Abstract
Long non-coding RNA (lncRNA) plays a contributory role in rheumatoid arthritis (RA). In this review, we summarized the current findings of lncRNAs in RA, including cellular function and the potential mechanisms. Serum lncRNA levels are associated with serum proinflammatory cytokines and disease activity. LncRNAs regulate proliferation, migration, invasion and apoptosis of RA fibroblast-like synoviocytes (FLSs), modulate the differentiation of T lymphocytes and macrophages, and affect bone formation-destruction balance of chondrocytes. Besides, lncRNAs are involved in inflammation and cell motivation signaling pathways. In-depth research on lncRNAs may help elucidate the pathogenesis of RA and provides clues for novel treatment targets.
Keywords: lncRNA, Rheumatoid arthritis, Fibroblast-like synoviocyte, Inflammation, Invasiveness
Introduction
Rheumatoid arthritis (RA) is a chronic autoimmune disease associated with progressive joint destruction, systemic complications and decreased life expectancy.[1] Due to the advances in understanding the pathogenesis of the disease, treatment for RA is improved greatly, emphasizing early intervention soon after diagnosis and escalating the therapy based on the assessment of disease activity and treatment response in pursuit of clinical remission.
However, the mechanisms of RA are not fully elucidated. Also, limitations exist in the current conventional and biological therapies. Joint destruction continues in some patients even after aggressive treatment.[2] Toxicity associated with immunosuppressive agents contributes to the high mortality of patients with RA.[3] Therefore, elucidating the pathogenesis that initiates and perpetuates RA may provide the promise of discovering novel treatment targets.
The exploration in the epigenetics has shed light on the regulatory roles of a variety of noncoding RNAs (ncRNAs) that are transcribed from what was previously thought to be junk DNA. NcRNAs are loosely categorized into two classes based on the transcript size: small ncRNA (<200 nt), such as microRNAs (miRNAs), and long ncRNAs (lncRNAs) (>200 nt).[4] In addition, circular RNAs (circRNAs), which are covalently closed circles of single-stranded RNA are emerging as a new class of ncRNAs.[5] Although the biosynthesis and biological function of miRNAs are explicitly elucidated, we are still in the infancy of unveiling the biological activities of lncRNAs. Referring to the GENCODE database (version 31), there are 17,904 lncRNA genes identified in human. LncRNAs are subclassified into five categories that include the long intergenic ncRNAs (lincRNAs), intronic lncRNAs, bidirectional lncRNAs, sense-overlapping lncRNAs, and natural antisense lncRNAs according to their positions relative to protein coding genes [Figure 1]. LncRNAs are involved in a wide range of biological activities from epigenetic regulation and chromatin remodeling to transcriptional and posttranscriptional modification [Figure 2].[6,7]
Figure 1.
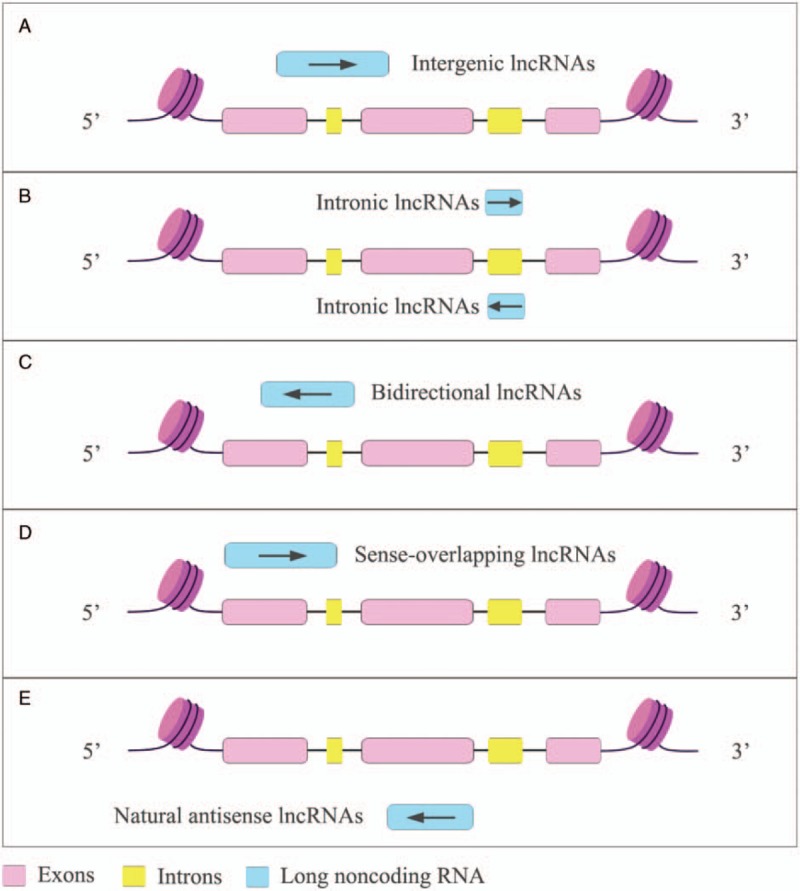
The classification of lncRNA. LncRNA can be subcategorized according to their relationship associated to the protein coding genes. (A) Intergenic lncRNAs are transcribed from the location between annotated protein-coding genes. (B) Intronic lncRNAs are RNA molecules that originate from the intron coding genes in either sense or antisense orientation. (C) Bidirectional lncRNAs are oriented from a location close to protein-coding genes but to an opposite direction. (D) Sense overlapping lncRNAs are transcribed from and overlapped with the sense strands of protein-coding genes. (E) Natural antisense lncRNAs are transcribed from the antisense strands and overlapped with one or several exons/introns. lncRNAs: Long noncoding RNAs.
Figure 2.
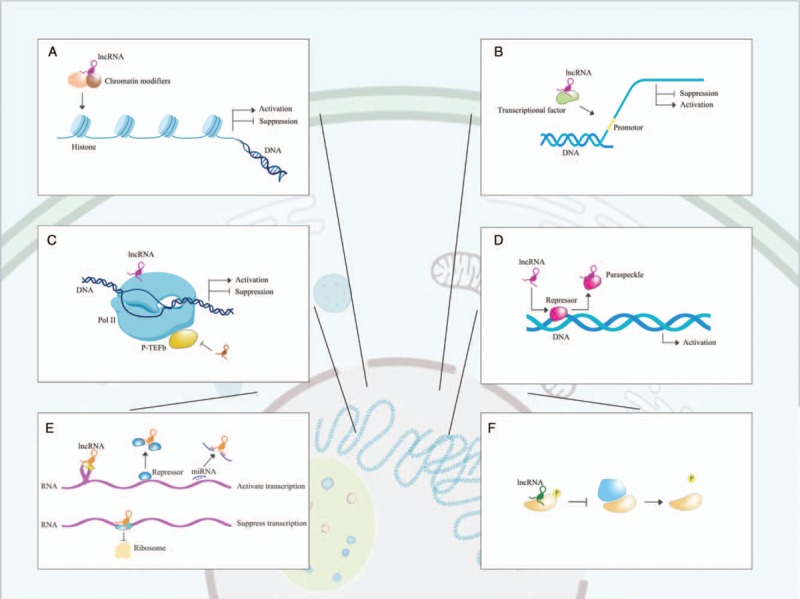
Cellular function of lncRNA. (A) LncRNAs recruit chromatin modifiers to regulate the epigenetic modification such as methylation and acetylation of the chromatin, altering the accessibility of genes to DNA-binding proteins and thus, interfering gene transcription. (B) LncRNAs can bind to the transcriptional factors to promote or inhibit their activity to interact with the promoter regions of certain genes. (C) LncRNAs interfere the transcriptional function of RNA polymerase II (Pol II). (D) LncRNAs are involved in the formation of some subnuclear structures such as paraspeckles to release their suppressive effect on DNA transcription. (E) The regulatory roles of lncRNA on mRNA translation are multifactorial. LncRNAs have impact on pre-mRNA splicing. LncRNAs modulate mRNA stability by associating with proteins involved in mRNA degradation. LncRNAs also function as sponge to miRNA and reverse their inhibitory effect on mRNA translation. LncRNA can recruit translational repressors to block the binding between mRNA and ribosome. (F) LncRNAs regulate protein phosphorylation at post-translation level in cytoplasm. lncRNAs: Long noncoding RNAs; miRNAs: microRNAs.
Emerging evidence suggests that lncRNAs are involved in RA development, and the current studies are listed in Table 1. Albeit a number of aberrant expressing lncRNAs are reported in RA,[8,9] only a few of them are functionally determined. We herein summarize the current findings of lncRNAs that could participate in the pathogenesis of RA, aiming to foster future research on this issue.
Table 1.
Summary of literatures of lncRNAs in RA
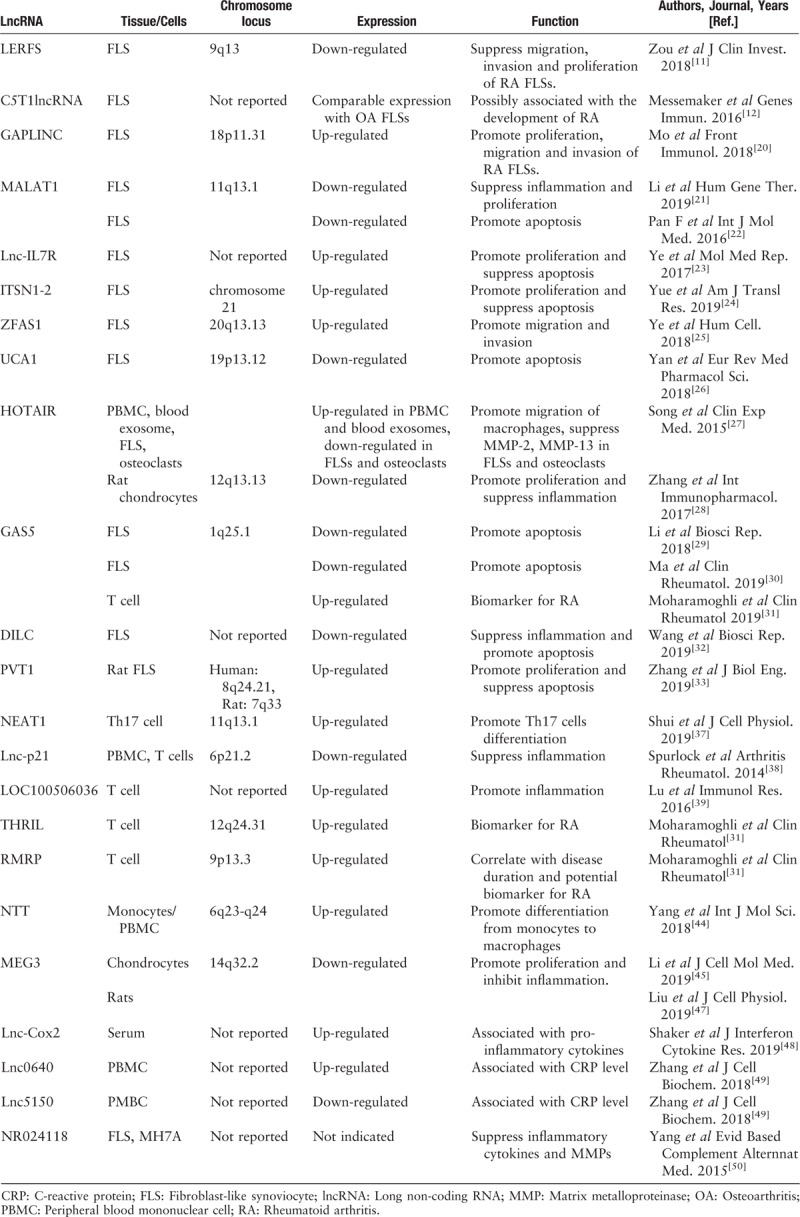
LncRNA in RA fibroblast-like synoviocytes (FLSs)
RA FLSs are the major components in synovial tissue. Different from normal FLSs, RA FLSs produce an abundant of cytokines to promote local inflammation, and proteolytic enzymes to degrade the extracellular matrix. Increasing proliferation, migration, invasion, and decreasing apoptosis is observed in RA FLSs, and reversing the aggressive phenotype could improve clinical outcomes without suppressing systemic immunity.[10] Recent studies suggest that lncRNAs are involved in the regulation of biological function of RA FLSs, and some of the major findings are shown as follows.
LncRNA LERFS
LERFS is a newly identified lncRNA in RA FLSs using microarray analysis.[11] The decreased expression of LERFS negatively regulates proliferation, migration and invasion of RA FLSs. The mechanistic analysis indicated that LERFS interacts with hnRNP Q, an RNA-binding protein, to form an lncRNA-protein complex which anchors to the mRNAs of RhoA, Rac1 and Cdc42 and reduces their stability or translation. This study provides evidence that LERFS is involved in synovial aggression and hyperplasia that characterize joint abnormalities in RA.
LncRNA C5T1lncRNA
C5T1lncRNA is identified recently within the TRAF1-C5 region, which is a susceptibility locus for RA discovered by genomewide study (GWAS).[12–14] In RA FLSs, C5T1lncRNA suppresses the mRNA of C5, a protein that has been detected in inflamed joints of patients with RA, and without which mice are resistant to the development of collagen-induced arthritis (CIA).[15,16] However, the expression of C5T1lncRNA and C5 in FLS obtained from patients with RA or OA is comparable. Whether C5T1lncRNA is inflammation-specific requires further validation and so does its functional role in RA.
LncRNA GAPLINC
GAPLINC was previously reported to be involved in proliferation and metastasis in tumor cells.[17–19] Its role in the regulation of aggressive phenotype of RA FLSs was recently revealed by Mo et al.[20] Increasing expression of GAPLINC promotes proliferation as well as in vitro migration and invasion of RA FLSs. Bioinformatics analysis suggested that GAPLINC could act as a molecular sponge of miR-382–5p and miR-575 since a negative correlation was observed between the expression of GAPLINC and the aforementioned miRNAs. Biological pathway enrichment analysis based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and Gene ontology (GO) enrichment analysis demonstrated that a part of the signaling pathway including MAPK signaling pathway, Ras signaling pathway and PI3K-Akt signaling pathway could be regulated by GAPLINC. However, the regulatory effect of miR-382–5p and miR-575 on the biological behavior of RA FLSs is only presumed based on the findings in cancer cells but not verified in this study.
LncRNA MALAT1
MALAT1 is involved in the regulation of inflammation and cell proliferation of RA FLSs.[21] MALAT1 might promote cell apoptosis at the same time as claimed by Pan et al, since a group of apoptosis-associated proteins including caspase-3, caspase-9, Bax, and Bcl-2 were suppressed in RA FLSs transfected with MALTA1 siRNA.[22] However, one of the defects in this study is the absence of the apoptotic rates in the control groups, which obscures the advantage of MALAT1. At least two signaling pathways were identified through which MALAT1 exerts biological function. On one hand, MALAT1 binds to catenin beta-1 (CTNNB1) promoter region, recruiting methyltransferase to increase CTNNB1 promoter methylation and inhibiting the transcription and expression of CTNNB1. β-catenin, as a transcriptional product of CTNNB1 and critical molecule in Wnt signaling pathway, is prevented from nucleation in the case of MALAT1 overexpression and so are the secretion of inflammatory cytokines such as interleukin (IL)-6. On the other hand, the activity of PI3K/AKT pathway increases when MALAT1 is down-regulated in RA FLSs.
LncRNA-IL7R
LncRNA-IL17R exerts pro-proliferation and anti-apoptosis effect on RA FLSs.[23] Analysis of mechanisms suggested that lncRNA-IL17R increases the binding of enhancer of zeste homolog 2 (EZH2) and H3K27me3 levels across cyclin dependent kinase inhibitor 2A (p16) and cyclin dependent kinase inhibitor 1A (p21) promoters, suppressing the transcription of p16 and p21.
LncRNA ITSN1-2
ITSN1-2 is up-regulated in both synovial tissue and FLSs obtained from patients with RA.[24] Knockdown of ITSN1-2 inhibits proliferation and promotes apoptosis of RA FLSs. ITSN1-2 is positively correlated with pro-inflammatory cytokines including tumor necrosis factor (TNF)-α and IL-17, and negatively correlated with IL-10 in RA FLSs and synovial tissue. GO and KEGG enrichment analysis found a total of 242 differentially expressed genes (DEGs) regulated by ITSN1-2 knockdown, all of which are associated with pathways related to RA. NOD2 is one of the top DEGs and proved to be associated with inflammation in synovial tissue in RA patients.
LncRNA ZFAS1
ZFAS1 contributes to the invasive phenotype of RA FLSs.[25] The activity of matrix metalloproteinases (MMP)-2 and MMP-9 is positively regulated by ZFAS1. MiR-27a, whose expression is decreased in synovial tissue and FLSs in RA, is recognized as a target of ZFAS1. By suppressing miR-27a, ZFAS1 promotes in vitro migration and invasion of RA FLSs.
LncRNA UCA1
Decreased expression of UCA1 is associated with the reduced activity of caspase-3 and therefore, increases cell viability of RA FLSs.[26] Wnt6 is one of the mediators through which UCA1 regulates apoptosis in RA FLSs.
LncRNA HOTAIR
The expression of HOTAIR in RA is context-dependent. Song et al found that the expression of HOTAIR increases in peripheral blood mononuclear cell (PBMC) and blood exosomes using lncRNA array analysis.[27] Stimulated by exosomes containing high level of HOTAIR, active macrophage displays enhanced in vitro migration. Meanwhile, the expression of HOTAIL is decreased in FLSs and osteoclasts isolated from patients with RA. Overexpression of HOTAIR suppresses the activation of MMP-2 and MMP-13. HOTAIR was also found to inhibit inflammation and promote proliferation in LPS-induced chondrocytes, probably by suppressing the nuclear factor (NF)-κB signaling pathway through miR-138.[28] Thus, HOTAIR seems to have widespread impact on RA and its biological function is complex.
LncRNA GAS5
Overexpression of GAS5 in RA FLSs promotes cell apoptosis partially by activating cleaved caspase-3 and caspase-9, and suppressing PI3K/AKT signaling pathway.[29,30] The expression of GAS5 in T cells and blood serum obtained from patients with RA, however, is inconsistent. An increased expression of GAS5 to approximately 3.3-fold was observed in T cells vis-à-vis a decreased serum level of GAS5 in patients with RA.[30,31] One of the limitations of these studies is that only the functional roles of GAS5 in RA were explored roughly and the underlying mechanisms were not specified. Much is needed to confirm the discriminative role of GAS5 in RA.
LncRNA DILC
Another lncRNA that is negatively correlated with serum IL-6 is DILC.[32] Similar to GAS5, DILC induces cell apoptosis of RA FLSs.
LncRNA PVT1
The role of PVT1 in RA was determined using CIA model in rats.[33] Established rat CIA model displays higher level of PVT1 in the synovial tissue compared with control rats. Knockdown of PVT1 in FLS isolated from CIA rats suppresses the production of pro-inflammatory cytokines including tumor necrosis factor (TNF)-α and IL-1β. Besides, cell proliferation is inhibited, and apoptosis increases in FLS transfected with PVT1 shRNA. Bioinformatics prediction and dual-luciferase reporter gene assay found sirt6, a gene which plays a contributory role in inflammation and bone destruction, as a target for PVT1.[34] Down-regulation of PVT1 hinders the methylation of sirt6. In this study, although increasing invasiveness of RA FLSs was presumed to be a result of PVT1 overexpression, it was not proven directly and could be secondary to increasing cell proliferation. Therefore, further research to validate the exact biological function of PVT1 in synovial tissue especially from RA patients is necessary.
LncRNA in lymphocytes from RA
The synovium of RA contains a great number of lymphocytes, and T lymphocytes especially type I helper T cells (Th1) and type 17 helper T cells (Th17) are considered to be major mediators to initiate RA. A shift of T lymphocytes towards inflammation facilitates the release of various cytokines that promote the accumulation of other inflammatory cells and activate the adjacent FLSs and chondrocytes.[35,36] Herein, we summarize four studies that were focusing lncRNA in T lymphocytes in the context of RA.
LncRNA NEAT1
The upregulation of NEAT1 induces the differentiation of Th17 cells in patients with RA.[37] The protective effect of NEAT1 knockdown on arthritis development is also demonstrated in vivo using mice CIA models. Overexpression of NEAT1 stabilizes the protein level of STAT3 and consequently, skews immune repertoire to Th17 cells.
LncRNA-p21
Methotrexate (MTX) is the cornerstone of RA treatment, and it ameliorates arthritis by multiple mechanisms. A recent literature disclosed the role of lncRNA-p21 in MTX-induced inhibition of NF-κB activity in T lymphocytes.[38] In RA, the expression of lncRNA-p21 is low and can be restored by the treatment of MTX. By sequestering RELA mRNA, lncRNA-p21 interferes the translation of RELA and thus, suppresses the activation of NF-κB.
LncRNA LOC100506036
Microarray analysis showed the expression of LOC100506036 is increased in peripheral T cells in patients with RA. LOC100506036 promotes the production of IFN-γ possibly by suppressing sphingomyelin phosphodiesterase 1 (SMPD1) protein.[39]
LncRNA THRIL and RMBP
Upward trends of THRIL and RMBP are detected in T cells from patients with RA.[31] Both of them are suggested as biomarkers for RA and RMRP is correlated with disease duration as well.
LncRNA in monocyte-derived macrophages from RA
The high number of macrophages contribute to the cytokine storm as well as cartilage and bone destruction in the synovium, and are considered as early hallmarks of active RA.[40–42] Macrophages in RA synovial tissue are partially derived from monocytes in response to inflammation.[43] A recent study reported that lncRNA is involved in this process.
LncRNA NTT
There is a dearth of knowledge of lncRNAs in monocyte/macrophage system in RA. Yang et al found an increased expression of NTT in PBMC derived from untreated early RA patients.[44] Overexpression of NTT enhances the expression of prostate and breast cancer overexpressed 1 (PBOV1), and facilitates the differentiation from monocytes to macrophages and the production of chemokines such as CXCL10. This study also identified the upstream regulator of NTT, that is, C/EBPβ, which binds to NTT promoter and regulates NTT expression. The activation of C/EBPβ/NTT/PBOV1 axis is correlated with high disease activity measured by DAS28 score.
LncRNA GAS5
The increased expression of GAS5 in T cells isolated from PBMC from patients with RA is discussed above.[31] The subsets of T cells, however, were not specified. And GAS5 shows no correlation with TNF-α or IL-17 in T cells in this study. One of the explanations for the discrepancy could be that when calculating the average expression of GAS5 in patients with RA, two outliers were included, and that might interfere the final results. Therefore, whether GAS5 is up-regulated in T cells and what subtypes of T cells from RA needs to be determined in larger sample size.
LncRNA in chondrocytes from RA
Chondrocytes are critical regulators to maintain cartilage matrix. Chondrocytes exert dual function as building up cartilage matrix in physiological condition and breaking it down under chronic inflammation. When stimulated by IL-1 and IL-17α, chondrocytes in RA synovial tissue generate abundant MMPs, resulting in increased aggrecanolysis and reduced proteoglycan synthesis. To decipher the molecular mechanisms of the turnover behavior of chondrocytes is in growing importance in RA.
LncRNA MEG3
MEG3 is involved in the regulation of inflammation in RA. The expression of MEG3 is suppressed in chondrocytes in the stimulation of lipopolysaccharide (LPS) and the overexpression of MEG3 inhibits the production of pro-inflammatory cytokines including IL-17 and IL-23 in RA.[45] The protective effect of MEG3 on chondrocytes is supported by another study, reporting that overexpression of MEG3 promotes proliferation and relieves the degradation of extracellular matrix in IL-1β-induced chondrocytes in osteoarthritis (OA).[46] The activation of AKT-mTOR signaling pathway is suppressed by MEG overexpression in chondrocytes.[45] By contrast, the role of MEG3 in RA FLSs seems to be controversial. It has found an increased expression of MEG3 in human synovial tissue and FLSs from RA.[45] However, another study[47] found the decreased expression of MEG3 in complete Freund adjuvant (CFA)-induced rat models for RA. Although Li et al also established rat model for RA, whether it was CFA-induced was not clarified. Besides, they did not detect the expression of MEG3 in synovial tissue or FLSs isolated from the animal models. Whether the cell phenotype in rat models is consistent with human beings remains to be determined. Thus, further research is still needed to determine the role of MEG3 in RA FLSs.
LncRNA HOTAIR
The involvement of HOTAIR in RA chondrocytes is discussed above.[28]
Other lncRNAs that are detected in RA PBMC or synovial cell lines include: (i) Lnc-COX2 isolated from blood serum from RA participants displays a positive correlation with serum levels of IL-6 and MMP-9.[48] (ii) The altered expression of lnc0640 and lnc5150 is observed in PBMC from patients with RA, and both of them are associated with C-reactive protein (CRP) levels.[49] Genetically, TT genotype of rs13039216 in lnc0640 gene is associated with a reduced risk of RA, implying that lnc0640 could contribute to the onset of RA. (iii) lncRNA NR024118 is involved in the regulation of inflammatory cytokines and MMPs in RA.[50] Inhibition of NR024118 suppresses the mRNA expression of IL-6, IL-8, MMP-1, and MMP-3 in MH7A synovial cell lines.
LncRNA in signaling pathways involved in RA
The aggressive behavior of RA is regulated by signal network. Due to the constraints of space and scope, two pathways as the representatives of inflammation and cell motility that were subjected to the regulation of lncRNA are highlighted, that is, NF-κB and RhoGTPases. The involvement of lncRNAs in other classical signaling pathways in RA was discussed in previous studies.[51]
NF-κB signaling pathway
NF-κB signaling pathway is pivotal in regulating inflammation in RA. NF-κB continues to be a master target in pursuit for controlling inflammation. Until recently, many lncRNAs are reported to be involved in the activation of NF-κB [Figure 3A], among which lncRNA-p21 and lncRNA-COX2 are implicated in RA.
Figure 3.
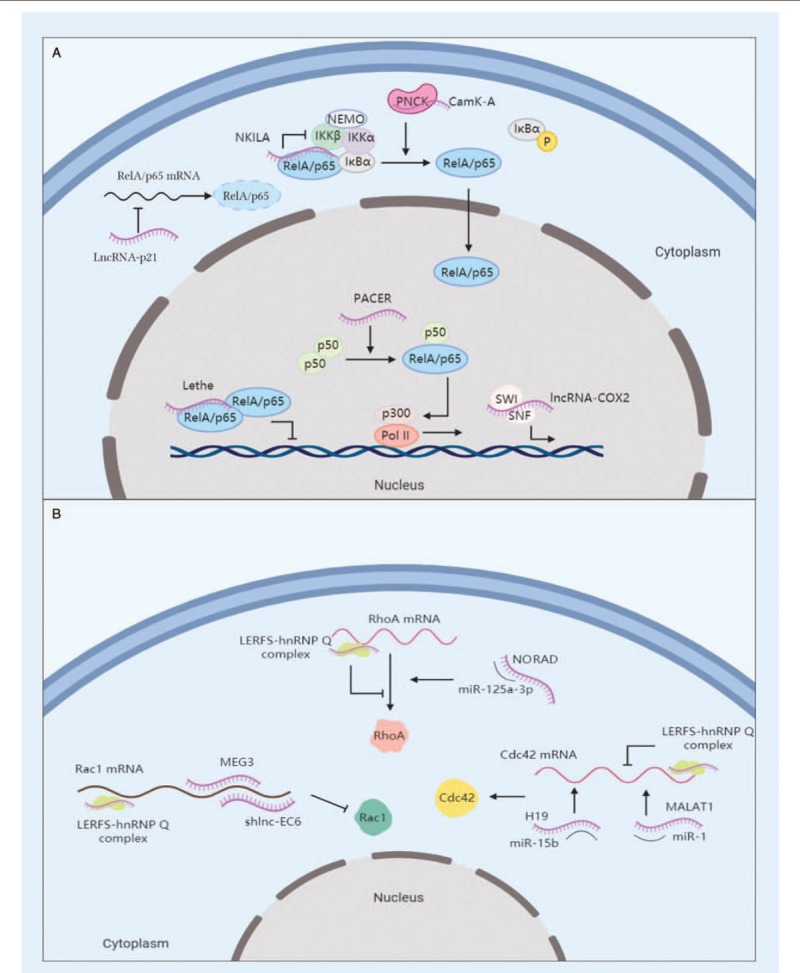
Functional lncRNAs in signaling pathway. (A) LncRNAs regulate NF-κB signaling pathways. NKILA masks the phosphorylating sites of IKKβ in IκBα-RelA complex and suppresses RelA activation. LncRNA-p21 sequesters RelA by suppressing its mRNA translation. Lethe blocks the DNA-binding activity of RelA homodimer. PACER sequesters p50 and facilitates the formation of Pol II transcriptional complex. LncRNA-Cox2 is integrated into the SWI/SNF complex and transactivates late-primary response genes regulated by NF-κB. CAMK-A, in accompany with calmodulin-dependent kinase PNCK, promotes IκBα phosphorylation and nuclear transportation of RelA. (B) LncRNAs participate in the regulation of RhoGTPases-mediating pathways. Newly-identified lncRNA LERFS forms a complex with hnRNP Q to interfere the translation of RhoA, Rac1, and Cdc42. NORAD, MEG3, shlnc-EC6, H19, and MALAT1 act as miRNA sponges to regulate the translation of the aforementioned RhoGTPases, respectively. IKK: Inhibitory-κB kinase; lncRNA: Long non-coding RNA; NEMO: NF-κB essential modulator; NF: Nuclear factor; NKILA: NF-κB interacting lncRNA; PNCK: Pregnancy upregulated non-ubiquitous calmodulin kinase.
LncRNA-p21 suppresses inflammation in RA through sequestering NF-κB. LncRNA-p21 is the mediator whereby MTX suppresses TNF-α-induced NF-κB activation.[38] Knockdown of lncRNA-p21 abrogates the inhibitory effect of MTX on NF-κB activation. Further experiment showed that treatment with MTX inhibits the protein expression of RelA and the phosphorylation of RelA. Since lncRNA-p21 was found to interact intensively with RelA transcripts, it is postulated that lncRNA-21 suppresses NF-κB activation by reducing RelA translation. However, it would be more persuasive if protein expression of RelA had been examined in cells with lncRNA-p21 knockdown directly.
The transcription of lncRNA-COX2 stimulated by proinflammatory cytokines is controlled by NF-κB signaling in macrophages.[52] LncRNA-COX2 develops a positive feedback loop by assembling into the SWItch/Sucrose NonFermentable (SWI/SNF) complex, triggering SWI/SNF-associated chromatin remodeling and transactivating late-primary response genes (ie, CCL2, CCL5, CXCL10, PELI1, TRAF1, SAA3, IFNβ1) following NF-κB activation. LncRNA-COX2 knockdown reduces histone H3 methylation in SAA3 and CCL5 promoters in RAW264.7 cells, and is considered to be a consequence of the impaired recruitment of SWI/SNF complex. Since molecular mechanisms are shared in many chronic inflammatory diseases, it is conceivable that lncRNAs involved in NF-κB signaling pathways in other diseases might have a role in RA. The hypothesis deserves in-depth surveys.
RhoGTPases signaling pathway
RhoGTPases are critical in the regulation of cell motility. However, the role of lncRNA in RhoGTPases-mediated signaling pathway is less discussed. We reviewed literatures focusing on the regulatory role of lncRNAs in RhoGTPases [Figure 3B] and highlighted three lncRNAs that are involved in RA.
The involvement of lncRNA in regulating RhoGTPases in RA was reported by Zou et al for the first time.[11] As indicated above, LERFS interferes the expression of RhoA, Rac1 and Cdc42 through hnRNP Q, which is the only protein found to be combined with LERFS by pull-down assay in RA FLSs. However, LERFS seems to exert biological function in different ways. Although both the mRNA and protein expression of Rac1 is inhibited by LERFS overexpression, only the protein levels of RhoA and Cdc42 are affected. Different from the common interaction between lncRNA and mRNA, LERFES does not bind to mRNA of the aforementioned proteins directly. Therefore, LERFS could act as a cofactor to facilitate hnRNP Q binding to mRNA and the underlying mechanism requires further exploration.
LncRNA MEG3 and lncRNA MALAT1 were found to be involved in the pathogenesis of RA. However, the underlying mechanism is not well-defined. Studies on tumor cells indicated that both of the lncRNAs participate in the regulation of Rho GTPases. Rac1 exhibits a specific target within the 3’UTR for MEG3, and the interaction was proven by dual luciferase reporter assay in TPC-1 and HTH83 thyroid cancer cell lines.[53] An inverse correlation is also observed between MEG3 and Rac1 in papillary thyroid carcinoma tissue. Overexpression of MEG3 strongly inhibits cell migration and invasion by suppressing protein expression of Rac1. MALAT1 accelerates migration and invasion of breast cancer cells MCF7 and MDA-MB-231.[54] The expression of MALAT1 is reversely correlated with miR-1. Bioinformatics prediction distinguishes a binding site in miR-1 shared by MALAT1 and Cdc42 3’UTR. The increased expression of Cdc42 induced by MALAT1 overexpression is hindered by miR-1 mimics. Therefore, it is suggested that MALAT1 serves as a competing endogenous RNA (ceRNA) to competitively bind to miR-1 and reverse the inhibitory effect of miR-1 on Cdc42 translation. Taken together, we assume that MEG3 and MALAT1 could exert regulatory function on RA FLSs through Rho GTPases-mediating signaling pathway. Further study to investigate the hypothesis will be helpful.
Conclusions
Emerging evidence suggests that lncRNAs are important regulators in RA. Continuing to explore the aberrant expression profile of lncRNAs in RA, to determine their functional roles and the mode of action would deepen our understanding of disease origin. A group of lncRNAs are found to be associated with clinical indicators such as CRP, ESR, serum proinflammatory cytokines and DAS28, suggesting that lncRNAs can serve as biomarkers to monitor disease activity. The validity of lncRNAs needs to be verified in a large RA population. Various lncRNAs are candidate central regulators in inflammatory signaling pathways, but only a few of them have been testified in RA. The sharing mechanisms of RA and other inflammatory diseases imply a similar role of lncRNAs guiding the onset of RA. It would be informative to dig out the function of these lncRNAs in the context of RA. The beneficial effect of lncRNAs manipulation in relieving arthritis is preliminarily examined in animal models in a few studies. It is conceivable that lncRNAs are promising treatment targets for RA and deserve in-depth investigation in vivo. Taken together, research on lncRNAs in RA is very much a nascent field. Deepening the understanding of lncRNAs will likely lead to novel mechanistic insights into RA pathogenesis and ultimately, to new avenues in therapeutic treatment.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81871275, 81671591, U1401222, 81601403).
Conflicts of interest
None.
Footnotes
How to cite this article: Lao MX, Xu HS. Involvement of long non-coding RNAs in the pathogenesis of rheumatoid arthritis. Chin Med J 2020;133:941–950. doi: 10.1097/CM9.0000000000000755
References
- 1.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature 2003; 423:356–361. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 2.Brown AK, Conaghan PG, Karim Z, Quinn MA, Ikeda K, Peterfy CG, et al. An explanation for the apparent dissociation between clinical remission and continued structural deterioration in rheumatoid arthritis. Arthritis Rheum 2008; 58:2958–2967. doi: 10.1002/art.23945. [DOI] [PubMed] [Google Scholar]
- 3.Rutherford AI, Subesinghe S, Hyrich KL, Galloway JB. Serious infection across biologic-treated patients with rheumatoid arthritis: results from the British Society for Rheumatology Biologics Register for Rheumatoid Arthritis. Ann Rheum Dis 2018; 77:905–910. doi: 10.1136/annrheumdis-2017-212825. [DOI] [PubMed] [Google Scholar]
- 4.Perkel JM. Visiting “noncodarnia”. Biotechniques 2013; 54:301.303-4. doi: 10.2144/000114037. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Li D, Luo H, Zhu X. Circular RNAs: the star molecules in cancer. Mol Aspects Med 2019; 70:141–152. doi: 10.1016/j.mam.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Schonrock N, Harvey RP, Mattick JS. Long noncoding RNAs in cardiac development and pathophysiology. Circ Res 2012; 111:1349–1362. doi: 10.1161/CIRCRESAHA.112.268953. [DOI] [PubMed] [Google Scholar]
- 7.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 2013; 339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Xu YZ, Sun N, Liu JH, Chen FF, Guan XL, et al. Long noncoding RNA expression profile in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Res Ther 2016; 18:227.doi: 10.1186/s13075-016-1129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan M, Wang S, Yu L, Qu B, Xu L, Liu L, et al. Long noncoding RNA profiling revealed differentially expressed lncRNAs associated with disease activity in PBMCs from patients with rheumatoid arthritis. PLoS One 2017; 12:e0186795.doi: 10.1371/journal.pone.0186795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev 2010; 233:233–255. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou Y, Xu S, Xiao Y, Qiu Q, Shi M, Wang J, et al. Long noncoding RNA LERFS negatively regulates rheumatoid synovial aggression and proliferation. J Clin Invest 2018; 128:4510–4524. doi: 10.1172/JCI97965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messemaker TC, Frank-Bertoncelj M, Marques RB, Adriaans A, Bakker AM, Daha N, et al. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes Immun 2016; 17:85–92. doi: 10.1038/gene.2015.54. [DOI] [PubMed] [Google Scholar]
- 13.Kurreeman FA, Padyukov L, Marques RB, Schrodi SJ, Seddighzadeh M, StoekenRijsbergen G, et al. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS Med 2007; 4:e278.doi: 10.1371/journal.pmed.0040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plenge RM, Seielstad M, Padyukov L, Lee T, Remmers EF, Ding B, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis-a genomewide study. N Engl J Med 2007; 357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooke TD, Hurd ER, Jasin HE, Bienenstock J, Ziff M. Identification of immunoglobulins and complement in rheumatoid articular collagenous tissues. Arthritis Rheum 1975; 18:541–551. doi: 10.1002/art.1780180603. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Kristan J, Hao L, Lenkoski CS, Shen Y, Matis LA. A role for complement in antibody-mediated inflammation: C5-deficient DBA/1 mice are resistant to collagen-induced arthritis. J Immunol 2000; 164:4340–4347. doi: 10.4049/jimmunol.164.8.4340. [DOI] [PubMed] [Google Scholar]
- 17.Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res 2014; 74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 18.Liu L, Zhao X, Zou H, Bai R, Yang K, Tian Z. Hypoxia promotes gastric cancer malignancy partly through the HIF-1alpha dependent transcriptional activation of the long non-coding RNA GAPLINC. Front Physiol 2016; 7:420.doi: 10.3389/fphys.2016.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang P, Chen T, Xu Z, Zhu H, Wang J, He Z. Long noncoding RNA GAPLINC promotes invasion in colorectal cancer by targeting SNAI2 through binding with PSF and NONO. Oncotarget 2016; 7:42183–42194. doi: 10.18632/oncotarget.9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mo BY, Guo XH, Yang MR, Liu F, Bi X, Liu Y, et al. Long non-coding RNA GAPLINC promotes tumor-like biologic behaviors of fibroblast-like synoviocytes as MicroRNA sponging in rheumatoid arthritis patients. Front Immunol 2018; 9:702.doi: 10.3389/fimmu.2018.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li GQ, Fang YX, Liu Y, Meng FR, Wu X, Zhang CW, et al. MALAT1-driven inhibition of Wnt signal impedes proliferation and inflammation in fibroblast-like synoviocytes through CTNNB1 promoter methylation in rheumatoid arthritis. Hum Gene Ther 2019; 30:1008–1022. doi: 10.1089/hum.2018.212. [DOI] [PubMed] [Google Scholar]
- 22.Pan F, Zhu L, Lv H, Pei C. Quercetin promotes the apoptosis of fibroblast-like synoviocytes in rheumatoid arthritis by upregulating lncRNA MALAT1. Int J Mol Med 2016; 38:1507–1514. doi: 10.3892/ijmm.2016.2755. [DOI] [PubMed] [Google Scholar]
- 23.Ye Z, Xu J, Li S, Cai C, Li T, Sun L, et al. Lnc-IL7R promotes the growth of fibroblast-like synoviocytes through interaction with enhancer of zeste homolog 2 in rheumatoid arthritis. Mol Med Rep 2017; 15:1412–1418. doi: 10.3892/mmr.2017.6150. [DOI] [PubMed] [Google Scholar]
- 24.Yue T, Fan X, Zhang Z, Liu Z, Guo M, Bai F, et al. Downregulation of lncRNA ITSN1-2 correlates with decreased disease risk and activity of rheumatoid arthritis (RA), and reduces RA fibroblast-like synoviocytes proliferation and inflammation via inhibiting NOD2/RIP2 signaling pathway. Am J Transl Res 2019; 11:4650–4666. [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Y, Gao X, Yang N. LncRNA ZFAS1 promotes cell migration and invasion of fibroblast-like synoviocytes by suppression of miR-27a in rheumatoid arthritis. Hum Cell 2018; 31:14–21. doi: 10.1007/s13577-017-0179-5. [DOI] [PubMed] [Google Scholar]
- 26.Yan ZF, Zhao XY, Liu W, Liu XP. UCA1 impacts progress of rheumatoid arthritis by inducing the apoptosis of fibroblast-like synoviocyte. Eur Rev Med Pharmacol Sci 2018; 22:914–920. doi: 10.26355/eurrev_201802_14370. [DOI] [PubMed] [Google Scholar]
- 27.Song J, Kim D, Han J, Kim Y, Lee M, Jin EJ. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med 2015; 15:121–126. doi: 10.1007/s10238-013-0271-4. [DOI] [PubMed] [Google Scholar]
- 28.Zhang HJ, Wei QF, Wang SJ, Zhang HJ, Zhang XY, Geng Q, et al. LncRNA HOTAIR alleviates rheumatoid arthritis by targeting miR-138 and inactivating NF-(B pathway. Int Immunopharmacol 2017; 50:283–290. doi: 10.1007/s10238-013-0271-4. [DOI] [PubMed] [Google Scholar]
- 29.Li G, Liu Y, Meng F, Xia Z, Wu X, Fang Y, et al. Tanshinone IIA promotes the apoptosis of fibroblast-like synoviocytes in rheumatoid arthritis by up-regulating lncRNA GAS5. Biosci Rep 2018; 38:pii: BSR20180626.doi: 10.1042/BSR20180626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma C, Wang W, Li P. LncRNA GAS5 overexpression downregulates IL-18 and induces the apoptosis of fibroblast-like synoviocytes. Clin Rheumatol 2019; 38:3275–3280. doi: 10.1007/s10067-019-04691-2. [DOI] [PubMed] [Google Scholar]
- 31.Moharamoghli M, Hassan-Zadeh V, Dolatshahi E, Alizadeh Z, Farazmand A. The expression of GAS5, THRIL, and RMRP lncRNAs is increased in T cells of patients with rheumatoid arthritis. Clin Rheumatol 2019; 38:3073–3080. doi: 10.1007/s10067-019-04694-z. [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Tang L, Zhang X, Li Y. LncRNA DILC participates in rheumatoid arthritis by inducing apoptosis of fibroblast-like synoviocytes and down-regulating IL-6. Biosci Rep 2019; 39:pii: BSR20182374.doi: 10.1042/BSR20182374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang CW, Wu X, Liu D, Zhou W, Tan W, Fang YX, et al. Long non-coding RNA PVT1 knockdown suppresses fibroblast-like synoviocyte inflammation and induces apoptosis in rheumatoid arthritis through demethylation of sirt6. J Biol Eng 2019; 13:60.doi: 10.1186/s13036-019-0184-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HS, Ka SO, Lee SM, Lee SI, Park JW, Park BH. Overexpression of sirtuin 6 suppresses inflammatory responses and bone destruction in mice with collagen-induced arthritis. Arthritis Rheum 2013; 65:1776–1785. doi: 10.1002/art.37963. [DOI] [PubMed] [Google Scholar]
- 35.Chabaud M, Fossiez F, Taupin JL, Miossec P. Enhancing effect of IL-17 on IL-1-induced IL-6 and leukemia inhibitory factor production by rheumatoid arthritis synoviocytes and its regulation by Th2 cytokines. J Immunol 1998; 161:409–414. [PubMed] [Google Scholar]
- 36.Miossec P, Korn T, Kuchroo VK. Interleukin-17 and type 17 helper T cells. N Engl J Med 2009; 361:888–898. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- 37.Shui X, Chen S, Lin J, Kong J, Zhou C, Wu J. Knockdown of lncRNA NEAT1 inhibits Th17/CD4+ T cell differentiation through reducing the STAT3 protein level. J Cell Physiol 2019; 234:22477–22484. doi: 10.1002/jcp.28811. [DOI] [PubMed] [Google Scholar]
- 38.Spurlock CF, 3rd, Tossberg JT, Matlock BK, Olsen NJ, Aune TM. Methotrexate inhibits NF-κB activity via long intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol 2014; 66:2947–2957. doi: 10.1002/art.38805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu MC, Yu HC, Yu CL, Huang HB, Koo M, Tung CH, et al. Increased expression of long noncoding RNAs LOC100652951 and LOC100506036 in T cells from patients with rheumatoid arthritis facilitates the inflammatory responses. Immunol Res 2016; 64:576–583. doi: 10.1007/s12026-015-8756-8. [DOI] [PubMed] [Google Scholar]
- 40.Chu CQ, Field M, Feldmann M, Maini RN. Localization of tumor necrosis factor α in synovial tissues and at the cartilage-pannus junction in patients with rheumatoid arthritis. Arthritis Rheum 1991; 34:1125–1132. doi: 10.1002/art.1780340908. [DOI] [PubMed] [Google Scholar]
- 41.Bertolini DR, Nedwin GE, Bringman TS, Smith DD, Mundy GR. Stimulation of bone resorption and inhibition of bone formation in vitro by human tumour necrosis factors. Nature 1986; 319:516–518. doi: 10.1038/319516a0. [DOI] [PubMed] [Google Scholar]
- 42.Lam J, Takeshita S, Barker JE, Kanagawa O, Ross FP, Teitelbaum SL. TNF-α induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J Clin Invest 2000; 106:1481–1488. doi: 10.1172/JCI11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merad M, Manz MG, Karsunky H, Wagers A, Peters W, Charo I, et al. Langerhans cells renew in the skin throughout life under steady-state conditions. Nat Immunol 2002; 3:1135–1141. doi: 10.1038/ni852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang CA, Li JP, Yen JC, Lai IL, Ho YC, Chen YC, et al. LncRNA NTT/PBOV1 axis promotes monocyte differentiation and is elevated in rheumatoid arthritis. Int J Mol Sci 2018; 19:pii: E2806.doi: 10.3390/ijms19092806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li G, Liu Y, Meng F, Xia Z, Wu X, Fang Y, et al. LncRNA MEG3 inhibits rheumatoid arthritis through miR-141 and inactivation of AKT/mTOR signaling pathway. J Cell Mol Med 2019; 23:7116–7120.doi: 10.1111/jcmm.14591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen K, Zhu H, Zheng MQ, Dong QR. LncRNA MEG3 inhibits the degradation of the extracellular matrix of chondrocytes in osteoarthritis via targeting miR-93/TGFBR2 axis. Cartilage 2019; 1947603519855759. doi: 10.1177/1947603519855759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu YR, Yang L, Xu QQ, Lu XY, Ma TT, Huang C, et al. Long noncoding RNA MEG3 regulates rheumatoid arthritis by targeting NLRC5. J Cell Physiol 2019; 234:14270–14284. doi: 10.1002/jcp.28126. [DOI] [PubMed] [Google Scholar]
- 48.Shaker OG, Mahmoud RH, Abdelaleem OO, Ahmed TI, Fouad NA, Hussein HA, et al. Expression profile of long noncoding RNAs, lnc-Cox2, and HOTAIR in rheumatoid arthritis patients. J Interferon Cytokine Res 2019; 39:174–180. doi: 10.1089/jir.2018.0117. [DOI] [PubMed] [Google Scholar]
- 49.Zhang TP, Zhang Q, Wu J, Zhao YL, Wang JB, Leng RX, et al. The expression levels of long noncoding RNAs lnc0640 and lnc5150 and its gene single-nucleotide polymorphisms in rheumatoid arthritis patients. J Cell Biochem 2018; 119:10095–10106. doi: 10.1002/jcb.27346. [DOI] [PubMed] [Google Scholar]
- 50.Yang KY, Chen DL. Shikonin inhibits inflammatory response in rheumatoid arthritis synovial fibroblasts via lncRNA-NR024118. Evid Based Complement Alternat Med 2015; 2015:631737.doi: 10.1155/2015/631737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pearson MJ, Jones SW. Long Noncoding RNAs in the regulation of inflammatory pathways in rheumatoid arthritis and osteoarthritis. Arthritis Rheumatol 2016; 68:2575–2583. doi: 10.1002/art.39759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu G, Gong AY, Wang Y, Ma S, Chen X, Chen J, et al. LincRNA-Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF-mediated chromatin remodeling. J Immunol 2016; 196:2799–2808. doi: 10.4049/jimmunol.1502146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C, Yan G, Zhang Y, Jia X, Bu P. Long non-coding RNA MEG3 suppresses migration and invasion of thyroid carcinoma by targeting of Rac1. Neoplasma 2015; 62:541–549. doi: 10.4149/neo_2015_065. [DOI] [PubMed] [Google Scholar]
- 54.Chou J, Wang B, Zheng T, Li X, Zheng L, Hu J, et al. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun 2016; 472:262–269. doi: 10.1016/j.bbrc.2016.02.102. [DOI] [PubMed] [Google Scholar]


