Abstract
The chemokine-like factor (CKLF)-like MARVEL transmembrane domain-containing family (CMTM) is widely expressed in the immune system. Abnormal expression of CMTM is associated with the development of various diseases. This article summarizes the relevant research on the role of the CMTM family in immune disorders. This information will increase our understanding of pathogenesis and identify promising targets for the diagnosis and treatment of autoimmune diseases. The CMTM family is highly expressed in peripheral blood mononuclear cells. CKLF1 may be involved in the development of arthritis through its interaction with C-C chemokine receptor 4. CKLF1 is associated with the pathogenesis of lupus nephritis and psoriasis. Both CMTM4 and CMTM5 are associated with the pathogenesis of systemic lupus erythematosus. CMTM1, CMTM2, CMTM3, and CMTM6 play a role in rheumatoid arthritis, systemic sclerosis, Sjögren syndrome, and anti-phospholipid syndrome, respectively. The CMTM family has been implicated in various autoimmune diseases. Further research on the mechanism of the action of CMTM family members may lead to the development of new treatment strategies for autoimmune diseases.
Keywords: CMTM, CKLFSF, Autoimmune Diseases, Immune system
Introduction
The chemokine-like factor super family (CKLFSF), a new class of chemokines, was discovered by Peking University Human Disease Gene Research Center. CKLF1, which was isolated from phytohemagglutinin (PHA)-stimulated U937 cells, was the initial member of the CKLFSF.[1] Subsequent studies identified CKLFSF1-8 using a combination of bioinformatics and reverse transcription polymerase chain reaction techniques. The CKLFSF consists of nine genes, CKLF and CKLFSF1-8.[2,3] The characteristics of their gene products lie somewhere between classical chemokines and members of the transmembrane 4 super family (TM4SF).[2] According to their structural features, the Human Gene Nomenclature Committe recommended denominating CKLFSF1-8 as CKLF-like MARVEL transmembrane (CMTM)1-8 [Figure 1].[4] The CMTM family members are widely expressed throughout the immune system and they are associated with autoimmune diseases.[2] Therefore, CMTM members may represent promising targets for the diagnosis and treatment in autoimmune diseases. This article summarizes the relevant research on the role of the CMTM family in the immune system and autoimmune diseases.
Figure 1.
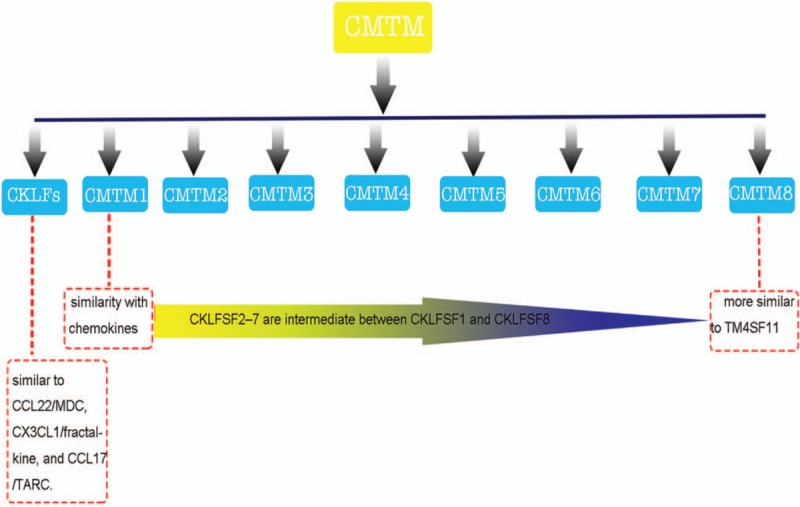
The composition of the CMTM family. The characteristics of CMTM members lie between that of classical chemokines and transmembrane 4 super family (TM4SF) members. CCL: C-C class chemokines; CMTM: Chemokine-like factor (CKLF)-like MARVEL transmembrane; CX3CL: CX3C chemokine ligand; MDC: Macrophage-derived chemokine; TARC: Thymus-and activation-regulated chemokine.
CMTM Family Members and the Immune System
CKLF1
CKLF1 is located on chromosome 16q 22.1 and has four exons and three introns. There are at least three alternative RNA splicing forms: CKLF2, CKLF3, and CKLF4. The expression of CKLFs in tissues is variable. CKLF1 expression is similar to that of CKLF2, but higher than that of CKLF4. CKLF3 expression is comparatively the lowest. CKLF1 and CKLF3 are secreted proteins, while CKLF2 and CKLF4 are transmembrane proteins. CKLF2 and CKLF4 are primarily located on the cell periphery.[1]
CKLF1 is different from classical chemokines. CKLF1 has a CC motif, but lacks the additional C-terminal cysteines as compared with the other classical CC sub-family members.[1] However, the key amino acids surrounding the CKLF1 motif are similar to those of thymus-and activation-regulated chemokine (TARC)/C-C class chemokines (CCL) 17 and macrophage-derived chemokine (MDC)/CCL22.[2] Previous studies have shown that both TARC/CCL17 and MDC/CCL22 are specific ligands for C-C chemokine receptor 4 (CCR4).[5,6] Several in vitro experiments found that CKLF1 induced calcium flux in CCR4-transfected HEK293 cells and desensitized CCR4 transfectants to subsequent TARC/CCL17 treatment. Meanwhile, the internalization of CCR4 receptors was significantly increased following treatment with CKLF1 in pCCR4-enhanced green fluorescent protein transfected cells.[7] CKLF1 is a novel functional ligand for CCR4.[8] CCR4 is a chemokine receptor that has been shown to promote recruitment, homing, and education of activated leukocytes (mainly CD4+ Th2 lymphocytes).[6,9] Moreover, CCR4 is expressed on dendritic cells (DCs), basophils, T cells, and platelets.[10] This suggests that the interaction of CKLF1 with CCR4 might play a role in autoimmune diseases.
CKLF1 has a broad spectrum of chemotactic activity for many cells including lymphocytes, macrophages, and neutrophils.[11] Li et al[12] reported that CKLF1 expression was increased in activated CD4+ and CD8+ lymphocytes. They further explored the kinetic expression of CKLF1 in PHA-stimulated (specific for T lymphocytes) peripheral blood lymphocytes and found that CKLF1 was significantly increased in a time-dependent manner. This was similar to the expression of CD25 (an activation marker of T cells).[12,13] These studies suggest that CKLF1 could be involved in T lymphocyte activation.[14] Previous studies have shown that CKLF may exhibit chemotactic activity and activate neutrophils through mitogen-activated protein kinase (MAPK) pathway. Anti-CKLF1 antibody decreased the production of the inflammatory factors tumor necrosis factor (TNF)-α, interleukin (IL)-1β, macrophage inflammatory protein-2 and IL-8 as well as the adhesion molecules, intercellular adhesion molecule 1 and vascular cell adhesion molecule 1.[15] In addition, CKLF1 was found to be highly expressed in monocytes. Shao et al[16] reported that, during DCs differentiation, CKLF1 was increased on day 2 and decreased on days 3 to 5. The expression of CKLF1 in DCs activated by stimuli was demonstrated to be lower compared with immature DCs.[16] DC maturation requires the activation of nuclear factor (NF)-κB transcription.[17] In a recent study, researchers demonstrated that IMM-H004 (a novel coumarin derivative screened from a CKLF1/C-C CCR4 system) protects against ischemic stroke-induced inflammation through a CKLF1 pathway in coordination with NF-κB.[18–20] Thus, the function of CKLF1 in DC maturation may involve the NF-κB pathway.[16] Furthermore, GC-Th cells were shown to significantly up-regulate the expression of C-X-C motif chemokine ligand (CXCL) 13, CKLF1 and inducible co-stimulator, which are important costimulatory molecules in the humoral immune response and germinal center formation.[21]
CKLF1 has two secreted forms at the C-terminus, known as C19 and C27.[22] The C19 peptide lacks eight amino acids at its N-terminus compared with the C27 peptide. This truncated region is important for receptor chemotaxis, calcium flux, and heparin binding.[23] Both C19 and C27 can interact with CCR4, whereas C27 exhibits more activity than C19. C27 acts as an agonist of CCR4, whereas C19 acts as an antagonist.[22] Furthermore, C19 and C27 inhibit stromal cell-derived factor-1 (SDF-1)-induced C-X-C motif chemokine receptor (CXCR) 4-mediated chemotaxis by binding to CCR4 and activating phosphatidyl inositol 3-kinase /protein kinase C pathway-mediated cross desensitization.[24] Both C19 and C27 may increase the capacity of immature DCs to induce proliferation and interferon (IFN)-γ production by T cells. C19 and C27 also up-regulate the expression of HLA-DR and the production of IL-12 in immature DCs.[25] Zheng et al[26] reported that intranasal treatment with large doses of C19 significantly reduced the numbers of Th2 cells. Finally, the expression of CKLF1 is up-regulated in various autoimmune diseases.[27]
CMTM1–5
CMTM1–4 is located in a gene cluster on chromosome 16q22.1.[28]CMTM1 consists of seven exons, six introns, and consists of 23 isoforms that are highly expressed in testis and tumor tissues. CMTM1 may play a role in spermatogenesis or testicular development and tumorigenesis.[29,30]CMTM2 is tightly linked with CMTM1, which is also highly expressed in testis, located in spermatogonia and secreted into the seminiferous tubules. Meanwhile, CMTM2 is expressed in bone marrow and peripheral cells, including CD4+T cells.[31] Intra-cellular CMTM2 can negatively regulate HIV-1 transcription in Jurkat (lymphoblastoid T-cell line) and U937 (human monocyte cell line) cells by targeting the AP-1 and cyclic adenosine monophosphate (AMP) response element binding (CREB) signaling pathways. CMTM2 may have unknown immune related functions.[32]
CMTM3 is highly expressed in the immune system and the male reproductive system.[4] Previous study reported that CMTM3 was highly expressed in peripheral blood mononuclear cells (PBMCs).[2] In a cDNA library prepared from peripheral blood, CMTM3 was predominantly expressed in resting B lymphocytes. CMTM3 is also highly expressed in CD4+ T lymphocytes and monocytes. These studies suggest that CMTM3 has a significant function in the immune system.[4] Moreover, CMTM3 can be released via exosomes.[33] CMTM4 has three transcript variants, CMTM4-v1, CMTM4-v2 and CMTM4-v3. CMTM4 is a regulator of the programmed cell death-ligand 1 (PD-L1) protein.[34]
CMTM5 is located on chromosome 14q11.2.[35] While studying granulopoietic gene expression, Liu et al[36] found that CMTM5 was expressed in the granulocyte system and is up-regulated during the process of differentiation. CMTM5 may have immune related functions. CMTM5-v1 is secreted via a vesicle-mediated secretory pathway.[37]
CMTM6–8
CMTM6–8 is located in a gene cluster on chromosome 3p22. CMTM6 is a widely expressed protein.[38] Studies have shown that CMTM6 regulates anti-tumor immunity by maintaining the expression of PD-L1.[39] Interference with CMTM6 expression resulted in impaired PD-L1 protein expression in all tested tumor cell types and primary human DCs.[39] Murine CMTM7 is highly expressed in immune cells.[40] Thus, CMTM7 may play an important role in the immune system. CMTM7 links B cell receptor (BCR) and B-cell linker protein (BLNK), and initiates BLNK-mediated signal transduction in B cells.[41] Zhang et al[40] demonstrated that CMTM7 plays a specific role in BCR expression and survival of B-1a cells. A recent study reported that Cmtm7 controls B-1a cell development at the transitional stage.[42] Moreover, increased numbers of B-1a cells are often associated with autoimmunity.[43] CMTM8 is more structurally similar to TM4SF11[2] and is a negative regulator of epidermal growth factor (EGF)-induced signaling.[44]
CMTM Members and Autoimmune Diseases
Autoimmune diseases represent a series of diseases, ranging from organ-specific diseases (antibodies and T cells react to localized autoantigens in specific tissues) to organ-non-specific or systemic diseases (characterized by reacting to antigens distributed throughout various tissues).[45] According to previous studies, CMTM members are highly expressed in the immune system.[2] Here, we discuss the relationship between the CMTM and autoimmune diseases.
CKLF1
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease characterized by an autoimmune response to self-antigens which can affect organs and tissues such as the brain, blood, and kidneys.[46,47] Lupus nephritis (LN) is a major risk factor for morbidity and mortality in SLE patients.[48] Previous studies have suggested that the pathogenesis of LN was associated with neutrophil activation. In a model of SLE, researchers reported that the disease may initiate pre-clinically with an IFN response followed by differentiation of B cells into plasmablasts.[49] As mentioned above, CKLF1 was associated with neutrophil activation, IFN production, and B cell function. CKLF1 may play a role in the pathogenesis of LN. The expression of the CKLF gene was found to be associated with the development of LN in a study of the molecular mechanism of renal inflammation during the progression, remission, and recurrence of LN in mice.[50] High expression of CKLF1 may be related to the excessive involvement of inflammatory cells in chemotaxis and CKLF1 may mediate the immune inflammatory reaction process observed in LN [Figure 2]. In this experiment, CMTM was transferred into SLE mice, resulting in high expression of CKLF1, which increased the urine protein level in SLE mice and aggravated the inflammatory reaction.[51] In a clinical study, the expression of CKLF1 in lupus patients was increased and positively correlated with the lupus active index compared with healthy controls.[52]
Figure 2.
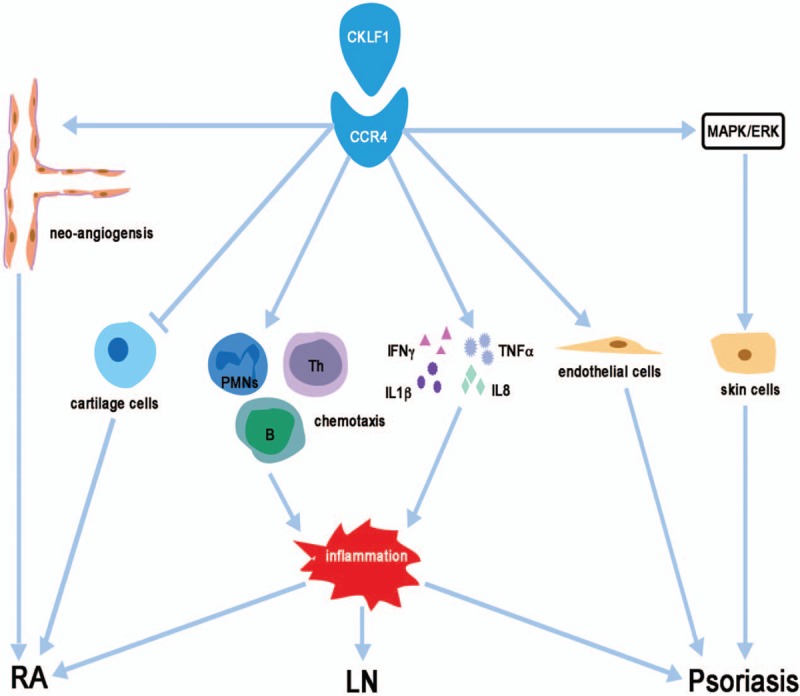
The major mechanisms regulated by CKLF1 in autoimmune diseases. CKLF1 may mediate the immune inflammatory reaction process in LN; CKLF1 may promote development and progression of the inflammatory response and neo-angiogenesis and inhibit cartilage cell proliferation in RA; CKLF1 may promote the proliferation of endothelial cells, participate in local inflammation and dysregulate the function of psoriatic skin cells. CCR4: C-C chemokine receptor 4; CKLF: Chemokine-like factor; ERK: Extracellular signal-regulated kinase; IFN: Interferon; IL: Interleukin: MAPK: Mitogen-activated protein kinase; PMNs: Polymorphonuclear leukocytes; LN: Lupus nephritis; Th: T helper cells; RA: Rheumatoid arthritis; TNF: Tumor necrosis factor.
Rheumatoid arthritis (RA) is a chronic inflammatory joint disease, which can lead to cartilage and bone damage. Early diagnosis is the key to optimal treatment.[53] T-cell response may be involved in the pathogenesis of RA.[54] An increase in citrulline-specific T-helper-1 cells was found in the circulation of patients with RA, especially in patients with early disease.[55] Leukocyte infiltration into the synovial and chemokine network play a role in the pathogenesis of RA.[56,57] Osteoarthritis (OA) is the most common joint disease that affects one or several diarthrodial joints.[58] OA is considered to be a disease of the whole joint, including articular cartilage, subchondral bone, ligament, articular capsule, and synovium, which eventually leads to joint failure.[59] Ankylosing spondylitis (AS) is a chronic inflammatory arthritis and the initiation of AS is unknown. T cells and TNF-α are involved in the pathogenesis of AS.[60] Chemokines play an important role in the pathogenesis of arthritis by inhibiting the synthesis and metabolism of articular cartilage and promoting the degradation of the matrix.[61] Rioja et al[62] reported that CKLF1, as measured by microarray analysis, was up-regulated in joints in a reactivation model of SCW-induced arthritis in Lewis (LEW/N) rats. A recent study found that the expression of CKLF1 was up-regulated in synovium from AS and RA patients, and CCR4 mRNA levels were increased in RA patients, but not in OA or AS patients. Meanwhile, this study also found that the expression levels of CKLF1 were correlated with C-reactive protein/erythrocyte sedimentation rate (ESR) and ESR in RA and AS patients, respectively.[63] Dysregulation of NF-κB plays an important role in the pathogenesis of arthritis.[64] Keith et al[65] reported that WAY-169916, a selective NF-κB transcriptional inhibitor, caused a marked decrease in CKLF1 expression in rat spleens. Consequently, CKLF may bind to CCR4 and promote development and progression of the inflammatory response and neo-angiogenesis in rheumatic diseases [Figure 2].[63] Furthermore, CKLF1 may inhibit cartilage cell proliferation, collagen, and protein polysaccharide synthesis.[66] Thus, CKLF1 may play an important role in the pathogenesis of arthritis.
Psoriasis is an immune-mediated genetic disease characterized by hyperproliferation of the epidermis, accumulation of inflammatory cells, and dilation of dermal papillary vessels.[67] CKLF1 is associated with the pathogenesis of psoriasis. A recent study has shown that CKLF1 and CCR4 are highly expressed in psoriatic lesions. CKLF1 may promote the proliferation of microvascular endothelial cells and contribute to local inflammation. It has been demonstrated that the MAPK/extracellular signal-regulated kinase (ERK) signaling pathway plays an important role in the pathophysiological function of psoriatic skin cells. Studies have shown that the C19 and C27 peptides bind to CCR4 and activate the MAPK/ERK pathway[68] [Figure 2].
CMTM1
Calcium (Ca2+) plays an important role in the pathogenesis of autoimmunity and inherited immunological dysregulation. Ca2+, as a second messenger, regulates many cell functions including gene transcription, apoptosis, and the immune response.[69] Previous studies have shown that intra-cellular Ca2+ signaling plays a role in the pathogenesis of RA.[70] Rheumatoid arthritis synovial fibroblasts (RASFs) are important effector cells that cause joint inflammation and deformities.[71] While studying the mechanism of celastrol in RA, Liu et al[72] found that the expression of CMTM1 was down-regulated in RASFs from patients treated with celastrol. They further found that celastrol treatment mobilized cytosolic Ca2+ in RASFs. As described, a regulatory role exists for celastrol-induced Ca2+ signaling in synovial fibroblasts of RA patients. Wong et al[73] also reported that CMTM1 may be suppressed by calmodulin.
CMTM2
In 2007, Kuttapitiya et al[74] used a detailed histologic and microarray analysis to investigate knee OA bone marrow lesions. They found that the expression of CMTM2 was down-regulated in OA patients compared with healthy controls.[74] Systemic sclerosis (SSc) is a chronic autoimmune disease characterized by progressive fibrosis of the skin and internal organs.[75] Previous studies indicated that SSc was associated with multiple susceptibility genes including HLA-classII, PTPN22, IRF5, and STAT4. While determining if polymorphisms in the C8orf13-BLK region were associated with SSc, Gourh et al[76] discovered that CMTM2 was differentially expressed in SSc peripheral blood cells between rs2736340- rs13277113 heterozygotes versus homozygotes. Pimentel-Santos et al[77] used a whole-genome microarray approach to identify candidate genes associated with AS and to measure gene expression changes occurring during the disease process. They discovered that CMTM2 was up-regulated in the peripheral blood of AS patients.[77]
CMTM3
Sjögren syndrome (SS) is an autoimmune disease characterized by lymphocytic infiltration of the exocrine glands resulting in significant reduction of saliva and tear production.[78] In 2011, Hu et al[79] utilized a protein microarray approach to identify salivary autoantibody biomarkers for primary SS (pSS). They reported that there was an elevated interaction of CMTM3 with autoantibodies in SLE and pSS patients compared with those from healthy control subjects.[79]
CMTM4
Encore is an open source network analysis pipeline for genome-wide association studies and rare variant data. To demonstrate Encore utility in the analysis of genetic sequencing data, Davis et al[80] analyzed the exome resequencing data from healthy individuals and those with SLE. They found a novel candidate gene, CMTM4, associated with their epistasis network model of the exome data.[80] Variability in DNA methylation levels contributes to recruitment and expression balance of inflammatory cytokines and to the development of autoimmune diseases. A better understanding of cytokine methylation variation among autoimmune diseases will help to identify potential epigenetic biomarkers and therapeutic targets.[81] Wang et al[81] reported that the methylation status of the CMTM4 promoter was significantly different in SLE, RA, and pSS.
CMTM5
The pathogenesis of lupus remains unclear. Dysregulation of gene expression, accompanied by accumulation of abnormal transcripts, may lead to apoptosis or increased expression of IFN-1.[82] In SLE patients, sense-antisense duplex RNA can drive IFN-1 expression and antisense transcripts can regulate cis transcription.[83] Shi et al[84] found approximately 5000 coding genes had antisense transcription strands in monocytes of SLE. The antisense transcriptional activity of CMTM5 was much higher compared with sense transcription in SLE patients.[84] CMTM5 may contribute to the immunologic dysregulation observed in SLE. As mentioned above, variabilities in methylation status of inflammatory cytokines play an important role in autoimmune diseases. Wang et al[81] also found that CMTM5 was hypermethylated in SLE and pSS, but hypomethylated in RA.
CMTM6
Anti-phospholipid syndrome (APS) is an autoimmune disorder characterized by arterial and/or venous thrombosis, pregnancy morbidity, and the presence of anti-phospholipid antibodies.[85] Previous studies have suggested that neutrophils are involved in the pathogenesis of APS. Neutrophil extracellular traps (NETs) play an important role in APS because neutrophils of APS patient are prone to spontaneous NET release.[86] Recently, Knight et al[87] found that CMTM6 was up-regulated in the neutrophils of APS patients.
CMTM7, CMTM8
At present, CMTM7 and CMTM8 have been identified primarily in tumor and cardiovascular diseases.[88–90] There are no relevant reports with respect to their involvement in immunologic diseases.
Several studies have indicated that CMTMs have a significant association with tumorigenesis and metastasis. For example, CMTM7 promotes the internalization of epidermal growth factor receptor (EGFR) and inhibits cell proliferation and metastasis. With respect to protein kinase (AKT) and ERK activation, Liu et al[91] found that CMTM7 down-regulated AKT phosphorylation and inhibited ERK activation in KYSE180 cells, thus inhibiting EGFR activation of downstream targets.[91] Previous studies concluded that cancer and autoimmune diseases represent fundamentally different pathological conditions. The fact that cancer and autoimmune diseases may sometimes occur in the same person indicates that there may be a connection between these two different clinical conditions.[92] The “internal pathway” occurring in tissue cells links cancer with inflammation, thus genetic events that activate oncogenes or inhibit tumor suppressor genes may also lead to the induction of inflammatory proteins. For example, EGFR activation can activate cyclooxygenase-2 by activation of transcription factors Sp1 and p38-MAPK.[93] Therefore, more studies are needed to define the relationship between CMTM and autoimmune diseases.
At present, a large number of studies have shown that CMTM is closely related to the function of the immune system. CMTM plays an important role in the pathogenesis of many autoimmune diseases. Previous studies have reported that CMTM is highly expressed in the immune system and participates in T cell and B cell activation. However, as a member of the chemokine superfamily, studies on the role of CMTM in autoimmune diseases are limited. The relationship between other family members and autoimmune diseases exists primarily at the level of expression, except for CKLF1 which has been studied in autoimmune diseases. The number of studies showing that CMTM is involved in the occurrence and development of autoimmune diseases has increased. CMTM may be useful as a new prognostic factor or therapeutic target in the future. This will require further understanding of how CMTM participates in the pathogenesis of autoimmune diseases.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (No. 81501390).
Conflicts of interest
None.
Footnotes
How to cite this article: Duan HJ, Li XY, Liu C, Deng XL. Chemokine-like factor-like MARVEL transmembrane domain-containing family in autoimmune diseases. Chin Med J 2020;133:951–958. doi: 10.1097/CM9.0000000000000747
References
- 1.Han W, Lou Y, Tang J, Zhang Y, Chen Y, Li Y, et al. Molecular cloning and characterization of chemokine-like factor 1 (CKLF1), a novel human cytokine with unique structure and potential chemotactic activity. Biochem J 2001; 357:127–135. doi: 10.1042/0264-6021:3570127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han W, Ding P, Xu M, Wang L, Rui M, Shi S, et al. Identification of eight genes encoding chemokine-like factor superfamily members 1-8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics 2003; 81:609–617. doi: 10.1016/s0888-7543(03)00095-8. [DOI] [PubMed] [Google Scholar]
- 3.Harris SL, Thorne LB, Seaman WT, Hayes DN, Couch ME, Kimple RJ. Association of p16(INK4a) overexpression with improved outcomes in young patients with squamous cell cancers of the oral tongue. Head Neck 2011; 33:1622–1627. doi: 10.1002/hed.21650. [DOI] [PubMed] [Google Scholar]
- 4.Zhong J, Wang Y, Qiu X, Mo X, Liu Y, Li T, et al. Characterization and expression profile of CMTM3/CKLFSF3. J Biochem Mol Biol 2006; 39:537–545. doi: 10.5483/bmbrep.2006.39.5.537. [DOI] [PubMed] [Google Scholar]
- 5.Imai T, Baba M, Nishimura M, Kakizaki M, Takagi S, Yoshie O. The T cell-directed CC chemokine TARC is a highly specific biological ligand for CC chemokine receptor 4. J Biol Chem 1997; 272:15036–15042. doi: 10.1074/jbc.272.23.15036. [DOI] [PubMed] [Google Scholar]
- 6.Imai T, Chantry D, Raport CJ, Wood CL, Nishimura M, Godiska R, et al. Macrophage-derived chemokine is a functional ligand for the CC chemokine receptor 4. J Biol Chem 1998; 273:1764–1768. doi: 10.1074/jbc.273.3.1764. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Zhang YM, Yang X, Han WL, Liu YN, Xu QM, et al. Chemokine-like factor 1 is a functional ligand for CC chemokine receptor 4 (CCR4). Life Sci 2006; 78:614–621. doi: 10.1016/j.lfs.2005.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Zhang Y, Yang X, Han W, Liu Y, Xu Q, et al. Chemokine-like factor 1 is a functional ligand for CC chemokine receptor 4 (CCR4). Life Sci 2006; 78:614–621. doi: 10.1016/j.lfs.2005.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark-Lewis I, Kim KS, Rajarathnam K, Gong JH, Dewald B, Moser B, et al. Structure-activity relationships of chemokines. J Leukoc Biol 1995; 57:703–711. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- 10.D’Ambrosio D, Panina-Bordignon P, Sinigaglia F. Chemokine receptors in inflammation: an overview. J Immunol Methods 2003; 273:3–13. doi: 10.1016/S0022-1759(02)00414-3. [DOI] [PubMed] [Google Scholar]
- 11.Ke X, Jia L, Jing H, Liu Y, Zhang Y, Di C. Effects of novel human chemokine-like factor 1 (CKLF1) on bone marrow hematopoietic stem cell/progenitor cell in vitro (in Chinese). Chin J Hematol 2002; 23:301–303. [PubMed] [Google Scholar]
- 12.Licastro F, Davis LJ, Morini MC. Lectins and superantigens - membrane interactions of these compounds with T-lymphocytes affect immune-responses. Int J Biochem 1993; 25:845–852. doi: 10.1016/0020-711x(93)90239-B. [DOI] [PubMed] [Google Scholar]
- 13.Fan J, Nishanian P, Breen EC, McDonald M, Fahey JL. Cytokine gene expression in normal human lymphocytes in response to stimulation. Clin Diagn Lab Immun 1998; 5:335–340. doi: 10.1089/aid.1998.14.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Zhong J, Chen YY, Qiu XY, Zhang T, Ma DL, et al. Expression of chemokine-like factor 1 is upregulated during T lymphocyte activation. Life Sci 2006; 79:519–524. doi: 10.1016/j.lfs.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 15.Kong LL, Wang ZY, Han N, Zhuang XM, Wang ZZ, Li H, et al. Neutralization of chemokine-like factor 1, a novel C-C chemokine, protects against focal cerebral ischemia by inhibiting neutrophil infiltration via MAPK pathways in rats. J Neuroinflammation 2014; 11:112.doi: 10.1186/1742-2094-11-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao L, Li T, Mo X, Majdic O, Zhang Y, Seyerl M, et al. Expressional and functional studies of CKLF1 during dendritic cell maturation. Cell Immunol 2010; 263:188–195. doi: 10.1016/j.cellimm.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 17.Dai SY, Nakagawa R, Itoh A, Murakami H, Kashio Y, Abe H, et al. Galectin-9 induces maturation of human monocyte-derived dendritic cells. J Immunol 2005; 175:2974–2981. doi: 10.4049/jimmunol.175.5.2974. [DOI] [PubMed] [Google Scholar]
- 18.Li G, Wang D, Sun M, Li G, Hu J, Zhang Y, et al. Discovery and optimization of novel 3-piperazinylcoumarin antagonist of chemokine-like factor 1 with oral antiasthma activity in mice. J Med Chem 2010; 53:1741–1754. doi: 10.1021/jm901652p. [DOI] [PubMed] [Google Scholar]
- 19.Sun M, Hu J, Song X, Wu D, Kong L, Sun Y, et al. Coumarin derivatives protect against ischemic brain injury in rats. Eur J Med Chem 2013; 67:39–53. doi: 10.1016/j.ejmech.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 20.Ai Q, Chen C, Chu S, Luo Y, Zhang Z, Zhang S, et al. IMM-H004 protects against cerebral ischemia injury and cardiopulmonary complications via CKLF1 mediated inflammation pathway in adult and aged rats. Int J Mol Sci 2019; 20:E1661.doi: 10.3390/ijms20071661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim CH, Lim HW, Kim JR, Rott L, Hillsamer P, Butcher EC. Unique gene expression program of human germinal center T helper cells. Blood 2004; 104:1952–1960. doi: 10.1182/blood-2004-03-1206. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Zhang Y, Han W, Li D, Tian L, Yin C, et al. Two C-terminal peptides of human CKLF1 interact with the chemokine receptor CCR4. Int J Biochem Cell Biol 2008; 40:909–919. doi: 10.1016/j.biocel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Zhang S, Ling X, Li Y, Zhang Y, Han W, et al. Analysis of the interactions between the peptides from secreted human CKLF1 and heparin using capillary zone electrophoresis. J Pept Sci 2008; 14:984–988. doi: 10.1002/psc.1028. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Tian L, Zheng Y, Qi H, Guo C, Sun Q, et al. C-terminal peptides of chemokine-like factor 1 signal through chemokine receptor CCR4 to cross-desensitize the CXCR4. Biochem Biophys Res Commun 2011; 409:356–361. doi: 10.1016/j.bbrc.2011.05.047. [DOI] [PubMed] [Google Scholar]
- 25.Shao LN, Li T, Mo XN, Majdic O, Zhang YF, Seyerl M, et al. Expressional and functional studies of CKLF1 during dendritic cell maturation. Cell Immunol 2010; 263:188–195. doi: 10.1016/j.cellimm.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Y, Guo CY, Zhang Y, Qi H, Sun QY, Xu EQ, et al. Alleviation of murine allergic rhinitis by C19, a C-terminal peptide of chemokine-like factor 1 (CKLF1). Int Immunopharmacol 2011; 11:2188–2193. doi: 10.1016/j.intimp.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Zhong J, Chen Y, Qiu X, Zhang T, Ma D, et al. Expression of chemokine-like factor 1 is upregulated during T lymphocyte activation. Life Sci 2006; 79:519–524. doi: 10.1016/j.lfs.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 28.Lu J, Wu QQ, Zhou YB, Zhang KH, Pang BX, Li L, et al. Cancer research advance in CKLF-like MARVEL transmembrane domain containing member family (review). Asian Pac J Cancer Prev 2016; 17:2741–2744. [PubMed] [Google Scholar]
- 29.Wang L, Wu C, Zheng Y, Qiu X, Wang L, Fan H, et al. Molecular cloning and characterization of chemokine-like factor super family member 1 (CKLFSF1), a novel human gene with at least 23 alternative splicing isoforms in testis tissue. Int J Biochem Cell Biol 2004; 36:1492–1501. doi: 10.1016/j.biocel.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 30.Si J, Zhang P, Tian D, Wang X, Ma Y, Zhang J, et al. CMTM1_v17 is associated with chemotherapy resistance and poor prognosis in non-small cell lung cancer. World J Surg Oncol 2017; 15:34.doi: 10.1186/s12957-016-1094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi S, Rui M, Han W, Wang Y, Qiu X, Ding P, et al. CKLFSF2 is highly expressed in testis and can be secreted into the seminiferous tubules. Int J Biochem Cell Biol 2005; 37:1633–1640. doi: 10.1016/j.biocel.2004.04.028. [DOI] [PubMed] [Google Scholar]
- 32.Song HS, Shi SA, Lu XZ, Gao F, Yan L, Wang Y, et al. Intracellular CMTM2 negatively regulates human immunodeficiency virus type-1 transcription through targeting the transcription factors AP-1 and CREB. Chin Med J 2010; 123:2440–2445. doi: 10.3760/cma.j.issn.0366-6999.2010.17.027. [PubMed] [Google Scholar]
- 33.Liu B, Li H, Fu W, Cheng Y, Yuan W, Liu W, et al. CMTM3 presents a secreted form released via exosomes. Acta Biochim Biophys Sin (Shanghai) 2016; 48:584–586. doi: 10.1093/abbs/gmw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 2017; 549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li P, Liu K, Li L, Yang MX, Gao WJ, Feng JB, et al. Reduced CMTM5 expression correlates with carcinogenesis in human epithelial ovarian cancer. Int J Gynecol Cancer 2011; 21:1248–1255. doi: 10.1097/IGC.0b013e3182259c31. [DOI] [PubMed] [Google Scholar]
- 36.Liu PC, Barb J, Woodhouse K, Taylor JG, Munson PJ, Raghavachari N. Transcriptome profiling and sequencing of differentiated human hematopoietic stem cells reveal lineage-specific expression and alternative splicing of genes. Physiol Genomics 2011; 43:1117–1134. doi: 10.1152/physiolgenomics.00099.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li HN, Guo XH, Shao LN, Plate M, Mo XN, Wang Y, et al. CMTM5-v1, a four-transmembrane protein, presents a secreted form released via a vesicle-mediated secretory pathway. BMB Rep 2010; 43:182–187. doi: 10.5483/BMBRep.2010.43.3.182. [DOI] [PubMed] [Google Scholar]
- 38.Yafune A, Kawai M, Itahashi M, Kimura M, Nakane F, Mitsumori K, et al. Global DNA methylation screening of liver in piperonyl butoxide-treated mice in a two-stage hepatocarcinogenesis model. Toxicol Lett 2013; 222:295–302. doi: 10.1016/j.toxlet.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 2017; 549:101–105. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Wang JY, Han W. A role for CMTM7 in BCR expression and survival in B-1a but not B-2 cells. Int Immunol 2014; 26:47–57. doi: 10.1093/intimm/dxt042. [DOI] [PubMed] [Google Scholar]
- 41.Miyazaki A, Yogosawa S, Murakami A, Kitamura D. Identification of CMTM7 as a transmembrane linker of BLNK and the B-Cell receptor. Plos One 2012; 7:e31829.doi: 10.1371/journal.pone.0031829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, Liu Y, Li T, Wang P, Mo X, Lv P, et al. Cmtm7 knockout inhibits B-1a cell development at the transitional (TrB-1a) stage. Int Immunol 2019; 31:715–728. doi: 10.1093/intimm/dxz041. [DOI] [PubMed] [Google Scholar]
- 43.Youinou P, Mackenzie L, Katsikis P, Merdrignac G, Isenberg DA, Tuaillon N, et al. The relationship between CD5-expressing B lymphocytes and serologic abnormalities in rheumatoid arthritis patients and their relatives. Arthritis Rheum 1990; 33:339–348. doi: 10.1002/art.1780330306. [DOI] [PubMed] [Google Scholar]
- 44.Jin C, Ding P, Wang Y, Ma D. Regulation of EGF receptor signaling by the MARVEL domain-containing protein CKLFSF8. FEBS Lett 2005; 579:6375–6382. doi: 10.1016/j.febslet.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Wang FS, Gershwin ME. Human autoimmune diseases: a comprehensive update. J Intern Med 2015; 278:369–395. doi: 10.1111/joim.12395. [DOI] [PubMed] [Google Scholar]
- 46.La Paglia GMC, Leone MC, Lepri G, Vagelli R, Valentini E, Alunno A, et al. One year in review 2017: systemic lupus erythematosus. Clin Exp Rheumatol 2017; 35:551–561. [PubMed] [Google Scholar]
- 47.Aringer M, Schneider M. Systemic lupus erythematosus. Dtsch Med Wochenschr 2016; 141:537–543. doi: 10.1055/s-0041-110604. [DOI] [PubMed] [Google Scholar]
- 48.Alarcon GS. Multiethnic lupus cohorts: what have they taught us? Reumatol Clin 2011; 7:3–6. doi: 10.1016/j.reuma.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Almaani S, Meara A, Rovin BH. Update on lupus nephritis. Clin J Am Soc Nephrol 2017; 12:825–835. doi: 10.2215/CJN.05780616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bethunaickan R, Berthier CC, Zhang W, Eksi R, Li HD, Guan YF, et al. Identification of stage-specific genes associated with lupus nephritis and response to remission induction in NZB/W and NZM2410 mice. Arthritis Rheumatol 2014; 66:2246–2258. doi: 10.1002/art.38679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vielhauer V, Anders HJ, Schlondorff D. Chemokines and chemokine receptors as therapeutic targets in lupus nephritis. Semin Nephrol 2007; 27:81–97. doi: 10.1016/j.semnephrol.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 52.Ji Y, Zhang H, Yuan H, Yang GP, Zhang K, Xie LH. Expression of chemokine like factor-1 in nephridial tissue of lupus nephritis (in Chinese). J Central South Univ (Med Ed) 2007; 32:490–493. [PubMed] [Google Scholar]
- 53.Scott DL, Wolfe F, Huizinga TW. Rheumatoid arthritis. Lancet 2010; 376:1094–1108. doi: 10.1016/S0140-6736(10)60826-4. [DOI] [PubMed] [Google Scholar]
- 54.Klarenbeek PL, de Hair MJ, Doorenspleet ME, van Schaik BD, Esveldt RE, van de Sande MG, et al. Inflamed target tissue provides a specific niche for highly expanded T-cell clones in early human autoimmune disease. Ann Rheum Dis 2012; 71:1088–1093. doi: 10.1136/annrheumdis-2011-200612. [DOI] [PubMed] [Google Scholar]
- 55.James EA, Rieck M, Pieper J, Gebe JA, Yue BB, Tatum M, et al. Citrulline-specific Th1 cells are increased in rheumatoid arthritis and their frequency is influenced by disease duration and therapy. Arthritis Rheumatol 2014; 66:1712–1722. doi: 10.1002/art.38637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med 2011; 365:2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 57.Feldmann M, Maini SR. Role of cytokines in rheumatoid arthritis: an education in pathophysiology and therapeutics. Immunol Rev 2008; 223:7–19. doi: 10.1111/j.1600-065X.2008.00626.x. [DOI] [PubMed] [Google Scholar]
- 58.Hunter DJ, Bierma-Zeinstra S. Osteoarthritis. Lancet 2019; 393:1745–1759. doi: 10.1016/S0140-6736(19)30417-9. [DOI] [PubMed] [Google Scholar]
- 59.Martel-Pelletier J, Wildi LM, Pelletier JP. Future therapeutics for osteoarthritis. Bone 2012; 51:297–311. doi: 10.1016/j.bone.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 60.Braun J, Bollow M, Neure L, Seipelt E, Seyrekbasan F, Herbst H, et al. Use of immunohistologic and in-situ hybridization techniques in the examination of sacroiliac joint biopsy specimens from patients with ankylosing-spondylitis. Arthritis Rheum 1995; 38:499–505. doi: 10.1002/art.1780380407. [DOI] [PubMed] [Google Scholar]
- 61.Borzi RM, Mazzetti I, Cattini L, Uguccioni M, Baggiolini M, Facchini A. Human chondrocytes express functional chemokine receptors and release matrix-degrading enzymes in response to C-X-C and C-C chemokines. Arthritis Rheum 2000; 43:1734–1741. doi: 10.1002/1529-0131(200008)43:8<1734::Aid-Anr9>3.0.Co;2-B. [DOI] [PubMed] [Google Scholar]
- 62.Rioja I, Clayton CL, Graham SJ, Life PF, Dickson MC. Gene expression profiles in the rat streptococcal cell wall-induced arthritis model identified using microarray analysis. Arthritis Res Ther 2005; 7:R101–R117. doi: 10.1186/ar1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao K, Tang X, Wang B, Li RJ, Zhang BQ, Lin JH, et al. Distinct expression of chemokine-like factor 1 in synovium of osteoarthritis, rheumatoid arthritis and ankylosing spondylitis. J Huazhong Univ Sci-Med 2016; 36:70–76. doi: 10.1007/s11596-016-1544-4. [DOI] [PubMed] [Google Scholar]
- 64.Tak PP, Firestein GS. NF-kappaB: a key role in inflammatory diseases. J Clin Invest 2001; 107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keith JC, Albert LM, Leathurby Y, Follettie M, Wang LL, Borges-Marcucci L, et al. The utility of pathway selective estrogen receptor ligands that inhibit nuclear factor-kappa B transcriptional activity in models of rheumatoid arthritis. Arthritis Res Ther 2005; 7:R427–R438. doi: 10.1186/ar1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng A, Han W, Ma D, Lou S. Effects of chemokine-like factor 1 (CKLF1) on proliferation and metabolism of chondrocytes (in Chinese). J Peking Univ (Healt Sci) 2003; 35:399–401. doi: 10.1142/S0252959903000104. [PubMed] [Google Scholar]
- 67.MacDonald A, Burden AD. Psoriasis: advances in pathophysiology and management. Postgrad Med J 2007; 83:690–697. doi: 10.1136/pgmj.2007.061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan Y, Wang Y, Li L, Xia J, Peng S, He Y. Chemokine-like factor 1-derived C-terminal peptides induce the proliferation of dermal microvascular endothelial cells in psoriasis. PLoS One 2015; 10:e0125073.doi: 10.1371/journal.pone.0125073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clapham DE. Calcium signaling. Cell 2007; 131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 70.Izquierdo JH, Bonilla-Abadia F, Canas CA, Tobon GJ. Calcium, channels, intracellular signaling and autoimmunity. Reumatol Clin 2014; 10:43–47. doi: 10.1016/j.reuma.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 71.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev 2010; 233:233–255. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Seabra Rodrigues Dias IR, Mok SWF, Gordillo-Martinez F, Khan I, Hsiao WWL, Law BYK, et al. The calcium-induced regulation in the molecular and transcriptional circuitry of human inflammatory response and autoimmunity. Front Pharmacol 2017; 8:962.doi: 10.3389/fphar.2017.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wong VKW, Qiu C, Xu SW, Law BYK, Zeng W, Wang H, et al. Ca(2+) signalling plays a role in celastrol-mediated suppression of synovial fibroblasts of rheumatoid arthritis patients and experimental arthritis in rats. Br J Pharmacol 2019; 176:2922–2944. doi: 10.1111/bph.14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuttapitiya A, Assi L, Laing K, Hing C, Mitchell P, Whitley G, et al. Microarray analysis of bone marrow lesions in osteoarthritis demonstrates upregulation of genes implicated in osteochondral turnover, neurogenesis and inflammation. Ann Rheum Dis 2017; 76:1764–1773. doi: 10.1136/annrheumdis-2017-211396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chizzolini C. T cells, B cells, and polarized immune response in the pathogenesis of fibrosis and systemic sclerosis. Curr Opin Rheumatol 2008; 20:707–712. doi: 10.1097/BOR.0b013e32830c45ae. [DOI] [PubMed] [Google Scholar]
- 76.Gourh P, Agarwal SK, Martin E, Divecha D, Rueda B, Bunting H, et al. Association of the C8orf13-BLK region with systemic sclerosis in North-American and European populations. J Autoimmun 2010; 34:155–162. doi: 10.1016/j.jaut.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pimentel-Santos FM, Ligeiro D, Matos M, Mourao AF, Costa J, Santos H, et al. Whole blood transcriptional profiling in ankylosing spondylitis identifies novel candidate genes that might contribute to the inflammatory and tissue-destructive disease aspects. Arthritis Res Ther 2011; 13:R57.doi: 5710.1186/ar3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fox RI. Sjogren's syndrome. Lancet 2005; 366:321–331. doi: 10.1016/S0140-6736(05)66990-5. [DOI] [PubMed] [Google Scholar]
- 79.Hu S, Vissink A, Arellano M, Roozendaal C, Zhou H, Kallenberg CGM, et al. Identification of autoantibody biomarkers for primary Sjogren's syndrome using protein microarrays. Proteomics 2011; 11:1499–1507. doi: 10.1002/pmic.201000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davis NA, Lareau CA, White BC, Pandey A, Wiley G, Montgomery CG, et al. Encore: genetic association interaction network centrality pipeline and application to SLE exome data. Genet Epidemiol 2013; 37:614–621. doi: 10.1002/gepi.21739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang XQ, Lei DY, Ding J, Liu S, Tao L, Zhang F, et al. A DNA-methylated sight on autoimmune inflammation network across RA, pSS, and SLE. J Immunol Res 2018; 2018:4390789.doi: 10.1155/2018/4390789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alexander JJ, Saxena AK, Bao LH, Jacob A, Haas M, Quigg RJ. Prominent renal expression of a murine leukemia retrovirus in experimental systemic lupus erythematosus. J Am Soc Nephrol 2002; 13:2869–2877. doi: 10.1097/01.Asn.0000036868.73317.7a. [DOI] [PubMed] [Google Scholar]
- 83.Tranter M, Helsley RN, Paulding WR, McGuinness M, Brokamp C, Haar L, et al. Coordinated post-transcriptional regulation of Hsp70.3 gene expression by microRNA and alternative polyadenylation. J Biol Chem 2011; 286:29828–29837. doi: 10.1074/jbc.M111.221796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi LH, Zhang Z, Yu AM, Wang W, Wei Z, Akhter E, et al. The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. Plos One 2014; 9:e93846.doi: 10.1371/journal.pone.0093846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyakis S, Lockshin MD, Atsumi T, Branch DW, Brey RL, Cervera R, et al. International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS). J Thromb Haemost 2006; 4:295–306. doi: 10.1111/j.1538-7836.2006.01753.x. [DOI] [PubMed] [Google Scholar]
- 86.Yalavarthi S, Gould TJ, Rao AN, Mazza LF, Morris AE, Nunez-Alvarez C, et al. Release of neutrophil extracellular traps by neutrophils stimulated with antiphospholipid antibodies: a newly identified mechanism of thrombosis in the antiphospholipid syndrome. Arthritis Rheumatol 2015; 67:2990–3003. doi: 10.1002/art.39247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Knight JS, Meng H, Coit P, Yalavarthi S, Sule G, Gandhi AA, et al. Activated signature of antiphospholipid syndrome neutrophils reveals potential therapeutic target. JCI Insight 2017; 2:93897.doi: 10.1172/jci.insight.93897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu B, Su Y, Li T, Yuan W, Mo X, Li H, et al. CMTM7 knockdown increases tumorigenicity of human non-small cell lung cancer cells and EGFR-AKT signaling by reducing Rab5 activation. Oncotarget 2015; 6:41092–41107. doi: 10.18632/oncotarget.5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morrison AC, Felix JF, Cupples LA, Glazer NL, Loehr LR, Dehghan A, et al. Genomic variation associated with mortality among adults of European and African Ancestry with heart failure the cohorts for heart and aging research in genomic epidemiology consortium. Circ-Cardiovasc Gene 2010; 3:248–264. doi: 10.1161/Circgenetics.109.895995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang WJ, Qi H, Mo XN, Sun QY, Li T, Song QS, et al. CMTM8 is frequently downregulated in multiple solid tumors. Appl Immunohisto Mol Morphol 2017; 25:122–128. doi: 10.1097/Pai.0000000000000274. [DOI] [PubMed] [Google Scholar]
- 91.Liu Q, Su Y, Jiang GC, Zhou ZL, Liu BC, Bu L, et al. Change of CMTM7 expression, a potential tumor suppressor, is associated with poor clinical outcome in human non-small cell lung cancer. Chin Med J 2013; 126:3006–3012. doi: 10.3760/cmaj.issn.0366-6999.20123625. [PubMed] [Google Scholar]
- 92.Rahat MA, Shakya J. Parallel aspects of the microenvironment in cancer and autoimmune disease. Mediat Inflamm 2016; 2016:4375120.doi: 10.1155/2016/4375120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Del Prete A, Allavena P, Santoro G, Fumarulo R, Corsi MM, Mantovani A. Molecular pathways in cancer-related inflammation. Biochem Medica 2011; 21:264–275. doi: 10.11613/bm.2011.036. [DOI] [PubMed] [Google Scholar]


