Abstract
HIV-1 replication in CD4-positive T lymphocytes requires counteraction of multiple different innate antiviral mechanisms. Macrophage cells are also thought to provide a reservoir for HIV-1 replication but less is known in this cell type about virus restriction and counteraction mechanisms. Many studies have combined to demonstrate roles for APOBEC3D, APOBEC3F, APOBEC3G and APOBEC3H in HIV-1 restriction and mutation in CD4-positive T lymphocytes, whereas the APOBEC enzymes involved in HIV-1 restriction in macrophages have yet to be delineated fully. We show that multiple APOBEC3 genes including APOBEC3G are expressed in myeloid cell lines such as THP-1. Vif-deficient HIV-1 produced from THP-1 is less infectious than Vif-proficient virus, and proviral DNA resulting from such Vif-deficient infections shows strong G to A mutation biases in the dinucleotide motif preferred by APOBEC3G. Moreover, Vif mutant viruses with selective sensitivity to APOBEC3G show Vif null-like infectivity levels and similarly strong APOBEC3G-biased mutation spectra. Importantly, APOBEC3G-null THP-1 cells yield Vif-deficient particles with significantly improved infectivities and proviral DNA with background levels of G to A hypermutation. These studies combine to indicate that APOBEC3G is the main HIV-1 restricting APOBEC3 family member in THP-1 cells.
Keywords: APOBEC3G, G to A hypermutation, HIV-1 restriction, myeloid cell line THP-1, Vif
Introduction
Human cells have the potential to encode up to seven different single-stranded DNA cytosine deaminase enzymes of the APOBEC3 (A3) subfamily, APOBEC3A-D and APOBEC3F-H (A3A-D, A3F-H). These enzymes have overlapping functions in providing innate immune protection against a broad number of parasitic DNA-based elements (reviewed by [1–5]). Due to reverse transcription having obligate single-stranded cDNA replication intermediates, retroviruses are particularly sensitive to attack by A3 enzymes as cDNA strand C to U deamination events result in genomic strand G to A mutations. The best-studied example to date is the mechanism of HIV-1 restriction in which A3D, A3F, A3G and A3H have the capacity to mutate viral cDNA replication intermediates, as well as interfere with reverse transcription by deaminase-independent mechanisms (reviewed by [1–5]). Due to the intrinsic preferences of A3 enzymes for cytosine bases in specific dinucleotide motifs, 5′-CC (A3G) or 5′-TC (A3D, A3F and A3H), viral cDNA deamination events typically manifest as genomic strand 5′-GG to AG mutations or 5′-GA to AA mutations.
HIV-1, HIV-2 and non-human lentiviruses counteract this restriction mechanism through the virus-encoded virion infectivity factor (Vif), which nucleates the formation of an E3 ubiquitin ligase complex that binds and degrades restrictive A3 enzymes (reviewed by [1–5]). At the heart of this complex is Vif heterodimerization with the transcription cofactor CBF-β [6, 7], which also interferes with expression of restrictive A3 genes by preventing formation of CBF-β/RUNX transcriptional activation complexes [8]. Although this Vif-mediated counterdefense mechanism is certainly effective, a large body of evidence also indicates that A3 enzymes frequently escape degradation in CD4-positive T cells, package into assembling viral particles, deaminate viral cDNA replication intermediates, and ultimately contribute to HIV-1 genetic diversification including mutations responsible for immune evasion and drug resistance (e.g. [9–13]).
HIV-1 also infects myeloid lineage cell types including macrophages, which may constitute an additional reservoir for virus replication and latency in vivo (reviewed by [14–17]). However, considerably less is known about A3 function in these cell types in comparison to the plethora of studies already done using T cells. Here we ask whether the A3 restriction mechanism works similarly or differently against Vif-deficient HIV-1 in the myeloid cell line THP-1. This cell line was selected for studies here because it has already proven to be a robust model system for prior HIV-1 studies including several on restriction factors (e.g. [18–21]). Interestingly, although multiple restrictive A3s are expressed in THP-1, infectivity data and G to A hypermutation patterns of a variety of different HIV-1 constructs in both endogenous A3G-expressing and A3G-null THP-1 lines combine to indicate that A3G is the predominant A3 family member capable of virus restriction in this system.
Results
Multiple APOBEC3 family members are expressed in THP-1 and other myeloid cell lines
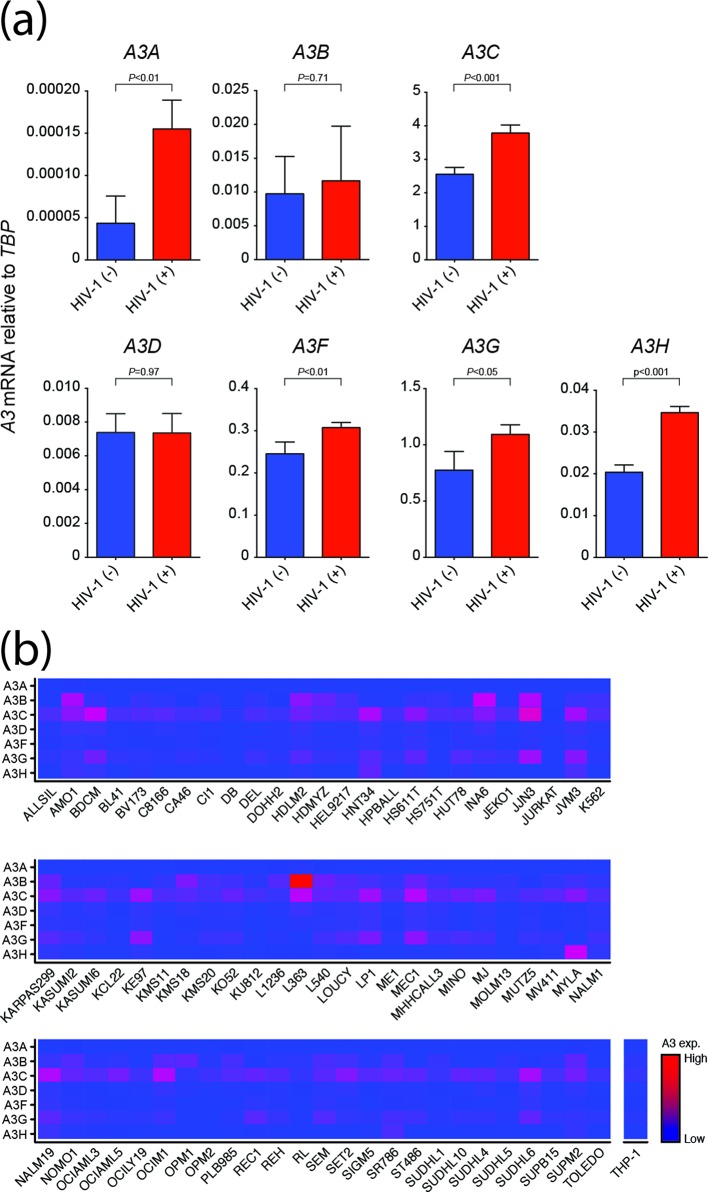
Previous studies have reported mRNA expression of multiple A3 family members including A3F, A3G and A3H in primary myeloid lineage cell types including macrophages and dendritic cells [18, 22–25]. To determine whether a similarly complex A3 repertoire is expressed in a more experimentally tractable model, we first used established reverse transcription-quantitative PCR (RT-qPCR) assays [23, 24, 26] to quantify the mRNA levels of each of the seven human A3 genes in the monocyte cell line THP-1. Relative to the housekeeping gene TATA-binding protein (TBP), significant levels of five different A3 family member mRNAs were evident – A3B, A3C, A3F, A3G and A3H (Fig. 1a). Moreover, infection by HIV-1IIIB caused a modest but statistically significant increase in mRNA levels for A3A, A3C, A3F, A3G and A3H, but to lesser extents than reported previously for HIV-1 infection of primary CD4-positive T lymphocytes [27, 28].
Fig. 1.
Multiple A3 genes are expressed in myeloid lineage cell lines. (a) A3 mRNA levels relative to the housekeeping gene TBP in THP-1 cells+/-HIV-1IIIB infection (m.o.i.=0.25). Each histogram bar shows the mean+/-sd of three independent experiments (P-values obtained by Student’s t-test). (b) A3 mRNA levels relative to the housekeeping gene TBP in 72 different myeloid cell lines (RNAseq data from CCLE). Red indicates high expression levels and blue lower levels.
To ask whether this mRNA expression profile is similar to those in other myeloid cell lines, we analysed A3 expression levels in RNAseq data sets representing 72 different myeloid cell lines available through the Cancer Cell Line Encyclopedia (CCLE) [29]. These analyses revealed a similar overall expression pattern for most of the cell lines with high levels of A3B, A3C and A3G and varying amounts of other A3 mRNAs (Fig. 1b). These gene expression studies combined to indicate that THP-1 may be a good model system for studies on A3 restriction in a myeloid lineage cell line.
Vif-deficient HIV-1 is restricted in THP-1 cells
Next, we wanted to determine if the A3 enzymes expressed in THP-1 could functionally restrict virus infectivity. VSV-G pseudotyped Vif-proficient and Vif-deficient HIV-1IIIB stocks were produced using 293T cells, and m.o.i. were determined by titring on CEM-GXR reporter cells [30]. Equivalent amounts of each virus were used to infect THP-1 cells (m.o.i.=0.25). As controls, parallel infections were done using the T cell line SupT11 expressing an empty vector or A3G. SupT11 does not express any A3 mRNA to significant levels [27] and, therefore, the empty vector line is expected to be fully permissive for replication of both viruses and the A3G expressing line will be non-permissive for Vif-deficient virus replication and permissive for Vif-proficient virus replication. In both cases, newly produced viruses were harvested after 48 h of incubation, and then infectivity was quantified using TZM-bl reporter cells. Short incubation durations of 48 h were chosen to enable estimates of the magnitude of virus restriction in a single round (or near single round) of virus replication.
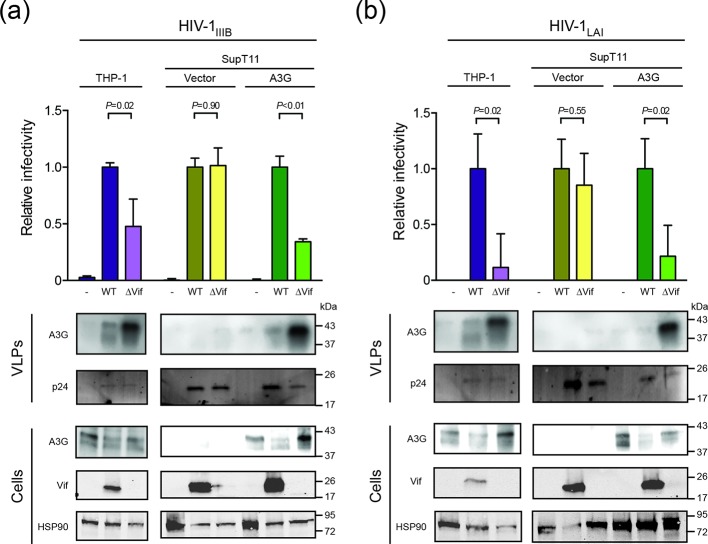
As expected for cell lines expressing multiple restrictive A3 enzymes, the infectivity of Vif-proficient virus was higher than that of the Vif-deficient virus following production in THP-1 cells (Fig. 2a). A similar fold-difference was observed for the same viruses produced in the SupT11-A3G cell line and, also as expected, no infectivity difference was seen for viruses produced in the SupT11-vector cell line (Fig. 2a). These results were corroborated by anti-A3G immunoblots of protein extracts from these infected cell lines and from the resulting viral particles. For both the THP-1 and SupT11-A3G cell lines, A3G was clearly expressed in cells and incorporated into viral particles. Importantly, Vif caused A3G to be degraded, which resulted in lower steady-state levels of A3G in cells, lower amounts of A3G incorporated into particles, and higher overall infectivity levels in comparison to Vif-deficient reaction conditions. Similar results were obtained for a different HIV-1 strain (LAI), which produces a Vif protein that more effectively degrades human A3H [31] (Fig. 2b). These results therefore combined to implicate at least one A3 family member in HIV-1 restriction in THP-1 cells.
Fig. 2.
Infectivity of Vif-proficient and Vif-deficient HIV-1 in THP-1 cells. (a) Infectivity of Vif-proficient and Vif-deficient HIV-1IIIB produced from THP-1 cells in comparison to the same viruses produced from SupT11 cells expressing either a control vector or A3G. Each histogram bar is the average of triplicate experiments +/-sd in which raw data are normalized to Vif-proficient virus infectivity (P-values obtained by Student’s t-test). Immunoblots of the indicated cellular and viral proteins in whole cell extracts and virus-like particles (VLPs) with HSP90 and p24 as loading controls, respectively. (b) Experiments similar to those in (a) except a different HIV-1 strain was used (LAI).
Base substitution mutation spectra for Vif-deficient and Vif-proficient HIV-1 produced from THP-1 cells
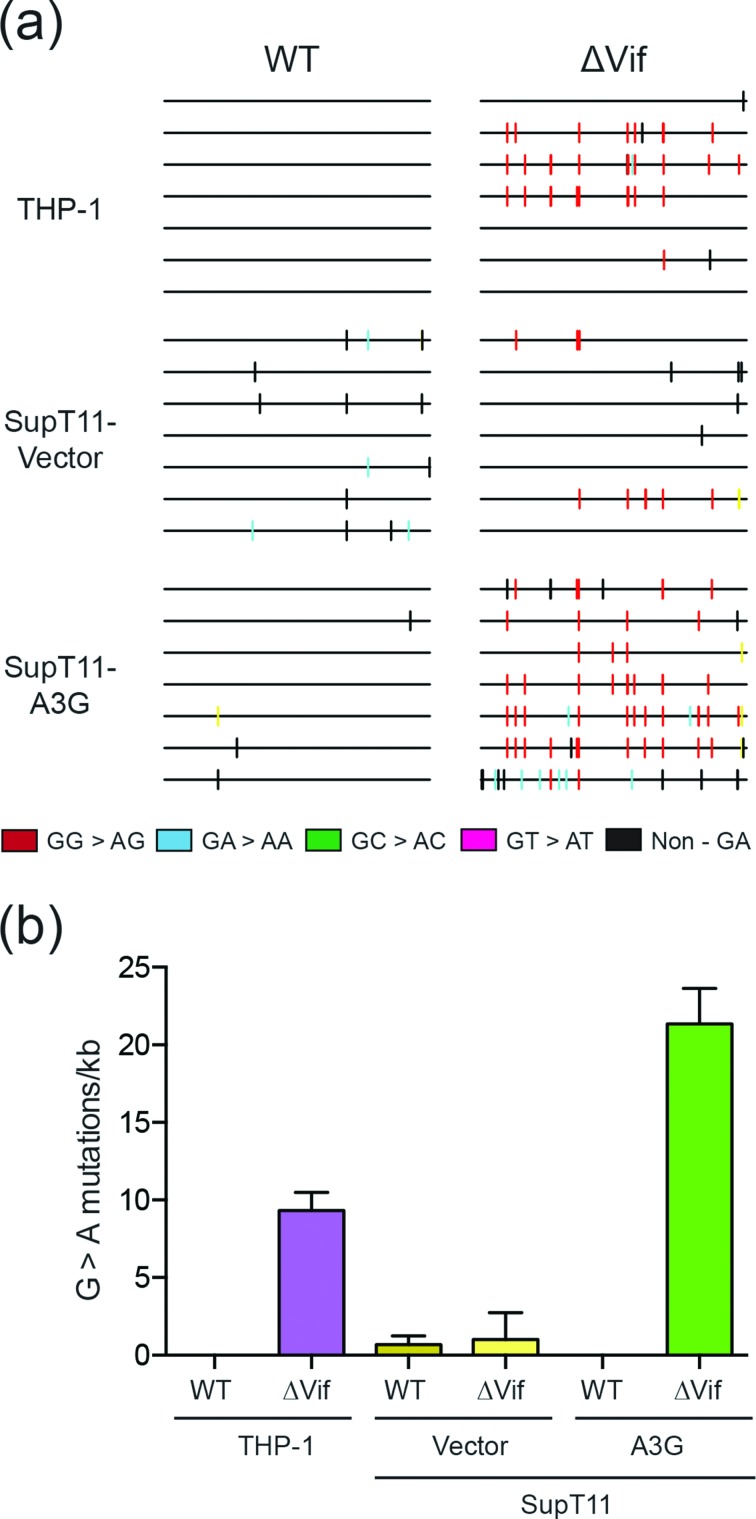
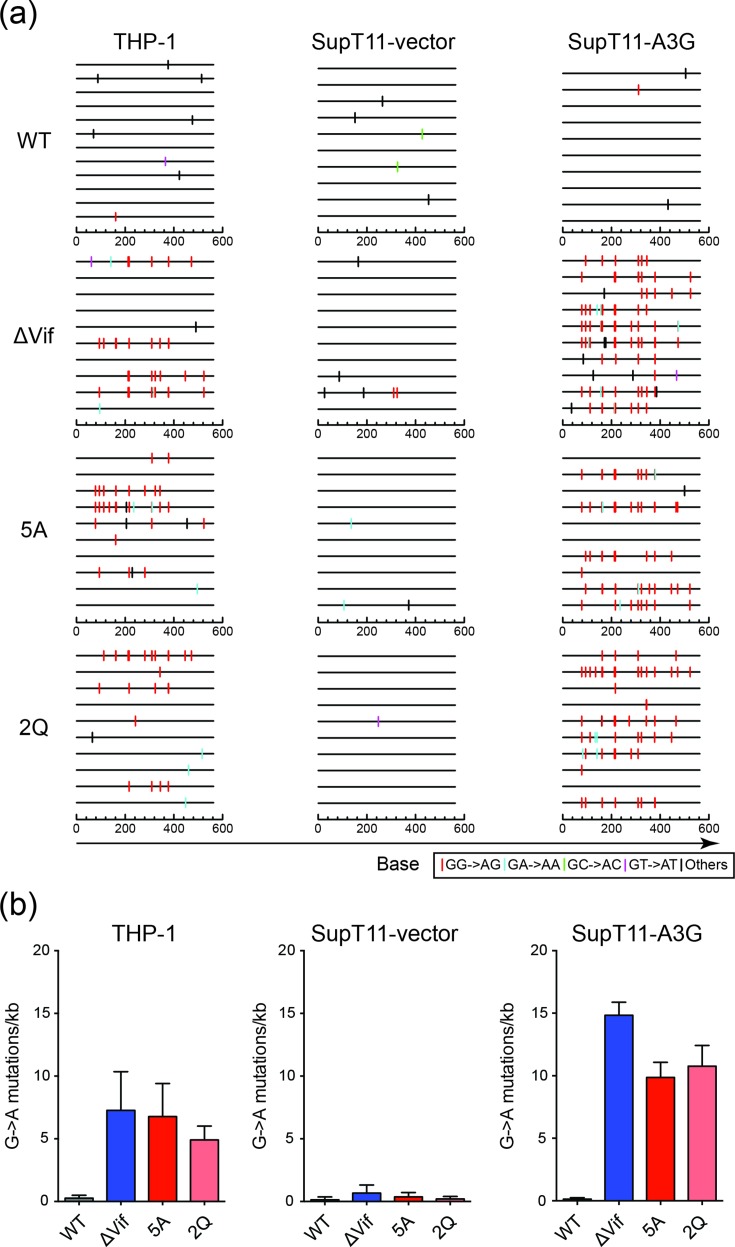
To examine the base substitution mutation spectra, and particularly the dinucleotide context of G to A mutations, the HIV-1IIIB proviral DNA from the infectivity experiments described above in Fig. 2 was cloned and sequenced. Specifically, total genomic DNA was prepared from infected TZM-bl reporter cells, and the pol region of the virus was amplified by high-fidelity PCR, cloned and Sanger sequenced. As expected, the mutation levels were low in all Vif-proficient experimental conditions, likely due to degradation of restrictive A3 enzymes (WT in Fig. 3a–b). Vif-deficient viruses produced in SupT11-A3G cells had high frequencies of G to A mutations in A3G-preferred motifs (88 % GG to AG on the genomic strand due to CC to CU deamination events on the cDNA strand; Fig. 3a–b). Similarly high frequencies of G to A mutations in A3G-preferred motifs were evident in proviral DNA derived from THP-1 infections (85 %; Fig. 3a–b). In both the SupT11-A3G and THP-1 systems, most of the remaining G to A mutations occurred in GA to AA dinucleotide motifs (11 and 3 %, respectively). Based on prior observations showing that A3G preferentially deaminates 5′-CC (~85 %) over 5′-TC (~15 %) [32–35], the distribution of G to A mutations in the THP-1 system may be attributable solely to enzymatic deamination by A3G; however, these sequencing results do not exclude the possibility of contributions from one or more A3 family members that prefer 5′-TC substrates.
Fig. 3.
Base substitution mutation spectra of Vif-proficient and Vif-deficient HIV-1 produced from THP-1 cells. (a) Dinucleotide contexts of G to A mutations in the pol region of the indicated HIV-1IIIB viruses produced from the indicated cell lines. Each G to A mutation is shown as a vertical line (tick) on top of individual sequences illustrated to scale on a 564 bp amplicon with colours corresponding to dinucleotide context. (b) Average G to A mutation loads in proviral DNA following Vif-proficient and Vif-deficient HIV-1IIIB production from THP-1 cells or SupT11 cells expressing either a control vector or A3G. Each histogram bar is the average +/-sd of triplicate infections.
APOBEC3G causes the majority of Vif-deficient HIV-1 restriction in THP-1
To more precisely gauge the importance of A3G in the Vif-deficient HIV-1 restriction phenotype in THP-1 cells, we used Vif separation-of-function mutants that are completely sensitive to A3G and simultaneously fully capable of counteracting the other restrictive A3 enzymes. Specifically, penta-alanine (5A) substitution of Vif residues 40–44 (YRHHY in IIIB and NL4-3) renders HIV-1 susceptible to restriction by A3G but not by A3D, A3F or A3H [36, 37]. Similarly, dual-glutamine (2Q) substitution of Vif residues 26 and 27 (i.e. KH to QQ) renders HIV-1 sensitive to restriction by A3G but not by other A3 enzymes [38]. These separation-of-function Vif mutant viruses were used as molecular probes, along with Vif-deficient and Vif-proficient viruses in pseudo-single cycle experiments with THP-1, SupT11-vector and SupT11-A3G cell lines as described above.
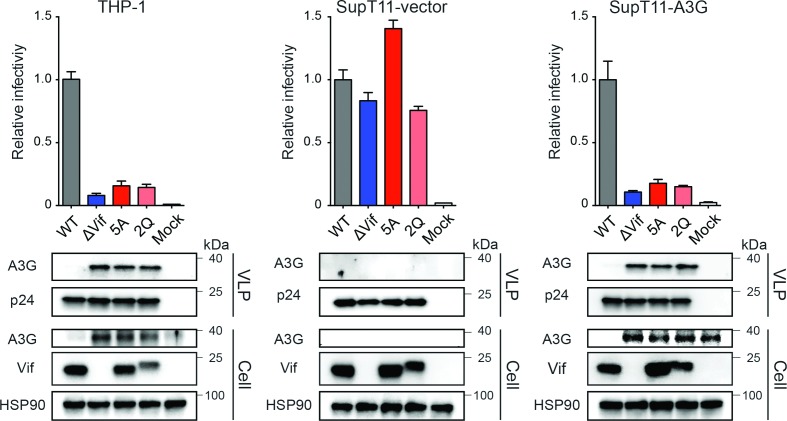
We were surprised to find that the declines in infectivity of the 5A and 2Q Vif mutant viruses produced in THP-1 cells approach that of the fully Vif-null virus produced in parallel (left data sets in Fig. 4). Together with the strong hypermutation bias discussed above, these results further supported the notion that A3G may be the dominant restrictive A3 family member in THP-1 cells. In support of this possibility and confirming that the 5A and 2Q viruses are truly deficient in counteracting A3G, immunoblots showed that levels of A3G incorporated into 5A, 2Q and Vif-null particles are similar. As expected, these three Vif mutants all showed similarly high levels of A3G encapsidation and similarly low levels of infectivity following production in SupT11-A3G cells (right data sets in Fig. 4). The otherwise fully functional nature of these Vif mutant HIV-1 constructs was confirmed through observation of uniformly high levels of infectivity following production in SupT11-vector cells (middle data sets in Fig. 4).
Fig. 4.
A3G is the main Vif-counteracted APOBEC family member in THP-1 cells. Histograms showing relative infectivity levels for the indicated HIV-1IIIB viruses produced from THP-1, SupT11-vector or SupT11-A3G cells. The 5A and 2Q viruses are Vif mutants that are selectively sensitive to restriction by A3G (see text for additional details). Each histogram bar is the mean+/-sd of three independent experiments. Immunoblots are shown below for the indicated viral and cellular proteins in viral particle and whole cell extracts with HSP90 and p24 as loading controls, respectively.
APOBEC3G signature mutations dominate the mutation spectra of Vif-mutant viruses
The infectivity data for the Vif separation-of-function viruses described above indicated that A3G might be the only source of A3 mutagenesis in THP-1 cells. To further investigate this possibility, the pol region of the proviruses from these infectivity studies was cloned and sequenced as described above. Although the mutation levels were relatively low in Vif mutant viruses produced in THP-1 cells, the observed G to A mutations were predominantly within the GG to AG signature motif of A3G (Fig. 5a–b). Importantly, the relative proportion of mutations within GG to AG motifs was similar for Vif-null virus and the two Vif separation-of-function mutants 5A and 2Q (94±2 %, 90±3 % and 89±5 %, respectively; Fig. 5b). Moreover, high levels of G to A mutations within GG motifs were observed for all three Vif mutant viruses produced in SupT11-A3G cells and the relative proportions were similar to those observed in viruses from THP-1 cells (93±2, 92±3 and 92±1 %, respectively; Fig. 5a–b). The similarity of the results from THP-1 cells with a complex A3 repertoire and SupT11-A3G cells with only a single A3G protein provided further support to the idea that A3G may be the only functionally restrictive A3 enzyme in THP-1 cells.
Fig. 5.
A3G is the primary source of G to A mutations in THP-1 cells. (a) Dinucleotide contexts of G to A mutations in the pol region of the indicated HIV-1IIIB viruses produced from the indicated cell lines. Each G to A mutation is shown as a vertical line (tick) on top of individual sequences illustrated to scale on a 564 bp amplicon with colours corresponding to dinucleotide context. (b) Average G to A mutation loads +/-sd for three independent experiments for the viruses and conditions described in (a).
A3G-null THP-1 cells are semi-permissive for Vif-deficient HIV-1 and fully permissive for 5A and 2Q Vif separation-of-function mutants
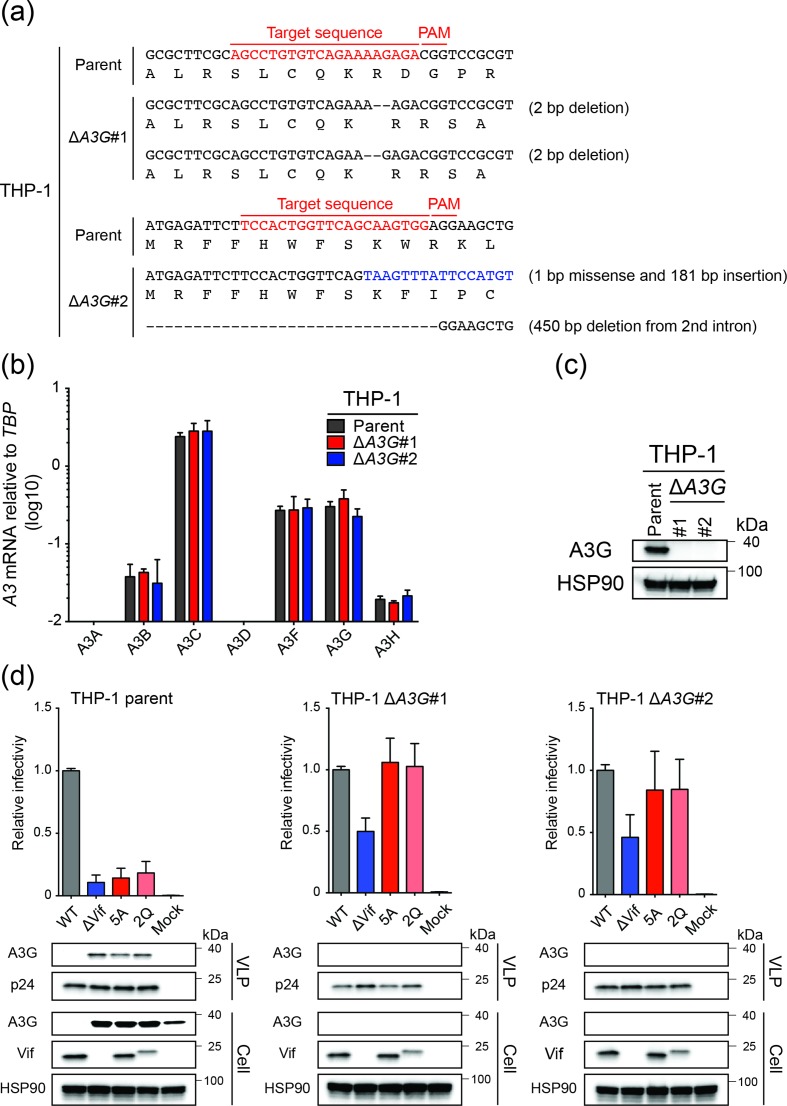
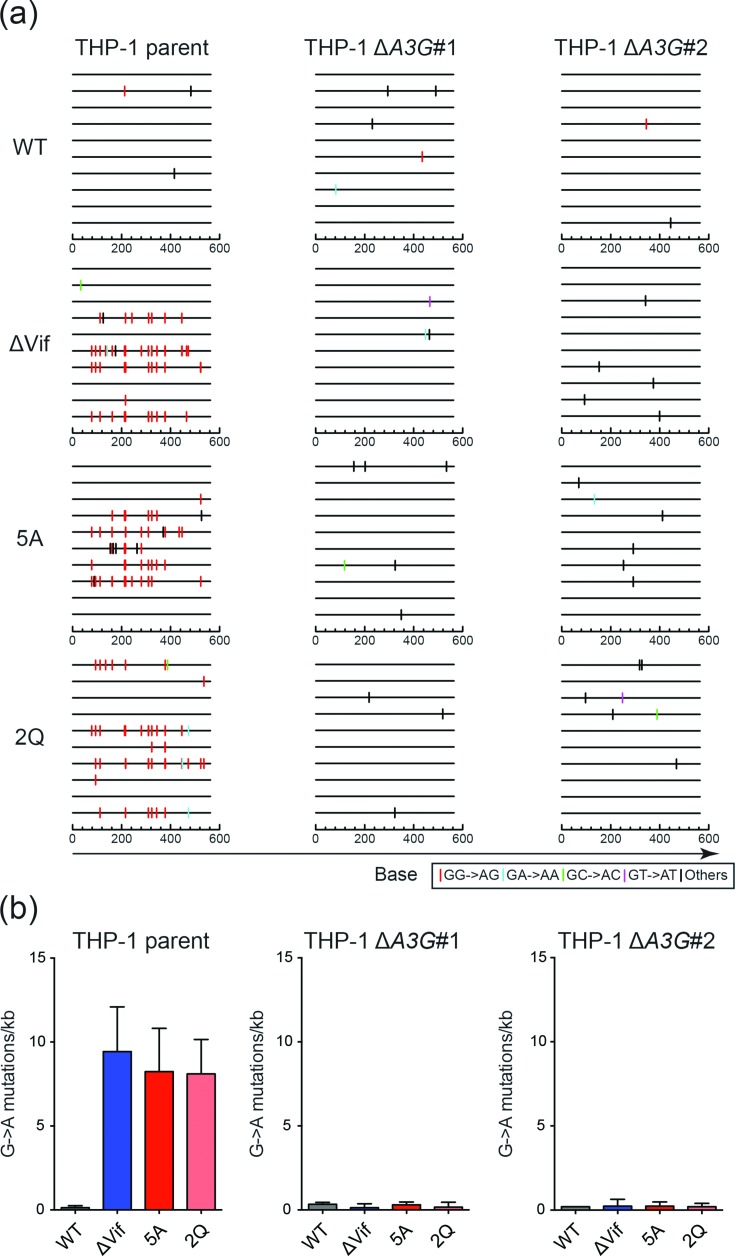
To unambiguously address whether A3G might be the only HIV-1 restrictive deaminase family member in THP-1 cells, we used CRISPR to knock-out both copies of the endogenous A3G gene and performed infectivity and proviral DNA sequencing studies as described above. Two independent A3G-null clones were obtained as evidenced by genomic DNA sequences spanning the Cas9/gRNA cleavage site, with one clone harbouring two different 2 bp deletion mutations adjacent to the PAM site and the other two different insertion/deletion alleles (Fig. 6a). In both instances, RT-qPCR analyses showed that neither A3G nor flanking A3 mRNA levels were affected by these mutations (Fig. 6b). However, also in both instances as predicted by the nature of the mutational events, A3G protein levels were undetectable by immunoblots indicating fully null alleles had been obtained (Fig. 6c). Next, pseudo-single cycle HIV-1 infectivity studies were conducted as described above with Vif-proficient, Vif-deficient, Vif-5A and Vif-2Q constructs. The THP-1 parental cell line yielded results similar to those described above in Fig. 4. In contrast, the A3G-null clones yielded two important results. First, the infectivity of the Vif-null virus increased from 10 % to approximately 50 %, but not completely to 100 % (Fig. 6d). Second, the infectivity of the 5A and the 2Q viruses recovered to 100 % in the A3G-null conditions (Fig. 6d). Moreover, for both the Vif-null and separation-of-function viruses, the A3G-biased hypermutation signature was no longer evident in proviral DNA sequences (Fig. 7). These results combined with the aforementioned data to indicate the existence of either a partial virus restriction activity from another A3 family member in this system (possibly a deamination-independent activity based on the near-exclusivity of the A3G-biased hypermutation data) or, alternatively, an A3 independent function for Vif that is genetically separable from amino acids required for A3G neutralization.
Fig. 6.
HIV-1 infectivity phenotypes of parental and A3G-null THP-1 cells. (a) A3G exon 3 DNA sequences encompassing the gRNA binding site for the two alleles in each A3G-null clone. (b) RT-qPCR analyses of A3 gene expression in parental and A3G-null THP-1 cells. Each histogram bar is the mean+/-sd of three independent experiments. (c) Immunoblot analysis of A3G in parental and A3G-null THP-1 cells. Anti-HSP90 was used to show equal loading of whole cell extracts from the indicated lines. (d) Histograms showing relative infectivities of the indicated HIV-1IIIB viruses produced from THP-1 or two independent A3G-null clonal derivatives. Each histogram bar is the mean+/-sd of three independent experiments. Immunoblots are shown below for the indicated viral and cellular proteins in viral particle and whole cell extracts with HSP90 and p24 as loading controls, respectively.
Fig. 7.
A3G disruption in THP-1 erases G to A mutation bias in proviral DNA. (a) Dinucleotide contexts of G to A mutations in the pol region of the indicated HIV-1IIIB viruses produced from the indicated cell lines. Each G to A mutation is shown as a vertical line (tick) on top of individual sequences illustrated to scale on a 564 bp amplicon with colours corresponding to dinucleotide context. (b) Average G to A mutation loads +/-sd for three independent experiments for the viruses and conditions described in (a).
Discussion
We initiated these studies to ask whether the ‘rules for HIV-1 restriction’ by A3 enzymes are similar in myeloid and T cell lines. Prior work from our group and others had combined to demonstrate that four different A3 enzymes, A3D, A3F, A3G and A3H (stable haplotypes only), contribute to Vif-deficient HIV-1 restriction and hypermutation in CD4-positive T cell lines and primary T-lymphocytes (e.g. [27, 31, 39]). Here, using the myeloid lineage cell line THP-1, we show that A3G is the main and possibly the only A3 family member capable of causing Vif-deficient HIV-1 restriction and G to A hypermutation. One approach revealed similar restriction and hypermutation phenotypes for Vif-null virus and two different Vif separation-of-function mutants (Figs 4 and 5). In particular, in all three of these conditions, emergent viruses showed similarly high levels of A3G signature GG to AG mutations and, importantly, similarly low levels of GA to AA mutations (i.e. low numbers that could be due to A3G or another family member). A complementary approach using A3G-null clones of the parental THP-1 cell line showed incomplete recovery of the infectivity of the Vif-null virus, yet full recovery of the infectivity of two Vif separation-of-function mutants (5A and 2Q) (Fig. 6). Moreover, proviral DNA sequences from the same experiments showed a near-complete erasure of the A3G-biased GG to AG hypermutation signature (Fig. 7). Taken together, we conclude that A3G is the dominant A3 family member capable of HIV-1 restriction and hypermutation in THP-1 cells. However, the reduced infectivity phenotype of the Vif-null virus produced in A3G-null clones indicated that THP-1 has at least one additional A3 enzyme exerting a largely deamination-independent restriction mechanism or that Vif may have an A3 independent function (that minimally does not require the A3G interaction surfaces defined by the 5A and 2Q amino acid substitutions).
Prior to the discovery of A3 enzymes in 2002 [40–42], multiple independent studies had demonstrated the importance of Vif for HIV-1 replication in primary macrophages [43–47]. Several studies also indicated a role for Vif in monocytic lineage cell lines [43, 45, 48–50]. However, since the discovery of A3 enzymes, the prime cellular targets of HIV-1 Vif, a disproportionate number of studies have addressed the Vif-A3 interaction using primary T lymphocytes and immortal T cell lines (reviewed by [1–5]). Comparatively fewer studies have focused on myeloid lineage cell types. First, multiple groups have shown that primary macrophages express a complex A3 repertoire invariably including A3G [24, 25, 51–53]. Second, the Cimarelli group has implicated A3A in HIV-1 restriction in THP-1 and primary macrophages [18]. However, A3A was not addressed by our studies here because interferon was not used to induce A3A transcription [22, 24, 25, 51, 52, 54, 55]. Lastly and perhaps most relevant to our studies, Pathak and coworkers showed that a Vif mutant virus defective for interaction with A3G (5A also used here) has attenuated replication kinetics in cultures of primary monocyte-derived macrophages, whereas a Vif mutant defective for interaction with A3D and A3F was largely unaffected [56]. Moreover, it is important to note that this study also observed A3G signature-biased G to A hypermutations but likely due to difficulties engineering primary macrophages did not demonstrate a genetic requirement for endogenous A3G. Therefore, our studies and particularly the CRISPR knock-outs are important because they provide direct support to the idea that A3G is the predominant HIV-1 restrictive deaminase family member in macrophages and indicate that THP-1 cells may be a powerful model system for additional mechanistic studies.
Future studies should also consider the A3 genotype of the host cells because, for instance, stable haplotypes of A3H are restrictive whereas unstable haplotypes are not [31, 39, 57, 58]. Moreover, the restriction capacity of the cellular A3 repertoire may be influenced by developmental stage and, especially, by the immune microenvironment including levels of various interferons, cytokines and chemokines [22–25, 51–55, 59–64]. For example, the overall antiviral state, including expression levels of several A3 family members, is dramatically strengthened through activation of the type I interferon response [18, 22–25, 51–55, 64, 65]. Such studies may ultimately contribute to a greater understanding of potential reservoirs for HIV-1 infection and latency in vivo, and possibly also to prospects of activating antiviral restriction factors to suppress and/or clear virus infection.
Methods
Cell lines and culture conditions
293T (CRL-3216) was obtained from American Type Culture Collection. TZM-bl (no. 8129) [66] was obtained from the NIH AIDS Reagent Program. CEM-GXR (CEM-GFP expressing CCR5) was provided by Dr Todd Allen (Harvard University, USA) [30]. THP-1 was provided by Dr Andrea Cimarelli (INSERM, France) [18]. Adherent cells were maintained in Dulbecco's modified Eagle medium supplemented with 10 % FBS and 0.5 % penicillin/streptomycin (P/S). Suspension cells were cultured in RPMI with 10 % FBS and 0.5 % P/S.
The creation and characterization of the permissive T cell line SupT11 has been reported [19, 35]. A transfer vector pTRPE-CCR5 was created by placing the coding region of CCR5 (AH005786.2) in pTRPE [67]. To create SupT11 stably expressing CCR5 (SupT11-CCR5), high titre lentiviral supernatant was generated as previously described [68] [kindly provided by Dr James L. Riley (University of Pennsylvania, USA)] and used for transduction. A single cell subclone of SupT11-CCR5 (clone no. 8) was isolated by limiting dilution and expanded. pcDNA3.1 and pcDNA3.1-A3G have been described [69]. To create SupT11-CCR5 cells stably expressing A3G, SupT11-CCR5 cells were electroporated with 20 µg of linearized pcDNA3.1-A3G or vector control, plated at several dilutions in 96-well plates and selected with 1 mg ml− 1 G418. Then, single cell clones of SupT11-CCR5-A3G and -vector were expanded and analysed by immunoblots for A3G expression (data now shown).
A3G specific guides for exon 3 (Fig. 6a) were designed using Synthego's CRISPR design tool (https://design.synthego.com/#/) and evaluated manually for specificity to A3G via an alignment with the most related members of the A3 family. Oligos with ends compatible with the Esp3I sites in pLentiCRISPR1000 were ordered from IDT. The targeting constructs were generated by annealing oligos [ΔA3G gRNA#1: (5′-CAC CGA GCC TGT GTC AGA AAA GAG A) and (5′-GAA CTC TCT TTT CTG ACA CAG GCT C). ΔA3G gRNA#2: (5′-CAC CGT CCA CTG GTT CAG CAA GTG G) and (5′-GAA CCC ACT TGC TGA ACC AGT GGA C)], and cloned by GoldenGate ligation into pLentiCRISPR1000 [70]. All constructs were confirmed by sequencing. 293T cells were transfected with the lentiCRISPR1000 targeting construct along with pΔ-NRF (HIV-1 gag, pol, rev, tat genes) and pMDG (VSV-G) expression vectors. After 48 h, virus-containing supernatants were filtered (0.45 µm, Millipore) and concentrated by centrifugation (26 200 g, 4 ˚C, 2 h). Then, viral pellets were resuspended in 10 % FBS/RPMI and incubated with cells for 48 h before being placed under drug selection (1 µg ml−1 puromycin). Single cell clones were isolated by limiting dilution of the drug-resistant cell pool, expanded and analysed for A3G expression by gnomic DNA sequencing, RT-qPCR and immunoblotting (Fig. 6a–c).
RT-qPCR
Total RNA was extracted from THP-1 cells using the High Pure RNA Isolation Kit (Roche), and cDNAs were made using qScript cDNA SuperMix XL (Quantabio). After RT-qPCR was performed as described using specific primer–probe combinations for each APOBEC [23, 26].
CCLE gene expression analysis
Gene expression information for relevant cell lines was obtained from the CCLE [29] (accessible online at https://portals.broadinstitute.org/ccle). Reads per kilobase per million (RPKM) values were obtained for all available haematopoietic and lymphoid cells lines from the latest CCLE preprocessing pipeline to quality control reads, align/assemble reads to a human reference genome, and quantify individual human gene expression RPKM values (CCLE_DepMap_18Q1_RNAseq_RPKM_20180214.gct). Data were formatted using the statistical analysis software R and RStudio, and visualized using the R package ggplot2 [71, 72].
HIV-1 infectivity assays
Vif-proficient and Vif-deficient (X26 and X27) HIV-1 IIIB C200 proviral expression constructs have been reported [73]. An HIV-1 IIIB C200 variant encoding Vif Q26 and Q27 has been characterized [38] (here called 2Q mutant). An HIV-1 IIIB C200 Vif 5A variant (40AAAAA44) was created by digesting pNLCSFV3-5A proviral DNA construct ([74]; kindly provided by Dr Kei Sato, University of Tokyo, Japan) at SwaI and SalI sites and similarly cloned into pIIIB C200 proviral construct. HIV-1 single-cycle assays using VSV-G pseudotyped viruses were performed as described previously with minor modifications [27, 75]. VSV-G pseudotyped viruses were generated by transfecting 2.4 µg of proviral DNA construct and 0.6 µg of VSV-G expression vector using TransIT-LT1 reagent (Mirus Bio) into 293T cells (3.0×106). Then, 48 h later, supernatants were harvested, filtered by 0.45 µm filters and used to infect into 2.5×104 CEM-GXR reporter cells for m.o.i. determinations. 5×105 target cells were infected with a m.o.i. of 0.05 (for SupT11-CCR5-vector and SupT11-CCR5-A3G) or 0.25 (for THP-1), washed with PBS after 24 h of the infection, and then incubated for an additional 24 h. Then, 24 h later, supernatants were collected and filtered. The resulting viral particles were normalized by p24 ELISA (ZeptoMetrix) and viral infectivity was measured using TZM-bl cells.
Hypermutation analysis
Hypermutation analyses were performed as described [27, 28]. Proviral DNAs were recovered by infecting virus produced in THP-1 or SupT11-CCR5 cells into TZM-bl or SupT11-CCR5 using Gentra Puregene Cell Kit (Qiagen) [75]. Following DpnI digestion, the viral pol region was amplified by nested PCR with outer primers (876 bp) [(5′-TCC ART ATT TRC CAT AAA RAA AAA) and (5′-TTY AGA TTT TTA AAT GGY TYT TGA)] and inner primers (564 bp) [(5′-AAT ATT CCA RTR TAR CAT RAC AAA AAT) and (5′-AAT GGY TYT TGA TAA ATT TGA TAT GT)]. Tdata-word-spacing="data-word-spacing="0.2w"0.2w"he resulting 564 bp amplicon was cloned into CloneJet (ThermoScientific). At least seven independent clones were Sanger sequenced for each condition. Clones with identical mutations were eliminated.
Immunoblots
Cells were lysed in 2.5×Laemmli sample buffer. Virions were dissolved in 2.5×Laemmli sample buffer after pelleting down using 20 % sucrose (26 200 g, 4 ˚C, 2 h). Proteins in cell and viral lysates were separated by SDS-PAGE and transferred to PVDF membranes (LI-COR Biosciences). Membranes were blocked with 5 % milk in PBS containing 0.1 % Tween 20 and incubated with primary antibodies: mouse anti-HSP90 (Thermo Fisher Scientific, PA3-012); rabbit anti-APOBEC3B (RSH#10.87.13; first described in [76]); rabbit anti-A3G (NARP, #10201); mouse anti-Vif (319, NARP, #6459); mouse anti-p24 (183-H12-5C, NARP, #1513). Subsequently, the membranes were incubated with horseradish peroxidase (HRP) or fluorescent dye-conjugated secondary antibodies: IRDye 680LT anti-mouse IgG (LI-COR Biosciences, 926–68020); anti-rabbit IgG-HRP (Jackson ImmunoResearch, 111-035-144); anti-mouse IgG-HRP (Jackson ImmunoResearch, 715-035-150). HyGlo chemiluminescent HRP detection reagent (Denville Scientific) was used for HRP detection. Bands were visualized by using film exposure or the Odyssey Fc Imager (LI-COR Biosciences).
Funding information
This work was supported by NIAID R37 AI064046 and NIGMS R01 GM118000. A. M. M. received partial salary support from NIAID T32 AI83196, M. C. J. from NCI T32 CA009138, and D. J. S. from NIDCR T90 DE022732.
Acknowledgements
We thank B. Anderson for advice early in the project, T. Allen, A. Cimarelli, J. Riley and K. Sato for providing reagents, and A. Cheng for proofreading this manuscript. We extend additional thanks for reagents obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. R. S. H. is the Margaret Harvey Schering Land Grant Chair for Cancer Research, a Distinguished McKnight University Professor, and an Investigator of the Howard Hughes Medical Institute.
Conflicts of interest
R. S. H. is a co-founder, shareholder and consultant of ApoGen Biotechnologies. The other authors have no conflicts of interest to declare.
Footnotes
Abbreviations: A3, APOBEC3, apolipoprotein mRNA editing catalytic subunit-like 3; gRNA, guide RNA; HIV-1, human immunodeficiency virus-type 1; m.o.i., multiplicity of infection; Vif, virion infectivity factor; VSV-G, vesicular stomatitis virus G protein.
References
- 1.Malim MH, Bieniasz PD. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moris A, Murray S, Cardinaud S. Aid and APOBECs span the gap between Innate and adaptive immunity. Front Microbiol. 2014;5:534. doi: 10.3389/fmicb.2014.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479-480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon V, Bloch N, Landau NR. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol. 2015;16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jäger S, Kim DY, Hultquist JF, Shindo K, LaRue RS, et al. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature. 2012;481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Du J, Evans SL, Yu Y, Yu XF. T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 8.Anderson BD, Harris RS. Transcriptional regulation of APOBEC3 antiviral immunity through the CBF-β/RUNX axis. Sci Adv. 2015;1:e1500296. doi: 10.1126/sciadv.1500296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulder LCF, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci U S A. 2008;105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jern P, Russell RA, Pathak VK, Coffin JM. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009;5:e1000367. doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim EY, Bhattacharya T, Kunstman K, Swantek P, Koning FA, et al. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. J Virol. 2010;84:10402–10405. doi: 10.1128/JVI.01223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim EY, Lorenzo-Redondo R, Little SJ, Chung Y-S, Phalora PK, et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog. 2014;10:e1004281. doi: 10.1371/journal.ppat.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noguera-Julian M, Cozzi-Lepri A, Di Giallonardo F, Schuurman R, Däumer M, et al. Contribution of APOBEC3G/F activity to the development of low-abundance drug-resistant human immunodeficiency virus type 1 variants. Clin Microbiol Infect. 2016;22:191–200. doi: 10.1016/j.cmi.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Stevenson M. Role of myeloid cells in HIV-1-host interplay. J Neurovirol. 2015;21:242–248. doi: 10.1007/s13365-014-0281-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sattentau QJ, Stevenson M. Macrophages and HIV-1: an unhealthy constellation. Cell Host Microbe. 2016;19:304–310. doi: 10.1016/j.chom.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wacleche V, Tremblay C, Routy JP, Ancuta P. The biology of monocytes and dendritic cells: contribution to HIV pathogenesis. Viruses. 2018;10:65. doi: 10.3390/v10020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herskovitz J, Gendelman HE. HIV and the macrophage: from cell reservoirs to drug delivery to viral eradication. J Neuroimmune Pharmacol. 2019;14:52–67. doi: 10.1007/s11481-018-9785-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berger G, Durand S, Fargier G, Nguyen XN, Cordeil S, et al. APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 2011;7:e1002221. doi: 10.1371/journal.ppat.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goujon C, Moncorgé O, Bauby H, Doyle T, Ward CC, et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature. 2013;502:559–562. doi: 10.1038/nature12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koning FA, Newman ENC, Kim EY, Kunstman KJ, Wolinsky SM, et al. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, et al. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thielen BK, McNevin JP, McElrath MJ, Hunt BVS, Klein KC, et al. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J Biol Chem. 2010;285:27753–27766. doi: 10.1074/jbc.M110.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Refsland EW, Hultquist JF, Harris RS. Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog. 2012;8:e1002800. doi: 10.1371/journal.ppat.1002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brockman MA, Tanzi GO, Walker BD, Allen TM. Use of a novel GFP reporter cell line to examine replication capacity of CXCR4- and CCR5-tropic HIV-1 by flow cytometry. J Virol Methods. 2006;131:134–142. doi: 10.1016/j.jviromet.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Refsland EW, Hultquist JF, Luengas EM, Ikeda T, Shaban NM, et al. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS Genet. 2014;10:e1004761. doi: 10.1371/journal.pgen.1004761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/S0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 33.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Q, König R, Pillai S, Chiles K, Kearney M, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 36.Russell RA, Smith J, Barr R, Bhattacharyya D, Pathak VK. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J Virol. 2009;83:1992–2003. doi: 10.1128/JVI.01621-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith JL, Pathak VK. Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J Virol. 2010;84:12599–12608. doi: 10.1128/JVI.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albin JS, Haché G, Hultquist JF, Brown WL, Harris RS. Long-term restriction by APOBEC3F selects human immunodeficiency virus type 1 variants with restored Vif function. J Virol. 2010;84:10209–10219. doi: 10.1128/JVI.00632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ooms M, Brayton B, Letko M, Maio SM, Pilcher CD, et al. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe. 2013;14:411–421. doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme Apobec1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/S1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 41.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 42.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 43.Gabuzda DH, Li H, Lawrence K, Vasir BS, Crawford K, et al. Essential role of Vif in establishing productive HIV-1 infection in peripheral blood T lymphocytes and monocyte/macrophages. J Acquir Immune Defic Syndr. 1994;7:908–915. [PubMed] [Google Scholar]
- 44.Kawamura M, Ishizaki T, Ishimoto A, Shioda T, Kitamura T, et al. Growth ability of human immunodeficiency virus type 1 auxiliary gene mutants in primary blood macrophage cultures. J Gen Virol. 1994;75:2427–2431. doi: 10.1099/0022-1317-75-9-2427. [DOI] [PubMed] [Google Scholar]
- 45.Michaels FH, Hattori N, Gallo RC, Franchini G. The human immunodeficiency virus type 1 (HIV-1) Vif protein is located in the cytoplasm of infected cells and its effect on viral replication is equivalent in HIV-2. AIDS Res Hum Retroviruses. 1993;9:1025–1030. doi: 10.1089/aid.1993.9.1025. [DOI] [PubMed] [Google Scholar]
- 46.von Schwedler U, Song J, Aiken C, Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowdhury IH, Chao W, Potash MJ, Sova P, Gendelman HE, et al. vif-negative human immunodeficiency virus type 1 persistently replicates in primary macrophages, producing attenuated progeny virus. J Virol. 1996;70:5336–5345. doi: 10.1128/jvi.70.8.5336-5345.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan L, Peden K. Cell-free transmission of Vif mutants of HIV-1. Virology. 1992;190:19–29. doi: 10.1016/0042-6822(92)91188-Z. [DOI] [PubMed] [Google Scholar]
- 49.Gargan S, Ahmed S, Mahony R, Bannan C, Napoletano S, et al. HIV-1 promotes the degradation of components of the type 1 IFN JAK/STAT pathway and blocks anti-viral ISG induction. EBioMedicine. 2018;30:203–216. doi: 10.1016/j.ebiom.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du J, Rui Y, Zheng W, Li P, Kang J, et al. Vif-CBFβ interaction is essential for Vif-induced cell cycle arrest. Biochem Biophys Res Commun. 2019;511:910–915. doi: 10.1016/j.bbrc.2019.02.136. [DOI] [PubMed] [Google Scholar]
- 51.Koning FA, Goujon C, Bauby H, Malim MH. Target cell-mediated editing of HIV-1 cDNA by APOBEC3 proteins in human macrophages. J Virol. 2011;85:13448–13452. doi: 10.1128/JVI.00775-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng G, Greenwell-Wild T, Nares S, Jin W, Lei KJ, et al. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood. 2007;110:393–400. doi: 10.1182/blood-2006-10-051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stopak KS, Chiu YL, Kropp J, Grant RM, Greene WC. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J Biol Chem. 2007;282:3539–3546. doi: 10.1074/jbc.M610138200. [DOI] [PubMed] [Google Scholar]
- 54.Land AM, Law EK, Carpenter MA, Lackey L, Brown WL, et al. Endogenous APOBEC3A DNA cytosine deaminase is cytoplasmic and nongenotoxic. J Biol Chem. 2013;288:17253–17260. doi: 10.1074/jbc.M113.458661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohanram V, Sköld AE, Bächle SM, Pathak SK, Spetz AL. IFN-α induces APOBEC3G, F, and A in immature dendritic cells and limits HIV-1 spread to CD4+ T cells. J Immunol. 2013;190:3346–3353. doi: 10.4049/jimmunol.1201184. [DOI] [PubMed] [Google Scholar]
- 56.Chaipan C, Smith JL, Hu WS, Pathak VK. APOBEC3G restricts HIV-1 to a greater extent than APOBEC3F and APOBEC3DE in human primary CD4+ T cells and macrophages. J Virol. 2013;87:444–453. doi: 10.1128/JVI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.OhAinle M, Kerns JA, Li MMH, Malik HS, Emerman M. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe. 2008;4:249–259. doi: 10.1016/j.chom.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pion M, Granelli-Piperno A, Mangeat B, Stalder R, Correa R, et al. APOBEC3G/3F mediates intrinsic resistance of monocyte-derived dendritic cells to HIV-1 infection. J Exp Med. 2006;203:2887–2893. doi: 10.1084/jem.20061519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen K, Huang J, Zhang C, Huang S, Nunnari G, et al. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J Virol. 2006;80:7645–7657. doi: 10.1128/JVI.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kreisberg JF, Yonemoto W, Greene WC. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J Exp Med. 2006;203:865–870. doi: 10.1084/jem.20051856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pido-Lopez J, Whittall T, Wang Y, Bergmeier LA, Babaahmady K, et al. Stimulation of cell surface CCR5 and CD40 molecules by their ligands or by HSP70 up-regulates APOBEC3G expression in CD4(+) T cells and dendritic cells. J Immunol. 2007;178:1671–1679. doi: 10.4049/jimmunol.178.3.1671. [DOI] [PubMed] [Google Scholar]
- 63.Lafferty MK, Sun L, DeMasi L, Lu W, Garzino-Demo A. CCR6 ligands inhibit HIV by inducing APOBEC3G. Blood. 2010;115:1564–1571. doi: 10.1182/blood-2009-06-226423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kane M, Zang TM, Rihn SJ, Zhang F, Kueck T, et al. Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe. 2016;20:392–405. doi: 10.1016/j.chom.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol. 2009;83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leibman RS, Richardson MW, Ellebrecht CT, Maldini CR, Glover JA, et al. Supraphysiologic control over HIV-1 replication mediated by CD8 T cells expressing a re-engineered CD4-based chimeric antigen receptor. PLoS Pathog. 2017;13:e1006613. doi: 10.1371/journal.ppat.1006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parry RV, Rumbley CA, Vandenberghe LH, June CH, Riley JL. CD28 and inducible costimulatory protein Src homology 2 binding domains show distinct regulation of phosphatidylinositol 3-kinase, Bcl-xL, and IL-2 expression in primary human CD4 T lymphocytes. J Immunol. 2003;171:166–174. doi: 10.4049/jimmunol.171.1.166. [DOI] [PubMed] [Google Scholar]
- 69.Haché G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 70.Carpenter MA, Law EK, Serebrenik A, Brown WL, Harris RS. A lentivirus-based system for Cas9/gRNA expression and subsequent removal by Cre-mediated recombination. Methods. 2019;156:79–84. doi: 10.1016/j.ymeth.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ito K, Murphy D. Application of ggplot2 to pharmacometric graphics. CPT: pharmacomet syst pharmacol. 2013;2:e79. doi: 10.1038/psp.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.RDC . R: a Language and ENvironmenral for Statistical Computing. Vienna, Austria: the R Foundation for Statistical Computing; 2011. http://www.r-project.org [Google Scholar]
- 73.Haché G, Shindo K, Albin JS, Harris RS. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr Biol. 2008;18:819–824. doi: 10.1016/j.cub.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sato K, Takeuchi JS, Misawa N, Izumi T, Kobayashi T, et al. APOBEC3D and APOBEC3F potently promote HIV-1 diversification and evolution in humanized mouse model. PLoS Pathog. 2014;10:e1004453. doi: 10.1371/journal.ppat.1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ikeda T, Symeonides M, Albin JS, Li M, Thali M, et al. HIV-1 adaptation studies reveal a novel Env-mediated homeostasis mechanism for evading lethal hypermutation by APOBEC3G. PLoS Pathog. 2018;14:e1007010. doi: 10.1371/journal.ppat.1007010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leonard B, McCann JL, Starrett GJ, Kosyakovsky L, Luengas EM, et al. The PKC/NF-κB signaling pathway induces APOBEC3B expression in multiple human cancers. Cancer Res. 2015;75:4538–4547. doi: 10.1158/0008-5472.CAN-15-2171-T. [DOI] [PMC free article] [PubMed] [Google Scholar]