Summary
Defining functions for the full complement of proteins is a grand challenge in the post-genomic era, and is essential for our understanding of basic biology and disease pathogenesis. In recent times, this endeavor has benefitted from a combination of modern large-scale and classical reductionist approaches—a process we refer to as ‘systems biochemistry’—that helps surmount traditional barriers to the characterization of poorly understood proteins. This strategy is proving to be particularly effective for mitochondria, whose well-defined proteome has enabled comprehensive analyses of the full mitochondrial system that can position understudied proteins for fruitful mechanistic investigations. Recent systems biochemistry approaches have accelerated the identification of new disease-related mitochondrial proteins and of long-sought ‘missing’ proteins that fulfill key functions. Collectively, these studies are moving us toward a more complete understanding of mitochondrial activities and providing a molecular framework for the investigation of mitochondrial pathogenesis.
Keywords: mitochondria, systems biochemistry, multi-omics, rare disease, orphan proteins
eTOC Blurb
In this review, Sung et al. describe the ‘systems biochemistry’ approach—a blend of large-scale, ‘omic’ methodologies and classical reductionist biochemistry—that has accelerated the study of mitochondrial proteins of unknown function over the past decade and moved us towards a more complete, molecular understanding of mitochondrial function.
Mitochondrial dark matter
The state of modern biological science is remarkable: Our research is performed at a time when ‘omics’ techniques enable us to measure nearly every biomolecule, revolutions in imaging and structural biology enable us to observe subcellular components at striking resolution, and gene editing technologies enable us to manipulate DNA seemingly without restriction. Yet, our ability to measure, observe, and manipulate biological systems has arguably outpaced our fundamental understanding of the basic gene functions that underlie them. Recent analyses have revealed that most research on human genes only concentrates on approximately 2,000 of the ~19,000 genes of the human genome (Stoeger et al. 2018), with just 100 genes accounting for more than one-quarter of the papers tagged by the National Library of Medicine (Dolgin 2017). Similarly, over 600 yeast proteins and 2000 human proteins have yet to be assigned any molecular function (Ellens et al. 2017), and even for the most well-studied model organisms such as Escherichia coli, more than a third of the genes have yet to be fully functionalized (Ghatak et al. 2019; Sevin et al. 2017). Relatedly, it is estimated that more than 1,000 known enzyme activities among the 5,000 entries of the Enzyme Commission (EC) classification still lack an associated protein (Sorokina et al. 2014).
The reasons why so many proteins remain poorly characterized are manifold. Many are simply hard to study because they may have redundant functions, lack essential functions under standard laboratory conditions, or have functions that affect multiple cellular processes (Oliver 1996; Hillenmeyer et al. 2008; Galperin and Koonin 2010). For others, progress is limited by the lack of tools and reagents (e.g., antibodies, mouse lines) available for more ‘popular’ proteins (Edwards et al. 2011). Additionally, this lingering focus on a small set of proteins may be rooted in the assumption that they are more relevant for human health and disease; however, this is likely misguided (Stoeger et al. 2018). A telling example in support of this can be seen in a recent analysis of human protein-protein interactions (Rolland et al. 2014). In this study, the authors found that highly studied proteins have a strikingly higher number of reported protein-protein interactions in the literature than uncharacterized proteins, giving the impression that that the former group is more connected to key biological processes. Yet, further analyses revealed the latter group to be comparably represented in genome-wide association studies (GWAS) and equally associated with Mendelian disorders. Such “inspection bias” risks erroneously assuming that well-studied proteins are more responsible for a given effect simply due to their familiarity.
This paradigm extends to mitochondria, whose iconic cellular “powerhouse” descriptor has mistakenly given the impression that this organelle is a fully defined system with a fully defined function. In reality, recent work has revealed hundreds of mitochondrial uncharacterized (x) proteins (MXPs) (Pagliarini and Rutter 2013), and new mitochondria-related processes continue to be identified (Nunnari and Suomalainen 2012). Progress toward defining the functions of MXPs has been accelerated by the advent of diverse large-scale methodologies. On their own, such ‘omics-level’ analyses risk being mere exercises in information gathering. Wielded properly, these experiments serve to generate sharper, more informed hypotheses into protein function. We refer to this blending of ‘systems’ analyses with classical mechanistic biochemical and bioenergetics approaches as ‘systems biochemistry.’ The well-established proteome of mitochondria, together with its manageable complexity and purifiability, seem to have made it particularly amenable to this approach. The goal of this review is to describe some of the successful applications of systems biochemistry that, over the past decade, have transformed mitochondrial research. We hope that this will provide a useful framework for surmounting some of the challenges in studying uncharacterized proteins. For those with further interest in works under the broader umbrella of mitochondrial ‘systems biology,’ we refer them to several other excellent recent reviews (Matilainen, Quiros, and Auwerx 2017; Hu, Go, and Jones 2020; Maldonado et al. 2019; Rahman and Rahman 2018).
Defining the system
A key first step in assigning functions to the parts of a biological system is to define its composition. The ability to separate discrete cellular fractions from much of the rest of the cell has been helpful in this regard, even before scientists knew what these fractions held. For example, early work by Otto Warburg with crude liver lysate fractions had shown that “succinoxidase” activity resided within subcellular “large granules” before it was clear that they contained mitochondria (Warburg 1913). Motivated by the mission to establish and quantify the “distribution of enzyme activities among the various cell components” (Claude 1946), Albert Claude and colleagues made subsequent advancements in ultracentrifugation and associated methods that allowed for the generation of a sample that was enriched for intact mitochondria (Claude 1946). Such samples were then used to show that succinate dehydrogenase and cytochrome oxidase are localized in the mitochondrial fraction (Hogeboom, Claude, and HotchKiss). Over the next several years, other enzymatic activities including the TCA cycle, fatty acid oxidation, and oxidative phosphorylation were localized to mitochondria (Kennedy and Lehninger 1949). Together, these studies solidified the role of mitochondria in energy metabolism, and cemented its designation as the “powerhouse of the cell” (Ernster and Schatz 1981).
After the core tenets of oxidative phosphorylation were established (Mitchell 1961; Jagendorf and Uribe 1966), the intense interest in mitochondria somewhat waned, marked by a significant decline in the percentage of papers related to mitochondrial function (Pagliarini and Rutter 2013). Nonetheless, pioneering work in subsequent decades led to the sequencing of the human mitochondrial genome (Anderson et al. 1981), the demonstration of maternal inheritance of mitochondria, and the direct link between mtDNA mutations and human disease (Wallace et al. 1988; Holt, Harding, and Morgan-Hughes 1988), undergirding the renaissance of interest that was soon to follow. By the late 1990s, further sparked by remarkable and unexpected new observations of mitochondrial functions and their association with human disease, broad enthusiasm in mitochondrial biology was rekindled (Pagliarini and Rutter 2013). Beyond ATP production, these efforts connected mitochondria to key roles in cell signaling, apoptosis, response to viral infection, aging, and many other processes (Wallace 1999; Nunnari and Suomalainen 2012). Mitochondrial dysfunction soon became associated with more than 150 unique diseases, including many inborn errors of metabolism, diabetes, and cancer (Nunnari and Suomalainen 2012; Vafai and Mootha 2012; Koopman, Willems, and Smeitink 2012). As such, it became important both to identify proteins of known function that localize to mitochondria (and thereby place their pathways within the organelle), and to identify new and uncharacterized mitochondrial proteins.
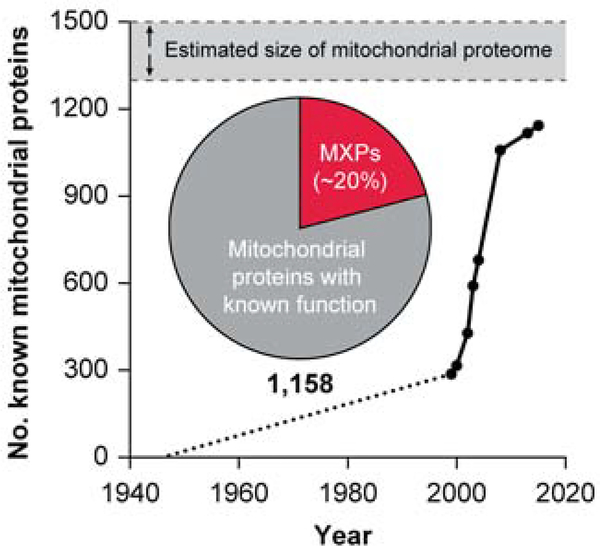
Fortunately, this renaissance in mitochondrial biology coincided with the development of new technologies, most notably mass spectrometry (MS)-based proteomics, that enabled scientists to explore the composition of mitochondria at a much greater depth (Aebersold and Mann 2003). These studies, covered in other reviews (Calvo and Mootha 2010; Gonczarowska-Jorge, Zahedi, and Sickmann 2017), rapidly increased the number of known mitochondrial proteins from yeast to human (Fig. 1). More recent advances in labeling and MS methods have further expanded the known mitochondrial proteome while simultaneously revealing the submitochondrial localization of many these proteins (Rhee et al. 2013; Hung et al. 2017; Hung et al. 2014; Geladaki et al. 2019; Vogtle et al. 2017). The current breadth of coverage stands at 1,158 unique mitochondrial proteins for mouse and human (Calvo, Clauser, and Mootha 2016). This number is consistent with earlier predictions that the true size of the mitochondrial proteome is approximately 1,300–1,500 proteins, as estimated based on 2-D gel and computational analyses (Pagliarini et al. 2008; Taylor et al. 2003; DiMauro and Schon 1998; Lopez et al. 2000).
Figure 1.
Understanding of the mammalian mitochondrial proteome has increased rapidly since 2000, yet ~20% of its proteins remain uncharacterized. The current list stands at 1,158 proteins, close to the early estimates of 1,300–1,500.
Systematic functional characterization of mitochondrial proteins
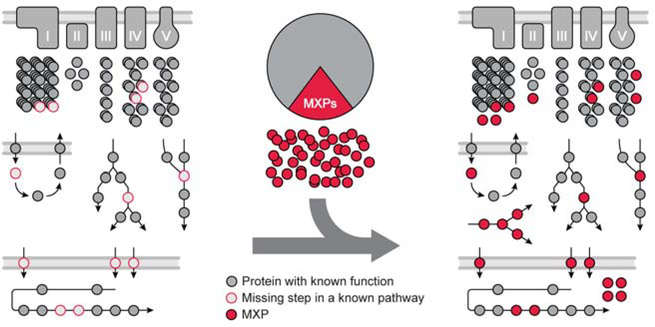
An established compendium of proteins for a biological system accomplishes two goals for the grand challenge noted above: It reveals those parts of the system that aren’t yet understood, and provides the basis for a systematic approach to assign functions to those parts (Fig. 2). For example, the MitoCarta study revealed that ~300 mammalian mitochondrial proteins had little to no functional characterization, and then leveraged this expanded compendium to discover new complex I disease-related genes (Pagliarini et al. 2008).
Figure 2.
A defined system catalyzes the assignment of orphan protein functions.
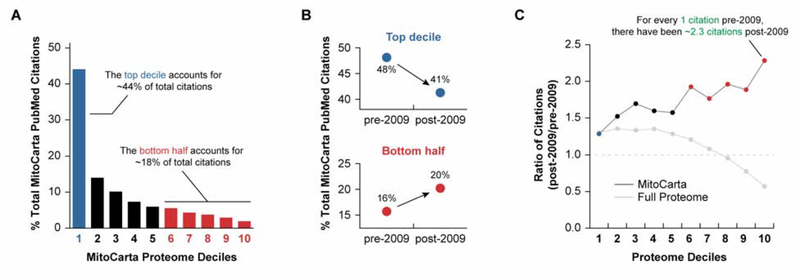
The continued influence of a defined mitochondrial system on accelerating the study of its understudied proteins is apparent from recent PubMed citation trends (Fig. 3). In this analysis, we determined the number of NCBI Pubmed citations attributed to the geneID of each human protein in MitoCarta 2.0 (Calvo, Clauser, and Mootha 2016). These proteins were then rank ordered based on the number of citations per protein and binned into deciles. In total, the top 10% of mitochondrial proteins (i.e., the ‘popular’ proteins) account for nearly triple the number of citations of the bottom half of the proteome (i.e., the ‘unpopular proteins’) (Fig. 3A). However, when the results of the analysis are separated into citations before and after the MitoCarta study (published in 2009) an encouraging pattern emerges. Prior to this study, nearly half of human mitochondria-related citations stemmed from the top decile of mitochondrial proteins, while the bottom half accounted for just 16% (Fig. 3B). During the subsequent decade, this distribution began to shift. The top decile decreased to ~40% of human mitochondria-related citations between 2009 and 2019, and the bottom increased to 20% (Fig. 3B). This pattern is even more striking when comparing the ratio of citations attributed to each decile in the past decade to all prior citations. Indeed, there have been more than twice as many citations for proteins in the bottom decile in the past decade than all citations in that decile prior to 2009 (Fig 3C). This trend is especially noteworthy given that the opposite trend is seen in the overall distribution of PubMed Citations for the whole human proteome, where the disparity between the top decile and bottom half of the proteome continues to grow (Fig 3C).
Figure 3.
Defining a biological system helps reverse the over focus on proteins of known function.
A, There is a striking disproportionality in citations across the mitochondrial proteome.
B, After defining a high-confidence mitochondrial proteome in 2009, the distribution in citations in the top decile and bottom half have begun to shift towards a more balanced distribution.
C, While this trend is true for the mitochondrial genes, research on the rest of the genome continues to be skewed towards the more ‘popular’ proteins.
The systems biochemistry approaches that have driven many of these studies have generally taken one of two forms: a top-down approach seeking to identify a missing component of an established process (known unknowns), or a bottom-up approach that begins with the intentional disruption of poorly characterized genes (unknown unknowns). In each case, the following demonstrate how a systematic analysis of a well-defined mitochondrial proteome led to clear hypotheses and new insight into protein function.
Top-down (known unknowns)
Analogous to the forward genetics approach, a top-down approach begins with a function or phenotype that is not well understood, and proceeds to identify the underlying genes/proteins. This approach has been successful in filling gaps in knowledge related to mitochondrial transporters and the formation of respiratory supercomplexes.
Mitochondrial Transporters
One powerful way that an established compendium facilitates the identification of protein function is by limiting the search space for a known missing activity. An illustrative example is the discovery of the mitochondrial calcium uniporter. Since the 1960s, it was documented that vertebrate mitochondria could take up calcium (Deluca and Engstrom 1961). Further studies showed that certain mitochondrial functions were activated by calcium. Yet, the machinery responsible for calcium import remained unidentified for decades (Mammucari et al. 2018). It had been established for some time that kinetoplastids exhibited rapid, uncoupler-sensitive Ca2+ import into mitochondria, but that this same activity was absent from S. cerevisiae. A simple phylogenetic analysis to identify genes present in the former but not the latter yielded too many candidates to be practical; however, when the list was filtered using mitochondrial proteome datasets, a manageable number of candidates emerged (Perocchi et al. 2010). Subsequent rigorous biochemistry and structural biology approaches have now established MICU1–2, MCU/MCUb, and EMRE as the full mitochondrial Ca2+ import machinery (Baughman et al. 2011; De Stefani et al. 2011; Sancak et al. 2013; Perocchi et al. 2010). Similarly, analysis of mitochondrial carrier-like proteins in these proteome lists helped prioritize candidates for the mitochondrial pyruvate carrier, which likewise went unidentified for ~50 years (Bricker et al. 2012; Herzig et al. 2012). Finally, a mitochondrial serine transporter was recently identified in part through the use of a custom CRISPR screen that focused only on genes encoding mitochondrial and other metabolism-related proteins, allowing for much deeper coverage of this prioritized set than would be achieved via a genome-wide screen (Kory et al. 2018). Together, these studies demonstrate the utility of a well-defined search space for identifying proteins responsible for missing steps in known pathways.
Respiratory Supercomplex Formation
Another example of a robust top-down systems biochemistry approach can be seen in the quantitative trait loci (QTL) mapping work by Williams et al. that helped establish a regulator of mitochondrial respiratory supercomplex (SC) formation (Williams et al. 2016). SCs are now recognized as key components of the electron transport chain, but their assembly is poorly understood (Milenkovic et al. 2017; Williams et al. 2016). In this work, the authors combined a targeted analysis of the mitochondrial proteome across hundreds of mice from the BXD genetic reference population (Andreux et al. 2012) with biochemical measurements of SC formation (among other genomic, metabolomic, and physiological measurements). Remarkably, the integration of the prominent SC QTL with mitochondrial protein QTLs nominated COX7A2L as a clear candidate regulator of SC formation, which was then confirmed with detailed follow-up analyses (Williams et al. 2016). Here again, a large-scale analysis guided by the well-defined mitochondrial proteome, coupled with detailed biochemical analyses, led to a robust hypothesis of protein function.
Bottom-up (unknown unknowns)
The surprisingly large number of MXPs comprises proteins that generally have not been studied and others that have been linked to a process (e.g., via genetic screens), but for unknown reasons. A number of approaches have been conducted that took direct aim at understanding what these proteins do without a specific knowledge gap in mind. These strategies parallel the classical reverse genetics paradigm in which genes or proteins of unknown function are disrupted in an effort to define their functions. Similarly, the goal of these bottom-up approaches has been to establish a more complete understanding of mitochondria by functionalizing their component parts and placing them within pre-existing pathways.
Deorphanizing MXPs through multi-omic analyses
In recent years, we have taken a multi-omics profiling approach to define uncharacterized mitochondrial protein function. These initiatives begin with the creation of a series of individual genetic perturbations in a common system, such as yeast strains or mammalian cell lines. These genes typically include many encoding MXPs and others encoding “sentinel” proteins of well-defined function. Each strain/cell line is then profiled using deep proteomic and metabolomic analyses, thereby providing a detailed cellular response signature unique to each disruption. The use of carefully select gene series is essential to this approach’s success because the effects of individual perturbations (e.g., drugs, gene deletions) are difficult to interpret in isolation due to cellular compensation and secondary responses.
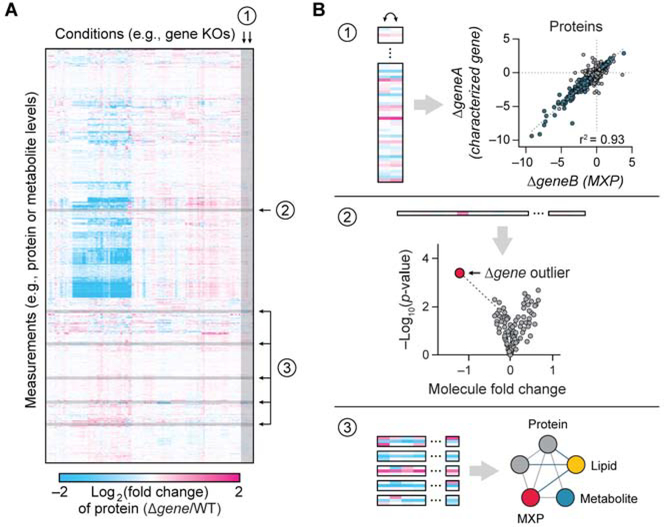
These rich datasets can be mined in many sophisticated ways, and we find that three straightforward analyses can quickly provide actionable new insights (Fig. 4). First, new connections can be made between distinct perturbations by virtue of their similar ‘omic’ responses using a regression analysis (Fig. 4B–1). For instance, if deletion of an uncharacterized protein yields a similar multi-omic signature across thousands of data points to that of a second gene deletion, it is likely that those genes have related functions. Second, the simultaneous analysis of many perturbations enables the identification of unique (i.e., “outlier”) responses for a given perturbation, even when the magnitude of change is modest (Fig. 4B–2). Third, co-regulated networks of proteins, lipids, and metabolites can be identified across the dataset, thus creating new functional hypotheses even when those molecules were not the focus of the initial perturbation (Fig. 4B–3).
Figure 4.
Multi-omics analyses of well-defined, contrasting biological states are a powerful approach for leveraging large-scale data to generate mechanistic hypotheses.
A, These large datasets can be mined in many ways.
B, Straightforward analyses include, 1) KO vs. KO regression analyses, 2) outlier analyses, and 3) molecular covariance network analyses.
Recently, we have used these multi-omic approaches to help establish mechanistic hypotheses for several yeast MXPs. First, we revealed that Hfd1p completes the conversion of tyrosine to 4-hydroxybenzoate (4-HB), the precursor to coenzyme Q biosynthesis (CoQ)—a step that had remained unresolved for more than 40 years (Stefely et al. 2016). The function of Hfd1 was concurrently identified by an independent genetic screen (Payet et al. 2016). We also used this strategy to systematically profile mitochondrial proteases, thereby connecting these key proteins to new processes and identifying new direct substrates (Veling et al. 2017), and to identify targets for an RNA binding protein that helps orchestrate mitochondrial biogenesis (Lapointe et al. 2018). In ongoing efforts, we have extended this strategy to a library of CRISPR-mediated knockouts in human cells. Overall, this multi-omic approach is providing molecular insight into mitochondrial protein function and disease and, more broadly, has established a powerful new high-throughput approach for defining gene function.
Coenzyme Q Biosynthesis - Structural Genomic Insights
Structural genomics is a bottom-up approach that aims to generate or model structural information for most proteins in nature. Targets for structural determination are often selected before their biochemical functions are understood, with the hope that structural information could lend insights into protein function and assist in the future prediction of protein structure from primary sequence (Skolnick, Fetrow, and Kolinski 2000). Recent efforts, such as the Protein Structure Initiative (Montelione 2012), have benefitted a number of mitochondrial proteins, including COQ8A, COQ9, and COQ4. These three proteins are required for coenzyme Q (CoQ) biosynthesis, but their biochemical activities remain incompletely defined (Stefely and Pagliarini 2017).
COQ8A (aka ADCK3) was originally speculated to be a protein kinase (Poon et al. 2000). However, subsequent structural work and biochemical work revealed that it more likely serves as an ATPase that supports the integrity of a biosynthetic complex of other CoQ-related proteins (Stefely et al. 2015; Reidenbach et al. 2018; He et al. 2014; Floyd et al. 2016). Similarly, determination of the COQ9 structure unexpectedly revealed that it adopts an ancient TetR (tetracycline resistance) fold typically used by bacteria for transcriptional regulation upon ligand binding. Further work revealed that COQ9 selectively binds prenyl lipids like CoQ, and possesses a strong physical and functional interaction with COQ7—the enzyme that catalyzes the penultimate step of CoQ biosynthesis (Lohman et al. 2014). Collectively, the COQ9 structural work accelerated its characterization as a novel prenyl lipid chaperone that helps surmount the biophysical challenges associated with synthesizing a highly hydrophobic molecule by binding CoQ intermediates and presenting them to other peripheral membrane-associated biosynthetic enzymes (Lohman et al. 2019). Finally, while the function of COQ4 remains unknown, a structure of a bacterial homolog, Alr8543, revealed a similar lipid-binding pocket and bound transition metals, motivating new mechanistic hypotheses into its function in the pathway (Rea et al. 2010). Overall, structural genomics is a prime example of the systems biochemistry approach, whereby the “screen” gives immediate information that helps to form specific new mechanistic hypotheses.
New Insights into Vitamin B12 Biology from Activity-Based Metabolic Profiling
Activity-based metabolomic profiling (ABMP) is an MS approach to assigning biochemical activity to proteins of unknown function. In a typical ABMP experiment, a recombinantly purified enzyme and its potential cofactors are first added to a cell/tissue metabolite extract. Then, liquid chromatography mass spectrometry is used to identify changes in the composition of the metabolite extract in the presence or absence of the recombinant protein. These changes are then used to infer specific biochemical activities of the protein in question (de Carvalho et al. 2010; Sevin et al. 2017).
The power of the ABMP approach can be seen in recent work by Shen et al. that deorphanized the mitochondrial protein CLYBL. CLYBL belongs to a small class of human genes that often harbor bi-allelic, loss-of-function mutations that appear to be well-tolerated. Homozygous mutations in CLYBL were linked with decreased circulating levels of vitamin B12 through GWAS, but the function of the protein remained unknown. ABMP was used to systematically probe substrates and products that could be acted on by wild-type CLYBL but not the catalytically deficient mutant, pointing to acetyl-CoA or citramalate as likely substrates. Subsequent biochemical follow-up established the role of CLYBL as a citromalyl-CoA lyase, loss of which leads to vitamin B12 inactivation through an accumulation of an upstream metabolite. Thus, through a bottom-up systems biochemistry approach, the investigators were able to define a function of the previously uncharacterized CLYBL protein and its effect on mitochondrial vitamin B12 levels (Shen et al. 2017).
Integrating Top-Down and Bottom-Up Approaches
On their own, the top-down and bottom-up approaches to systems biochemistry offer powerful insight into previously uncharacterized proteins and pathways. While the above vignettes have been focused on one approach or the other, these two approaches are of course not mutually exclusive. Rather, they are quite complementary and have jointly propelled progress in two areas of mitochondrial biology in particular: complex I assembly and the mitochondrial contact site and cristae organizing system.
Complex I Assembly
Isolated complex I (CI) deficiency is the most common inborn error of metabolism worldwide (Kirby et al. 1999). While the structure and biochemical mechanism of mature CI have been extensively studied (Agip et al. 2018; Zhu, Vinothkumar, and Hirst 2016; Vinothkumar, Zhu, and Hirst 2014; Friedrich 2014), a large percentage of diagnosed CI disorders are not associated with mutations in the structural CI subunits, but are instead due to disruption of genes that assist in the assembly and/or functionality of the mature complex — so-called CI assembly factors (CIAFs) (McKenzie et al. 2011; Calvo et al. 2010). Because CIAFs are not part of the final complex, they have been challenging to identify (Guerrero-Castillo et al. 2017). Systems biochemistry approaches have proved instrumental in spotting these elusive proteins.
A number of recent studies have employed a top-down systems biochemistry approach to identify novel CIAFs. First, in the original MitoCarta study, a phylogenetic approach was used to identify mitochondrial proteins whose presence/absence patterns across evolution matched those of the CI subunits conserved to bacteria. Six of the top 19 candidates from this analysis have now been validated as bona fide CIAFs that harbor mutations in human disease (Pagliarini et al. 2008; Sugiana et al. 2008; Formosa et al. 2018). Second, Stroud et al. identified two CIAFs by performing MS-based proteomics on cells each lacking one of either 25 accessory CI subunits or seven assembly factors (Stroud et al. 2016). Third, Guarani et al. employed an interaction proteomics approach to identify a novel CIAF through its reciprocal association with several core CI subunits as well as other known CIAFs (Guarani et al. 2014). Fourth, Heide et al. established a ‘complexome’ profiling approach that combined native gel electrophoresis and mass spectrometry to identify proteins that co-migrated with CI subunits during the CI assembly process, leading to the discovery of an additional assembly factor (Heide et al. 2012). This technique was then expanded upon by Guerrero-Castillo et al. to define the assembly intermediates of CI and infer the stepwise pathway by which they assemble into mature CI (Guerrero-Castillo et al. 2017). In each case, these large-scale efforts leveraged the established mitochondrial proteome and ‘omic’ technologies to motivate mechanistic biochemistry efforts to define new protein functions.
Similarly, bottom-up approaches have also been key in identifying novel CIAFs and for adding mechanistic depth to previously identified CIAFs. An example of the former is through the study of protein-protein interaction (PPI) as a means to infer functions for uncharacterized proteins. Floyd et al. used deep affinity-enrichment mass spectrometry analyses to map the PPIs of 50 MXPs. Among the very top hits was an interaction between an established assembly factor and the orphan protein, C17orf89, which itself was then validated as a CIAF and renamed NDUFAF8 (Floyd et al. 2016). An example of the latter is the work on NDUFAF7 by Rhein et al. The investigators used a mass spectrometric approach to compare the effects of knockdown of NDUFAF7 on the methylation status of its putative substrate, NDUFS2. Their work showed that NDUFAF7 was required for the symmetric demethylation of NDUFS2, defining its role in the post-translational maturation of NDUFS2 (Rhein et al. 2013). The combination of top-down and bottom-up systems biochemistry have thus added both breadth and depth to the understanding of CI assembly, with over 15 CIAFs identified to date (Formosa et al. 2018).
Mitochondrial Contact Site and Cristae Organizing System
The “mitochondrial contact site and cristae organizing system” (MICOS) is conserved from yeast to humans and plays crucial roles in the maintenance of cristae junctions, inner membrane architecture, and formation of contact sites between the inner and outer membranes. In 2011 and 2012, four independent studies converged upon the subunit definition of MICOS in S. cerevisiae (Pfanner et al. 2014). Here, we highlight two systems biochemistry approaches, one bottom-up and one top-down, used to identify the constituents of the MICOS complex in yeast.
In a seminal 2011 study, Hoppins et al. constructed a bottom-up quantitative genetic interaction map that focused specifically on genes encoding mitochondrial proteins, along with select genes whose functions likely to impact mitochondria (Hoppins et al. 2011). Systematic genetic interaction maps can be used to infer related gene function by measuring pairwise growth defects in double mutant cells (Schuldiner et al. 2005; Kornmann et al. 2009). The resulting MITO-MAP was particularly powerful in linking MXPs to roles in establishing physical and functional organization of mitochondrial membranes. Indeed, their work identified a cluster of genes that encode six proteins that form a large heterooligomeric protein complex on the inner mitochondrial membrane. At the same time, Harner et al. arrived at the same conclusion through an orthogonal top-down approach. Based on observations that the inner and outer membranes remain tethered at discrete sites during mitochondrial shrinking and swelling experiments, the investigators set out to identify the proteins responsible for forming these junctions. They first generated a marker protein that spanned both the outer and inner mitochondrial membranes at these contact sites. Then, using a mass spectrometry-based proteomic technique in conjunction with rigorous biochemical analyses, Harner et al. identified six previously uncharacterized proteins that shared the same distribution as the marker protein (Harner et al. 2011). In both cases, systematic analyses enabled focused biochemical experiments to define the composition of MICOS complex, and set a foundation for further systems biochemistry-based discovery of MICOS constituents, including the mammalian MICOS subunits APOOL/Mic27 (Weber et al. 2013) and QIL1/Mic13 (Guarani et al. 2015; Anand et al. 2016).
Systems Biochemistry and Mitochondrial Disease
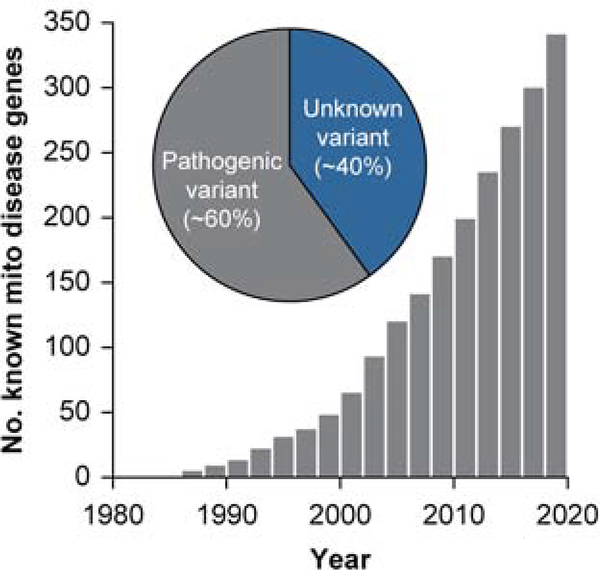
Since the first genetic cause of human mitochondrial disease was discovered in the late 1980s (Wallace et al. 1988; Holt, Harding, and Morgan-Hughes 1988), the number of unique human diseases with known causal mutations in mitochondrial proteins has increased exponentially (Frazier, Thorburn, and Compton 2019). Recent analysis of the OMIM database (Amberger et al. 2015) shows that 341 of the genes in MitoCarta 2.0 are linked to at least one OMIM disease phenotypes (Figure 5). With our expanded knowledge of mitochondrial functions at large and of their component parts, we might have expected to see substantially increased rates of successful molecular diagnosis, yet success rates have stalled at around 30–60% (Wortmann et al. 2015; Scharfe et al. 2000; Frazier, Thorburn, and Compton 2019; Stenton et al. 2019). Indeed, the current number of known disease genes still only account for ~60% of suspected cases of mitochondrial disease (Figure 5). Though new disease gene candidates continue to be uncovered through massively parallel sequencing and associated analyses, identification of genetic variants alone is insufficient for molecular diagnosis (Frazier, Thorburn, and Compton 2019). In the post-genomic era, the systems biochemistry approach offers a framework with which to bridge this gap between genotype and phenotype and thus improve the diagnosis and treatment of mitochondrial disorders.
Figure 5.
The identification of mitochondrial disease genes has increased markedly since the 1980s when the first mitochondrial disease was characterized. However, ~40% of diagnosed mitochondrial disorders have no identified genetic cause, and overall diagnostic success rates remain surprisingly low. Data from OMIM (Amberger et al. 2015) and (Frazier, Thorburn, and Compton 2019).
To better understand the pathogenesis of a disease, one must first pinpoint the cellular process being disrupted (Frazier, Thorburn, and Compton 2019). To that end, characterizing MXPs to understand protein function and then placing them within cellular pathways, akin to bottom-up systems biochemistry, can prioritize genes for deeper diagnostic analyses. For instance, characterization of the CIAF NDUFAF8 led to the subsequent identification of a deep intronic variant in a pediatric patient that was initially missed by whole exome sequencing (Alston et al. 2020).
Conversely, one may also take a top-down approach to the diagnosis of mitochondrial disease. Over the years, diagnostic yield for Mendelian diseases through genomic sequencing approaches alone has stagnated at ~40%. However, these limitations can be overcome by integrating transcriptomic and proteomic data from patient samples with existing genomic data, achieving a molecular diagnosis in >10% of unsolved cases (Stenton et al. 2019). Inclusion of other ‘omic’ measurements, such as metabolomics and lipidomics, will likely boost the diagnostic power of this approach. Ultimately, we anticipate that a robust portfolio of multi-omic profiles for various pathophysiologic states can be used to rapidly and accurately diagnose mitochondrial diseases based on a patient’s multi-omic ‘signature’ in the same way that these signatures empowered protein characterization in the examples above. Consistent with this, a recent review of work in model systems (Khan et al. 2020) identified ten multi-omic profiling studies that integrated genome, transcriptome, proteome, and metabolome data to identify mitochondrial drivers of metabolic phenotypes. While still a nascent field, this approach is being actively tailored to the clinic and is yielding promising early results (Rahman and Rahman 2018).
Beyond challenges in diagnosis, the vast majority of mitochondrial disorders lack effective therapies (Frazier, Thorburn, and Compton 2019; Pfeffer et al. 2012; Maldonado et al. 2019). Current treatment regimens are mostly palliative and generally fail to halt progression of the disease (Pfeffer et al. 2012). Nevertheless, progress towards understanding protein function at a molecular level has led to the development of small-molecule based interventions that are now in preclinical and clinical trials (Frazier, Thorburn, and Compton 2019). Further systematic efforts to define protein function and elucidate the biochemical underpinnings of mitochondrial dysfunction are thus needed to drive development of targeted therapeutics.
Conclusions and future directions
The human genome was fully sequenced in 2001 (Lander et al. 2001; Venter et al. 2001), yet progress towards characterizing its ~19,000 genes has been much slower than anticipated. This is in part due to a disproportionate amount of effort being focused on a small subset of genes at the expense of a broader understanding of gene/protein function (Edwards et al. 2011; Stoeger et al. 2018; Dolgin 2017). Fortunately, the advent of large-scale systems methodologies is accelerating the pace at which we can gather surface-level information on gene function, facilitating research on the unknown. However, such information-gathering alone is insufficient for properly deorphanizing genes of unknown functions. The systems biochemistry approach — a marrying of systems-level analyses with rigorous classical biochemistry — is a means to harness the power of large-scale, ‘omic’ methodologies to guide mechanistic, hypothesis-driven inquiry into the molecular function of uncharacterized proteins. While this approach holds great promise for addressing the disparity in research between under- and over-studied genes, its application genome-wide is often not yet feasible due to the daunting complexity of higher eukaryotic cells. In recent years, subcellular components, most notably mitochondria, have become more well-defined systems that match the capacity of the systems biochemistry technologies. The systems biochemistry approach has, in turn, spurred tremendous progress in the field of mitochondrial biology through deorphanizing MXPs and providing systematic functional annotation of mitochondrial pathways.
Moving forward, we see systems biochemistry maturing in a number of ways. First, new screening and computational methodologies will expand the scope of this approach. CRISPR-Cas9 technology has already revolutionized our ability to manipulate the genomes of higher-order model organisms (Doench 2018). Coupled with advances in metabolomic and massively parallel sequencing technologies, and the implementation of new computational and machine learning methods, this will enable larger and more precise screens to link additional genes to known processes and position them for in-depth biochemical follow-up. Second, further efforts to define cellular and subcellular systems (e.g., Human Cell Atlas (Regev et al. 2017), LOPIT (Geladaki et al. 2019)), inter-organellar interactions (Lackner 2019), and tissue-specific systems (e.g., GTEx Project (Consortium 2013), Human Protein Atlas (Uhlen et al. 2015)) will enable the application of systems biochemistry to increasingly complex systems. Ultimately, more studies such as these will pave the way for a genome-wide systems biochemistry effort to cast light on the ‘dark matter’ of the proteome. These efforts will facilitate ‘omics’ analyses of patient samples to identify genes variants causative for disease and enable intervention via precision medicine.
The recent heightened awareness that a surprisingly large percentage of our genomes have been experimentally neglected coupled with rapidly advancing technologies may now be ushering in the post-genomic era of comprehensive gene/protein characterization envisioned by the sequencing pioneers decades ago. Notably, as more proteins are functionalized, the more they power, via a bootstrapping effect, further understanding of other uncharacterized proteins in the systems biochemistry paradigm. Nonetheless, systematic approaches alone will never suffice to clarify complex biology, and these efforts must continue to be framed by elegant questions on the front end, and a dedication to rigorous, quantitative experimentation on the back. That, along with some well-timed serendipity, should continue to mold our understanding of mitochondria and beyond.
Acknowledgments
This work was supported by NIH grants R35GM131795 (to D.J.P.); T32AG00213 (to A.Y.S.); T32DK007665 (to B.J.F.); and T32GM008692 MSTP (to A.Y.S. and B.J.F.).
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aebersold R, and Mann M. 2003. ‘Mass spectrometry-based proteomics’, Nature, 422: 198–207. [DOI] [PubMed] [Google Scholar]
- Agip AA, Blaza JN, Bridges HR, Viscomi C, Rawson S, Muench SP, and Hirst J. 2018. ‘Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states’, Nat Struct Mol Biol, 25: 548–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alston CL, Veling MT, Heidler J, Taylor LS, Alaimo JT, Sung AY, He L, Hopton S, Broomfield A, Pavaine J, Diaz J, Leon E, Wolf P, McFarland R, Prokisch H, Wortmann SB, Bonnen PE, Wittig I, Pagliarini DJ, and Taylor RW. 2020. ‘Pathogenic Bi-allelic Mutations in NDUFAF8 Cause Leigh Syndrome with an Isolated Complex I Deficiency’, American journal of human genetics, 106: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, and Hamosh A. 2015. ‘OMIM.org: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders’, Nucleic Acids Res, 43: D789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R, Strecker V, Urbach J, Wittig I, and Reichert AS. 2016. ‘Mic13 Is Essential for Formation of Crista Junctions in Mammalian Cells’, PLoS One, 11: e0160258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, and Young IG. 1981. ‘Sequence and organization of the human mitochondrial genome’, Nature, 290: 457–65. [DOI] [PubMed] [Google Scholar]
- Andreux PA, Williams EG, Koutnikova H, Houtkooper RH, Champy MF, Henry H, Schoonjans K, Williams RW, and Auwerx J. 2012. ‘Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits’, Cell, 150: 1287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL, Koteliansky V, and Mootha VK. 2011. ‘Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter’, Nature, 476: 341–5. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker DK, Taylor EB, Schell JC, Orsak T, Boutron A, Chen YC, Cox JE, Cardon CM, Van Vranken JG, Dephoure N, Redin C, Boudina S, Gygi SP, Brivet M, Thummel CS, and Rutter J. 2012. ‘A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans’, Science, 337: 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, and Mootha VK. 2016. ‘MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins’, Nucleic Acids Res, 44: D1251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, and Mootha VK. 2010. ‘The mitochondrial proteome and human disease’, Annu Rev Genomics Hum Genet, 11: 25–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Tucker EJ, Compton AG, Kirby DM, Crawford G, Burtt NP, Rivas M, Guiducci C, Bruno DL, Goldberger OA, Redman MC, Wiltshire E, Wilson CJ, Altshuler D, Gabriel SB, Daly MJ, Thorburn DR, and Mootha VK. 2010. ‘Highthroughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency’, Nat Genet, 42: 851–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude A 1946. ‘Fractionation of Mammalian Liver Cells by Differential Centrifugation : I. Problems, Methods, and Preparation of Extract’, J Exp Med, 84: 51–9. [PubMed] [Google Scholar]
- Consortium G TEx 2013. ‘The Genotype-Tissue Expression (GTEx) project’, Nat Genet, 45: 580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho LP, Zhao H, Dickinson CE, Arango NM, Lima CD, Fischer SM, Ouerfelli O, Nathan C, and Rhee KY. 2010. ‘Activity-based metabolomic profiling of enzymatic function: identification of Rv1248c as a mycobacterial 2-hydroxy-3-oxoadipate synthase’, Chem Biol, 17: 323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani D, Raffaello A, Teardo E, Szabo I, and Rizzuto R. 2011. ‘A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter’, Nature, 476: 336–40. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca HF, and Engstrom GW. 1961. ‘Calcium uptake by rat kidney mitochondria’, Proc Natl Acad Sci U S A, 47: 1744–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, and Schon EA. 1998. ‘Nuclear power and mitochondrial disease’, Nat Genet, 19: 214–5. [DOI] [PubMed] [Google Scholar]
- Doench JG 2018. ‘Am I ready for CRISPR? A user’s guide to genetic screens’, Nat Rev Genet, 19: 67–80. [DOI] [PubMed] [Google Scholar]
- Dolgin E 2017. ‘The most popular genes in the human genome’, Nature, 551: 427–31. [DOI] [PubMed] [Google Scholar]
- Edwards AM, Isserlin R, Bader GD, Frye SV, Willson TM, and Yu FH. 2011. ‘Too many roads not taken’, Nature, 470: 163–5. [DOI] [PubMed] [Google Scholar]
- Ellens KW, Christian N, Singh C, Satagopam VP, May P, and Linster CL. 2017. ‘Confronting the catalytic dark matter encoded by sequenced genomes’, Nucleic Acids Res, 45: 11495–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernster L, and Schatz G. 1981. ‘Mitochondria: a historical review’, J Cell Biol, 91: 227s–55s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd BJ, Wilkerson EM, Veling MT, Minogue CE, Xia C, Beebe ET, Wrobel RL, Cho H, Kremer LS, Alston CL, Gromek KA, Dolan BK, Ulbrich A, Stefely JA, Bohl SL, Werner KM, Jochem A, Westphall MS, Rensvold JW, Taylor RW, Prokisch H, Kim JP, Coon JJ, and Pagliarini DJ. 2016. ‘Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function’, Mol Cell, 63: 621–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa LE, Dibley MG, Stroud DA, and Ryan MT. 2018. ‘Building a complex complex: Assembly of mitochondrial respiratory chain complex I’, Semin Cell Dev Biol, 76: 154–62. [DOI] [PubMed] [Google Scholar]
- Frazier AE, Thorburn DR, and Compton AG. 2019. ‘Mitochondrial energy generation disorders: genes, mechanisms, and clues to pathology’, J Biol Chem, 294: 5386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T 2014. ‘On the mechanism of respiratory complex I’, J Bioenerg Biomembr, 46: 255–68. [DOI] [PubMed] [Google Scholar]
- Galperin MY, and Koonin EV. 2010. ‘From complete genome sequence to ‘complete’ understanding?’, Trends Biotechnol, 28: 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geladaki A, Kocevar Britovsek N, Breckels LM, Smith TS, Vennard OL, Mulvey CM, Crook OM, Gatto L, and Lilley KS. 2019. ‘Combining LOPIT with differential ultracentrifugation for high-resolution spatial proteomics’, Nat Commun, 10: 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghatak S, King ZA, Sastry A, and Palsson BO. 2019. ‘The y-ome defines the 35% of Escherichia coli genes that lack experimental evidence of function’, Nucleic Acids Res, 47: 2446–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczarowska-Jorge H, Zahedi RP, and Sickmann A. 2017. ‘The proteome of baker’s yeast mitochondria’, Mitochondrion, 33: 15–21. [DOI] [PubMed] [Google Scholar]
- Guarani V, McNeill EM, Paulo JA, Huttlin EL, Frohlich F, Gygi SP, Van Vactor D, and Harper JW. 2015. ‘QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology’, Elife, 4: e06265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarani V, Paulo J, Zhai B, Huttlin EL, Gygi SP, and Harper JW. 2014. ‘TIMMDC1/C3orf1 functions as a membrane-embedded mitochondrial complex I assembly factor through association with the MCIA complex’, Mol Cell Biol, 34: 847–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Castillo S, Baertling F, Kownatzki D, Wessels HJ, Arnold S, Brandt U, and Nijtmans L. 2017. ‘The Assembly Pathway of Mitochondrial Respiratory Chain Complex I’, Cell Metab, 25: 128–39. [DOI] [PubMed] [Google Scholar]
- Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, and Neupert W. 2011. ‘The mitochondrial contact site complex, a determinant of mitochondrial architecture’, EMBO J, 30: 4356–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CH, Xie LX, Allan CM, Tran UC, and Clarke CF. 2014. ‘Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multisubunit Coq polypeptide complexes in yeast coq null mutants’, Biochim Biophys Acta, 1841: 630–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heide H, Bleier L, Steger M, Ackermann J, Drose S, Schwamb B, Zornig M, Reichert AS, Koch I, Wittig I, and Brandt U. 2012. ‘Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex’, Cell Metab, 16: 538–49. [DOI] [PubMed] [Google Scholar]
- Herzig S, Raemy E, Montessuit S, Veuthey JL, Zamboni N, Westermann B, Kunji ER, and Martinou JC. 2012. ‘Identification and functional expression of the mitochondrial pyruvate carrier’, Science, 337: 93–6. [DOI] [PubMed] [Google Scholar]
- Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D, Altman RB, Davis RW, Nislow C, and Giaever G. 2008. ‘The chemical genomic portrait of yeast: uncovering a phenotype for all genes’, Science, 320: 362–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogeboom GH, Claude A, and Hotch-Kiss RD. 1946. ‘The distribution of cytochrome oxidase and succinoxidase in the cytoplasm of the mammalian liver cell’, J Biol Chem, 165: 615–29. [PubMed] [Google Scholar]
- Holt IJ, Harding AE, and Morgan-Hughes JA. 1988. ‘Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies’, Nature, 331: 717–9. [DOI] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, and Nunnari J. 2011. ‘A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria’, J Cell Biol, 195: 323–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Go YM, and Jones DP. 2020. ‘Omics Integration for Mitochondria Systems Biology’, Antioxidants & redox signaling: 10.1089/ars.2019.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V, Lam SS, Udeshi ND, Svinkina T, Guzman G, Mootha VK, Carr SA, and Ting AY. 2017. ‘Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation’, Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V, Zou P, Rhee HW, Udeshi ND, Cracan V, Svinkina T, Carr SA, Mootha VK, and Ting AY. 2014. ‘Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging’, Mol Cell, 55: 332–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagendorf AT, and Uribe E. 1966. ‘ATP formation caused by acid-base transition of spinach chloroplasts’, Proc Natl Acad Sci U S A, 55: 170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EP, and Lehninger AL. 1949. ‘Oxidation of fatty acids and tricarboxylic acid cycle intermediates by isolated rat liver mitochondria’, J Biol Chem, 179: 957–72. [PubMed] [Google Scholar]
- Khan S, Ince-Dunn G, Suomalainen A, and Elo LL. 2020. ‘Integrative omics approaches provide biological and clinical insights: examples from mitochondrial diseases’, J Clin Invest, 130: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby DM, Crawford M, Cleary MA, Dahl HH, Dennett X, and Thorburn DR. 1999. ‘Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder’, Neurology, 52: 1255–64. [DOI] [PubMed] [Google Scholar]
- Koopman WJ, Willems PH, and Smeitink JA. 2012. ‘Monogenic mitochondrial disorders’, N Engl J Med, 366: 1132–41. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, and Walter P. 2009. ‘An ER-mitochondria tethering complex revealed by a synthetic biology screen’, Science, 325: 477–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N, Wyant GA, Prakash G, Uit de Bos J, Bottanelli F, Pacold ME, Chan SH, Lewis CA, Wang T, Keys HR, Guo YE, and Sabatini DM. 2018. ‘SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism’, Science, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL 2019. ‘The Expanding and Unexpected Functions of Mitochondria Contact Sites’, Trends Cell Biol, 29: 580–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J, and Consortium International Human Genome Sequencing. 2001. ‘Initial sequencing and analysis of the human genome’, Nature, 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lapointe CP, Stefely JA, Jochem A, Hutchins PD, Wilson GM, Kwiecien NW, Coon JJ, Wickens M, and Pagliarini DJ. 2018. ‘Multi-omics Reveal Specific Targets of the RNA-Binding Protein Puf3p and Its Orchestration of Mitochondrial Biogenesis’, Cell Syst, 6: 125–35 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman DC, Aydin D, Von Bank HC, Smith RW, Linke V, Weisenhorn E, McDevitt MT, Hutchins P, Wilkerson EM, Wancewicz B, Russell J, Stefely MS, Beebe ET, Jochem A, Coon JJ, Bingman CA, Dal Peraro M, and Pagliarini DJ. 2019. ‘An Isoprene Lipid-Binding Protein Promotes Eukaryotic Coenzyme Q Biosynthesis’, Mol Cell, 73: 763–74 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman DC, Forouhar F, Beebe ET, Stefely MS, Minogue CE, Ulbrich A, Stefely JA, Sukumar S, Luna-Sanchez M, Jochem A, Lew S, Seetharaman J, Xiao R, Wang H, Westphall MS, Wrobel RL, Everett JK, Mitchell JC, Lopez LC, Coon JJ, Tong L, and Pagliarini DJ. 2014. ‘Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis’, Proc Natl Acad Sci U S A, 111: E4697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez MF, Kristal BS, Chernokalskaya E, Lazarev A, Shestopalov AI, Bogdanova A, and Robinson M. 2000. ‘High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation’, Electrophoresis, 21: 3427–40. [DOI] [PubMed] [Google Scholar]
- Maldonado EM, Taha F, Rahman J, and Rahman S. 2019. ‘Systems Biology Approaches Toward Understanding Primary Mitochondrial Diseases’, Front Genet, 10: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C, Raffaello A, Vecellio Reane D, Gherardi G, De Mario A, and Rizzuto R. 2018. ‘Mitochondrial calcium uptake in organ physiology: from molecular mechanism to animal models’, Pflugers Archiv : European journal of physiology, 470: 1165–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilainen O, Quiros PM, and Auwerx J. 2017. ‘Mitochondria and Epigenetics - Crosstalk in Homeostasis and Stress’, Trends Cell Biol, 27: 453–63. [DOI] [PubMed] [Google Scholar]
- McKenzie M, Tucker EJ, Compton AG, Lazarou M, George C, Thorburn DR, and Ryan MT. 2011. ‘Mutations in the gene encoding C8orf38 block complex I assembly by inhibiting production of the mitochondria-encoded subunit ND1’, J Mol Biol, 414: 413–26. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Blaza JN, Larsson NG, and Hirst J. 2017. ‘The Enigma of the Respiratory Chain Supercomplex’, Cell Metab, 25: 765–76. [DOI] [PubMed] [Google Scholar]
- Mitchell P 1961. ‘Coupling of phosphorylation to electron and hydrogen transfer by a chemiosmotic type of mechanism’, Nature, 191: 144–8. [DOI] [PubMed] [Google Scholar]
- Montelione GT 2012. ‘The Protein Structure Initiative: achievements and visions for the future’, F1000 biology reports, 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, and Suomalainen A. 2012. ‘Mitochondria: in sickness and in health’, Cell, 148: 1145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver SG 1996. ‘From DNA sequence to biological function’, Nature, 379: 597–600. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, and Mootha VK. 2008. ‘A mitochondrial protein compendium elucidates complex I disease biology’, Cell, 134: 112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, and Rutter J. 2013. ‘Hallmarks of a new era in mitochondrial biochemistry’, Genes Dev, 27: 2615–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payet LA, Leroux M, Willison JC, Kihara A, Pelosi L, and Pierrel F. 2016. ‘Mechanistic Details of Early Steps in Coenzyme Q Biosynthesis Pathway in Yeast’, Cell Chem Biol, 23: 1241–50. [DOI] [PubMed] [Google Scholar]
- Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, and Mootha VK. 2010. ‘MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake’, Nature, 467: 291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N, van der Laan M, Amati P, Capaldi RA, Caudy AA, Chacinska A, Darshi M, Deckers M, Hoppins S, Icho T, Jakobs S, Ji J, Kozjak-Pavlovic V, Meisinger C, Odgren PR, Park SK, Rehling P, Reichert AS, Sheikh MS, Taylor SS, Tsuchida N, van der Bliek AM, van der Klei IJ, Weissman JS, Westermann B, Zha J, Neupert W, and Nunnari J. 2014. ‘Uniform nomenclature for the mitochondrial contact site and cristae organizing system’, J Cell Biol, 204: 1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer G, Majamaa K, Turnbull DM, Thorburn D, and Chinnery PF. 2012. ‘Treatment for mitochondrial disorders’, Cochrane Database Syst Rev: CD004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon WW, Davis DE, Ha HT, Jonassen T, Rather PN, and Clarke CF. 2000. ‘Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis’, J Bacteriol, 182: 5139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman J, and Rahman S. 2018. ‘Mitochondrial medicine in the omics era’, Lancet (London, England), 391: 2560–74. [DOI] [PubMed] [Google Scholar]
- Rea SL, Graham BH, Nakamaru-Ogiso E, Kar A, and Falk MJ. 2010. ‘Bacteria, yeast, worms, and flies: exploiting simple model organisms to investigate human mitochondrial diseases’, Dev Disabil Res Rev, 16: 200–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, Clevers H, Deplancke B, Dunham I, Eberwine J, Eils R, Enard W, Farmer A, Fugger L, Gottgens B, Hacohen N, Haniffa M, Hemberg M, Kim S, Klenerman P, Kriegstein A, Lein E, Linnarsson S, Lundberg E, Lundeberg J, Majumder P, Marioni JC, Merad M, Mhlanga M, Nawijn M, Netea M, Nolan G, Pe’er D, Phillipakis A, Ponting CP, Quake S, Reik W, Rozenblatt-Rosen O, Sanes J, Satija R, Schumacher TN, Shalek A, Shapiro E, Sharma P, Shin JW, Stegle O, Stratton M, Stubbington MJT, Theis FJ, Uhlen M, van Oudenaarden A, Wagner A, Watt F, Weissman J, Wold B, Xavier R, Yosef N, and Participants Human Cell Atlas Meeting. 2017. ‘The Human Cell Atlas’, Elife, 6: e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidenbach AG, Kemmerer ZA, Aydin D, Jochem A, McDevitt MT, Hutchins PD, Stark JL, Stefely JA, Reddy T, Hebert AS, Wilkerson EM, Johnson IE, Bingman CA, Markley JL, Coon JJ, Dal Peraro M, and Pagliarini DJ. 2018. ‘Conserved Lipid and Small-Molecule Modulation of COQ8 Reveals Regulation of the Ancient Kinase-like UbiB Family’, Cell Chem Biol, 25: 154–65 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, and Ting AY. 2013. ‘Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging’, Science, 339: 1328–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhein VF, Carroll J, Ding S, Fearnley IM, and Walker JE. 2013. ‘NDUFAF7 methylates arginine 85 in the NDUFS2 subunit of human complex I’, J Biol Chem, 288: 33016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, Kamburov A, Ghiassian SD, Yang X, Ghamsari L, Balcha D, Begg BE, Braun P, Brehme M, Broly MP, Carvunis AR, Convery-Zupan D, Corominas R, Coulombe-Huntington J, Dann E, Dreze M, Dricot A, Fan C, Franzosa E, Gebreab F, Gutierrez BJ, Hardy MF, Jin M, Kang S, Kiros R, Lin GN, Luck K, MacWilliams A, Menche J, Murray RR, Palagi A, Poulin MM, Rambout X, Rasla J, Reichert P, Romero V, Ruyssinck E, Sahalie JM, Scholz A, Shah AA, Sharma A, Shen Y, Spirohn K, Tam S, Tejeda AO, Trigg SA, Twizere JC, Vega K, Walsh J, Cusick ME, Xia Y, Barabasi AL, Iakoucheva LM, Aloy P, De Las Rivas J, Tavernier J, Calderwood MA, Hill DE, Hao T, Roth FP, and Vidal M. 2014. ‘A proteome-scale map of the human interactome network’, Cell, 159: 1212–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Markhard AL, Kitami T, Kovacs-Bogdan E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA, Calvo SE, Goldberger O, and Mootha VK. 2013. ‘EMRE is an essential component of the mitochondrial calcium uniporter complex’, Science, 342: 1379–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfe C, Zaccaria P, Hoertnagel K, Jaksch M, Klopstock T, Dembowski M, Lill R, Prokisch H, Gerbitz KD, Neupert W, Mewes HW, and Meitinger T. 2000. ‘MITOP, the mitochondrial proteome database: 2000 update’, Nucleic Acids Res, 28: 155–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, and Krogan NJ. 2005. ‘Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile’, Cell, 123: 507–19. [DOI] [PubMed] [Google Scholar]
- Sevin DC, Fuhrer T, Zamboni N, and Sauer U. 2017. ‘Nontargeted in vitro metabolomics for high-throughput identification of novel enzymes in Escherichia coli’, Nat Methods, 14: 187–94. [DOI] [PubMed] [Google Scholar]
- Shen H, Campanello GC, Flicker D, Grabarek Z, Hu J, Luo C, Banerjee R, and Mootha VK. 2017. ‘The Human Knockout Gene CLYBL Connects Itaconate to Vitamin B12’, Cell, 171: 771–82 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnick J, Fetrow JS, and Kolinski A. 2000. ‘Structural genomics and its importance for gene function analysis’, Nat Biotechnol, 18: 283–7. [DOI] [PubMed] [Google Scholar]
- Sorokina M, Stam M, Medigue C, Lespinet O, and Vallenet D. 2014. ‘Profiling the orphan enzymes’, Biol Direct, 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely JA, Kwiecien NW, Freiberger EC, Richards AL, Jochem A, Rush MJP, Ulbrich A, Robinson KP, Hutchins PD, Veling MT, Guo X, Kemmerer ZA, Connors KJ, Trujillo EA, Sokol J, Marx H, Westphall MS, Hebert AS, Pagliarini DJ, and Coon JJ. 2016. ‘Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling’, Nat Biotechnol, 34: 1191–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely JA, and Pagliarini DJ. 2017. ‘Biochemistry of Mitochondrial Coenzyme Q Biosynthesis’, Trends Biochem Sci, 42: 824–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely JA, Reidenbach AG, Ulbrich A, Oruganty K, Floyd BJ, Jochem A, Saunders JM, Johnson IE, Minogue CE, Wrobel RL, Barber GE, Lee D, Li S, Kannan N, Coon JJ, Bingman CA, and Pagliarini DJ. 2015. ‘Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis’, Mol Cell, 57: 83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenton SL, Kremer LS, Kopajtich R, Ludwig C, and Prokisch H. 2019. ‘The diagnosis of inborn errors of metabolism by an integrative “multi-omics” approach: A perspective encompassing genomics, transcriptomics, and proteomics’, J Inherit Metab Dis. [DOI] [PubMed] [Google Scholar]
- Stoeger T, Gerlach M, Morimoto RI, and Nunes Amaral LA. 2018. ‘Large-scale investigation of the reasons why potentially important genes are ignored’, PLoS Biol, 16: e2006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud DA, Surgenor EE, Formosa LE, Reljic B, Frazier AE, Dibley MG, Osellame LD, Stait T, Beilharz TH, Thorburn DR, Salim A, and Ryan MT. 2016. ‘Accessory subunits are integral for assembly and function of human mitochondrial complex I’, Nature, 538: 123–26. [DOI] [PubMed] [Google Scholar]
- Sugiana C, Pagliarini DJ, McKenzie M, Kirby DM, Salemi R, Abu-Amero KK, Dahl HH, Hutchison WM, Vascotto KA, Smith SM, Newbold RF, Christodoulou J, Calvo S, Mootha VK, Ryan MT, and Thorburn DR. 2008. ‘Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease’, Am J Hum Genet, 83: 468–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SW, Fahy E, Zhang B, Glenn GM, Warnock DE, Wiley S, Murphy AN, Gaucher SP, Capaldi RA, Gibson BW, and Ghosh SS. 2003. ‘Characterization of the human heart mitochondrial proteome’, Nat Biotechnol, 21: 281–6. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, and Ponten F. 2015. ‘Proteomics. Tissue-based map of the human proteome’, Science, 347: 1260419. [DOI] [PubMed] [Google Scholar]
- Vafai SB, and Mootha VK. 2012. ‘Mitochondrial disorders as windows into an ancient organelle’, Nature, 491: 374–83. [DOI] [PubMed] [Google Scholar]
- Veling MT, Reidenbach AG, Freiberger EC, Kwiecien NW, Hutchins PD, Drahnak MJ, Jochem A, Ulbrich A, Rush MJP, Russell JD, Coon JJ, and Pagliarini DJ. 2017. ‘Multi-omic Mitoprotease Profiling Defines a Role for Oct1p in Coenzyme Q Production’, Mol Cell, 68: 970–77 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, and Zhu X. 2001. ‘The sequence of the human genome’, Science, 291: 1304–51. [DOI] [PubMed] [Google Scholar]
- Vinothkumar KR, Zhu J, and Hirst J. 2014. ‘Architecture of mammalian respiratory complex I’, Nature, 515: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogtle FN, Burkhart JM, Gonczarowska-Jorge H, Kucukkose C, Taskin AA, Kopczynski D, Ahrends R, Mossmann D, Sickmann A, Zahedi RP, and Meisinger C. 2017. ‘Landscape of submitochondrial protein distribution’, Nat Commun, 8: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC 1999. ‘Mitochondrial diseases in man and mouse’, Science, 283: 1482–8. [DOI] [PubMed] [Google Scholar]
- Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, Lezza AM, Elsas LJ 2nd, and Nikoskelainen EK. 1988. ‘Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy’, Science, 242: 1427–30. [DOI] [PubMed] [Google Scholar]
- Warburg O 1913. ‘Über sauerstoffatmende Körnchen aus Leberzellen und über Sauerstoffatmung in Berkefeld-Filtraten wässriger Leberextrakte’, Arch. ges. Physiol, 154: 599. [Google Scholar]
- Weber TA, Koob S, Heide H, Wittig I, Head B, van der Bliek A, Brandt U, Mittelbronn M, and Reichert AS. 2013. ‘APOOL is a cardiolipin-binding constituent of the Mitofilin/MINOS protein complex determining cristae morphology in mammalian mitochondria’, PLoS One, 8: e63683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EG, Wu Y, Jha P, Dubuis S, Blattmann P, Argmann CA, Houten SM, Amariuta T, Wolski W, Zamboni N, Aebersold R, and Auwerx J. 2016. ‘Systems proteomics of liver mitochondria function’, Science, 352: aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortmann SB, Koolen DA, Smeitink JA, van den Heuvel L, and Rodenburg RJ. 2015. ‘Whole exome sequencing of suspected mitochondrial patients in clinical practice’, J Inherit Metab Dis, 38: 437–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Vinothkumar KR, and Hirst J. 2016. ‘Structure of mammalian respiratory complex I’, Nature, 536: 354–58. [DOI] [PMC free article] [PubMed] [Google Scholar]