Abstract
Background
Gastric cancer (GC) is one of the most globally prevalent cancers in the world. The pathogenesis of GC has not been fully elucidated, and there still lacks effective targeted therapeutics. The influence of altered kinesin superfamily protein 22 (KIF22) expression in GC progression is still unclearly. The aim of this study was to investigate the KIF22 effects on GC and related mechanisms.
Methods
Gastric carcinoma tissues and matching non-cancerous tissues were collected from patients with GC who have accepted a radical gastrectomy in Lanzhou University Second Hospital from May 2013 to December 2014. The expression of KIF22 was examined in GC of 67 patients and 20 para-carcinoma tissues by immunochemical staining. The relationship between the expression of KIF22 and clinicopathologic characteristics was next investigated in the remaining 52 patients except for 15 patients who did not complete follow-up for 5 years. Cell viability was performed via 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) test and colony formation assay in the MGC-803 and BGC-823 GC cells. Cell scratch and trans-well invasion assay was performed to assess migration ability in the MGC-803 and BGC-823 GC cells. Gene set enrichment analysis (GSEA) pathway enrichment analysis was performed to explore the potential functions. Cell cycle was detected by flow cytometry. In addition, the two GC cell lines were used to elucidate the underlying mechanism of KIF22 in GC in vitro via assessing the effects on mitogen-activated protein kinase and extracellular regulated protein kinases (MAPK/ERK) signal transduction pathway-related expressions by Western blotting assays. The differences were compared by t tests, one-way analysis of variance, and Chi-squared tests.
Results
The study showed that KIF22 was up-regulated in GC, and KIF22 high expression was significantly related to differentiation degree (χ2 = 12.842, P = 0.002) and poorly overall survivals. GSEA pathway enrichment analysis showed that KIF22 was correlated with the cell cycle. Silence of KIF22 decreased the ability of the proliferation and migration in gastric cells, induced G1/S phase cell cycle arrest via regulating the MAPK-ERK pathways.
Conclusions
KIF22 protein level was negatively correlated with prognosis. KIF22 knockdown might inhibit proliferation and metastasis of GC cells via the MAPK-ERK signaling pathway.
Keywords: Kinesin superfamily protein 22, Gastric cancer, MAPK-ERK
Introduction
As one of the most globally prevalent cancers, gastric cancer (GC) has an incidence in the world ranking sixth and a mortality ranking fourth in all kinds of cancers according to “National Cancer Report 2019” published by the National Cancer Center.[1] However, the pathogenesis of GC has not been fully elucidated, and there still lacks effective targeted therapeutics.[2] Chemotherapy is currently widely used to treat all kinds of cancers, including GC, by targeting the mitotic spindle in mitosis such as taxanes and vinca alkaloids.[3–5] However, these drugs with spindle poison have potent side effects, such as dose-dependent toxicities on many target organs,[6] and innate or acquired drug resistance.[7] It is urgent to find new chemicals to target the mitotic spindle with fewer side effects. As a type of molecular motor proteins dependent on the microtubule, the kinesin superfamily proteins (KIFs) are crucial for mitosis. These proteins play an important role in regulating some important functional molecules.[8,9] Current studies have shown that alterations of their expression are related to carcinogenesis for various cancer.[10–13] Thus, we have been attempting to screen a latent molecular target to diagnose and treat advanced GC by analyzing KIF gene expressions based on profiles of Gene Expression Profilling Interactive Analysis (GEPIA) database. Throughout these screenings in this study, we identified member 22 of the kinesin family, that is, kinesin-like DNA-binding protein (Kid), as potential candidate target genes for the treatment of GC. KIF22 is encoded by the Kinesin-like 4 (KNSL4) gene and localizes to the mitotic microtubules of cells. It is a positive microtubule-driven protein and essential for cell mitosis. It is also a crucial protein during the process of cell division, centrosome separation, as well as spindle formation. Given that KIF22 is vital in assembling and maintaining the mitotic spindle, it should be as a critical role in the cancer cellular mitosis and proliferation. Although there are some reports on the microtubule dynamics and biochemical characterization of KIF22 mentioned above, its role remains largely unknown in GC's development and progression.
By using immunohistochemistry, the KIF22's expression and distribution in human GC were examined in this study; also, the relationship of KIF22 positivity and clinicopathologic features was examined. Furthermore, the impact of suppressing KIF22 expression with RNA interference (RNAi) on the GC cells’ proliferation and migration was analyzed; its mechanism in promoting the GC occurrence was then illuminated. Collectively, KIF22 was indicated by these data that might have an important effect on cancer pathogenesis.
Methods
Gene expression data and enrichment pathway analysis
The KIF22 gene expression analysis of different tumors used from GEPIA database (http://gepia.cancer-pku.cn/index.html) is publically available. Gene set enrichment analysis (GSEA) pathway enrichment analysis was performed to reveal the potential functions.
Samples and ethical approval
Human tissue paraffin-embedded samples were collected from patients with GC who obtained the treatment of surgical resection in Lanzhou University Second Hospital from May 2013 to December 2014. The Medical Ethics Committee of Lanzhou University Second Hospital approved this study, and the informed consent was signed by all patients in this study. A total of 67 patients with primary GC were involved in this study, among which we excluded 15 patients for failing to follow-up at least 5 years. The remaining 52 patients including 38 males and 14 females were followed up for 5 years. None of the 52 patients was pretreated with chemotherapy or radiotherapy prior to surgery. All of the 52 specimens were confirmed through pathological examination, as well as staged primary tumor/regional lymph nodes/distant metastasis (TNM) according to The Union for International Cancer Control (UICC) classification (TNM 2010).
Cell culture
The MGC-803 and BGC-823 GC cells were purchased from and identified by the Shanghai Cell Bank of Type Culture Collection of Chinese Academy of Sciences (China). Short tandem repeat typing was used to previously authenticated the two cell lines. All the two cells were cultured at room temperature (RT, 37°C) and 5% CO2 in a humidified incubator, in which Dulbecco's modified Eagle's medium (Gibco, Gaithersburg, MD, USA) with a supplement of heat-inactivated fetal bovine serum (10%; FBS; Hyclone, Logan, UT, USA) was contained. Additionally, the KIF22 expression was detected through Western blotting. Beta-actin was set as a reference.
siRNA transfection
Small interfering RNA (siRNA) was employed to inhibit KIF22 expression. KIF22 and siRNA were synthesized using GenePharma (Shanghai, China), and the following sequences were: siRNA-1, sense, 5′-GGUCCAAGGAGGUGAUCAATT-3′, antisense, 5′-UUGAUCACCUCCUUGGACCTT-3′; siRNA 2, sense, 5′-AGAGAAGGACCUAGAGAUUTT-3′, antisense, 5′-AAUCUCUAGGUCCUUCUCUTT-3′; siRNA 3, sense, 5′-CACCAGGAGACUCUCAAAUTT-3′, antisense, 5′-AUUUGAGAGUCUCCUGGUGTT-3′. Control: sense, 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. After Western blotting identification, the final two target sequence after competition we selected with best inhibition effect for the KIF22 were the siRNA 1 and siRNA 2 and a corresponding negative control. Assays for evaluating gene silencing efficiency were performed 60 h after transfection by Western blotting. After 60 h, cells were harvested for analysis.
Cellular proliferation and colony formation assay
The two GC cells (MGC-803 and BGC-823) with an infection of negative control or siRNA targeting KIF22 were seeded at a density of 2000 cells in 100 μL/well in a 96-well culture plate. They were then placed at RT (37°C) under 5% CO2 in an incubator. Cell proliferation rates were determined using 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) test. At 490 nm, the absorbance values were read. siRNA targeting KIF22 or negative control cells were seeded for the colony formation assay in the culture plates of 6-well tissue. Paraformaldehyde (10%) was used to fix the cell colonies after 14 days of incubation, and then they were stained by Giemsa staining. By using a light microscope, the colonies, including over 50 cells were counted.
Cell scratch and trans-well invasion assay
By a cell scratch assay, the migration ability was evaluated. Six-well cell culture plates were prepared, into which 3 × 105 cells were seeded. The generation of three parallel and linear wounds was available in each plate with a Micro Scratch Tester. The migration was then measured after an extra 24-h culture. The invasive ability was assessed by an invasion assay of trans well which was coated with matrix. With a suspension in cell culture medium (500 μL), 1 × 105 cells were added to the top chamber of a 24-well plate which was coated with matrix and then incubated for 24 h. The membranes were fixed, and then a crystal violet solution was used to fix stain the membranes for 10 min. An investigation was conducted to count the number of cells with an invasion into the membrane.
Flow cytometry assay of the cell cycle
Flow cytometry was applied to detect cell cycle. Cells were collected by centrifugation (1000 r/min, 5 min), then fixed with 75% ethanol for 2 h at 4°C, and stained with propidium (PI). Cells were analyzed by flow cytometry at a speed of 300 to 500 cells/s.
Western blotting
In a radio-immunoprecipitation assay (RIPA) lysis buffer, the proteins were extracted from GC cells. Ten percents of polyacrylamide sodium dodecyl sulfate (SDS) gels (SDS-PAGE) were used to separate equivalent amounts of proteins. The proteins were then blotted onto membranes of polyvinylidene fluoride (Amersham, UK). After membranes were blocked at RT (37°C) in bovine serum albumin (5%) for 1 h, primary antibodies were used to culture the membranes overnight. After being cultured, they were placed in secondary antibodies conjugated with human resource planning (Perbio Science, Belgium), and incubated at RT (37°C) for 1 h. The system of enhanced chemiluminescence immunodetection (Immobilon, USA) was used to visualize immunoreactive proteins. The antibodies for Western blotting were as follows: anti-KIF22 (13403-1-AP; Proteintech Group, USA), anti-Phospho-MEK1/2 (CST: #9154; Proteintech Group), anti-MEK (CST: #8727; Cell Signaling Technology, USA), anti-Phospho-ERK1/2 (CST: #4370; Proteintech Group), anti-ERK (16443-1-AP; Proteintech Group), anti-Cyclin A2 (18202-1-AP; Proteintech Group), anti-Cyclin B1 (55004-1-AP; Proteintech Group), anti-Cyclin D1 (26939-1-AP; Proteintech Group), anti-P21 (10355-1-AP; Proteintech Group), and β-actin (20536-1-AP; Proteintech Group).
Immunohistochemistry and scoring
Sections of 5-μm-thick para-carcinoma tissues and gastric tumors were washed in xylene twice, 15 min for each time. Then, they were washed for sequential 10 min: xylene (50%)/ethanol mixture (50%), ethanol (100%, 95%, 85%, and 75%), double distilled H2O (ddH2O), and H2O2 (3%). Sections were blocked for 30 min in serum (10%) after antigen retrieval. Anti-KIF22 (13403-1-AP; Proteintech Group) was used to incubate the sections in a humidified chamber overnight at 4°C. An UltraSensitive™ SP kit (Fuzhou Maixin Biotech. Co., Ltd., China) was then used to incubate them in humidified chamber at RT (37°C) for 1 h. An inverted microscope (XDS-100; Shanghai Cai Kang Optical Instrument Co., Ltd., China) was applied to take photos after 3,3’ diaminobenzidine tetrahydrochloride (DAB) and hematoxylin staining. As described previously, two pathologists conducted the scores of KIF22 immunostaining in accordance with the stained cell proportion and staining strength of each section. The intensity was scored as “zero,” “one,” “two,” and “three”; that was negative, weak, moderate, and strong staining, respectively. The positively stained cells percentage was scored as “zero,” “one,” “two,” “three,” and “four,” representing 0%, 1% to 25%, 26% to 50%, 51% to 75%, and 76% to 100%. Then, the two scores were multiplied, and the final score was obtained. The median of KIF22 immunohistochemistry (IHC) score (6.0) was regarded as the cutoff value to classify the scores into two groups (low and high expression groups).
Statistical analysis
Unless otherwise stated, the results of continuous variables are expressed in the present study as mean ± standard deviation (SD). A comparison of variables with independent sample Chi-squared test, t tests, and one-way analysis of variance (ANOVA) was carried out. A P value <0.05 was considered to have statistical significance. IBM SPSS Statistics software version 23.0 (Chicago, IL, USA) was used to carry out all analyses.
Results
KIF22 gene expression and enrichment pathways analysis
A public GEPIA database was employed to collect information about KIF22 mRNA expression in different tissues of cancer, so the data of KIF22 mRNA expression from the tissues of GC could be further investigated. These different cancer tissues included esophageal carcinoma (n = 182) and normal esophageal tissues (n = 13), GC (n = 408) and normal gastric tissues (n = 36), breast cancer (n = 1085) and normal breast tissues (n = 112), bladder cancer (n = 404) and normal bladder tissues (n = 19), uterine corpus endometrial carcinoma (n = 174) and normal tissues (n = 13) [Figure 1A]. From these data, KIF22 was indicated to have an over-expression in various human cancer, suggesting that KIF22 might contribute to malignant tumor development. Then, a further gene set enrichment analysis (GSEA) pathway enrichment analysis was performed to reveal the potential functions. The results showed that KIF22 predominantly enriched in cell cycle, cell mitosis, DNA replication [Figure 2A–D].
Figure 1.
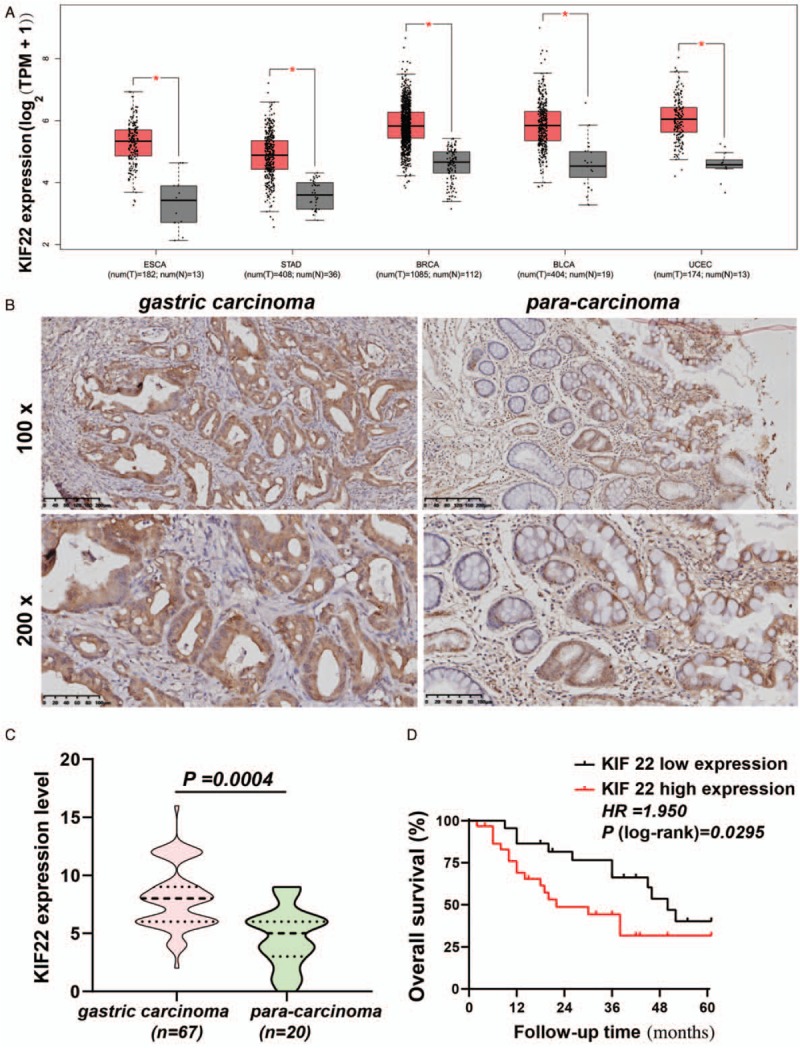
Expression of KIF22 and its association with prognosis in gastric cancer patients. (A) KIF22 mRNA expression in different cancer tissues by Gene Expression Profilling Interactive Analysis (GEPIA) database. (B) Representative images of immunohistochemical staining for KIF22 in gastric cancer tissues and para-carcinoma tissues. (C) Gastric cancer tissues had significantly higher expression levels of KIF22 than para-carcinoma tissues. (D) High levels of KIF22 predicted better overall survival (OS) in patients with gastric cancer. KIF22: Kinesin superfamily protein 22; ESCA: Esophageal carcinoma; STAD: Stomach adenocarcinoma; BRCA: Breast invasive carcinoma; BLCA: Bladder urothelial carcinoma; UCEC: Uterine corpus endometrial carcinoma.
Figure 2.
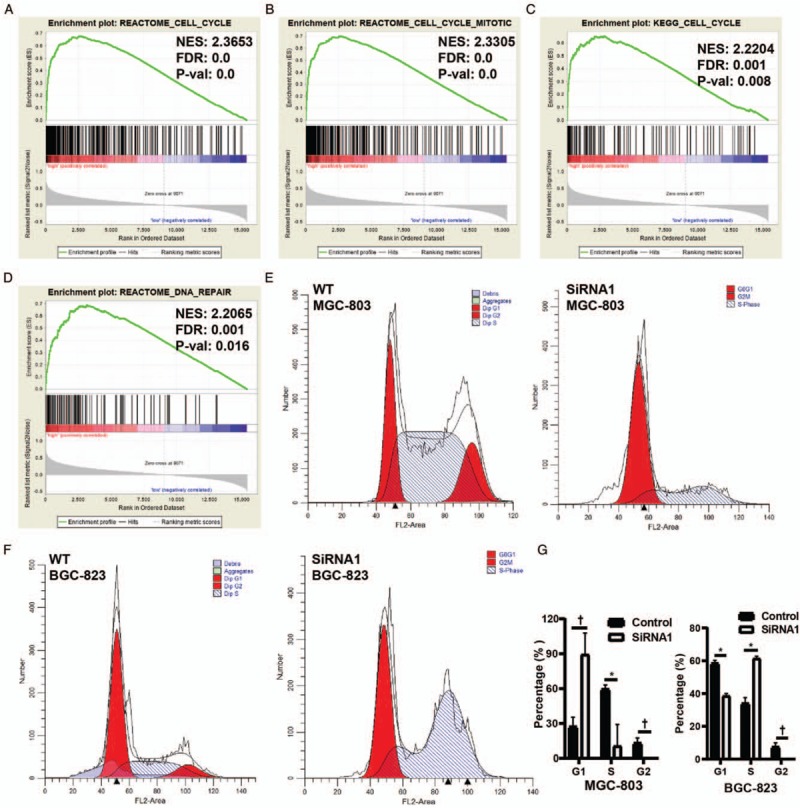
The gene set enrichment analysis (GSEA) showed that Kinesin superfamily protein 22 predominantly enriched in cell cycle (A and C), cell mitosis (B), DNA replication (D). (E–G) The percentages of cells in the cell cycle were detected using flow cytometry. G1 and S phase populations significantly increased while the G2/M phase population obviously decreased in MGC-803 and BGC-823. Data were expressed as mean ± standard deviation. ∗P < 0.05 and †P < 0.01 vs. the control group, respectively.
KIF22 up-regulation in GC tissues is correlated with poor patient prognosis
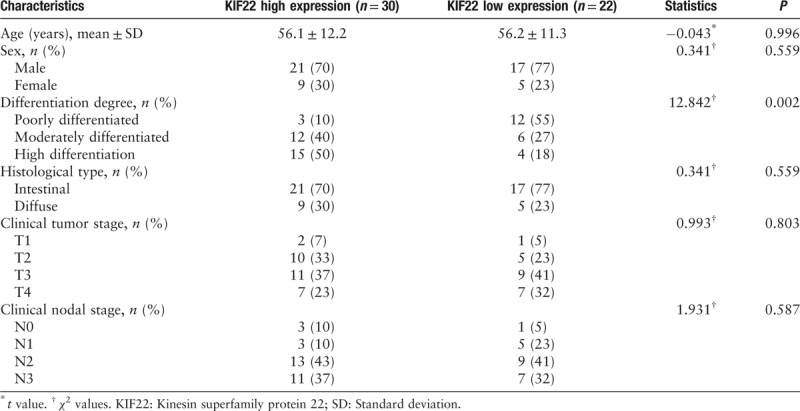
To study the effect of KIF22 on GC progression, we examined the KIF22 expression in the 67 samples from 67 patients with GC, and in 20 para-carcinoma tissues by immunochemical staining. Figure 1B shows that KIF22 was mainly localized to the human GC cells cytoplasm from immunohistochemical staining of tissue microarrays. Gastric tumor samples with high expression of KIF22 were significantly more than para-carcinoma samples [Figure 1C]. The clinicopathologic role that KIF22 expression plays in 52 patients with GC was then investigated. In accordance with the median of KIF22 immunohistochemistry score, we divided 52 patients with GC into two groups (low or high KIF22). The correlation between KIF22 expression and clinicopathologic characteristics is shown in Table 1. The analysis indicated that KIF22 high expression in GC was significantly related to differentiation degree (χ2 = 12.842, P = 0.002), whereas no significant difference with age, gender, and Lauren's classification as shown in Table 1 was observed.
Table 1.
Relationship between KIF22 expression and clinicopathologic characteristics of patients with gastric cancer.
It was revealed from Kaplan-Meier analyses that the KIF22-high patients have worse overall survival rate than the KIF22-low patients [Figure 1D].
By taking all mentioned into account, increased KIF22 expression is revealed to imply poor prognosis in patients with GC.
KIF22 promoted cellular growth and GC cells colony formation
To observe how KIF22 regulates gastric tumor development, we examined KIF22 expression by Western blotting in six human GC cell lines: AGS, BGC-823, HGC-27, MGC-803, MKN-45, SGC-7901, and the non-tumorigenic cell line GES [Figure 3A]. The results indicated an extremely low KIF22 expression in GES. We used KIF22 siRNA to knockdown its expression in BGC-823 cells and MGC-803 which have high expression. After 48 h, the green fluorescent protein (GFP) was also obviously observed by fluorescence microscopy. Furthermore, to confirm the efficiency of transfection, Western blotting was used. By Western blotting, the confirmation of the transfection efficiency at protein level was shown in Figure 3B. Group of cells with inhibited KIF22 expression was named as siKIF22, while siRNA control group was used to name the native control transfected with empty vector. MTT assay was conducted. As shown in Figure 3C and 3D, after inhibiting its expression using KIF22 siRNA, it indicated a significant decrease of GC cell proliferation in BGC-823 and MGC-803 cells. Colony formation assays were used to further validate the impact of KIF22 in cell growth. As can be seen from Figure 3E, results showed that KIF22 promotes colony formation in GC cells.
Figure 3.
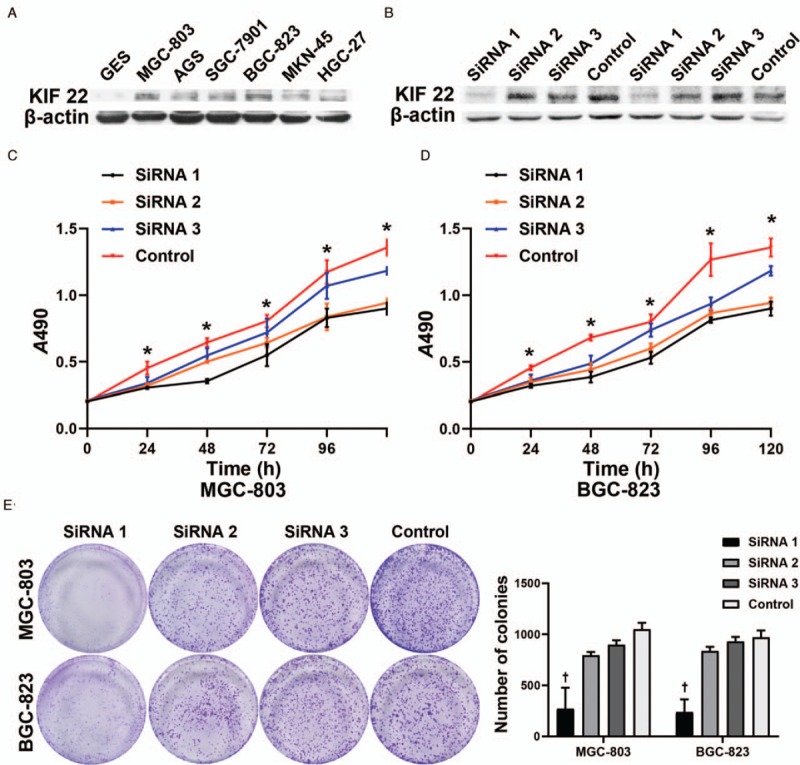
SiRNA-mediated inhibition of KIF22. (A) The expression of KIF22 in six GC cell lines was examined by Western blotting compared with one non-tumorigenic cell line. (B) KIF22 knockdown efficiencies were determined by Western blotting. (C) MTT assays displays decreased proliferation in MGC-803 after using KIF22 siRNA. (D) MTT assays display decreased proliferation in BGC-823 after using KIF22 siRNA. (E) Suppression of KIF22 decreased the colony formation ability of MGC-803 and BGC-823 cells. ∗P < 0.05 and +P < 0.01 for the SiRNA1 group vs. the control group, respectively. SiRNA: Small interfering RNA; KIF22: Kinesin superfamily protein 22; MTT: 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide.
KIF22 promoted tumor invasion and metastatic ability
Figure 4A and 4B showed that via the cell scratch test, migration rates in siKIF22 groups (MGC-803 cells and BGC-823 cells) were significantly reduced in comparison with shCtrl groups (all P < 0.01). By matrix-coated trans-well assays, both the MGC-803 and BGC-823 siKIF22 groups had a strongly decrease in cell invasion in comparison with the shCtrl groups (P < 0.01) [Figure 4C]. Therefore, the GC cell migration and its invasion could be promoted by KIF22.
Figure 4.
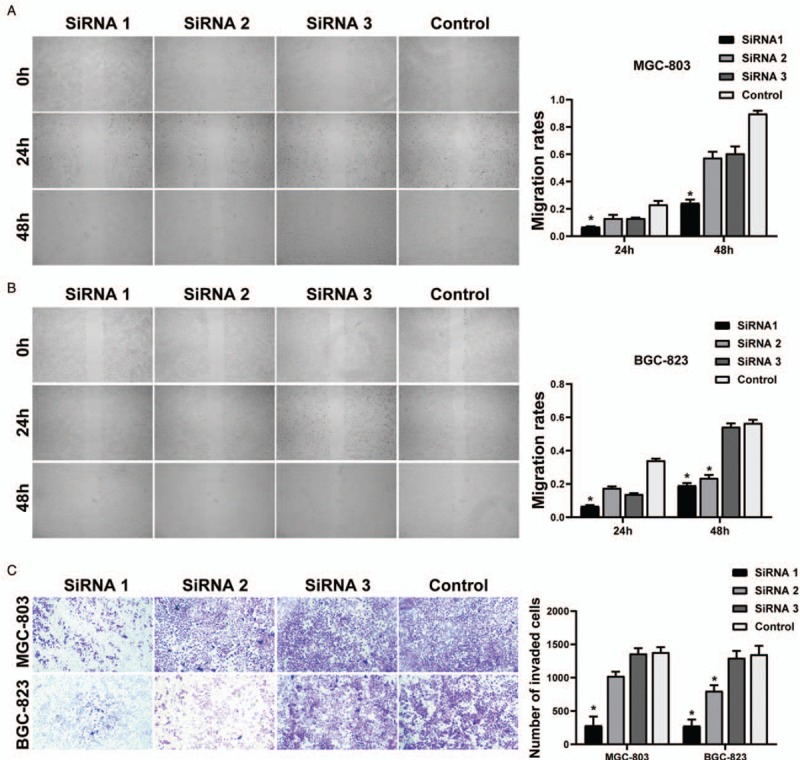
KIF22 promoted tumor invasion and metastatic ability. (A) Representative and quantification of wound-healing assay in MGC-803 cells. (B) Representative and quantification of wound-healing assay in BGC-823 cells. (C) The matrix-coated trans-well assay showed that the SiRNA groups had significant invasiveness. ∗P < 0.01 vs. the control group. KIF22: Kinesin superfamily protein 22.
KIF22-promoted cell cycle
As shown in Figure 2 E–G, suppression of KIF22-induced G1/S-phase cell cycle arrest. The SiRNA1-KIF22 group has significantly higher percentages of cells in G1 and S phases than control group, whereas the G2 phase has obviously decreased in siRNA1-KIF22 group.
KIF22-promoted GC cell proliferation, invasion via the activity of MAPK-ERK pathways
The underlying mechanism which KIF22 contributes to progression and worse clinical outcome of GC was next explored. As shown in Figure 2, KIF22 was showed to be significantly enriched in cell cycle by GSEA pathway enrichment analysis, and it played a vital role in cell cycle. By Western blotting analysis, KIF22 knockdown was shown to up-regulate cyclin B1, while cyclin A2 and cyclin D1 decreased [Figure 5].
Figure 5.
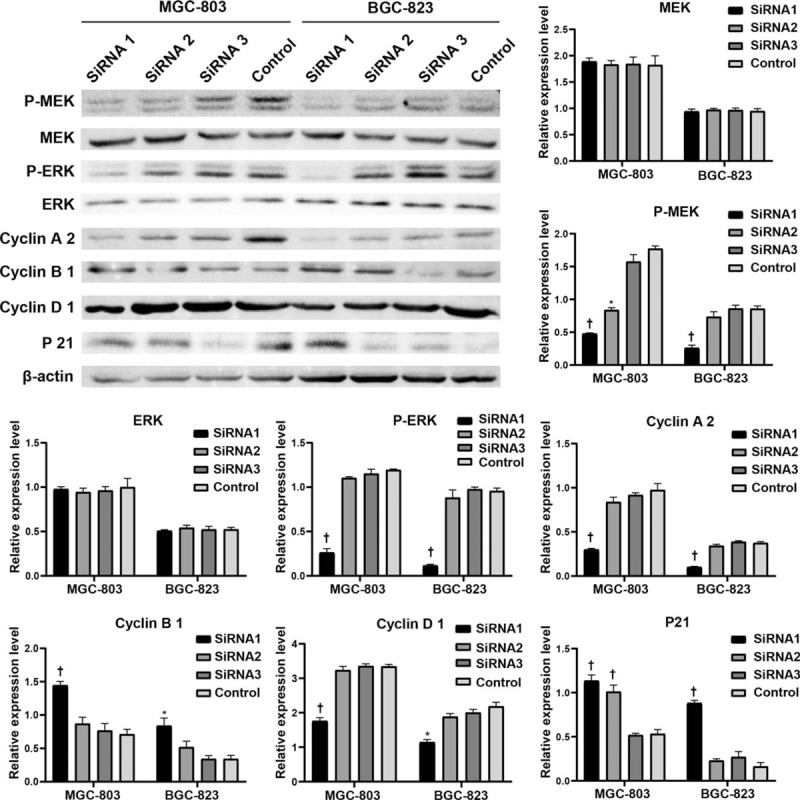
Knockdown of KIF22 suppressed the activation of MAPK-ERK signaling pathway. The expression of cycle related protein including cyclin B1, cyclin A2, and p21 and was significantly down-regulated in the MGC-803 and BGC-823 cells by kif22 silence. ∗P < 0.05 and †P < 0.01 vs. the control group, respectively. KIF22: Kinesin superfamily protein 22.
Previous studies indicated that the kinesin family KIF15 is closely related to the MAPK-ERK pathways. Thus, we further elucidate whether the activation of MEK-ERK signaling pathway could be affected by the changes in KIF22 expression. By Western blotting analysis, KIF22 siRNA was showed to inhibit phospho-MEK and ERK levels in both of the MGC-803 cells and BGC-823 cells. However, the total MEK and ERK protein levels were little affected. By Western blotting analysis, we further examined the down-stream genes of MEK-ERK signaling pathway including p21. The results revealed that in the MGC-803 and BGC-823 cells, the p21 expression was significantly up-regulated by KIF22 silence [Figure 5]. These data suggest that the MAPK-ERK pathways were inhibited when KIF22 was knockdowned in the two cells.
Discussion
In the present study, the expression pattern of KIF22 and its clinical significance by immunohistochemical analysis were examined in GC. Our study demonstrated a clearly increase of KIF22 in GCs in comparison with para-carcinoma tissues. Furthermore, the results also showed that the increase of the KIF22 expression was associated with differentiation degree, T classification, and N classification. While no significant difference appeared in age, gender, and Lauren's classification. What is more, the results showed that KIF22 could be the factor that independently influences the GC prognosis.
Current understanding suggest that mitosis is strictly regulated while abnormal mitosis such as cell cycle checkpoint deficiency, multipolar mitosis, and supernumerary centrosomes are often observed in various cancers.[14–17] Interestingly, previous studies indicated the important role of most kinesins over-expression in GC development, such as KIF2A,[18] KIFC1,[19] KIF11,[20] KIF20A,[21] and Xenopus kinesin-like protein 2 (TPX2).[22] Few kinesins such as KIF4 may play the opposite effect.[23] In kinesin family, KIF11 (also known as kinesin-5, EG5) is well studied as the target with the most clinical trials. However, limited efficiency caused by the cancer cells resistance could be found in many studies, resulting in the discontinued clinical trials (stop at phase I) such as AZD4877, MK-0731, and EMD534085. It was observed that KIF22, that is, Kid, plays an important role as a motor protein in cell mitosis. More importantly, in line with our study, previous research has also shown that the KIF22 was over-expressed in various human tumors and played an important role in carcinogenesis.[24–25]
As previously reported,[26–28] most kinesins promotes cancer cell proliferation and migration. To further characterize the regulation effect of KIF22 in the GC cells, we investigate how siRNA-mediated suppression of KIF22 could have a significant impact on tumor proliferation, invasion, and metastatic abilities. As expected, the results showed that KIF22 was observed to promote cellular growth and increase migration in GC cells. GSEA pathway enrichment analysis was used to further expound the potential mechanisms by which KIF22 promoted GC, and KIF22 was found to be enriched mainly in cell cycle, cell mitosis, and DNA replication. The previous researches also indicated that cyclin D1 transcription and the cell cycle process could be influenced by the MAPK-ERK signaling pathway.[29,30] We performed Western blotting to examine whether KIF22 knockdown inhibited the activation of MAPK-ERK. Our data indicate that knockdown of KIF22 obviously decreased the protein levels of phosphorylated MEK and ERK. However, there is no obvious change in total MEK and ERK protein expression. In addition, the studies also demonstrated that the protein related with cycle (cyclin D1, cyclin A2, and p21) has an expression which was significantly down-regulated in the MGC-803 and BGC-823 cells by KIF22 silence. Furthermore, the flow cytometry assay also indicated that suppression of KIF22 induced G1/S phase cell cycle arrest. Therefore, we concluded that by the inactivation of the MAPK-ERK signaling pathway, KIF22 knockdown decreased gastric cell proliferation and migration, induced G1/S phase cell cycle arrest. However, how KIF22 affected down-stream regulatory factors requires further exploration and the mechanism of KIF22 in promoting GC development by activating the MAPK-ERK signaling pathway will be illuminated in our future work.
In conclusion, KIF22 might act as tumor oncogene in promoting GC proliferation and metastasis via the MAPK-ERK signaling pathway. KIF22 could be a molecular marker in the progression of GC, and thereby is a potential therapeutic target in the future.
Funding
This research was funded by grants from Gansu Provincial Youth Science and Technology Fund Program (No. 18JR3RA330), and Cuiying Scientific and Technological Innovation Program of Lanzhou University Second Hospital (No. CY2018-QN11).
Conflicts of interest
None.
Footnotes
How to cite this article: Yu ZY, Jiang XY, Zhao RR, Qin JJ, Luo CJ, Ren YX, Ren W, Ma ZJ, Jiao ZY. Effect of KIF22 on promoting proliferation and migration of gastric cancer cells via MAPK-ERK pathways. Chin Med J 2020;133:919–928. doi: 10.1097/CM9.0000000000000742
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Aprile G, Giampieri R, Bonotto M, Bittoni A, Ongaro E, Cardellino GG, et al. The challenge of targeted therapies for gastric cancer patients: the beginning of a long journey. Expert Opin Investig Drugs 2014; 23:925–942. doi: 10.1517/13543784.2014.912631. [DOI] [PubMed] [Google Scholar]
- 3.Van Vuuren RJ, Visagie MH, Theron AE, Joubert AM. Antimitotic drugs in the treatment of cancer. Cancer Chemother Pharmacol 2015; 76:1101–1112. doi: 10.1007/s00280-015-2903-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marzo I, Naval J. Antimitotic drugs in cancer chemotherapy: promises and pitfalls. Biochem Pharmacol 2013; 86:703–710. doi: 10.1016/j.bcp.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Stanton RA, Gernert KM, Nettles JH, Aneja R. Drugs that target dynamic microtubules: a new molecular perspective. Med Res Rev 2011; 31:443–481. doi: 10.1002/med.20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azim HA, de Azambuja E, Colozza M, Bines J, Piccart MJ. Long-term toxic effects of adjuvant chemotherapy in breast cancer. Ann Oncol 2011; 22:1939–1947. doi: 10.1093/annonc/mdq683. [DOI] [PubMed] [Google Scholar]
- 7.Gordon RR, Nelson PS. Cellular senescence and cancer chemotherapy resistance. Drug Resist Updat 2012; 15:123–131. doi: 10.1016/j.drup.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirokawa N, Noda, Yasuko, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 2009; 10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 9.Wordeman L. How kinesin motor proteins drive mitotic spindle function: Lessons from molecular assays. Semin Cell Dev Biol 2010; 21:260–268. doi: 10.1016/j.semcdb.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheng J, Jiang K, Xue X. Knockdown of kinase family 15 inhibits cancer cell proliferation in vitro and its clinical relevance in triple-negative breast cancer. Curr Mol Med 2019; 19:147–155. doi: 10.2174/1566524019666190308122108. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Xie H, Zhu S, Chen X, Yu J, Shen T, et al. High expression of KIF22/Kinesin-like DNA binding protein (Kid) as a poor prognostic factor in prostate cancer patients. Med Sci Monit 2018; 24:8190–8197. doi: 10.12659/MSM.912643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q, Cao B, Nan N, Wang YU, Zhai XU, Li Y, et al. Elevated expression of KIF18A enhances cell proliferation and predicts poor survival in human clear cell renal carcinoma. Exp Ther Med 2016; 12:377–383. doi: 10.3892/etm.2016.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Wang H, Lian Y, Wu X, Zhou L, Wang J, et al. Upregulation of kinesin family member 4A enhanced cell proliferation via activation of Akt signaling and predicted a poor prognosis in hepatocellular carcinoma. Cell Death Dis 2018; 9:141.doi: 10.1038/s41419-017-0114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokai N, Fujimoto Nishiyama A, Toyoshima Y, Yonemura S, Tsukita S, Inoue J, et al. Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO J 1996; 15:457–467. doi: 10.1002/j.1460-2075.1996.tb00378.x. [PMC free article] [PubMed] [Google Scholar]
- 15.Gisselsson D. Mitotic instability in cancer: is there method in the madness? Cell Cycle 2005; 4:1007–1010. doi: 10.4161/cc.4.8.1884. [DOI] [PubMed] [Google Scholar]
- 16.Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev 2009; 28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- 17.Duensing A, Duensing S. Centrosomes, polyploidy and cancer. Adv Exp Med Biol 2010; 676:93–103. doi: 10.1007/978-1-4419-6199-0_6. [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Huang F, Wang Y, Song Q, Yang X, Wu H. KIF2A overexpression and its association with clinicopathologic characteristics and poor prognoses in patients with gastric cancer. Dis Markers 2016; 2016:7484516.doi: 10.1155/2016/7484516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oue N, Mukai S, Imai T, Song Q, Yang X, Wu H. Induction of KIFC1 expression in gastric cancer spheroids. Oncol Rep 2016; 36:349–355. doi: 10.3892/or.2016.4781. [DOI] [PubMed] [Google Scholar]
- 20.Imai T, Oue N, Nishioka M, Mukai S, Oshima T, Sakamoto N, et al. Overexpression of KIF11 in gastric cancer with intestinal mucin phenotype. Pathobiology 2017; 84:16–24. doi: 10.1159/000447303. [DOI] [PubMed] [Google Scholar]
- 21.Yan GR, Zou FY, Dang BL, Zhang Y, Yu G, Liu X, et al. Genistein-induced mitotic arrest of gastric cancer cells by downregulating KIF20A, a proteomics study. Proteomics 2012; 12:2391–2399. doi: 10.1002/pmic.201100652. [DOI] [PubMed] [Google Scholar]
- 22.Shao C, Duan C, Wang J, Luan S, Gao Y, Jin D, et al. Expression of microtubule-associated protein TPX2 in human gastric carcinoma and its prognostic significance. Cancer Cell Int 2016; 16:79.doi: 10.1186/s12935-016-0357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Sai N, Wang C, Sheng X, Shao Q, Zhou C, et al. Overexpression of chromokinesin KIF4 inhibits proliferation of human gastric carcinoma cells both in vitro and in vivo. Tumour Biol 2011; 32:53–61. doi: 10.1007/s13277-010-0090-0. [DOI] [PubMed] [Google Scholar]
- 24.Yu Y, Wang XY, Sun L, Wang YL, Wan YF, Li XQ, et al. Inhibition of KIF22 suppresses cancer cell proliferation by delaying mitotic exit through upregulating CDC25C expression. Carcinogenesis 2014; 35:1416–1425. doi: 10.1093/carcin/bgu065. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Xie H, Zhu S, Chen X, Yu J, Shen T, et al. High expression of KIF22/kinesin-like DNA binding protein (Kid) as a poor prognostic factor in prostate cancer patients. Med Sci Monit 2018; 24:8190–8197. doi: 10.12659/MSM.912643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Guo X, Xie C, Jiang J. KIF15 promotes pancreatic cancer proliferation via the MEK–ERK signalling pathway. Br J Cancer 2017; 117:245–255. doi: 10.1038/bjc.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z, Rebowe Re, Wang Z, Li Y, Wang Z, DePaolo JS, et al. KIF3A promotes prostate cancer cell proliferation, migration, and invasion via activation of DvI2 and the Wnt signaling pathway. Mol Cancer Res 2013; 73:4937–14937. doi: 10.1158/1538-7445.AM2013-4937. [Google Scholar]
- 28.Tian DW, Wu ZL, Jiang LM, Gao J, Wu CL, Hu HL, et al. KIF5A promotes bladder cancer proliferation in vitro and in vivo. Dis Markers 2019; 2019:4824902.doi: 10.1155/2019/4824902. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Modi PK, Komaravelli N, Singh N, Sharma P. Interplay between MEK-ERK signaling, cyclin D1, and cyclin-dependent kinase 5 regulates cell cycle reentry and apoptosis of neurons. Mol Biol Cell 2012; 23:3722–3730. doi: 10.1091/mbc.E12-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daksis JI, Lu RY, Facchini LM, Marhin WW, Penn LJ. Myc induces cyclin D1 expression in the absence of de novo protein synthesis and links mitogen-stimulated signal transduction to the cell cycle. Oncogene 1994; 9:3635–3645. doi: 10.1016/0197-0186(94)90157-0. [PubMed] [Google Scholar]


