Abstract
A growing body of clinical and experimental data supports the view that the efficacy of probiotics is strain-specific and restricted to particular pathological conditions, which means that newly isolated probiotic strains need to be targeted to a specific disease. Following national and international guidelines, we used a conventional in vitro experimental approach to characterize a novel strain of Lactobacillus reuteri, LMG P-27481, for safety (sensitivity to antibiotics and genome analysis) and putative efficacy (resistance to gastro-intestinal transit, adhesiveness, induction of cytokines, and release of antimicrobial metabolites). In vitro assays, which were carried out to examine the probiotic’s effect on diarrhea (lactose utilization, inhibition of pathogens such as bacteria and Rotavirus), showed that it was more efficacious with respect to well-known reference strains in antagonizing Clostridioides difficile (CD). Data confirming that the probiotic can effectively treat CD colitis was gained from in vivo trials involving mice conditioned with large spectrum antibiotics before they were subjected to CD challenge. Two out of the three antibiotic-treated groups received daily LMG P-27481 for different time durations in order to simulate a preventive approach (LMG P-27481 administered prior to CD challenge) or an antagonistic one (LMG P-27481 administered after CD challenge). Both approaches significantly reduced, with respect to the untreated controls, CD DNA concentrations in caecum and C. difficile toxin titers in the gut lumen. In addition, LMG P-27481 supplementation significantly mitigated body weight loss and the extent of inflammatory infiltrate and tissue damage. The study results, which need to be confirmed by in vivo clinical trials, have demonstrated that the L. reuteri LMG P-27481 strain is a promising probiotic candidate for the treatment of CD infection.
Keywords: probiotic, lactobacilli, pathogen, intestine, gut, diarrhea
Introduction
A variety of studies have evaluated the efficacy of probiotics in treating diarrhea, which represents a potentially serious health problem with more than 4 billion cases reported world-wide every year. McFarland et al. (2018) carried out an extensive systematic meta-analysis of the results of randomized controlled trials examining the use of probiotics to treat different types of diseases. In accordance with Ouwehand (2017), the investigators confirmed the strain- and disease-specificity of probiotics in treating diarrhea. Despite a wide heterogeneity across clinical studies, some probiotics have been found to be effective in treating antibiotic-associated adult diarrhea, Clostridium difficile diarrhea (CDAD), acute Rotavirus diarrhea, radiation therapy-related diarrhea, and enteral nutrition-related diarrhea. Their efficacy in treating traveler’s diarrhea (TD) has not, however, been supported by clinical data (Williams and Adcock, 2018). Hypothetical mechanisms explaining probiotics’ effects include competitive inhibition of pathogens, stabilization of the resident microbiota, and attenuation of increased gut barrier permeability. Their beneficial effects in terms of reducing the duration and intensity of fever has been demonstrated in children although they did not seem to affect the number of daily evacuations and accompanying symptoms such as vomiting (Salari et al., 2012).
The efficacy of probiotics in adults for the different types of diarrhea listed above has not yet been convincingly established and still requires further investigation (Ritchie and Romanuk, 2012; Salari et al., 2012). When Szajewska et al. (2019) examined in a systematic review 15 randomized controlled trials investigating the efficacy of Lactobacillus rhamnosus GG (LGG) in children affected by acute gastroenteritis, they concluded that the probiotic reduced the duration of diarrhea and hospitalization. Conflicting findings, which may be explained by the heterogeneity of the patients studied and methodological limitations, have nevertheless been reported. Capurso (2019) likewise reviewed data examining the use of L. rhamnosus GG supplementation to treat gastrointestinal infections and diarrhea, antibiotic and C. difficile associated diarrhea as well as its use for other pathological conditions.
Belonging to the Lactobacillus genus, Lactobacillus reuteri represents a natural inhabitant of the human and animal gastro-intestinal tract. It is an obligatory heterofermentative Lactobacillus that in particular conditions leads to the formation of carbon dioxide, ethanol, acetic acid and lactic acid following sugar fermentation. L. reuteri can also produce 3-hydroxypropionaldehyde (reuterin), a bacteriocin with antimicrobial properties, as well as folate and cobalamin (Mu et al., 2018). Some L. reuteri strains, such as DSM 17938 and RC-14, are already present in many commercially available food supplements. Evidence confirming the efficacy of L. reuteri in treating gastrointestinal disorders such as infantile colic, regurgitation, functional constipation, abdominal pain, and necrotizing enterocolitis (Srinivasan et al., 2018) has been extensively outlined in the literature.
Using the FAO/WHO (2002) for the evaluation of probiotics in food, whose timeliness has been confirmed by an expert panel (Hill et al., 2014; Shewale et al., 2014), the current study set out to evaluate the efficacy of a new probiotic strain in treating C. difficile-associated diarrhea. The new strain, named LMG P-27481, meets the conventional safety criteria for probiotics which is based on their survival in the GI tract as well as their ability to adhere to gut mucosa, to counteract the activity of pathogens and to boost the immune system. Its safety profile for human use was also assessed following the most recent international guidelines (EFSA Scientific Opinion, 2018). Characterization to assess the strain’s efficacy was, moreover, performed using an in vivo murine model of CD-associated diarrhea to investigate its ability to counteract the pathogen.
Materials and Methods
Identification of L. reuteri LMG P-27481
The LMG P-27481 L. reuteri strain was isolated in 2013 from a human fecal sample of a 9-month old vaginally delivered healthy infant. Written informed consent was obtained from the parents. The strain was identified at the genus level and then as a distinct L. reuteri species. LMG P-27481 was compared to other L. reuteri strains available on the Italian market to assess its originality using strain-specific REP-PCR amplification (Versalovic et al., 1994). Other relevant well-known commercial strains such as L. reuteri RC-14, L. reuteri DSM 17938, and L. rhamnosus ATCC 53103 were used as references for the assays conducted during the study (Mogna et al., 2012).
Safety Assessment
Minimum inhibitory concentrations (MICs) of 15 different antibiotics were determined for the strain under investigation in accordance with ISO 10932:2010 (IDF 223:2010). MIC values were compared with European Food Safety Authority (EFSA) breakpoints reported in their Scientific Opinions 2012 and 2018. The tests were carried out using antibiotics and doses listed in Table 1. Contemporaneously, we excluded the presence of plasmids within the cell using the method of plasmid extraction outlined by Anderson and McKay (1983) and by sequencing the entire genome of LMG P-27481 strain in order to check its safety profile.
TABLE 1.
The range of antibiotic concentrations used to determine the minimum inhibitory concentrations.
| Antibiotic | Concentration range (μg/ml) |
| Gentamicin | 0.5–256 |
| Kanamycin | 2–1024 |
| Streptomycin | 0.5–256 |
| Neomycin | 0.5–256 |
| Tetracycline | 0.12–64 |
| Erythromycin | 0.016–8 |
| Clindamycin | 0.03–16 |
| Chloramphenicol | 0.12–64 |
| Ampicillin | 0.03–16 |
| Vancomycin | 0.25–128 |
| Virginiamycin (Quinupristin-Dalfopristin) | 0.016–8 |
| Linezolid | 0.03–16 |
| Trimethoprim | 0.12–64 |
| Ciprofloxacin | 0.25–128 |
| Rifampin | 0.12–64 |
Complete Genome Sequencing of L. reuteri LMG P-27481 Strain
Genomic DNA from an overnight culture of LMG P-27481 L. reuteri, which was extracted using the MasterPure Gram Positive DNA Purification kit (Epicenter, Cambio, United Kingdom) following the manufacturer’s instructions, was quantified utilizing NanoDropTM spectrophotometer A280/260 (Thermo Fisher Scientific, Milan, Italy). Total DNA sequencing was determined by GenProbio Srl (Parma, Italy) using a MiSeq Illumina platform. Genomic libraries were constructed employing the TruSeq DNA PCR-Free LT Kit (Illumina) using 2.5 μg of genomic DNA, which was fragmented using a Bioruptor NGS ultrasonicator (Diagenode, United States), and size was assessed using Agilent 2200Tape Station (Agilent Technologies). Library samples were loaded onto a Flow Cell V3 600 cycles (Illumina) following the technical support guide instructions. Five hundred sequencing cycles resulted in an average reading length of approximately 250 nucleotides for both paired-end sequences.
Bioinformatic Analysis of L. reuteri LMG P-27481 Genome
The version 4.0.2 MIRA program (Chevreux et al., 1999, Computer Science and Biology: Proceedings of the German Conference on Bioinformatics) was used for de-novo assembly of the LMG P-27481 L. reuteri genome sequence. Assembly resulted in 134 contigs with a total genome size of 2,016,419 bp using a MIRA v4.0.2 assembler (Chevreux et al., 1999), with a coverage of 70.1X, GC content of 38.8%, N50 29849. Open Reading Frames (ORFs) prediction and subsequent GeneBank file construction were performed using Prodigal v2.6 software (Hyatt et al., 2010). Automatic annotation of the ORFs was carried out using a custom script performing a BLASTp search against the NCBI database (Milani et al., 2014). Ribosomal RNA gene prediction was performed using RNAmmer v1.2 (Lagesen et al., 2007), and transfer RNA gene prediction was carried out using tRNAscan-SE v1.21 (Lowe and Eddy, 1997).
Reordering of the final contigs comparable to NCBI template genomes L. reuteri DSM 20016 was performed using Mauve v2.3.1 (Darling et al., 2004). Burrows-Wheeler Alignment with SAMtools software (Li and Durbin, 2009) and VarScan v2.2.3 (Koboldt et al., 2013) were carried out to optimize the assembly to obtain final contigs. To confirm the results obtained from the MIC evaluation of the strain, the antibiotic resistome of LMG P-27481 L. reuteri was identified using Rapsearch (Zhao et al., 2012) against a custom database and CARD (McArthur et al., 2013) and Transporter Classification Database (TCDB) (Saier et al., 2014). Prediction of the mobilome was carried out using the PHAge Search Tool PHAST (Zhou et al., 2011) and the ISfinder database (Siguier, 2006). Prediction of the putative virulence genes was carried out using the Virulence Factors Database (VFDB) (Chen et al., 2005) and that of the bacteriocin encoding genes was performed using the Bagel3 software package (van Heel et al., 2013). Other relevant factors with a putative adhesion to probiotic functions, such as mucus and fibronectin-binding proteins and S-layer proteins, pilus encoding genes and those involved in Extracellular polysaccharide synthesis (EPS), were predicted using a custom database based on NCBI RefSeq (Pruitt et al., 2007).
Conventional Characterization of L. reuteri LMG P-27481
The procedures described by Charteris et al. (1998) and Gilliland et al. (1984) were followed to investigate the ability of the newly isolated and reference L. reuteri DSM 17938 and L. reuteri RC-14 strains to survive the hostile conditions of the gastrointestinal environment including the high pH levels of gastric and duodenal juices. Their ability to adhere to the human mucosa was verified and quantified using cultured monolayers of HT-29 cells (Jensen et al., 2012). All the strains were cultured in MRS medium at 37°C in anaerobiosis and collected by centrifugation. Bacteria were resuspended in sterile DMEM at 108 CFU/ml and added to HT-29 monolayers (MOI 1:50/cells to probiotic). The medium was incubated for 60 min at 37°C. Then, it was removed and discarded and sterile DMEM was added to the monolayers. The medium was checked three times to verify that all unbound bacteria were removed. One ml of sterile MRS was then added to each monolayer and the cells were collected using a cell scraper. The material was collected using a sterile syringe and passed three times through an 18-needle gauge, and following decimal dilutions, seeded on MRS agar to quantify the live bacteria. The adhesion was expressed as the percentage of viable bacterial cells adhering to HT-29. The experiments were performed in triplicate. L. paracasei ATCC 344 was used as the technical control.
The in vitro Effect on Human Dendritic Cells
Following international guidelines for the characterization of the properties of probiotics, the immune modulatory potential of the LMG P-27481 strain was verified on human-derived dendritic cells (DCs). The DCs were generated from monocytes obtained from the peripheral blood of healthy donors and cultured in complete RPMI medium containing granulocyte-macrophage colony-stimulating factor (GM-CSF, 50 ng/mL; Peprotech) and interleukin-4 (20 ng/mL; Peprotech, Milan, Italy) for 6 days (Mileti et al., 2009). On the day of the experiment, the DCs were collected, washed by centrifugation, counted, and either incubated in medium alone or in medium without antibiotics containing the specified probiotic strain (MOI 1:10/DC to probiotic) obtained from a fresh culture in the logarithmic growth phase. The dendritic cells were incubated 1 h at 37°C, then collected, pelleted by centrifugation, resuspended in 1 ml of sterile RPMI and subjected to centrifugation (800 rpm × 10 min). The cells were then incubated in fresh complete medium containing gentamicin (100 μg/ml). After 24 h, the DCs were collected by centrifugation, IL-10 and IL12p70 were quantified in the culture supernatants by ELISA (eBioscience), and the anti-inflammatory index was calculated (IL-10/IL12p70 concentration ratio).
The Release of Hydrogen Peroxide
The ability to release H2O2 in the surrounding environment confers antimicrobial and preservative properties to some lactobacilli. Bacterial strains were grown in MRS broth to assess the LMG P-27481 strain’s ability to release H2O2. The cultures were then centrifuged, washed twice, and finally resuspended with sodium phosphate buffer containing glucose 5 mM. After being incubated in refrigerated conditions for a maximum of 24 h, the cultures were centrifuged, and the supernatants were assayed for H2O2. Hydrogen peroxide was quantified following the protocol of Yap and Gilliland (2000). The RC-14 and DSM 17938 strains were used as positive and negative controls, respectively.
Reuterin Secretion
Some strains of L. reuteri are known to produce broad-spectrum antimicrobial substance, reuterin (Talarico et al., 1988), a product of the glycerol metabolism that is active against several intestinal pathogens. The ability to produce this bacteriocin was verified using the colorimetric test described by Cadieux et al. (2008) on a 24-h culture of the strain grown in MRS medium supplemented with glycerol (250 mM final concentration). The principle behind the test is that of transforming glycerol into 3-hydroxypropionaldehyde (reuterin) and to quantify it using a spectrophotometric method at 560 nm (Doleyres et al., 2005; Ortiz-Rivera et al., 2017).
Lactose Metabolism
The ability to use lactose as the growth substrate was tested by inoculating washed pure suspensions of L. reuteri DSM 17938, L. rhamnosus ATCC 53103, and L. reuteri LMG P-2748 in MRS medium without dextrose, containing either 1% glucose or 1% lactose as the sole carbon source (Arnold et al., 2018). After being incubated for 18 h in microaerophilic conditions at 37°C, live bacteria were enumerated by seeding decimal dilution of the cultures on MRS agarose plates. T18 results were compared to the initial counts at T0 for the different carbohydrate sources.
The in vitro Effect on Intestinal Pathogen Growth
These experiments were designed to assess the ability of the LMG P-27481 strain to exert a direct inhibitory effect on some of the most common intestinal pathogens. The pathogen strains used for these tests were obtained from ATCC and from the Culture Collection of the University of Göteborg (Sweden). The two L. reuteri strains (LMG P-27481 and DSM 17938) and L. rhamnosus ATCC 53103 were cultured in MRS medium, E. coli and Salmonella enterica in Luria Bertani (LB) broth, and C. difficile in Brain Heart Infusion (BHI) broth. To test the antimicrobial activity, the lactobacilli were separately inoculated at 107 CFU/ml and grown in MRS broth anaerobically at 37°C for 24 h. Once the bacteria had grown, the cultures were centrifuged, the supernatants were sterile filtered (0.2 μm), and the pH corrected to 7.0 before being inoculated with E. coli, Salmonella or C. difficile (107 CFU/ml) and incubated at 37°C. The aliquots of bacterial cultures were collected and plated 6, 12, and 24 h after inoculation onto appropriate agar media to quantify the viable pathogens. Each experiment was performed three times with triplicate determinations.
The in vitro Effect on the Rotavirus Infection
Rotavirus (ATCC VR-2018) was expanded and quantified on MA-104 (rhesus monkey kidney) cells. The efficacy of the L. reuteri LMG P-27481 strain in treating the Rotavirus infection was assessed utilizing two in vitro HT-29 cells protocols mimicking the human intestinal epithelium. In the pre-treatment in vitro model the strains were placed in contact with HT-29 cells (MOI 1:50/cells to probiotic) prior to exposure to trypsin activated human rotavirus (Guerrero et al., 2000). Seventy-two hours later, the cells were collected, and total RNA was extracted. In the competition protocol, trypsin activated human rotavirus was pre-incubated with the different probiotic strains for 90 min. The bacteria + virus mixture was briefly centrifuged, and then the clear supernatant was used to infect confluent monolayers of human intestinal epithelial HT-29 cells. Seventy-two hours later, the cells were collected, total RNA was extracted using the SV Total RNA Isolation System (Promega), and reverse-transcribed into complementary DNA. Viral genome copies were quantified with SYBR Green PCR Master Mix in an ABI PRISM 7000 Sequence Detection System (Applied Biosystems) using specific primers (van Maarseveen et al., 2010).
The in vivo Inhibitory Effect on C. difficile Infection
LMG P-27481‘s ability to inhibit C. difficile infection was assessed in vivo by verifying intestinal colonization and inflammation in mice treated with cefoperazone (Theriot et al., 2011). Twenty-four adult C57Bl/6 mice, purchased from Envigo Srl (Udine, Italy), were randomly allocated to one of four experimental groups. Experimental group 1 was not subjected to antibiotic treatment and did not receive LMG P-27481; experimental groups 2, 3, and 4 received a broad-spectrum antibiotic (cefoperazone 0.5 mg/ml) in drinking water for 10 days. Two days after the antibiotic was discontinued mice received an intragastric suspension of C. difficile VPI strain 10463 (105 CFUs) grown in BHI broth in anaerobic conditions. Experimental group 2 received only the C. difficile challenge without LMG P-27481 treatment. Experimental group 3 was administered an intragastric suspension of L. reuteri LMG P-27481 (109 CFU/day) 2 days before the antibiotic administration, which lasted for 9 days. Experimental group 4 received L. reuteri LMG P-27481 (109 CFU/day) beginning on day 1 after C. difficile challenge, that continued for 4 days (Figure 1).
FIGURE 1.
The experimental protocol of L. reuteri LMG P-27481 treatment of C. difficile colitis in mice.
Body weight was monitored daily until the animals were sacrificed 5 days after the C. difficile challenge. The caecum content was collected to assess the presence of C. difficile toxins, diluted 1:1 (vol/weight) with sterile PBS, and centrifuged at 10,000 × g for 5 min. The clear supernatant was then sterile filtered (0.2 μm) and serially diluted on Vero cell monolayers to determine the cytotoxin titer, which was defined as the highest dilution needed to cause >50% rounding of all cells. C. difficile colonization was evaluated by seeding aliquots of caecal content on C. difficile selective Agar (CDSA) plates (Becton Dickinson) incubated at 37°C in anaerobiosis. In addition, total DNA was extracted from 200 mg of caecal content using QIAmp DNA Stool mini extraction kit (Qiagen), and purified DNA was used as a template for qPCR using C. difficile 16S specific primers (Truong et al., 2017). Data were quantified using the ΔΔCt method and 16S rDNA as the reference gene. Tissue specimens from the colon of the mice were collected immediately after they were sacrificed and either fixed in 4% buffered formalin or snap frozen in liquid nitrogen. After the tissues had been kept in formalin for 24 h, they were paraffin embedded. At that time, 6 μm thick sections were cut, subjected to standard hematoxylin & eosin staining and scored for inflammatory damage by a pathologist (A.P.) (Castagliuolo et al., 1994). The frozen specimens were used to quantify myeloperoxidase activity (MPO) as a marker of neutrophil infiltration, as described elsewhere (Castagliuolo et al., 2005). All the experimental protocols were approved by the Animal Care and Use Ethics Committee of the University of Padua under license from the Italian Ministry of Health, and they were compliant with national and European guidelines for handling experimental animals and using them in experiments.
Statistical Analysis
Data are reported as mean ± standard error of the mean (SEM) or mean ± standard deviation (SD). Statistical analysis was performed using GraphPad Prism Software 6.0 (GraphPad Software Inc., La Jolla, CA, United States). Student’s t-test was used to compare two independent groups; One-way ANOVA was used to compare more than two experimental group. p < 0.05 values were considered statistically significant.
Results
Identification and Probiotic Features of L. reuteri LMG P-27481
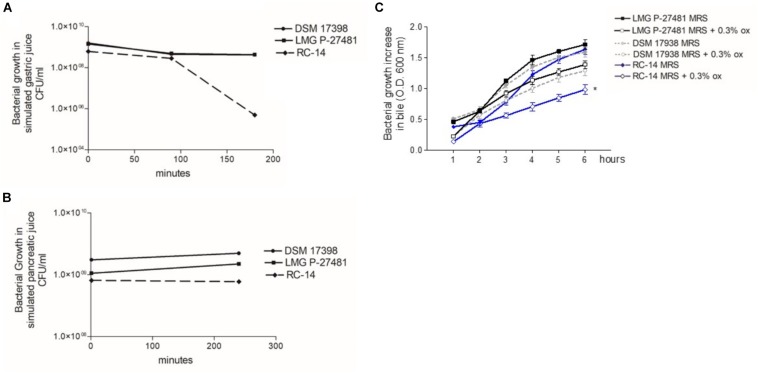
The LMG P-27481 strain was selected from other human isolates belonging to the Lactobacillus reuteri species in view of its susceptibility to antibiotics according to the breakpoint values for L. reuteri suggested by EFSA’s Scientific Opinion for 2018 (EFSA Scientific Opinion, 2018). L. reuteri species can be found, in fact, in the list of qualified presumption of safety (QPS) species, and our investigations using phenotypic and genotypic methods have confirmed the safety of LMG P-27481 strain. The strain was compared to other well-known, commercially available probiotic strains, such as L. reuteri DSM 17938, RC-14 and L. rhamnosus ATCC 53103 during our assessment and convincingly resulted a new potential probiotic product. We found that strain LMG P-27481 was able to survive exposure to the pH of the simulated gastric juice; it was comparable to DSM 17938 and higher than RC-14 strain (Figure 2A). Moreover, all the tested strains showed a marked resistance to exposure to simulated pancreatic juice (Figure 2B). After 6 h of incubation at 37°C, the presence of bile in the culture medium caused a comparable reduction in optical density in strains DSM 17938 and LMG P-27481; RC-14 strain showed instead a greater susceptibility (Figure 2C). LMG P-27481’s ability to produce oxygen peroxide placed it an intermediate level of the reference strains. In fact, 2.58 μg/109 CFU was detected after 1-h of incubation and 3.96 μg/109 CFU after 24 h (Figure 3A). The data obtained during the course of two different tests to determine the presence of reuterin in the culture medium were in agreement with previous studies uncovering reuterin release by DSM 17938 (positive control) and RC-14 (negative control) strains and the absence of reuterin in LMG P-27481 culture medium (data not shown).
FIGURE 2.
Lactobacillus reuteri LMG P-27481’s tolerance to in vitro gut transit gastric acid (A), pancreatic juice (B) and bile (C) exposure. (A) The probiotic’s growth, expressed in CFU/ml, following 0, 90, and 180 min of exposure to simulated gastric juice (n = 3 replicates for each strain). (B) The probiotic’s growth, expressed in CFU/ml, following 0 and 240 min of exposure to simulated pancreatic juice (n = 3 replicates for each strain). (C) Growth increase of probiotics, expressed as optical density OD 600 nm, following exposure for 6 h to bile salts. There are two curves for each strain tested: the reference growth standard on conventional laboratory medium MRS and the test curve reflecting the same medium supplemented with 0.3% vol/vol Oxgall.
FIGURE 3.
Lactobacillus reuteri LMG P-27481 hydrogen peroxide release and adherence to human HT-29 monolayers. (A) Production of H2O2 by probiotics expressed as μg/ml of metabolite released by 109 viable CFUs after 24 h of incubation. (B) Antibiotic resistance image obtained by CARD showed the presence of nonperfect and strict and 111 loose hits. (C) Confluent HT-29 monolayers were washed and incubated with the probiotic strain (MOI 1:10) for 1 h at 37°C. To quantify adhering bacterial cells, the monolayers were extensively washed, lysed and aliquots seeded on MRS agar plates. Following 48 h of incubation in anaerobiosis, the colonies were enumerated and adhering bacteria expressed as the percentage of the total initial population of viable probiotic cells (n = 3 different experiments in duplicate). Data are presented as mean ± SD.
Genome Sequencing: The Safety and Functional Profile of the LMG P-27481 Strain
As reported by Saulnier et al. (2011) genome analysis of L. reuteri LMG P-27481 strain revealed features that are compatible with those of other L. reuteri strains. The average genome size for the L. reuteri species is approximately 2.0 Mbp with a total average number of 1.600 genes, and G+C content of 39%. Genome sequencing has been deposited and is available (WHOJ00000000 release public date 2020-02-03). The “unique” genes of L. reuteri LMG P-27481 are listed in Table 2. With regard to resistome detection, 15 putative antibiotic resistance genes with enzymatic function and 13 putative antibiotic resistance genes with efflux/transport function were identified. Graphical representation of antibiotic resistances obtained by CARD indicated the presence of 111 loose with the absence of perfect or strict hits underlying the safety of the L. reuteri LMG P-27481 (Figure 3B). Most of the sequences identified belonged to ß-lactamase enzymes, but those gene(s) in L. reuteri were considered non-transferable by several authors because of their chromosomal location. No other antibiotic resistance mechanisms were identified. L. reuteri LMG P-27481 MIC values are listed in Table 3. During the investigation for possible virulence genes, 38 sequences were sorted out by bioinformatic tools as predictors of virulence factors. However, based on strict BLASTP standard (90% identity cutoff) none of the sequences raised from the VFDB analysis were considered as significant reporting identity at most of 70.4% (Li et al., 2019). The investigation confirmed that the LMG P-27481 strain was safe given the absence of significant sequence identity with any well-known virulence genes. No S-layer putative proteins were found in the L. reuteri LMG P-27481 genome. We performed comparative genomics between the LMG P-27481 strain and 18 other L. reuteri genomes available in public databases (Table 4). The analysis uncovered 188 unique sequences for strain LMG P-27481; 81 of these were classified as hypothetical proteins of unknown function. Most of the remaining putative genes, such as mucus-binding proteins, EPS biosynthesis and cell wall anchor domains, seemed to be involved in cell surface design and therefore in the interaction with the host.
TABLE 2.
“Unique” genes of L. reuteri LMG P-27481.
| LMG_P-27481 ORFs | LMG_P-27481 annotation |
| LMG_P-27481_0045 | Hypothetical protein |
| LMG_P-27481_0048 | Hypothetical protein |
| LMG_P-27481_0205 | Hypothetical protein |
| LMG_P-27481_0214 | Type III restriction protein, res subunit |
| LMG_P-27481_0286 | YSIRK signal domain/LPXTG anchor domain surface protein |
| LMG_P-27481_0293 | Cytochrome C554 |
| LMG_P-27481_0387 | Transposase |
| LMG_P-27481_0432 | Acetylornithine deacetylase |
| LMG_P-27481_0511 | Hypothetical protein |
| LMG_P-27481_0514 | Hypothetical protein |
| LMG_P-27481_0520 | Cro/C1-type HTH DNA-binding domain-containing protein |
| LMG_P-27481_0523,LMG_P-27481_0840 | Hypothetical protein |
| LMG_P-27481_0583 | Oxalyl-CoA decarboxylase |
| LMG_P-27481_0650 | Hypothetical protein |
| LMG_P-27481_0666 | Competence protein CoiA |
| LMG_P-27481_0754 | Hypothetical protein |
| LMG_P-27481_0846 | Integrase |
| LMG_P-27481_0849 | Hypothetical protein |
| LMG_P-27481_0850 | Hypothetical protein |
| LMG_P-27481_0851 | Alpha-amylase |
| LMG_P-27481_0852 | Hypothetical protein |
| LMG_P-27481_0853 | Hypothetical protein |
| LMG_P-27481_0854 | Hypothetical protein |
| LMG_P-27481_0856 | Glycosyl hydrolase family 25 |
| LMG_P-27481_0869 | Peptidoglycan-binding protein LysM |
| LMG_P-27481_0908 | Hypothetical protein |
| LMG_P-27481_0909 | Hypothetical protein |
| LMG_P-27481_0910 | PHAGE protein, ATPase |
| LMG_P-27481_0914 | Cro/Cl family transcriptional regulator |
| LMG_P-27481_0915 | Hypothetical protein |
| LMG_P-27481_0919 | Hypothetical protein |
| LMG_P-27481_0920 | Hypothetical protein |
| LMG_P-27481_0921 | Hypothetical protein |
| LMG_P-27481_0922 | Hypothetical protein |
| LMG_P-27481_0923 | Hypothetical protein |
| LMG_P-27481_0924 | Hypothetical protein |
| LMG_P-27481_0927 | Hypothetical protein |
| LMG_P-27481_0928 | Phage protein |
| LMG_P-27481_0936 | Hypothetical protein |
| LMG_P-27481_0937 | DNA replication protein |
| LMG_P-27481_0938 | Hypothetical protein |
| LMG_P-27481_0942 | Hypothetical protein |
| LMG_P-27481_0943 | Hypothetical protein |
| LMG_P-27481_0944 | Chorismate synthase |
| LMG_P-27481_0946,LMG_P-27481_1265 | Hypothetical protein |
| LMG_P-27481_0947 | Hypothetical protein |
| LMG_P-27481_0948 | Hypothetical protein |
| LMG_P-27481_0950 | Hypothetical protein |
| LMG_P-27481_0952 | Hypothetical protein |
| LMG_P-27481_0969 | NLP P60 protein |
| LMG_P-27481_0970 | Hypothetical protein |
| LMG_P-27481_0971 | Hypothetical protein |
| LMG_P-27481_0972 | Tail protein |
| LMG_P-27481_0973 | Hypothetical protein |
| LMG_P-27481_0974 | Hypothetical protein |
| LMG_P-27481_0975 | Hypothetical protein |
| LMG_P-27481_0976 | Hypothetical protein |
| LMG_P-27481_1014 | Hypothetical protein |
| LMG_P-27481_1098 | Cell wall anchor |
| LMG_P-27481_1185 | Hypothetical protein |
| LMG_P-27481_1187,LMG_P-27481_2156 | N-acetylmuramoyl-L-alanine amidase |
| LMG_P-27481_1193 | Hypothetical protein |
| LMG_P-27481_1194 | Polysaccharide biosynthesis family protein |
| LMG_P-27481_1195 | Exopolysaccharide biosynthesis protein |
| LMG_P-27481_1196 | Transposase |
| LMG_P-27481_1197 | O-antigen polysaccharide polymerase Wzy |
| LMG_P-27481_1239 | PTS sugar transporter subunit IIA |
| LMG_P-27481_1249 | Chromosome segregation ATPase, partial |
| LMG_P-27481_1250 | NLP/P60 protein |
| LMG_P-27481_1251 | Phage tail protein |
| LMG_P-27481_1252 | Tape measure protein |
| LMG_P-27481_1253 | Hypothetical protein |
| LMG_P-27481_1254 | Small major structural protein |
| LMG_P-27481_1255 | Tail component protein |
| LMG_P-27481_1256 | Tail component protein |
| LMG_P-27481_1257 | Phage head-tail adaptor |
| LMG_P-27481_1297 | Hypothetical protein |
| LMG_P-27481_1304 | Hypothetical protein |
| LMG_P-27481_1455 | Hypothetical protein |
| LMG_P-27481_1458 | Hypothetical protein |
| LMG_P-27481_1459 | Hypothetical protein |
| LMG_P-27481_1460 | Hypothetical protein |
| LMG_P-27481_1461 | Hypothetical protein |
| LMG_P-27481_1462 | Acyltransferase |
| LMG_P-27481_1472 | CDP-diacylglycerol diphosphatase |
| LMG_P-27481_1479 | N-acetylmuramoyl-L-alanine amidase |
| LMG_P-27481_1491 | Amino acid permease |
| LMG_P-27481_1525 | Hypothetical protein |
| LMG_P-27481_1526 | Transposase |
| LMG_P-27481_1527 | Uracil-DNA glycosylase |
| LMG_P-27481_1541 | Hypothetical protein |
| LMG_P-27481_1543 | Hypothetical protein |
| LMG_P-27481_1651 | NAD-dependent malic enzyme |
| LMG_P-27481_1671 | ABC transporter related |
| LMG_P-27481_1735 | MFS transporter |
| LMG_P-27481_1744 | Amino acid permease |
| LMG_P-27481_1745 | Decarboxylase |
| LMG_P-27481_1755 | Hypothetical protein |
| LMG_P-27481_1762 | Hypothetical protein |
| LMG_P-27481_1763 | Hypothetical protein |
| LMG_P-27481_1764 | Hypothetical protein |
| LMG_P-27481_1765 | Short-chain dehydrogenase |
| LMG_P-27481_1767 | Acetyltransferase |
| LMG_P-27481_1772 | Hypothetical protein |
| LMG_P-27481_1785 | CsbD family protein |
| LMG_P-27481_1846 | Hypothetical protein |
| LMG_P-27481_1847 | Hypothetical protein |
| LMG_P-27481_1855,LMG_P-27481_2303,LMG_P-27481_1454,LMG_P-27481_2232,LMG_P-27481_2259 | Transposase |
| LMG_P-27481_1871 | Hemolysin |
| LMG_P-27481_2037 | Transcriptional regulator |
| LMG_P-27481_2073,LMG_P-27481_1453 | Integrase, catalytic region |
| LMG_P-27481_2075 | hypothetical protein |
| LMG_P-27481_2077 | integrase |
| LMG_P-27481_2078 | Hypothetical protein |
| LMG_P-27481_2079 | Hypothetical protein |
| LMG_P-27481_2080 | Hypothetical protein |
| LMG_P-27481_2081 | DNA polymerase III |
| LMG_P-27481_2084 | Hypothetical protein |
| LMG_P-27481_2085 | Hypothetical protein |
| LMG_P-27481_2086 | Hypothetical protein |
| LMG_P-27481_2087 | Hypothetical protein |
| LMG_P-27481_2088 | Hypothetical protein |
| LMG_P-27481_2089 | XRE family transcriptional regulator |
| LMG_P-27481_2095 | Hypothetical protein |
| LMG_P-27481_2101 | Integrase |
| LMG_P-27481_2102 | Transposase |
| LMG_P-27481_2103 | LysR family transcriptional regulator |
| LMG_P-27481_2104 | 5-methyltetrahydropteroyltriglutamate–homocyste ine methyltransferase |
| LMG_P-27481_2105 | 5,10-methylenetetrahydrofolate reductase |
| LMG_P-27481_2107 | Cell wall anchor protein |
| LMG_P-27481_2108 | Mucus-binding protein |
| LMG_P-27481_2137 | Hypothetical protein |
| LMG_P-27481_2138 | DNA-binding protein |
| LMG_P-27481_2139 | Restriction endonuclease |
| LMG_P-27481_2140 | DNA methylase N-4/N-6 family protein |
| LMG_P-27481_2141 | Hypothetical protein |
| LMG_P-27481_2142 | Hypothetical protein |
| LMG_P-27481_2143 | Hypothetical protein |
| LMG_P-27481_2144 | Hypothetical protein |
| LMG_P-27481_2145 | Hypothetical protein |
| LMG_P-27481_2146 | ATPase para family protein |
| LMG_P-27481_2147 | Phage integrase |
| LMG_P-27481_2161 | MFS transporter, partial |
| LMG_P-27481_2163 | K02314 replicative DNA helicase |
| LMG_P-27481_2165 | hypothetical protein |
| LMG_P-27481_2166 | Guanine permease |
| LMG_P-27481_2167 | Adenine phosphoribosyltransferase |
| LMG_P-27481_2168 | Hypothetical protein |
| LMG_P-27481_2173 | Glycerol-3-phosphate transporter |
| LMG_P-27481_2186 | Hypothetical protein |
| LMG_P-27481_2187 | Restriction endonuclease |
| LMG_P-27481_2189,LMG_P-27481_2317,LMG_P-27481_0403,LMG_P-27481_2213,LMG_P-27481_0570,LMG_P-27481_0092,LMG_P-27481_0775,LMG_P-27481_0839,LMG_P-27481_2016,LMG_P-27481_2134,LMG_P-27481_1225,LMG_P-27481_2282,LMG_P-27481_1076,LMG_P-27481_1045,LMG_P-27481_1996,LMG_P-27481_0115,LMG_P-27481_1661,LMG_P-27481_0752,LMG_P-27481_0105,LMG_P-27481_2185,LMG_P-27481_0344,LMG_P-27481_0170,LMG_P-27481_0119,LMG_P-27481_0522,LMG_P- | |
| 27481_1775,LMG_P-27481_1107,LMG_P-27481_2274,LMG_P-27481_1908,LMG_P-27481_1770,LMG_P-27481_0532,LMG_P-27481_2288,LMG_P-27481_1190,LMG_P-27481_1863,LMG_P-27481_1243,LMG_P-27481_2300,LMG_P-27481_1487,LMG_P-27481_1710,LMG_P-27481_0409,LMG_P-27481_2319,LMG_P-27481_0645,LMG_P-27481_0196 | Hypothetical protein |
| LMG_P-27481_2195 | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridin e pyrophosphokinase |
| LMG_P-27481_2198 | Bifunctional folylpolyglutamate synthase/dihydrofolate synthase |
| LMG_P-27481_2204 | Phage integrase |
| LMG_P-27481_2206 | Hypothetical protein |
| LMG_P-27481_2208 | Glycerol-3-phosphate transporter |
| LMG_P-27481_2212 | AI-2E family transporter |
| LMG_P-27481_2222 | 2,5-diketo-D-gluconic acid reductase |
| LMG_P-27481_2223 | Restriction endonuclease |
| LMG_P-27481_2225,LMG_P-27481_2159 | Restriction endonuclease |
| LMG_P-27481_2228 | Multidrug ABC transporter permease |
| LMG_P-27481_2239,LMG_P-27481_2183 | YSIRK signal domain/LPXTG anchor domain surface protein |
| LMG_P-27481_2240 | Acetylornithine deacetylase |
| LMG_P-27481_2242 | Glycerol-3-phosphate transporter |
| LMG_P-27481_2243,LMG_P-27481_2162 | hypothetical protein |
| LMG_P-27481_2244,LMG_P-27481_2205 | K02314 replicative DNA helicase |
| LMG_P-27481_2245 | Phospholipase |
| LMG_P-27481_2248 | MazF family toxin-antitoxin system |
| LMG_P-27481_2250 | 3-beta-hydroxysteroid dehydrogenase |
| LMG_P-27481_2253 | Glycerol-3-phosphate transporter |
| LMG_P-27481_2257 | Ribonuclease HII |
| LMG_P-27481_2272 | Bacterial lipoprotein |
| LMG_P-27481_2276 | Integrase |
| LMG_P-27481_2278 | Carbamoyl-phosphate synthase large chain |
| LMG_P-27481_2286 | Guanine permease |
| LMG_P-27481_2287 | Hypothetical protein |
| LMG_P-27481_2292 | DNA polymerase III subunit gamma/tau |
| LMG_P-27481_2293 | Two-component system sensor histidine kinase |
| LMG_P-27481_2296 | Farnesyl pyrophosphate synthetase |
| LMG_P-27481_2297 | Non-canonical purine NTP pyrophosphatase |
| LMG_P-27481_2305 | Glutamine amidotransferase |
| LMG_P-27481_2307 | 30S ribosomal protein S4 |
| LMG_P-27481_2313 | UDP-glucose 4-epimerase |
| LMG_P-27481_2315 | Asparagine–tRNA ligase |
| LMG_P-27481_2325 | Transposase |
| LMG_P-27481_2327 | Peptide ABC transporter substrate-binding protein |
| LMG_P-27481_2338 | Transposase |
TABLE 3.
Lactobacillus reuteri LMG P-27481 Minimum inhibitory concentration values.
| Antibiotic | L. reuteri LMG P-27481 (μg/ml) | EFSA cut-off 2018 for L. reuteri (μg/ml) |
| Gentamycin | 4 | 16 |
| Kanamycin | 64 | 64 |
| Streptomycin | 32 | 64 |
| Tetracycline | 16 | 32 |
| Erythromycin | 0,5 | 1 |
| Clindamycin | 0,063 | 4 |
| Chloramphenicol | 4 | 4 |
| Ampicillin | 1 | 2 |
| Vancomycin | >128 | n.r. |
| Neomycin | 4 | / |
| Virginiamycin | 1 | / |
| Ciprofloxacin | 64 | / |
| Linezolid | 4 | / |
| Rifampicin | 0,125 | / |
| Trimethoprim | 64 | / |
n.r. not requested, / not reported.
TABLE 4.
Comparative genomics analyses performed with 18 L. reuteri publicly available strains. The percentages of identity with L. reuteri LMG P-27481 are reported.
| L. reuteri ATCC53608 | L. reuteri JCM1112 | L. reuteri LTH5448 | L. reuteri I5007 | L. reuteri ZLR003 | L. reuteri TMW1.656 | L. reuteri TD1 | L. reuteri MM2-3 | L. reuteri DSM20016 | L. reuteri mlc3 | L. reuteri 100-23 | L. reuteri TMW1.112 | L. reuteri lpuph | L. reuteri IRT | L. reuteri LTH2584 | L. reuteri SD2112 | L. reuteri MM4-1A | L. reuteri CF48-3A | |
| L. reuteri LMG P-27481 | 96,67 | 98,88 | 96,65 | 96,76 | 96,69 | 96,2 | 96,53 | 98,86 | 98,86 | 95,68 | 96,35 | 96,14 | 96,53 | 98,92 | 96,18 | 96,33 | 98,87 | 96,12 |
L. reuteri LMG P-27481 Efficiently Adheres to Human Intestinal Epithelial Cells
In terms of adhesive capacity to the HT-29 cell line, a high percentage of viable cells of the reference strains adhered to human enterocytes although with a slightly different aptitude (Figure 3C). As expected, the L. rhamnosus ATCC 53103 and DSM 17938 reference strains showed the highest robust adhesion ability (79 and 78% on average, respectively); L. paracasei ATCC 334 was the least effective (adhesion ranging between 67 and 71%). Indeed, L. reuteri strain LMG P-27481 adhered efficiently to human intestinal epithelial cells since it was only slightly less adhesive (73%) with respect to the most efficient strains.
L. reuteri LMG P-27481 Induces a Distinctive Cytokine Profile in Human DC
We exposed human immature DCs obtained from peripheral blood monocytes to viable logarithmic-phase lactobacilli and quantified the cytokines in the culture medium to determine the effect of our strain on DC phenotype. After 1 h of incubation with either L. reuteri LMG P-27481, L. reuteri DSM 17938 or L. rhamnosus ATCC 53103, the cells were washed thoroughly, the medium was changed to one containing antibiotic and 23 h later the cytokine released was quantified by ELISA. The three lactobacilli tested induced IL-10 and IL-12p70 release, although with a different profile (Figures 4A,B). Both L. reuteri strains were more effective than L. rhamnosus ATCC 53103 in stimulating cytokine secretion. Moreover, L. reuteri LMG P-27481 appeared more efficient than L. reuteri DSM 17938 in simulating IL-10 secretion. The anti-inflammatory index, calculated as the IL-10/IL-12p70 ratio, revealed that viable L. reuteri LMG P-27481 was the most effective strain in inducing an anti-inflammatory response in the DCs (Figure 4C). The same experiments were also conducted after the DCs were challenged with Salmonella. Overall, all the probiotic strains were significantly less efficient than Salmonella in inducing cytokine release (data not shown).
FIGURE 4.
Lactobacillus reuteri LMG P-27481 induces an anti-inflammatory cytokine profile in human dendritic cells. The DCs obtained from differentiated peripheral blood monocytes were incubated (1:10 ratio) with the reported live bacterial strains for 1 h in medium without antibiotics. The cells were then collected, washed by centrifugation and incubated for an additional 23-h period in complete medium with antibiotics. Culture supernatants were collected. (A) IL-10 content quantified by ELISA. (B) IL-12p70 concentration assessed by ELISA. (C) Shows the anti-inflammatory index calculated as the ratio between IL-10 and IL-12p70 listed in Panels (A,B). Data are reported as mean ± SD on values obtained on 3 different donors. *P < 0.05 versus control and °P < 0.05 versus LMG P-27481.
L. reuteri LMG P-27481 Efficiently Metabolizes Lactose
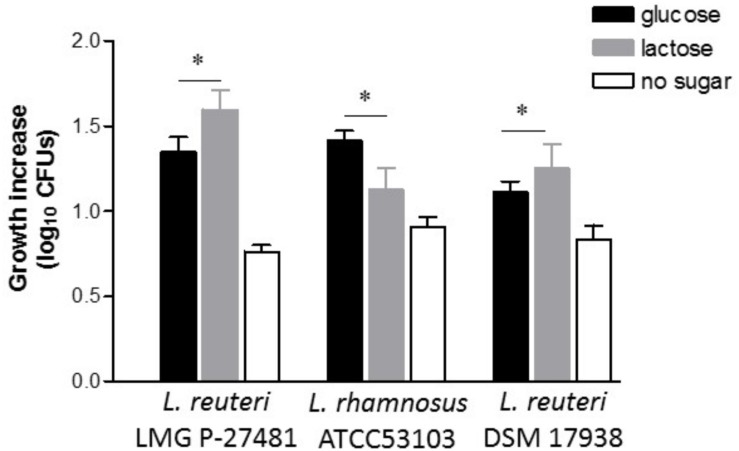
The ability to counteract or at least mitigate the negative effects of various etiological agents of a chemical and/or microbial origin that are able to cause diarrhea is a highly desirable feature for a probiotic strain. Since lactose intolerance is one of the most common causes of diarrhea, probiotic strains were tested for their ability to hydrolyze lactose and thus, potentially, to limit the symptoms associated with lactose intolerance. Two well-known probiotic strains, L. reuteri DSM 17938 and L. rhamnosus ATCC 53103, were selected as our reference of comparison with the LMG P-27481 strain tested.
Bacterial growth in the presence of glucose or lactose was compared. L. reuteri DSM 17938 seemed more active in the presence of lactose in the growth medium, indicating its ability to vigorously metabolize this carbon source. L. rhamnosus ATCC 53103 was instead clearly more active in presence of glucose with respect to lactose in the culture medium (Figure 5). The LMG P-27481 strain produced in the end the most significant growth in the presence of lactose with respect to the other strains (Figure 5). All strains tested showed a negligible growth in MRS broth without sugars (Figure 5).
FIGURE 5.
Lactobacillus reuteri LMG P-27481 preferentially utilizes lactose as a carbon source. The effect of the presence of only glucose (black bars), only lactose (gray bars) or no sugars (open bars) as carbon sources, on growth performance of L. reuteri LMG P-27481, L. reuteri DSM 17938 or L. rhamnosus ATCC 53103, was calculated as growth increase in log10 CFUs. Bacteria were incubated for 18 h with 1% of each type of sugar and growth was recorded by decimal counts, (n = 3 replicates for each condition). Data are presented as mean ± SD. * indicates P < 0.05 versus growth in MRS containing glucose.
L. reuteri LMG P-27481 Antagonizes Pathogen Growth in vitro
Intestinal pathogens, such as E. coli, Salmonella and C. difficile, were inoculated into supernatants of the probiotic strains tested and the inhibitory effects were verified. Figure 6 illustrates the growth patterns of the pathogens 6, 12, and 24 h following incubation in the probiotic supernatants. Although the probiotics tested were unable to significantly affect the growth of Salmonella, they all, however, slightly reduced pathogen growth as early as 6 h after incubation (0.2–0.3 log10 CFUs reduction compared to the control). L. reuteri DSM 17938 was the most effective in reducing Salmonella growth 24 h after incubation. Similarly, both L. reuteri strains hindered E. coli growth 6 h after incubation, and growth inhibition was maintained for the 24 h following co-incubation. L. rhamnosus, instead, did not modify the pathogen’s growth pattern. More interestingly, L. reuteri LMG P-27481 significantly inhibited C. difficile growth 24 h after incubation as the pathogen growth was reduced by more than 0.5 log10 CFUs. All the other strains produced a growth increase similar to that in the controls.
FIGURE 6.
Lactobacillus reuteri LMG P-27481 inhibits pathogen growth in vitro. MRS broth was inoculated with 107 CFU/ml of the strain and incubated in anerobiosis for 24 h. MRS conditioned broths were sterile filtered, pH adjusted to 7 and inoculated at 37°C with either E. coli (107 CFU/ml) (A), Salmonella (107 CFU/ml) (B) or C. difficile (107 CFU/ml) (C). Aliquots of culture were collected after 6, 12 or 24 h and live bacteria quantified by seeding on proper Agar medium incubated at 37°C in aerobiosis for E. coli, Salmonella and anaerobiosis for C. difficile. Data are reported as mean ± SD on values obtained on three different determinations. *P < 0.05 versus not treated pathogen.
L. reuteri LMG P-27481 Antagonizes Rotavirus in the in vitro Infection Model
The probiotics were then tested to verify their ability to protect human gut cells from Rotavirus infection. Two different experimental designs were used to mimic the probiotics’ potential protective action: in the first case by pre-treating HT-29 cells monolayers with probiotics before exposure to Rotavirus, and in the second by co-incubating probiotics and Rotavirus with human enterocytes. All the strains tested showed inhibitory effects against Rotavirus infection in vitro. The L. rhamnosus ATCC 53103 showed the highest inhibitory activity in the pre-treatment and competition protocols (86 and 91%, respectively) The L. reuteri DSM 17938 (50 and 89%, respectively) and L. reuteri LMG P-27481 (66 and 79%, respectively) also significantly reduced the copies of Rotavirus produced following the infection of the human intestinal cells (Table 5).
TABLE 5.
The average number of copies of viral genome in the presence of the probiotics tested during the study (raw data expressed as DNA copies/5 × 105 cells ± SEM) in the two experimental designs.
| Probiotic | N. copies viral genome |
|
| Pre-treatment | Competition | |
| Rotavirus alone | 4371 ± 660 × 103 | 4630 ± 720 × 103 |
| L. reuteri DSM 17938 | 2198 ± 450 × 103* | 531 ± 85 × 103* |
| L. reuteri LMG P-27481 | 1528 ± 390 × 103* | 1012 ± 410 × 103* |
| L. rhamnosus ATCC 53103 | 627 ± 90 × 103* | 421 ± 83 × 103* |
*p < 0.05 versus Rotavirus alone.
L. reuteri LMG P-27481 Protects From C. difficile Infection in vivo
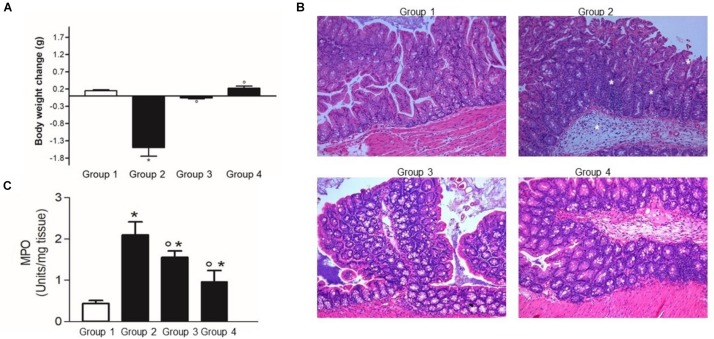
Following the challenge with C. difficile, mice with no probiotic supplementation (group 2) showed a significant body weight loss over the next 5 days (Figure 7A). As expected, at the histological examination the mice challenged with C. difficile showed a significant inflammatory infiltrate with massive edema and loss of epithelial cells in the colonic mucosa (Figure 7B). Indeed, myeloperoxidase (MPO) activity in colonic mucosa, a marker of neutrophils infiltration, was significantly higher in Experimental group 2 with respect to the control mice (Figure 7C).
FIGURE 7.
Lactobacillus reuteri LMG P-27481 reduces the severity of C. difficile colitis in a murine model. The mice were randomly divided in four experimental groups (Figure 1). Five days after intragastric (IG) challenge with C. difficile (105 CFU) the animals were sacrificed, and the colon content collected. (A) Body weight was measured at baseline (IG challenge) and every day for the following 5 days. Weight change was calculated as the variation in weight at sacrifice compared with that at the baseline. (B) Full thickness colonic specimens were fixed in 4% formalin, paraffin embedded and 8 μm thick sections stained with H&E. Tissue edema, inflammatory infiltrate and mucosal ulcers were evaluated and indicated with * in the Group 2 image. (C) Thickness colonic specimens were homogenized and used to quantify MPO activity as a marker of neutrophils infiltration. Data are reported as mean ± SD on values obtained on six animals for each experimental group. *P < 0.01 versus group 1 and °P < 0.05 versus group 2.
By seeding the caecal content on selective media, C. difficile was isolated from the caecal content of the mice challenged with the pathogen, but not from the control group (data not shown). qRT-PCR uncovered a low ΔΔCt value only in the mice challenged with the pathogen confirming that C. difficile was present in those mice (Figure 8A). Finally, there was a high cytotoxin titer (>50% Vero cells rounding at 10–4 dilution) in the caecal content of the mice challenged with C. difficile (Figure 8B).
FIGURE 8.
Lactobacillus reuteri LMG P-27481 reduces colonization in an antibiotic-induced C. difficile murine model. The mice were randomly divided into four experimental groups (Figure 1). Five days after intragastric (IG) challenge with C. difficile (105 CFU) the animals were sacrificed, and the caecum content collected. (A) Total DNA was extracted, and C. difficile specific DNA was quantified by qPCR. The data were quantified by the ΔΔCt method using 16S rDNA as the reference gene. (B) The caecal content was centrifuged, the clear supernatant sterile filtered and added to Vero cells monolayers to determine cytotoxicity. (C) Serial 1:2 dilutions of caecal content were added to Vero cells monolayers; the titer of C. difficile toxins was defined as the highest dilution causing >50% rounding 24 h after incubation. Data are reported as mean ± SD on values obtained on six animals for each experimental group. *P < 0.05 versus group 2.
The L. reuteri LMG P-27481 supplementation effectively reduced C. difficile colonization and colitis (Figures 7, 8), and it significantly diminished C. difficile induced body weight loss following pathogen challenge (Figure 7A). Moreover, histologic analysis and quantification of MPO activity demonstrated that it significantly reduced the inflammatory damage of the colonic mucosa (Figures 7B,C). Likewise, the amount of C. difficile DNA as well as toxins in the caecal content was significantly diminished in Experimental Groups 3 and 4 with respect to Group 2 (Figures 8A–C).
Discussion
Lactobacilli are important members of the gut microbiota community that are able to influence host health through a variety of mechanisms (Oh et al., 2010). L. reuteri, a separate species within the Lactobacillus species, was chosen for isolation because it has been shown to exhibit probiotic efficacy and to confer broad-spectrum protection against disease in humans (Casas and Dobrogosz, 2000). Beyond its ability to survive the hostile conditions of the gastrointestinal environment including the high pH level of gastric juices and to tolerate bile salts, it shows resistance to antibiotics. It is, in addition, able to metabolize lactose, meaning that it can potentially alleviate the symptoms of intolerant subjects. Like some control strains (L. reuteri DSM 17938 and L. rhamnosus ATCC 53103), L. reuteri LMG P-27481 has been shown to produce a marked inhibitory effect on several pathogens (E. coli, Salmonella and Rotavirus) in different in vitro models. But as opposed to the control strains, L. reuteri LMG P-27481 has also been shown to diminish C. difficile growth in vitro and to significantly reduce C. difficile colonization and inflammatory colitis in mice.
Its safety, effectiveness, and genetic stability were investigated by means of genome sequencing, which uncovered the presence of two amino acid sequences similar to the mucus-binding MUB domains as well as a sequence coding for the typical N-terminal signal peptide (YSIRK motif) targeting the protein for secretion, and a C-terminal sortase recognition site (LPxTG), targeting the protein for covalent attachment to the peptidoglycan layer on the outside of the bacterial cell (Sengupta et al., 2013). As described by Etzold et al. (2014) and Bath et al. (2005), our experiments showed that the strain carried five sequences with a significant homology to mucus-binding proteins, cell wall anchor domains, and signal peptides characterizing the surface-exposed adhesins from L. reuteri. Moreover, the presence of putative EPS-coding sequences, which are known to interfere with the adhesion of enteropathogenic E. coli (EPEC) to pig erythrocytes, was also confirmed in the LMG P-27481 genome (Chen et al., 2014). L. reuteri is, in fact, one of the lactobacilli species known to produce EPS, whose chemical features seem to depend on the type of sugar substrate metabolized by the strain. Wang et al. (2010) demonstrated that EPS from L. reuteri was able to interfere with the in vitro adhesion ability of EPEC to pig erythrocytes (hemagglutination). Peptidoglycan hydrolases and EPS production could enhance the strain’s potential to exert antimicrobial activities against gut pathogens. Our experiments confirmed the safety of the LMG P-27481 strain because they demonstrated that sequences of well-known virulence genes were absent. The unforeseen finding regarding the anti-inflammatory potential of the LMG P-27481 strain that emerged here was the higher stimulation of IL-10 cytokine with respect to IL-12p70, as demonstrated by the resulting anti–inflammatory index (IL-10/IL-12 ratio). The differences in the expression of key markers of DC activation and polarization for the L. reuteri DSM 17938 and LMG P-27481 and L. rhamnosus ATCC 53103 strains may reflect putative synergy between strains possessing complementary beneficial features (Mileti et al., 2009). That synergy, or putative antagonism, should be carefully tested using a scientific approach, for example, by comparing the efficacy of single pure strains versus blended ones in treating diarrhea caused by different physio-pathological mechanisms.
Lactobacillus reuteri LMG P-27481 significantly reduced C. difficile growth in vitro, and, following oral challenge in mice, it reduced colonization of pathogen and colitis in vivo. In the authors’ opinion, the probiotic’s beneficial effects in the C. difficile colitis setting depended on the strain’s anti-microbial as well as anti-inflammatory action. It is well known that all stress or inflammatory mechanisms have a detrimental effect on gut barrier integrity, and other exacerbations induced by cyclic mechanisms disrupt the gut barrier function (Marjoram et al., 2015; Gil-Cardoso et al., 2016; Cui et al., 2017). The probiotic’s ability to induce anti-inflammatory effects in dendritic cells may have contributed to reducing inflammatory mucosal damage given the active role of these cells during C. difficile infection (Ausiello et al., 2006; Lee et al., 2009).
The inhibitory effects noted during our in vitro assays demonstrated that LMG P-27481 supplementation was able to significantly reduce in vivo C. difficile intestinal colonization. The reduced cytotoxic load in the colon lumen observed in the LMG P-27481-supplemented mice could have been the direct result of the probiotic’s inhibitory action against the pathogen. Since C. difficile toxins mediate colonic inflammatory damage (Abt et al., 2016), the reduced toxic load could have ameliorated the clinical outcome.
The mechanisms underlying the in vivo efficacy of L. reuteri LMG P-27481 observed require further investigation, but some observations can be made with regard to our genome analysis. The study, in fact, uncovered some distinctive genetic features associated to the presence of a unique pool of traits with respect to other L. reuteri strains whose genomes are publicly available. L. reuteri LMG P-27481 can adhere to and interact with gut epithelial cells and host cells and enhance epithelial barrier function; it also has putative but reliable beneficial effects on impaired gut mucosal functions. Moreover, peptidoglycan hydrolases and EPS production could further support the strain’s antimicrobial activities against gut pathogens.
Conclusion
Taken together, the results of our work characterizing a new probiotic lactobacillus lead to the conclusion that L. reuteri LMG P-27481 represents a promising strain by virtue of its ability to adhere to human enterocytes and to stimulate anti-inflammatory cytokine secretion. After an initial assessment of the probiotic’s unique properties carried out following conventional protocols and international guidelines, the strain was subjected to a battery of assays leading to target-selection and characterization. The probiotic not only showed conventional probiotic features, which are essential for its successful activity, but also the ability to counteract the growth and colonization in vitro and in vivo of C. difficile. Further studies are warranted given the strain’s efficacy even in those cases in which it is administrated in association with antibiotic therapy. Its clinical application to treat C. difficile infection and forms of diarrhea caused by other conditions (i.e. lactose intolerance) also warrant further investigation.
Data Availability Statement
The datasets generated for this study can be found in the WHOJ00000000.
Ethics Statement
The strain involving human subjects was isolated following the written informed consent of the parents. Approval by an Ethical Committee was not required by local legislation at the time the strain was isolated. The animal study was carried out in accordance with the principles of the Basel Declaration and the recommendations of the Animal Care and Use Ethics Committee of the University of Padua under license from the Italian Ministry of Health; they were in compliance with national and European guidelines for handling and use of experimental animals. Experimental protocols were approved by the Animal Care and Use Ethics Committee of the University of Padua under license from the Italian Ministry of Health.
Author Contributions
VS, FU, GB, AP, PB, IC, and ME performed the experiments. LB, LR, GM, PB, IC, and ME designed and interpreted the experiments. VS, FU, IC, and ME wrote the manuscript. LM critically reviewed the manuscript.
Conflict of Interest
LB was employed by the Moviscom S.r.l. company that funded the study and owns the strain. GM and LR are employed by the Nòos S.r.l., the company that hold the license for the strain’s commercialization. ME, FU, and VS are employed by the AAT S.r.l. company, the contract laboratory that performed the assays. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Linda Inverso for reading and checking the language of the manuscript.
Footnotes
Funding. The authors declare that this study received funding from the Moviscom Srl. The funder had the following involvement with the study: ownership of the strain isolated.
References
- Abt M. C., McKenney P. T., Pamer E. G. (2016). Clostridium difficile colitis: pathogenesis and host defence. Nat. Rev. Microbiol. 14 609–620. 10.1038/nrmicro.2016.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D., McKay L. L. (1983). Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46 549–552. 10.1128/aem.46.3.549-552.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J. W., Simpson J. B., Roach J., Bruno-Barcena J. M., Azcarate-Peril M. A. (2018). Prebiotics for lactose intolerance: variability in galacto-oligosaccharide utilization by intestinal Lactobacillus rhamnosus. Nutrients 10:1517. 10.3390/nu10101517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausiello C. M., Cerquetti M., Fedele G., Spensieri F., Palazzo R., Nasso M., et al. (2006). Surface layer proteins from Clostridium difficile induce inflammatory and regulatory cytokines in human monocytes and dendritic cells. Microbes Infect. 8 2640–2646. 10.1016/j.micinf.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Bath K., Roos S., Wall T., Jonsson H. (2005). The cell surface of Lactobacillus reuteri ATCC 55730 highlighted by identification of 126 extracellular proteins from the genome sequence. FEMS Microbiol. Lett. 253 75–82. 10.1016/j.femsle.2005.09.042 [DOI] [PubMed] [Google Scholar]
- Cadieux P., Wind A., Sommer P., Schaefer L., Crowley K., Britton R. A., et al. (2008). Evaluation of reuterin production in urogenital probiotic Lactobacillus reuteri RC-14. Appl. Environ. Microbiol. 74 4645–4649. 10.1128/AEM.00139-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurso L. (2019). Thirty years of Lactobacillus rhamnosus GG: a review. J. Clin. Gastroenterol. 53(Suppl. 1) S1–S41. 10.1097/MCG.0000000000001170 [DOI] [PubMed] [Google Scholar]
- Casas I. A., Dobrogosz W. J. (2000). Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microb. Ecol. Health Dis. 12 247–285. 10.1080/08910600050216246-1 [DOI] [Google Scholar]
- Castagliuolo I., Galeazzi F., Ferrari S., Elli M., Tormen D., Brun P., et al. (2005). Beneficial effect of Lactobacillus crispatus on experimentally induced colitis in mice: effect of different phenotypes. FEMS Immunol. Med. Microbiol. 43 197–204. 10.1016/j.femsim.2004.08.011 [DOI] [PubMed] [Google Scholar]
- Castagliuolo I., LaMont J. T., Letourneau R., Kelly C., O’Keane J. C., Jaffer A., et al. (1994). Neuronal involvement in the intestinal effects of Clostridium difficile toxin A and Vibrio cholerae enterotoxin in rat ileum. Gastroenterol 107 657–665. 10.1016/0016-5085(94)90112-0 [DOI] [PubMed] [Google Scholar]
- Charteris W. P., Kelly P. M., Morelli L., Collins J. K. (1998). Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J. Appl. Microbiol. 84 759–768. 10.1046/j.1365-2672.1998.00407.x [DOI] [PubMed] [Google Scholar]
- Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33 D325–D328. 10.1093/nar/gki008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. Y., Woodward A., Zijlstra R. T., Ganzle M. G. (2014). Exopolysaccharides synthesized by Lactobacillus reuteri protect against Enterotoxigenic Escherichia coli in Piglets. Appl. Environ. Microbiol. 80 5752–5760. 10.1128/AEM.01782-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B., Wetter T., Suhai S. (1999). Genome sequence assembly using trace signals and additional sequence information computer science and biology. Proc. German Conference Bioinform. (GCB) 99 45–56 [Google Scholar]
- Cui Y., Liu L., Dou X., Wang C., Zhang W., Gao K., et al. (2017). Lactobacillus reuteri ZJ617 maintains intestinal integrity via regulating tight junction, autophagy and apoptosis in mice challenged with lipopolysaccharide. Oncotarget 24 77489–77499. 10.18632/oncotarget.20536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. C. E., Mau B., Blattner F. R., Perna N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14 1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doleyres Y., Beck P., Vollenweider S., Lacroix C. (2005). Production of 3-hydroxypropionaldehyde using a two-step process with Lactobacillus reuteri. Appl. Microbiol. Biotechnol. 68 467–474. 10.1007/s00253-005-1895-4 [DOI] [PubMed] [Google Scholar]
- EFSA Scientific Opinion (2012). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA Journal 10:2740. [Google Scholar]
- EFSA Scientific Opinion (2018). Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 16:5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzold S., MacKenzie D. A., Jeffers F., Walshaw J., Roos S., Hemmings A. M., et al. (2014). Structural and molecular insights into novel surface-exposed mucus adhesins from Lactobacillus reuteri human strains. Mol. Microbiol. 92 543–556. 10.1111/mmi.12574 [DOI] [PubMed] [Google Scholar]
- FAO/WHO (2002). Joint FAO/WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food. London, Ontario, Canada, April 30 and May 1. Rome: FAO. [Google Scholar]
- Gil-Cardoso K., Ginés I., Pinent M., Ardévol A., Blay M., Terra X. (2016). Effects of flavonoids on intestinal inflammation, barrier integrity and changes in gut microbiota during diet-induced obesity. Nutr. Res. Rev. 29 234–248. 10.1017/s0954422416000159 [DOI] [PubMed] [Google Scholar]
- Gilliland S. E., Staley T. E., Bush L. J. (1984). Importance of bile tolerance of Lactobacillus acidophilus used as dietary adjunct. J. Dairy Sci. 67 3045–3051. 10.3168/jds.s0022-0302(84)81670-7 [DOI] [PubMed] [Google Scholar]
- Guerrero C. A., Zarate S., Corkidi G., Lopez S., Arias C. F. (2000). Biochemical characterization of rotavirus receptors in MA104 cells. J. Virol. 74 9362–9371. 10.1128/jvi.74.20.9362-9371.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Guarner F., Reid G., Gibson G. R., Merenstein D. J., Pot B., et al. (2014). The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11 506–514. 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- Hyatt D., Chen G. L., LoCascio P. F., Land M. L., Larimer F. W., Hauser L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen H., Grimmer S., Naterstad K., Axelsson L. (2012). In vitro testing of commercial and potential probiotic lactic acid bacteria. Int. J. Food Microbiol. 153 216–222. 10.1016/j.ijfoodmicro.2011.11.020 [DOI] [PubMed] [Google Scholar]
- Koboldt D. C., Larson D. E., Wilson R. K. (2013). Using varscan 2 for germline variant calling and somatic mutation detection. Curr. Protoc. Bioinformatics 44 15.4.1–15.4.17. 10.1002/0471250953.bi1504s44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K., Hallin P., Rødland E. A., Stærfeldt H. H., Rognes T., Ussery D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35 3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Kim H., Cha M. Y., Park H. G., Kim Y. J., Kim I. Y., et al. (2009). Clostridium difficile toxin A promotes dendritic cell maturation and chemokine CXCL2 expression through p38, IKK, and the NF-kappaB signaling pathway. J. Mol. Med. (Berl) 87 169–180. 10.1007/s00109-008-0415-2 [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pang B., Wang D., Li J., Xu J., Fang Y., et al. (2019). Expanding dynamics of the virulence-related gene variations in the toxigenic Vibrio cholerae serogroup O1. BMC Genomics 20:360. 10.1186/s12864-019-5725-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25 955–964. 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjoram L., Alvers A., Deerhake M. E., Bagwell J., Mankiewicz J., Cocchiaro J. L., et al. (2015). Epigenetic control of intestinal barrier function and inflammation in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 112 2770–2775. 10.1073/pnas.1424089112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur A. G., Waglechner N., Nizam F., Yan A., Azad M. A., Baylay A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother 57 3348–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland L. V., Evans C. T., Goldstein E. J. C. (2018). Strain-specificity and disease-specificity of probiotic efficacy: a systematic review and meta-analysis. Front. Med. 5:124. 10.3389/fmed.2018.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C., Lugli G. A., Turroni F., Mancabelli L., Duranti S., Viappiani A., et al. (2014). Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol. Ecol. 90 493–503. 10.1111/1574-6941.12410 [DOI] [PubMed] [Google Scholar]
- Mileti E., Matteoli G., Iliev I. D., Rescigno M. (2009). Comparison of the immunomodulatory properties of three probiotic strains of Lactobacilli using complex culture systems: prediction for in vivo efficacy. PLoS One. 4:e7056. 10.1371/journal.pone.0007056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogna L., Del Piano M., Deidda F., Nicola S., Soattini L., Debiaggi R., et al. (2012). Assessment of the in vitro inhibitory activity of specific probiotic bacteria against different Escherichia coli strains. J. Clin. Gastroenterol. 46 S29–S32. 10.1097/MCG.0b013e31826852b7 [DOI] [PubMed] [Google Scholar]
- Mu Q., Tavella V. J., Luo X. M. (2018). Role of Lactobacillus reuteri in human health and diseases. Front. Microbiol. 2018:757 10.3389/fmicb.2018.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh P. L., Benson A. K., Peterson D. A., Patil P. B., Moriyama E. N., Roos S., et al. (2010). Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 4 377–387. 10.1038/ismej.2009.123 [DOI] [PubMed] [Google Scholar]
- Ortiz-Rivera Y., Sánchez-Vega R., Gutiérrez-Méndez N., León-Félix J., Acosta-Muñiz C., Sepulveda D. R. (2017). Production of reuterin in a fermented milk product by Lactobacillus reuteri: inhibition of pathogens, spoilage microorganisms, and lactic acid bacteria. J. Dairy Sci. 100 4258–4268. 10.3168/jds.2016-11534 [DOI] [PubMed] [Google Scholar]
- Ouwehand A. C. (2017). A review of dose-responses of probiotics in human studies. Benef. Microbes 8 143–151. 10.3920/BM2016.0140 [DOI] [PubMed] [Google Scholar]
- Pruitt K. D., Tatusova T., Maglott D. R. (2007). NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35 D61–D65. 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. L., Romanuk T. N. (2012). A meta-analysis of probiotic efficacy for gastrointestinal diseases. PLoS One 7:e34938. 10.1371/journal.pone.0034938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M. H., Reddy V. S., Tamang D. G., Västermark Å. (2014). The transporter classification database. Nucleic Acids Res. 42 D251–D258. 10.1093/nar/gkt1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salari P., Nikfar S., Abdollahi M. (2012). A meta-analysis and systematic review on the effect of probiotics in acute diarrhea. Inflamm. Allergy Drug Targets 11 3–14. 10.2174/187152812798889394 [DOI] [PubMed] [Google Scholar]
- Saulnier D. M., Santos F., Roos S., Mistretta T. A., Spinler J. K., Molenaar D., et al. (2011). Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features. PLoS One 6:e18783. 10.1371/journal.pone.0018783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta R., Altermann E., Anderson R. C., McNabb W. C., Moughan P. J., Roy N. C. (2013). The role of cell surface architecture of Lactobacilli in host-microbe interactions in the gastrointestinal tract. Mediators Inflamm. 2013:237921. 10.1155/2013/237921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewale R. N., Sawale P. D., Khedkar C. D., Singh A. (2014). Selection criteria for probiotics; a review. Int. J. Prob. Preb. 9 17–22. [Google Scholar]
- Siguier P. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 D32–D36. 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R., Kesavelu D., Veligandla K. C., Muni S. K., Mehta S. C. (2018). Lactobacillus reuteri DSM 17938: review of evidence in functional gastrointestinal disorders. Pediatr. Ther. 8:350 10.4172/2161-0665.1000350 [DOI] [Google Scholar]
- Szajewska H., Kolodziej M., Gieruszczak-Bialek D., Skorka A., Ruszczynski M., Shamir R. (2019). Systematic review with meta−analysis: Lactobacillus rhamnosus GG for treating acute gastroenteritis in children – a 2019 update. Aliment. Pharmacol. Ther. 49 1376–1384. 10.1111/apt.15267 [DOI] [PubMed] [Google Scholar]
- Talarico T. L., Casas I. A., Chung T. C., Dobrogoszi W. J. (1988). Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemoter 32 1854–1858. 10.1128/aac.32.12.1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot C. M., Koumpouras C. C., Carlson P. E., Bergin I. L., Aronoff D. M., Young B. V. (2011). Cefoperazone-treated mice as an experimental platform to assess differential virulence of Clostridium difficile strains. Gut Microbes 2 326–334. 10.4161/gmic.19142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong C., Schoreder L. F., Gaur R., Anikst V. E., Komo I., Watters C., et al. (2017). Clostridium difficile rates in asymptomatic and symptomatic hospitalized patients using nucleic acid testing. Diagn. Microbiol. Infect. Dis. 87 365–370. 10.1016/j.diagmicrobio.2016.12.014 [DOI] [PubMed] [Google Scholar]
- van Heel A. J., de Jong A., Montalbán-López M., Kok J., Kuipers O. P. (2013). BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 41 W448–W453. 10.1093/nar/gkt391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Maarseveen N. M., Wessels E., de Brouwer C. S., Vossen A. C., Claas E. C. (2010). Diagnosis of viral gastroenteritis by simultaneous detection of Adenovirus group F, Astrovirus, Rotavirus group A, Norovirus genogroups I and II, and Sapovirus in two internally controlled multiplex real-time PCR assays. J. Clin. Virol. 49 205–210. 10.1016/j.jcv.2010.07.019 [DOI] [PubMed] [Google Scholar]
- Versalovic J., Schneider M., de Bruijn F. J., Lupski J. R. (1994). Genomic fingerprinting of bacteria with repetitive sequence based polymerase chain reaction. Methods Mol. Cell Biol. 5 25–40. [Google Scholar]
- Wang Y., Gänzle M. G., Schwab C. (2010). Exopolysaccharide synthesized by Lactobacillus reuteri decreases the ability of enterotoxigenic Escherichia coli to bind to porcine erythrocytes. Appl. Environ. Microbiol. 76 4863–4866. 10.1128/AEM.03137-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D., Adcock L. (2018). Probiotics for Antibiotic-Associated Diarrhea and Clostridium Difficile Infection: A Review of Clinical Effectiveness. Ottawa, ON: Canadian Agency for Drugs and Technologies in Health. [PubMed] [Google Scholar]
- Yap P. S., Gilliland S. E. (2000). Comparison of newly isolated strains of Lactobacillus delbrueckii subsp. lactis for hydrogen peroxide production at 5°C. J. Dairy Sci. 83 628–632. 10.3168/jds.s0022-0302(00)74922-8 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Tang H., Ye Y. (2012). RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 28 125–126. 10.1093/bioinformatics/btr595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liang Y., Lynch K. H., Dennis J. J., Wishart D. S. (2011). PHAST: a fast phage search tool. Nucleic Acids Res. 39(Suppl. 2) W347–W352. 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study can be found in the WHOJ00000000.