular Life Sciences 2011, 68 (3), 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H; Yang J; Low PS; Cheng JX, Cholesterol level regulates endosome motility via Rab proteins. Biophys J 2008, 94 (4), 1508–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Millard EE; Gale SE; Dudley N; Zhang J; Schaffer JE; Ory DS, The sterol-sensing domain of the Niemann-Pick C1 (NPC1) protein regulates trafficking of low density lipoprotein cholesterol. J Biol Chem 2005, 280 (31), 28581–28590. [DOI] [PubMed] [Google Scholar]
- 50.Li X; Lu F; Trinh MN; Schmiege P; Seemann J; Wang J; Blobel G, 3.3 A structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport. Proc Natl Acad Sci U S A 2017, 114 (34), 9116–9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Online Methods
- 51.Mukherjee S; Maxfield FR, Trafficking of lipid analogs with varying tail lengths and unsaturations in Niemann Pick C fibroblasts. Mol Biol Cell 2000, 11, 325a–325a.10637311 [Google Scholar]
- 52.Devaraj NK; Upadhyay R; Hatin JB; Hilderbrand SA; Weissleder R, Fast and Sensitive Pretargeted Labeling of Cancer Cells through a Tetrazine/trans-Cyclooctene Cycloaddition. Angewandte Chemie-International Edition 2009, 48 (38), 7013–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts RL; Barbieri MA; Pryse KM; Chua M; Stahl PD, Endosome fusion in living cells overexpressing GFP-rab5. Journal of Cell Science 1999, 112 (21), 3667–3675. [DOI] [PubMed] [Google Scholar]
- 54.Humphries WH; Szymanski CJ; Payne CK, Endo-Lysosomal Vesicles Positive for Rab7 and LAMP1 Are Terminal Vesicles for the Transport of Dextran. Plos One 2011, 6 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinauer A; LaRochelle JR; Knox SL; Wissner RF; Berry S; Schepartz A, HOPS-dependent endosomal fusion required for efficient cytosolic delivery of therapeutic peptides and small proteins. P Natl Acad Sci USA 2019, 116 (2), 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aits S; Kricker J; Liu B; Ellegaard AM; Hamalisto S; Tvingsholm S; Corcelle-Termeau E; Hogh S; Farkas T; Jonassen AH; Gromova I; Mortensen M; Jaattela M, Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 2015, 11 (8), 1408–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ibach J; Radon Y; Gelleri M; Sonntag MH; Brunsveld L; Bastiaens PIH; Verveer PJ, Single Particle Tracking Reveals that EGFR Signaling Activity Is Amplified in Clathrin-Coated Pits. Plos One 2015, 10 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roepstorff K; Grandal MV; Henriksen L; Knudsen SLJ; Lerdrup M; Grovdal L; Willumsen BM; van Deurs B, Differential Effects of EGFR Ligands on Endocytic Sorting of the Receptor. Traffic 2009, 10 (8), 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tinevez JY; Perry N; Schindelin J; Hoopes GM; Reynolds GD; Laplantine E; Bednarek SY; Shorte SL; Eliceiri KW, TrackMate: An open and extensible platform for single-particle tracking. Methods 2017, 115, 80–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The materials and data reported in this study are available upon reasonable request from the corresponding author.