To the editor,
Asthma, one of the most common chronic diseases in childhood, is caused by interactions between genes and environmental factors. The mainstay of treatment is daily use of inhaled corticosteroids (ICS), which are the most effective medication for controlling asthma symptoms and preventing (severe) exacerbations. ICS use reduces both hospitalizations and mortality rates1 and improves asthma control; reflected in forced expiratory volume in 1 second (FEV1) levels and fraction of exhaled nitric oxide (FeNO). These effects are particularly observed in asthma patients with eosinophilic, type 2 airway inflammation.2 However, responses to ICS are heterogeneous, which while controversial, possibly reflect genetic associations.3, 4
Genome‐wide association studies (GWAS) have reproducibly found the Interleukin 1 receptor like 1 (IL1RL1, ST2) gene to be associated with asthma susceptibility.5 IL1RL1 single‐nucleotide polymorphisms (SNPs) and IL1RL1 expression levels have been associated with blood eosinophils and markers of Th2 type inflammation.6, 7 However, the influence of IL1RL1 SNPs on the effectiveness of asthma treatment has not been investigated. Since the IL‐33/IL1RL1 pathway has been associated with eosinophilic, type 2, inflammation, we hypothesized that IL1RL1 SNPs may affect corticosteroid treatment response in asthma patients. Since IL1RL1‐a functions as a decoy receptor to dampen IL‐33‐induced signaling, genetically determined low levels of IL1RL1‐a may predispose to enhanced IL‐33‐induced inflammation with consequently more exacerbations.
In the current study, we investigated whether IL1RL1 gene variants are associated with asthma exacerbations (based on ER visits/hospitalizations and courses of oral corticosteroid [OCS] use), questionnaire‐based asthma control and FeNO levels in asthma patients using ICS. Furthermore, we aimed to identify whether there is a pharmacogenetic effect of IL1RL1 variants on change in FeNO levels and FEV1% predicted in asthma patients after 4‐6 weeks of ICS treatment.
After close inspection of the Linkage Disequilibrium structure of IL1RL1, we selected 6 IL1RL1 SNPs that tag important LD blocks in IL1RL1 (r 2 > .8) with SNPs previously found to be associated with asthma5; rs13431828, rs1041973, rs1420101, rs1946131, rs1921622, and rs10204137 (Table S1). Cross‐sectional IL1RL1 SNP discovery analysis was performed in ICS treated asthmatic children, mainly of European ancestry, from the Pharmacogenetics of Asthma Medication in Children: Medication with Anti‐inflammatory effects (PACMAN) cohort (N = 820) using logistic and linear regression models. We replicated FDR corrected significant findings (P < .05) in four different cohorts collaborating within the Pharmacogenomics in Childhood Asthma (PiCA) consortium,8 one Hispanic/Latino study; Genes‐Environment and Admixture in Latino Americans (GALA II, N = 876) study, one African American population; Study of African Americans, Asthma, Genes, and Environments (SAGE, N = 525), and two European studies (≥96% European ancestry); the Effectiveness and Safety of Treatment with Asthma Therapy in children (ESTATe, N = 197) and SLOVENIA (N = 104). In addition, we performed a meta‐analysis (N = 2412). The longitudinal effect of IL1RL1 on FeNO levels and FEV1% predicted upon ICS treatment in asthmatic children and adults was assessed in the SLOVENIA cohort. Conditional analysis was performed in PACMAN to assess the independent effects of the IL1RL1 SNPs.
A detailed representation of the included cohorts and the allele frequencies of the IL1RL1 SNPs are provided in Tables S2 and S3, respectively. In PACMAN, we found a significant association between four of the six SNPs (rs13431828, rs1420101, rs1921622, and rs10204137) with ER visits and “any exacerbation” (Table 1A‐C), which were selected for the replication study. Sensitivity analyses on Dutch ethnicity, atopy, and medication adherence did not change these results. We did not observe an association with questionnaire‐based asthma control or FeNO measurements (Table S4A‐B).
Table 1.
Results of associations of IL1RL1 SNPs with ER visits/hospitalizations, OCS use, and “any exacerbation” per study and meta‐analysis
| A. | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Allele (R/E)a | ER visits/hospitalisations | ||||||||||||||||||
| PACMAN (n = 698) | GALA II (n = 876) | SAGE (n = 525) | SLOVENIA (n = 187) | ESTATe (n = 104) | Meta‐analysis (n = 2421) | |||||||||||||||
| OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | N | ||
| rs13431828 | T/C | 2.78 (1.11‐6.94) | .02 | .04 | 1.45 (1.04‐2.03) | .03 | .04 | 1.18 (0.88‐1.58) | .26 | .52 | 1.20 (0.62‐2.35) | .57 | .77 | 1.01 (0.18‐5.72) | .99 | .99 | 1.32 (1.08‐1.62) | .005 | .02 | 2412 |
| rs1041973 | A/C | 1.35 (0.77‐2.37) | .30 | .30 | ||||||||||||||||
| rs1420101 | G/A | 1.61 (1.05‐2.47) | .02 | .04 | 1.28 (1.04‐1.58) | .02 | .04 | 0.90 (0.69‐1.17) | .45 | .60 | 1.09 (0.67‐1.76) | .72 | .77 | 1.35 (0.56‐3.25) | .51 | .94 | 1.16 (1.01‐1.34) | .03 | .06 | 2412 |
| rs1946131 | G/A | 1.47 (0.81‐2.68) | .20 | .24 | ||||||||||||||||
| rs1921622 | G/A | 1.89 (1.18‐3.03) | .01 | .04 | 1.30 (1.06‐1.59) | .01 | .04 | 0.74 (0.55‐0.99) | .05 | .20 | 0.93 (0.57‐1.51) | .77 | .77 | 1.13 (0.97‐1.31) | .13 | .17 | 2308 | |||
| rs10204137 | G/A | 1.37 (0.87‐2.16) | .18 | .24 | 1.24 (0.99‐1.56) | .06 | .06 | 1.01 (0.76‐1.35) | .92 | .92 | 0.75 (0.46‐1.22) | .24 | .77 | 0.81 (0.34‐1.94) | .63 | .94 | 1.10 (0.95‐1.29) | .18 | .18 | 2412 |
| B. | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Allele (R/E)a | OCS use | ||||||||||||||||||
| PACMAN (n = 720) | GALA II (n = 876) | SAGE (n = 525) | SLOVENIA (n = 187) | ESTATe (n = 104) | Meta‐analysis (n = 2421) | |||||||||||||||
| OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | N | ||
| rs13431828 | T/C | 2.70 (1.08‐6.79) | .03 | .09 | 1.25 (0.87‐1.79) | .23 | .65 | 0.93 (0.68‐1.27) | .65 | .70 | 1.60 (0.57‐4.47) | .36 | .90 | 1.15 (0.46‐2.85) | .77 | .77 | 1.13 (0.91‐1.41) | .24 | .78 | 2412 |
| rs1041973 | A/C | 1.52 (0.86‐2.66) | .15 | .27 | ||||||||||||||||
| rs1420101 | G/A | 1.32 (0.88‐1.98) | .18 | .27 | 1.10 (0.86‐1.40) | .49 | .65 | 0.78 (0.57‐1.05) | .09 | .20 | 0.95 (0.49‐1.85) | .89 | .90 | 0.68 (0.34‐1.36) | .28 | .77 | 0.98 (0.84‐1.16) | .90 | .90 | 2412 |
| rs1946131 | G/A | 1.08 (0.57‐2.02) | .83 | .83 | ||||||||||||||||
| rs1921622 | G/A | 1.20 (0.78‐1.86) | .41 | .49 | 1.10 (0.88‐1.38) | .44 | .65 | 0.76 (0.54‐1.08) | .10 | .10 | 0.81 (0.41‐1.59) | .54 | .90 | 1.00 (0.84‐1.18) | .96 | .96 | 2308 | |||
| rs10204137 | G/A | 1.69 (1.05‐2.73) | .03 | .09 | 1.03 (0.81‐1.32) | .80 | .80 | 0.94 (0.69‐1.28) | .70 | .70 | 1.04 (0.52‐2.05) | .90 | .90 | 1.15 (0.58‐2.27) | .70 | .70 | 1.07 (0.90‐1.27) | .39 | .39 | 2412 |
| C. | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Allele (R/E)a | Any exacerbation | ||||||||||||||||||
| PACMAN (n = 720) | GALA II (n = 876) | SAGE (n = 525) | SLOVENIA (n = 187) | ESTATe (n = 104) | Meta‐analysis (n = 2421) | |||||||||||||||
| OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | OR (95% CI) | P | P b | N | ||
| rs13431828c | T/C | 2.63 (1.33‐5.18) | .006 | .03 | 1.63 (1.14‐2.32) | .009 | .01 | 1.04 (0.78‐1.39) | .80 | .80 | 1.19 (0.66‐2.12) | .73 | .83 | 1.09 (0.47‐2.56) | .83 | .86 | 1.31 (1.07‐1.59) | .007 | .02 | 2412 |
| rs1041973 | A/C | 1.28 (0.84‐1.96) | .26 | .26 | ||||||||||||||||
| rs1420101 | G/A | 1.52 (1.08‐2.13) | .01 | .03 | 1.35 (1.06‐1.72) | .01 | .01 | 0.83 (0.63‐1.10) | .18 | .36 | 1.05 (0.65‐1.67) | .83 | .83 | 0.85 (0.48‐1.53) | .60 | .60 | 1.14 (0.98‐1.32) | .07 | .14 | 2412 |
| rs1946131 | G/A | 1.37 (0.83‐2.25) | .22 | .26 | ||||||||||||||||
| rs1921622 | G/A | 1.45 (1.03‐2.04) | .03 | .04 | 1.35 (1.08‐1.70) | .009 | .01 | 0.67 (0.50‐0.90) | .009 | .03 | 0.84 (0.52‐1.35) | .47 | .83 | 1.08 (0.92‐1.25) | .31 | .31 | 2308 | |||
| rs10204137d | G/A | 1.52 (1.05‐2.18) | .02 | .04 | 1.29 (1.00‐1.66) | .04 | .04 | 0.91 (0.68‐1.21) | .50 | .66 | 0.81 (0.50‐1.30) | .38 | .83 | 0.95 (0.53‐1.70) | .86 | .86 | 1.11 (0.96‐1.30) | .14 | .18 | 2412 |
Bold‐faced results are FDR corrected significant results (P < .05). Missing values mean the SNP was not present in the study.
Abbreviations: CI, confidence interval; ER. Emergency room; OCS, oral corticosteroid; OR, odds ratio; SNP, single‐nucleotide polymorphism.
R = reference allele, E = effect allele.
FDR corrected P value.
rs13431828 was not present in ESTATe, and rs3771180 was used as a surrogate marker (LD r 2 = 1).
rs10204137 was not present in ESTATe, and rs4988956 was used as a surrogate marker (LD r 2 = 1).
In GALA II, we replicated our findings with significant results with the same direction of effect for rs13431828, rs1420101, and rs1921622 on ER visits/hospitalizations and “any exacerbation.” Rs10204137 showed a significant association with “any exacerbation” (Table 1A‐C). In SAGE, rs1921622 was associated with “any exacerbation” but the direction of the effect differed when compared to PACMAN. No association between IL1RL1 and questionnaire‐based asthma control was found. In the smaller SLOVENIA and ESTATe studies, no significant cross‐sectional or longitudinal associations were found (Table S5).
Meta‐analysis of the 4 IL1RL1 SNPs carried through to replication showed statistically significant results for rs13431828. The C allele of rs13431828 was associated with ER visits/hospitalizations (OR = 1.32, P = .02) and increased risk of “any exacerbations” (1.31, P = .02; Table 1A‐C, Figure 1). No evidence of heterogeneity was found (Q = 3.6, P = .33).
Figure 1.
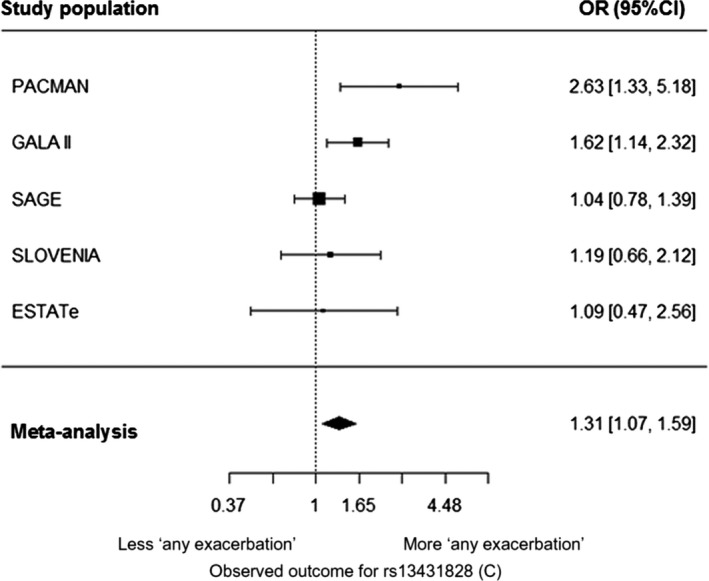
Forest plot showing the meta‐analysis result of the association between the IL1RL1 SNP rs13431828 (C) and ‘any exacerbation’ (P = 0.02). Included cohorts are PACMAN, GALA II, SAGE, SLOVENIA and ESTATe. Odds ratio (OR) and 95% confidence intervals (CI) are shown for the effect alleles (additive model). ‘Any exacerbation’ was defined as ER visits/hospitalizations and/or OCS use
Conditional analysis in PACMAN on rs13431828, rs142010, rs1921622, and rs10204137 for “any exacerbation” indicated that rs13431828 was the most independently associated SNP (Table S6).
These results provide new evidence that children and adolescents with the IL1RL1 risk alleles are prone to more exacerbations than children with the protective genotypes, while using ICS. This extends previous findings that SNPs in IL1RL1 are important in different asthma phenotypes, with more prominent effect in studies investigating childhood‐onset asthma.5 Rs1420101 has been specifically linked to the type 2‐high asthma phenotype,6 as well as to increased eosinophil numbers in peripheral blood.9
We observed replicable associations of the same IL1RL1 risk alleles in the Caucasian (PACMAN) and Hispanic/Latino (GALA II) population, but not in the African American study population (SAGE). This could be due to differences in ethnicity between study groups and LD patterns in this gene, suggested by the observed differences in allele frequency between the cohorts (see Table S3). It is possible that our results may have been influenced by factors other than currently included in the model such as inhalation technique or respiratory infections, but as such data were not available in all cohorts these were not considered.
Different mechanisms may explain our findings. Firstly, IL1RL1 SNPs may modify the asthma phenotype into a more severe phenotype, with more severe exacerbations, which are insufficiently treated with the ICS dosages prescribed to the children in this study. The risk alleles described in our study for rs13431828 (C), rs1420101 (T), rs1921622 (A), and rs10204137 (A) were previously associated with lower IL1RL1 blood methylation levels and lower serum IL1RL1‐a levels,7 indicating that the associated SNPs are important for regulation of IL1RL1 expression. Another mechanism to explain our results is that IL1RL1 may have a direct pharmacogenetic interaction with steroids resulting in reduced efficacy of the steroids. Rs10204137 is a missense mutation and has been associated with increased IL1RL1‐a expression, which induces IL‐33 expression and enhances IL‐33 responsiveness.10 Moreover, rs10204137 tags an LD block that contains 5 nonsynonymous coding SNPs that result in changes to four amino acids in the intracellular domain of IL1RL1‐b. These coding changes affect the Toll/interleukin‐1 receptor (TIR) domain of the intracellular part of the IL1RL1 protein, which plays an important role in IL‐33 induced signal transduction by IL1RL1. This triggers a signaling cascade that eventually results in the activation of downstream mitogen‐activated protein kinases and transcription factors, such as nuclear factor kB (NF‐kB) and activator protein‐1.5 Through this pathway, asthmatic children carrying the risk allele of rs10204137 may be more sensitive to IL‐33. As IL1RL1 is expressed on effector cells of the type‐2 immune response such as mast cells, eosinophils, basophils, Th2 cells and ILC2 cells,11 an increased sensitivity to IL33 will contribute to an exaggerated type‐2 inflammatory response after viral or allergen exposure.
Secondly, IL1RL1 may have a direct pharmacogenetic interaction with steroids resulting in reduced efficacy of the steroids. A recent study on ulcerative colitis found an association between dexamethasone and upregulation of soluble IL1RL1 transcription mediated via interaction of the steroid with the glucocorticoid‐responsive element in the IL1RL1 promotor patients carrying polymorphisms.12 To gain more insight into the mechanism underlying our finding, future studies should be performed in larger cohorts or with the use of biobank data.
This study shows that an IL1RL1 SNP effect is present in asthmatic children using ICS. This highlights the potential investigating if novel treatment strategies targeting the IL33/IL1RL1 pathway could be used as add‐on asthma treatment in patients using ICS.
CONFLICT OF INTEREST
N. Hernandez‐Pacheco received a grant from Instituto de Salud Carlos III (ISCIII) and was co‐funded by the European Social Funds from the European Union (ESF) “ESF invests in your future.” MC Nawijn received a grant from GSK during the conduct of this study and outside the submitted work. ME Engelkes received a grant from Zonmw. K. M. Verhamme received a grant from ZonMw and she works for a research group who in the past received unconditional grants from: Yamanouchi, Pfizer/Boehringer Ingelheim, Novartis and GSK. M Pino‐Yanes received a grant from the Spanish Ministry of Economy, Industry and Competitiveness and a grant from Instituto de Salud Carlos III (ISCIII). AH Maitland‐van der Zee received an unrestricted research grant from GSK, and Boehringer Ingelheim. She also received a grant from ERANET ERACOSYSMED, and she participated in an advisory board for Astra Zeneca. DS Postma declares that the University of Groningen has received money for DS Postma regarding a grant for research from Astra Zeneca, Chiesi, Genentec, GSK and Roche. Fees for consultancies were given to the University of Groningen by Astra Zeneca, Chiesi, and GSK. GH Koppelman received grants from the Lung Foundation of the Netherlands, the Ubbo Emmius Foundation, during the conduct of the study; and he received grants from Lung Foundation of the Netherlands, GSK, Tetri Foundation, Vertex, TEVA the Netherlands, outside the submitted work. GHK participated in an advisory board meeting of GSK. The rest of the authors declare that they have no relevant conflict of interests.
Funding information
The PACMAN study was supported by an unrestricted grant from GlaxoSmithKline (GSK), whereas genetic analysis for the present study was supported by a Lung Foundation of the Netherlands grant no. AF3.2.09.081JU. FND was supported by the Ubbo Emmius Foundation. The GALA II and SAGE studies were funded by the Sandler Family Foundation, the American Asthma Foundation, the RWJF Amos Medical Faculty Development Program, Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II, National Institutes of Health (1R01HL117004, R01Hl128439, R01HL135156, and 1X01HL134589), National Institute of Health and Environmental Health Sciences (R01ES015794 and R21ES24844), the National Institute on Minority Health and Health Disparities (1P60MD006902, U54MD009523, and 1R01MD010443), and the Tobacco‐Related Disease Research Program under Award Number 24RT‐0025 to EGB. This work was also funded by Instituto de Salud Carlos III (AC15/00015), through Strategic action for Health Research (AES) and European Community (EC) within the Active and Assisted Living (ALL) Programme framework, and by the SysPharmPedia grant from the ERACoSysMed first joint Transnational cCall from the European Union under the Horizon 2020. NH‐P was funded by a fellowship (FI16/00136) from Instituto de Salud Carlos III (ISCIII) and co‐funded by the European Social Funds from the European Union (ESF) “ESF invests in your future” and MP‐Y was supported by the Ramón y Cajal Program (RYC‐2015‐17205) by the Spanish Ministry of Economy, Industry, and Competitiveness. For the SLOVENIA study, the authors acknowledge the financial support from the Slovenian Research Agency (research core funding No. P3‐0067) and from SysPharmPedia grant, co‐financed by Ministry of Education, Science and Sport of the Republic of Slovenia. The ESTATe project was supported by a ZonMw Grant No 113201006. The PiCA study was overall supported by ERACoSysMed 1st Joint Transnational Call (SysPharmPedia).
Supporting information
ACKNOWLEDGMENTS
We would like to thank the participants and their parents of the studied cohorts for their participation. We also would like to acknowledge the field workers, data managers, and scientific collaborators dedicated to these cohorts. The authors acknowledge the GALA II and SAGE investigators (Kelley Meade, Harold J. Farber, Pedro C. Avila, Denise Serebrisky, Shannon M. Thyne, Emerita Brigino‐Buenaventura, William Rodriguez‐Cintron, Saunak Sen, Rajesh Kumar, Michael Lenoir, Luisa N. Borrell, and Jose R. Rodriguez‐Santana), the recruiters, participants, and the study coordinator Sandra Salazar. We thank as well the ESTATe investigators (Pharmo: Ron Herings, Annemarie Janse, Jettie Overbeek, Josine Kuiper. IPCI: Katia Verhamme, Hettie Janssens, Johan de Jongste, Miriam Sturkenboom).
Maitland‐van der Zee and Koppelman contributed equally.
REFERENCES
- 1. Global Initiative for Asthma . Global strategy for asthma management and prevention. 2018. http://www.ginasthma.org
- 2. Woodruff PG, Modrek B, Choy DF, et al. T‐helper Type 2 – driven inflammation defines major subphenotypes of asthma. Am J Respir CritCare Med. 2009;180:388‐395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kersten ETG, Koppelman GH. Pharmacogenetics of asthma: toward precision medicine. Curr Opin Pulm Med. 2017;23:12‐20. [DOI] [PubMed] [Google Scholar]
- 4. Mosteller M, Hosking L, Murphy K, et al. No evidence of large genetic effects on steroid response in asthma patients. J Allergy Clin Immunol. 2017;139:797‐803. [DOI] [PubMed] [Google Scholar]
- 5. Grotenboer NS, Ketelaar ME, Koppelman GH, Nawijn MC. Decoding asthma: translating genetic variation in IL33 and IL1RL1 into disease pathophysiology. J Allergy Clin Immunol. 2013;131:856‐865. [DOI] [PubMed] [Google Scholar]
- 6. Gordon ED, Palandra J, Wesolowska‐Andersen A, et al. IL1RL1 asthma risk variants regulate airway type 2 inflammation. JCI Insight. 2016;1:e87871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dijk FN, Xu C, Melén E, et al. Genetic regulation of IL1RL1 methylation and IL1RL1‐a protein levels in asthma. Eur Respir J. 2018;51:1701377. [DOI] [PubMed] [Google Scholar]
- 8. Farzan N, Vijverberg SJ, Andiappan AK, et al. Rationale and design of the multiethnic pharmacogenomics in childhood asthmaconsortium. Pharmacogenomics. 2017;18:931‐943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gudbjartsson DF, Bjornsdottir US, Halapi E, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342‐347. [DOI] [PubMed] [Google Scholar]
- 10. Brusselle GG, Maes T, Bracke KR. Eosinophils in the spotlight: eosinophilic airway inflammation in nonallergic asthma. Nat Med. 2013;19:977‐979. [DOI] [PubMed] [Google Scholar]
- 11. Saluja R, Ketelaar ME, Hawro T, Church MK, Maurer M, Nawijn MC. The role of the IL‐33/IL‐1RL1 axis in mast cell and basophil activation in allergic disorders. Mol Immunol. 2015;63:80‐85. [DOI] [PubMed] [Google Scholar]
- 12. Díaz‐Jiménez D, Núñez L, De La Fuente M, et al. A functional IL1RL1 variant regulates corticosteroid‐induced sST2 expression in ulcerative colitis. Sci Rep. 2017;7:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


